Abstract
BACKGROUND:
The X-chromosome gene USP9X encodes a deubiquitylating enzyme that has been associated with neurodevelopmental disorders (NDDs) primarily in females. USP9X escapes X-inactivation, and in females de novo heterozygous copy number loss or truncating mutations cause haploinsufficiency culminating in a recognisable syndrome with intellectual disability (ID), signature brain and congenital abnormalities. In contrast, the involvement of USP9X in male NDDs remains tentative.
METHODS:
We collected and interrogated the pathogenicity of 44 male-ascertained USP9X variants associated with NDDs using clinically recommended guidelines. Functional studies in patient derived cell lines and mice were used to determine mechanisms of pathology.
RESULTS:
Twelve missense variants showed strong evidence of pathogenicity. We define a characteristic phenotype of the CNS (white matter disturbances, thin corpus callosum and widened ventricles), global delay with significant alteration of speech, language and behaviour, hypotonia, joint hypermobility, visual system defects and other common congenital and dysmorphic features. Comparison of in silico and phenotypical features align additional variants of unknown significance with likely pathogenicity. In support of partial loss-of-function mechanisms, using patient derived cell lines we show loss of only specific USP9X substrates which regulate neurodevelopmental signalling pathways and a united defect in TGF signalling. In addition, we find correlates of the male phenotype in Usp9x brain-specific knockout mice, and further resolve loss of hippocampal dependent learning and memory.
CONCLUSION:
Our data demonstrate the involvement of USP9X variants in a distinctive neurodevelopmental and behavioural syndrome in males and identify plausible mechanisms of pathogenesis centred on disrupted TGFβ signalling and hippocampal function.
Keywords: USP9X, Deubiquitylating Enzyme, TGFβ, Hippocampus, Brain Malformation, Neurodevelopmental Disorder
INTRODUCTION
USP9X is a highly conserved X-chromosome gene encoding a substrate specific deubiquitylating enzyme (1). Complete Usp9x loss of function (LOF) is embryonic lethal in mouse (2), and homo- or hemizygous complete LOF germline mutations have never been identified in human. We previously reported the identification of 17 females with neurodevelopmental disorders (NDDs) due to USP9X de novo heterozygous complete LOF mutations (predominately early frame shift / stop gain mutations) (3). USP9X escapes X-inactivation and in these subjects the mRNA and protein levels are significantly reduced. The phenotype is recognisable, and involves intellectual disability (ID), structural brain abnormalities, characteristic facial features, and distinctive congenital malformations (3). We also reported two missense variants and a truncating frame shift variant (escaping NMD) associated with male intellectual disability (4). These variant proteins retained core enzymatic activity, and instead impaired specific USP9X ‘brain functions’ including neuronal migration and growth (4). Two additional novel missense variants were also implicated in epilepsy, one de novo and likely pathogenic, and another of unknown significance (5). Thus the involvement of USP9X remains only tentatively associated to nonspecific male NDDs.
USP9X has a central deubiquitylating catalytic domain and long N- and C-terminal extensions used to mediate substrate recognition (1). USP9X interacts with at least 53 proteins, each in a tissue and context dependent manner. USP9X deubiquitylates substrates, typically antagonising their proteasomal degradation and as such stabilising their levels (1). In brain, many USP9X substrates are encoded by NDD-associated genes (1), whilst others regulate neurodevelopmental signalling pathways including TGFβ, Notch, Wnt and mTOR (6–15). Conditional deletion of Usp9x in the embryonic forebrain alters these signalling pathways, and causes defective neural progenitor cell function, neuronal cell growth and maturation (4, 7, 12, 16–18). Prominent anatomical features of these mice include agenesis of the corpus collosum, and loss of post-natal hippocampal growth (17, 18). Establishing behavioural phenotypes of these mice is critical to establish models of human NDDs involving USP9X.
Here we interrogate 44 additional USP9X missense variants in males with NDDs, establish a characteristic clinical phenotype and resolve key features in knockout mice. We use patient derived cell lines to discover molecular mechanisms involving neurodevelopmental signalling pathways. Our data underscore the relevance of partial LOF effect of human USP9X variants and point to a loss of TGFβ signalling and hippocampal function as major contributors to pathology.
METHODS
Subjects
This study was approved by the Women’s and Children’s Health Network Human Research Ethics Committee, South Australia, Australia (HREC786-07-2020). All subject information was provided following informed parental consent (Table S7).
Cell Culture
Primary fibroblasts were maintained as previously described (3). TGFβ luciferase assays were conducted as previously described (18) in biological quadruplicate using 20ng/ml TGFβ (R&D Systems, In Vitro Technologies, Australia). Scratch migration assays were conducted as previously described (19). Number of cells migrating into the scratch were quantified using ImageJ (NIH, USA). Assays were blinded to genotype, and biological triplicates were assayed in technical triplicate. TGFβ-SMAD4 localisation assay were performed by incubating cells without serum for 8 hours prior to addition of 20ng/ml TGFβ. Assay was conducted blinded to genotype and conducted across 5 experiments.
Immunofluorescence
Immunofluorescence was performed as previously described (20). List of antibodies is provided in Table S6.
Biochemical Analysis
Protein isolation and western blots were performed as previously described (3). List of antibodies is provided in Table S6. RNA isolation and qPCR was described previously (3).
Proteomics
Immunoprecipitation was as previously described (4). Rabbit IgG or anti-USP9X antibody (5 μg/treatment; A301–350A, Bethyl Laboratories, USA) were used. Proteins were identified and quantified using tandem-mass-tag 1D-liquid-chromatography Electron-Spray-Ionisation tandem-mass-spectrometry by Australian Proteome Analysis Facility, Sydney Australia. Raw data was searched using Proteome Discoverer v2.1 to identify proteins. Raw quantitative values were mean normalized to handle batch effects and log transformed. Paired t-tests identified proteins significantly enriched (adjusted p<0.05 and fold change>0.5) in USP9X (wildtype and mutant) IPs compared to IgG controls.
Mouse husbandry
Usp9xLoxP/LoxP female mice (129SvJ / C57Bl6 mixed background) and Emx1-Cre male mice (C57Bl6 background) were crossed as previously described (17, 18). As Usp9x is located on the X chromosome, male offspring that inherit the Emx1-Cre allele lacked Usp9x in the telencephalon and derived cortex and hippocampal structures (referred to as Usp9x−/Y; Emx1-Cre or simply knockout mice). Cre-negative males were used as controls (referred to as Usp9xloxP/Y or simply wildtype). Female mice were not analysed.
Open Field Test
Locomotor behaviour was assessed in adult mice as previously described (21). Ethovision XT software (Noldus Information Technology, NLD) recorded distance travelled over a 30 minute test period and data were assessed in six 5 minute time-bins.
Primary SHIRPA screen
Adult mice were screened for gross neurological deficits using a primary SHIRPA (SmithKline Beecham Pharmaceuticals; Harwell, MRC Mouse Genome Centre and Mammalian Genetics Unit; Imperial College School of Medicine at St. Mary’s; Royal London Hospital, St. Bartholomew’s and the Royal London School of Medicine; Phenotype Assessment) screen (22). Mice were observed in a cylindrical viewing jar for 5 min, transferred to an arena (45 cm × 45 cm), followed by a series of anatomical and neurological measures, including assessments of muscular, spinocerebellar, sensory, neuropsychiatric and autonomic functions.
Active place avoidance task
Adult animals included (6–7 months) Usp9x −/Y; Emx1-Cre (n = 17) knockout and Usp9xloxP/Y control (n=16) mice. Littermates were raised together regardless of genotype. Test mice were placed onto a rotating platform arena within a room marked by visual cues (23). Upon entering the stationary shock zone, mice received electric shocks (0.5 milliamps at 15 ms intervals) until they exited. Habituation consisted of exploration without shock. The following 5 consecutive days, mice were placed on the rotating platform for 10 minutes with active shocks. Data was acquired using Ethovision™XT software. Testing and analysis was performed blind to genotype. Two-way ANOVA was performed involving two independent variables, with repeated measures if applicable. Multiple comparisons were adjusted (Bonferroni correction). Statistical significance was set at p<0.05.
Histology
Histology and immunofluorescence was conducted as previously described (17).
RESULTS
Identification of de novo and inherited USP9X missense variants in affected males
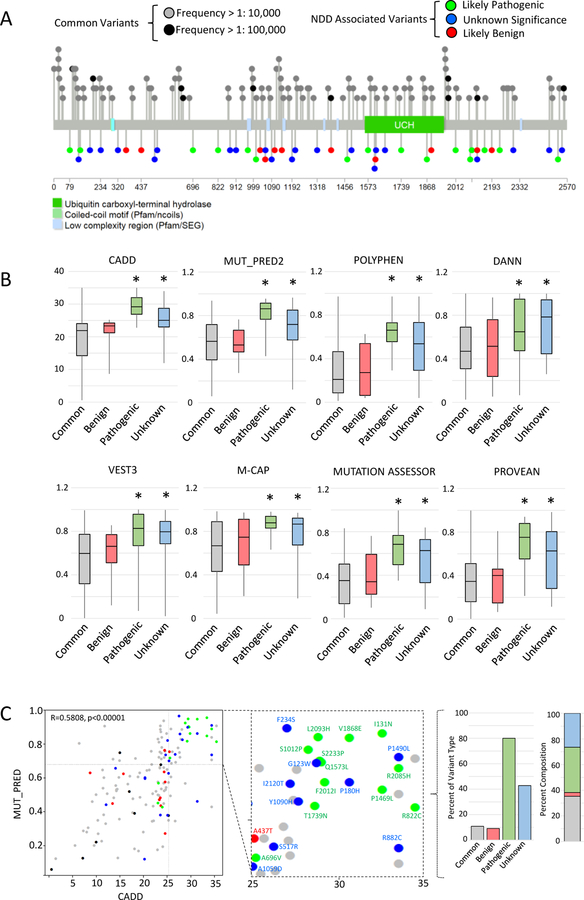
Targeting males with NDDs, we discovered 48 cases with 44 unique USP9X variants (3 were recurrent), primarily through trio-based exome sequencing (Table S1). Two of these subjects were obtained via DECIPHER (UK1 = Decipher Patient: 260068; and Netherlands 2 = Decipher Patient: 323395; (24)). The clinical history of each case is provided in Supplemental Information. We classified each variant’s pathogenicity in accordance with the American College of Medical Genetics and Genomics (ACMG) guidelines (Figure S1); (25). Eleven variants (13 families) were classified as likely benign (Table S1). The remaining 33 variants (in 35 families) were considered the most plausible genetic cause, either being the sole genetic finding, or prioritised over other variants of unknown/unlikely significance (Table S2). Nine de novo variants were classified as likely pathogenic (Table S1 and Figures S1–S2). Three of these located in the catalytic domain, however structural homology modelling mapped these variants to positions outside of the core catalytic site (Figure S3 and S4); (26). The remaining 24 variants (26 families) were maternally inherited, of which segregation beyond trio analysis was able to be performed in 11 cases (Table S1). We conducted functional studies to provide evidence of pathogenicity of three such inherited variants (see below) (Figure S2). Another variant p.Ala2481fs*17 was recurrent in 3 unrelated cases (Table S1). However, this variant impacted only the longer of two alternative coding USP9X isoforms, and was intronic in the short (Figure S5). Interpretation was further complicated by the presence of four hemizygous alleles in gnomAD (Table S1). Preferential isoform usage could underlie a variable penetrance, and rare SNPs (dbSNP and gnomAD) affecting the 5’ donor splice sites exist, but we were unable to classify its involvement beyond VUS (Figure S5). Thus from our collection of 44 variants, 12 were likely pathogenic (9 de novo, 3 maternally inherited), 11 likely benign, and 21 VUS (Table S1, Figure S1). The likely pathogenic variants altered highly conserved residues and were distributed throughout the protein (Figure S2).
USP9X variants are associated with a spectrum of neurodevelopmental features in males.
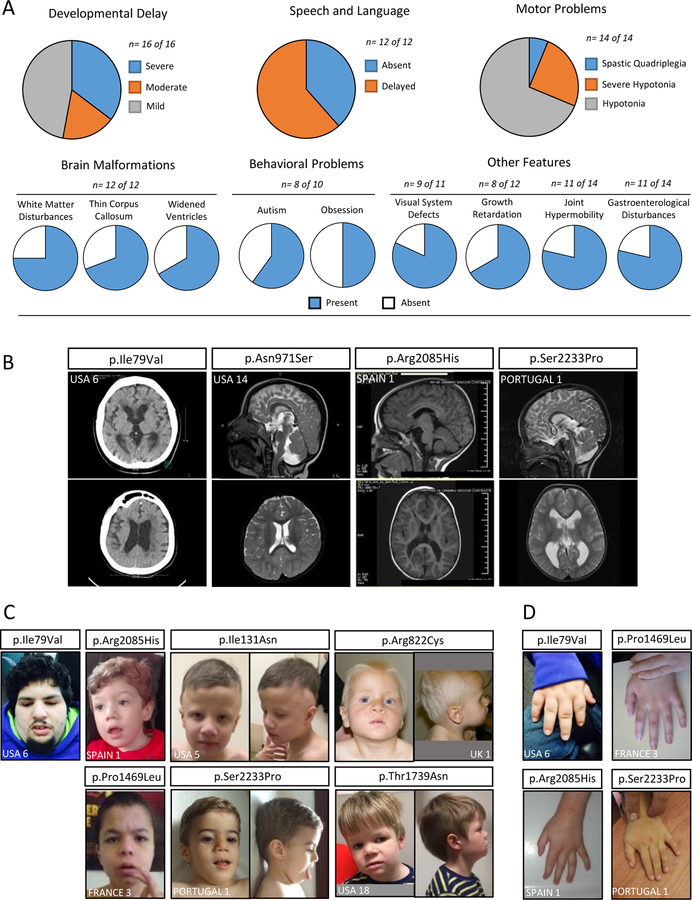
We collated clinical information of subjects with the 12 likely pathogenic variants and from the four previously published male likely pathogenic variants (Table S3); (4, 5). Global developmental delay / ID was reported in all cases varying from mild to severe (Figure 1A). Speech and language problems and motor disability were also found in all cases (where reported). Subjects also presented with behavioural issues, predominantly autistic and obsessive behaviours, but also attention deficit hyperactivity disorder (ADHD), anxiety and aggression (Figure 1A). Ophthalmic abnormalities, in particular strabismus, were also prevalent. In all cases where neuro-imaging was performed, brain malformations were present that included (but not limited to) white matter disturbances, hypoplastic corpus callosum, widened ventricles and cerebellar defects (Figure 1A–B). Outside of the brain, the affected individuals’ most common features included joint hypermobility, a range of gastroenterological problems (feeding difficulties, reflux and constipation in particular) and growth defects of pre- and post-natal onset (Figure 1A). All affected males presented with dysmorphic facial features, although variable in nature across the cohort (Figure 1C). Digital defects were also frequently reported, mainly tapered and pointed fingers (Figure 1D). Collectively, we associate several anomalies of the central nervous system, global delay with significant alteration of speech, language and behaviour, hypotonia, joint hypermobility, strabismus and some common dysmorphic features with missense USP9X variants in males. The prominent neurological features reported in female USP9X subjects (3) are frequent in this male cohort, however other major congenital features of females are infrequent or absent in males (Figure S6).
Figure 1. Likely pathogenic USP9X variants cause a characteristic NDD in males.
A. Constellation and penetrance of defining clinical features. n = the number of subjects whose information contributed to the data. B. Magnetic Resonance Imaging of the brains of individuals with likely pathogenic USP9X variants. Examples highlight evidence of white matter loss and ventricular widening in all, and in particular peri-ventricular leukomalacia (p.Ile79Val), loss of myelination / gliosis of posterior peri-ventricular white matter (p.Asn971Ser), cerebellar vermis hypoplasia (p.Arg2085His) and hypoplastic corpus callosum (p.Ser2233Pro). C-D. Photographs of individuals with USP9X variants. Note short, tapered fingers.
USP9X variants of unknown significance exhibit features of pathogenicity
We leveraged our resources of likely pathogenic and benign variants (Table S1) and common USP9X variants (gnomAD hemizygous variants with allele frequency > 1:100,000) to comparatively assess VUS. There was no clear difference in spatial distribution of variant types across the protein (Figure 2A). Three VUS located in the catalytic domain were also shown to lie outside of the catalytic site (Figure S4). All variants were then compared using ANNOVAR predictive tools (27) to discover algorithms with discriminatory power for USP9X. Eight predictive tools validated for which (1) common and benign variant scores were similar, and (2) common and pathogenic variants score were significantly different (Figure 2B). These validated tools aligned VUS more closely with likely pathogenic variants (Figure 2B). CADD and MutPred2 tools employ independent algorithms (28, 29), and respective variant scores were moderately correlated (Figure 2C). Combining these scores to predict pathogenicity (CADD >25 and MutPred2 >0.7) enriched for likely pathogenic variants (80% of all likely pathogenic variants), and were accompanied by ~half of VUS (Figure 2C). Similar results were obtained by combining CADD and PROVEAN scores (Figure S7).
Figure 2. USP9X VUS share in silico signatures with likely pathogenic variants.
A. Protein location of USP9X variants and common variants extracted from gnomAD data base. B. Bulk comparison of common, benign, likely pathogenic and variants of unknown significance by a suite of in silico prediction tools. *significantly different from common variants p<0.05 by Student’s t-test. C. Comparison of CADD and MUT_PRED2 scores reveal clustering of variants of unknown significance with likely pathogenic variants in upper-right quadrant consistent with pathogenicity (CADD >25, MUT_PRED >0.7). Scores are significantly correlated (Pearson’s correlation given). Colour scheme as in A and B. Inset identifies each variant in the ‘pathogenic quadrant’. Graphs show percent of each type of variant, and the overall composition of variant types within the pathogenic quadrant.
We also compared the prevalence of the characteristic clinical features in subjects with VUS. Developmental delay, speech and language problems and behavioural problems were frequent, whilst other features including motor problems, brain malformation and gastroenterological problems (among others) were observed at reduced frequency (Figure S7 and Table S4). In aggregate, these data reveal in silico and clinical overlap between USP9X VUS with likely pathogenic variants.
USP9X missense variants affect levels of USP9X and its substrates.
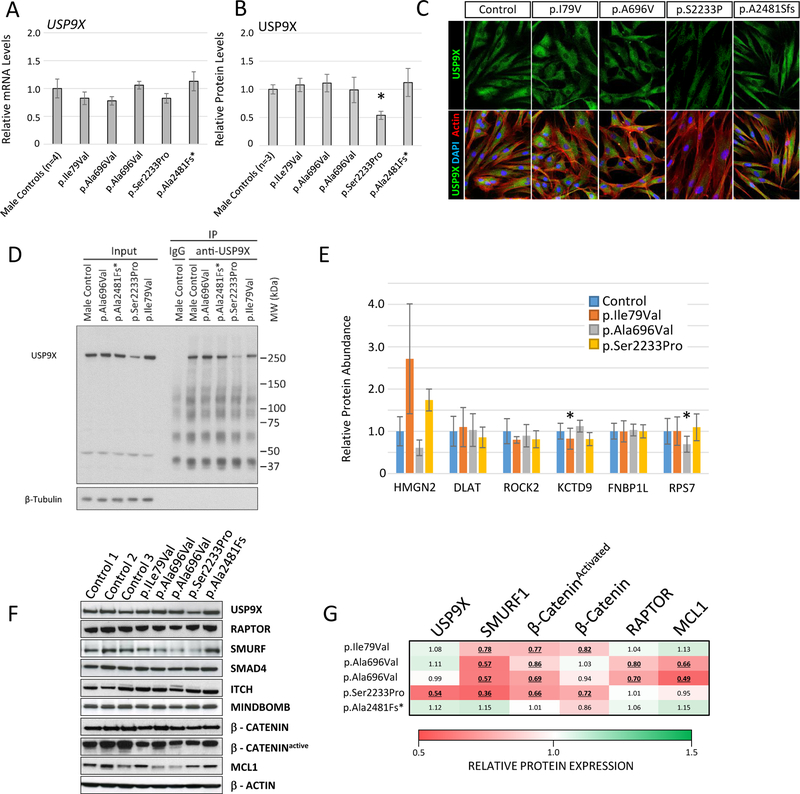
For four maternally inherited variants, we generated patient derived skin fibroblast cell lines and performed functional studies: USA 6 (p.Ile79Val); France 2 (p.Ala696Val); Portugal 1 (p.Ser2233Pro); and the recurrent frameshift variant from Netherlands 3 (p.Ala2481fs*17). Studies on the p.Ala2481fs*17 variant were uninformative, as the long isoform was barely expressed (Figure S5). We investigated the steady-state levels of USP9X mRNA and protein (Figure 3). The p.Ser2233Pro variant line showed a significant (~50%) reduction of USP9X protein level (Figures 3A–B, Figure S8). Subcellular localisation of USP9X variants was not overtly affected (Figure 3C and Figure S9).
Figure 3. USP9X variants impact substrates that regulate neurodevelopmental signalling pathways.
A. qRT-PCR of USP9X mRNA expression in male control and patient derived fibroblasts. B. Quantitation of n=3 western blot experiments analysing USP9X protein expression (See Figure S8). *p<0.05 Student’s t-test. C. Representative immunofluorescence images from control and USP9X variant fibroblast cell lines. D. Western-blot of representative USP9X immunoprecipitation (IP) experiment from control and USP9X variant fibroblast lysates. Immunoprecipitated proteins from n=3 independent experiments (See Figure S10) were analysed by tandem mass tag mass spectroscopy for quantitation. E. Relative protein quantities of significantly enriched USP9X interactors (enriched in USP9X IPs compared to IgG IPs in control cells) in variant USP9X IP experiments. *p<0.05 paired Student’s t-test. F. Representative western blot analysis of USP9X substrates implicated in neurodevelopmental signalling pathways in control and variant USP9X fibroblast cell lines. G. Quantitation of western-blots in C and replicates experiments (Figure S8; n=3 experiments). Values represent relative abundance compared to controls (n=3 cell lines); values underlined are significantly reduced (p<0.05 Student’s t-test).
As complete Usp9x LOF is embryonic lethal, we hypothesised a molecular mechanism of USP9X missense variants consisting of disruption of only specific subsets of USP9X protein-protein interactions, rather than all. To test this, we immunoprecipitated (IP) USP9X and interacting proteins from control and variant fibroblast cell lines, and subjected them to Tandem-Mass-Tag based quantitative proteomic analysis (Figure 3D–E and Figure S10). We identified 6 proteins (HMGN2, DLAT, ROCK2, KCTD9, FNBP1L and RPS7) in addition to USP9X statistically enriched in control USP9X IPs over IgG (Figure S10). Of these interactors, only KCTD9 was significantly depleted (by 20%) in the p.Ile79Val IPs; and RPS7 was significantly depleted (~40%) in the p.Ala697Val IPs (Figure 3E). We conclude that the variants did not overtly impact the majority of USP9X interactions detectable by IP coupled proteomics.
As a deubiquitylating enzyme, USP9X-substrate interactions are rapid and transient, which can render vigorous detection of interactions refractory to IP. We therefore took a targeted western-blot approach to study the protein expression levels of USP9X substrates. We studied substrates specifically involved in neurodevelopmental signalling pathways (Figure 3F–G and Figure S8); (1, 4, 7, 12). All USP9X missense variant cell lines had reduced levels of substrates SMURF1, a regulator of TGFβ signalling (30), and the activated (hypo-phosphorylated) form of CTNNB1 (aka β-CATENIN), a regulator of Wnt signalling (31). In addition, total β-CATENIN was significantly reduced in the p.Ile79Val and p.Ser2233Pro cell lines (Figure 3 F–G and Figure S8). We also found significant reduction of RAPTOR (mTOR pathway) and MCL1 (apoptotic pathway) levels in the p.Ala696Val cell lines (n=2 brothers) (32, 33). Other substrates including ITCH, MINDBOMB, and SMAD4 (regulators of EGF, NOTCH and TGFβ pathways respectively) were unchanged (Figure 3F and Figure S8). Ubiquitylation can also direct the nuclear localisation of SMAD4 and β-CATENIN, but we failed to identify any major difference in localisation under standard culture conditions (Figures S11 and S12). Taken together, USP9X missense variants lead to reduced levels of substrates specifically involved in neurodevelopmental signalling pathways, whilst other (more stable / robust interactions) identified via immunoprecipitation were largely unaffected.
USP9X missense variants lead to a loss of TGFβ signalling.
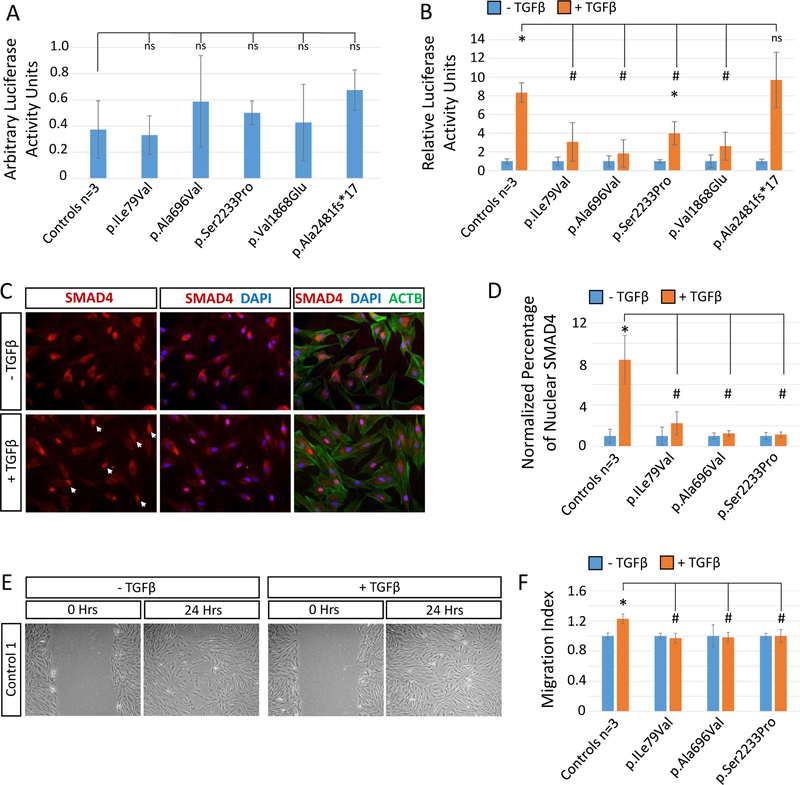
As SMURF1 levels were reduced across USP9X variant fibroblast lines, we assessed TGFβ signalling capacity. Basal levels of signalling assessed by TGFβ luciferase reporter assays were not affected (Figure 4A and Figure S13). The addition of TGFβ resulted in ~8-fold increase in luciferase activity in control cells, and in the p.Ala2481fs*17 cell line in which the variant isoform is barely expressed (Figure 4B and Figure S5 and S13). In contrast, only a 2–4 fold induction observed was observed in the remaining inherited variant cell lines. A similar result was obtained from a cell line derived from subject USA1 harbouring the de-novo likely pathogenic variant p.Val1868Glu (Figure 4B and Figure S13). We tested TGFβ signalling further using the inherited variant cell lines. SMAD4 is translocated into the nucleus during TGFβ signalling, and an ~8-fold increase in nuclear SMAD4 was identified in control cells following TGFβ stimulation, significantly greater than in the variant cell lines tested (Figure 4C–D and Figure S14). Lastly, we conducted a scratch migration assay to determine if variant cell lines are induced to migrate in response to TGFβ (19, 34). The TGFβ stimulated migration in control cells (~20% increase) was not observed in variant cells (Figure 4 E–F and Figure S15). In addition, we tested mTOR signalling capacity specifically in the p.Ala696Val cell lines (n=2 brothers) in which RAPTOR levels were reduced (Figure 4 C–D and Figure S8). Across two independent assays involving either a standard or serum stimulated cell culture protocol, the p.Ala696Val variant cell lines displayed evidence of a reduced mTOR response as assessed by reduced phospho-S6 (pS6) levels and reduced pS6:S6 ratio (Figure S16). We await additional cell lines with this variant/phenotype for more rigorous testing.
Figure 4. TGFβ signalling is disrupted in USP9X variant fibroblast cell lines.
Cells were serum starved (0.2% serum) for 16 hours prior to addition of TGFβ and assayed 24 hours later. A. In the absence of added TGFβ, cells display similar basal levels of signalling as assessed by TGFβ luciferase reporter assay. B. Relative increase of TGFβ signalling following addition of ligand as assessed by TGFβ luciferase reporter assays. Experiment done in quadruplicate. C. Representative immunofluorescent images of SMAD4 localisation before (time = 0 hr) and after (time = 24 hours) addition of TGFβ. Arrow heads indicate nuclear localisation. D. Quantitation of SMAD4 nuclear translocation following addition of TGFβ. n=5 replicates. E. Representative images of scratch migration assay. F. Quantitation of the relative stimulation of migration of cells into the scratch area following addition of TGFβ. n=3 technical x 3 biological replicates. * statistical difference between +/− TGFβ. # statistical difference between controls and USP9X variant cell lines. n.s.: non-significant difference between controls and USP9X variant cell lines. #* p<0.05 Student’s t-test.
Collectively, these data provide functional support for pathogenicity of three inherited USP9X missense variants p.Ile79Val, p.Ala696Val and p.Ser2233Pro, and reveal the strongest impact of these variants was on neurodevelopmental signalling pathways.
Loss of Usp9x function causes learning and memory deficits in mice
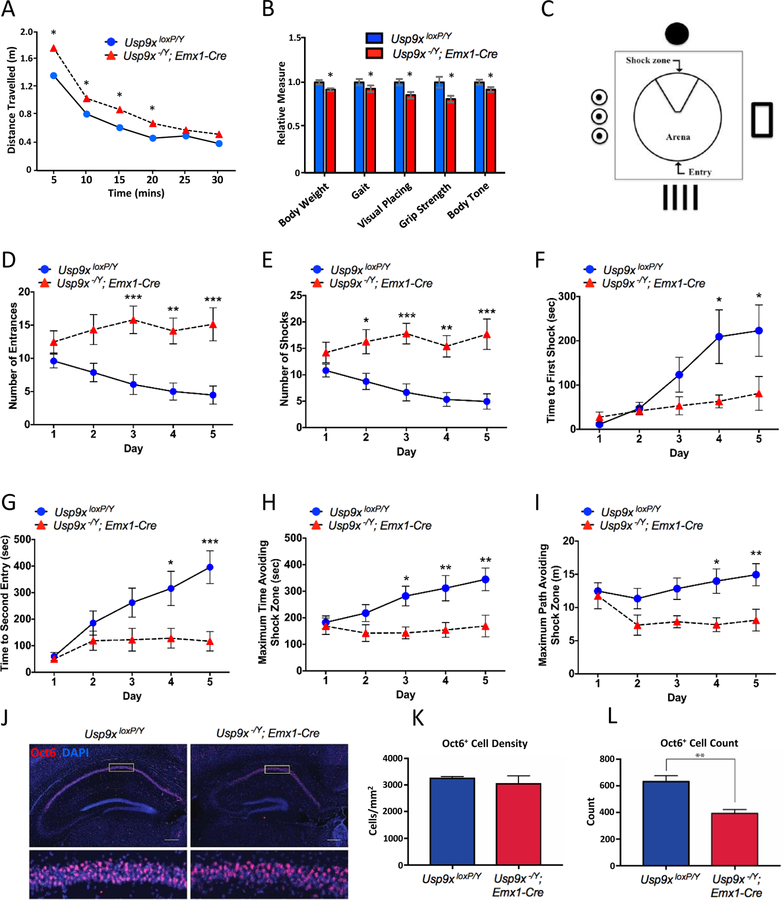
Our collective data on USP9X variants to date in males and females to date suggest partial LOF as the initial molecular driver of the associated pathology. To support this hypothesis, we interrogated the phenotypic consequence of Usp9x deficiency in mice. We mated floxed Usp9x allelic mice with Emx1-Cre driver mice to delete Usp9x in the embryonic forebrain as previously described (17, 18). Unlike in human, Usp9x is subjected to X-inactivation in mouse (35), and as such we forwent studies in heterozygous female offspring and studied hemizygous deletion in males (compared to wildtype male littermates). These mice, herein referred to as knockout mice, survive and provide opportunity to study behaviour. At postnatal day 60, we subjected knockout mice to a broad modified SHIRPA phenotype screen (See Methods, Figure 5A–B and Table S5); (22). Knockout mice displayed a significant increase in distance travelled in an open field test (Figure 5A), and exhibited deficits in weight, gait, grip strength, and visual placing (Figure 5B). Body position was also altered (less likely to be rearing or jumping), whilst no significant difference between control and knockout mice was identified for all other tests (Table S5).
Figure 5. Behavioral deficits in Usp9x knockout mice.
A. Adult Usp9x forebrain-specific knockout mice (Usp9x−/Y; Emx1-Cre) travel further than wildtype littermate controls (Usp9xLoxP/Y) in an open field test. B. Knockout mice also exhibited significant differences in various parameters of the modified SHIRPA neurological screening protocol (also see Supplementary Table 4); * p < 0.05; 2-tailed unpaired t test. C. Schematic of the active place avoidance (APA) arena. D-I. Knockout mice exhibited significantly reduced performance on different aspects of the APA task. Statistics relate to comparisons between wild-type and knockout animals on individual days of the five day test. * p < 0.05, ** p < 0.01, *** p < 0.001; two way ANOVA (also see Figure S16). J-L. Coronal sections of adult wild-type (left 2 panels) and mutant (right two panels) at the level of the hippocampus. OCT6 (red) was used a marker for CA1 hippocampal neurons, and DAPI (blue) was used to label nuclei. Whereas the density of OCT6-expressing neurons was not different between control and mutant animals (K), the total number of OCT6-expressing neurons per CA1 region was reduced within the hippocampus of mutant animals. * p < 0.05; t test. Scale bars in J; 250 μm in low magnification images; 30 μm in high magnification images.
Next we interrogated hippocampal-dependent cognitive function using the Active Place Avoidance (APA) test (23). This test assesses the capacity to learn and remember the position of a fixed shock zone within a rotating platform, using visual cues (Figure 5C). No significant differences in behaviour were observed during the APA habituation Phase (Figure S17). Total distance travelled, and average speed were also comparable over the test period (Figure S17B, C). Knockout mice did however display significantly reduced performance across a variety of test parameters in the APA task, including number of entrances into the shock zone (Figure 5D), total number of shocks (Figure 5E), latency to first shock (a measure of long-term memory; Figure 5F), latency to second entry to the shock zone (a measure of short-term memory; Figure 5G), maximum time and path avoiding the shock zone (Figure 5H–I). Moreover, intra-genotype analyses revealed that wild-type mice showed significant improvements in learning the avoidance task, whilst knockouts did not (Figure S17D–I). As the APA test is highly dependent on the CA1 region of the hippocampus, we assessed CA1 cellular architecture. Analysis revealed reduced total numbers of CA1 neurons in knockout mice, albeit at equivalent density (Figure 5J–L). Collectively, these data show that complete Usp9x LOF severely impacts hippocampal-dependent learning and memory, together with additional (CNS derived) motor, muscular and visual defects in adult mice.
DISCUSSION
Our study redefines the molecular and clinical effect of rare, predicted to be deleterious USP9X variants in males. Through integrated studies of patient-derived cell lines, and with evidence of learning and memory deficits of knockout mice, we conclude that DNA variation in USP9X leading to (partial) LOF has detrimental effects on normal brain development.
Prior to this study, only four male USP9X variants had been associated with pathogenicity in subjects with limited clinical information (4, 5). Here we report an additional 12 likely pathogenic cases. Although further clinically actionable information cannot be solely provided by in silico predictive tools, we discovered and utilised the best USP9X-centric tools to provide support of pathogenicity to around half of our cohort of VUS. This proportion aligned with the prevalence of the clinical attributes as defined by our likely pathogenic cohort. Taken together, our work provides incentive and framework for ongoing clinical and genetic studies towards resolving these cases.
We characterise the USP9X clinical presentation in males. Some features overlap with the more clearly defined female USP9X syndrome, including global developmental delay, ID, hypotonia, motor and speech delay and brain abnormalities including thin corpus callosum and cerebellar defects (3); however, consistent congenital features found in females were rare or absent in males. This difference is likely due to differing molecular consequences acting downstream of the mutation type: In females, mutations cause loss of USP9X dosage (with potential to impact all substrates) compared to missense mutations in males (including inherited through apparently asymptomatic mothers), which involve disruption to particular subsets of substrates (see below). The shared core neurological features between male and female cases do however suggest a convergent mechanism of pathology despite the differing mutation types. Whilst it remains to be tested in females, we speculate disruption to TGFβ as a prime candidate, whereby either reduced USP9X dosage (females) or missense mutation (males) may both culminate in a loss of TGFβ signalling in brain, stemming from a loss of key USP9X substrate(s) involved in signal transduction. Disrupted TGFβ has been implicated in several NDDs (36), and is involved in multiple aspects of brain development and function (37–40). USP9X joins an emerging group of X-linked genes including DDX3X, IQSEC2, KDM5C, SMC1A, ALG13 and OFD1 which (1) escape X-inactivation (2) feature de novo heterozygous LOF mutations in female NDDs, and (3) feature missense variants with milder allelic impact (e.g. partial LOF) in male NDDs, often maternally inherited (41–46).
The hippocampus plays significant roles in learning and memory, and human ID and NDDs, and we discovered hippocampal-dependent learning and memory deficits in Usp9x knockout mice (47–50). Reductions in grip strength, body tone, gait and visual placement also phenocopies hypotonia, motor deficits and visual defects seen in humans. Previous studies revealed several brain malformations in the mouse, which we now show are frequent in male subjects, including agenesis of the corpus callosum, dilated ventricles (ventriculomegaly) and other brain malformations (18). The remarkable phenotypic similarities between human and mouse models have two implications. Firstly they align the mechanism of USP9X missense variation in males with partial LOF. We appreciate that the mouse is a complete LOF model with a comparatively severe phenotype, and analogous germline compete LOF mutations in humans (i.e. hemizygous or homozygous) are not likely compatible with life (2). Nevertheless, the similarities between human and the knockout mice suggest the variants hinder specific USP9X brain functions. Secondly, because a brain LOF mechanism is suggested, we speculate that the cellular and molecular mechanisms resolved in knockout mice may be indeed relevant to human pathology.
Using this same Usp9x knockout mice model, we previously established that loss of Usp9x results in decreased TGFβ-mediated axonogenesis (18), decreased mTOR-mediated neural stem cell proliferation (7) and differentiation defects associated with defective Wnt and Notch signalling (12). As USP9X variant cell lines were refractory to TGFβ stimulation, it’s plausible that loss of axonal tracts (e.g. agenesis of the corpus callosum) stems in part from defective TGFβ signalling. USP9X has several substrates involved in regulating the TGFβ pathway, including SMURF1, SMAD4 and PJA1 (6, 9, 15). In our male subjects, we observed evidence of both down regulation of SMURF1 and loss of nuclear localisation of SMAD4. Both phenomena are consistent with a loss of USP9X interaction and may drive defective TGFβ signalling, but they may alternatively reflect loss of TGFβ signalling stemming from other molecular calamities downstream of USP9X. Indeed, it is intriguing that the variants tested in these assays are located in divergent regions of the protein that are predicted to mediate distinct protein-protein interactions. Thus whilst all tested variants caused a loss of TGFβ signalling, the key substrates driving this effect may be different. We provide evidence that different variants can uniquely impact various USP9X substrate interactions. For example, only the p.Ala696Val variant cell lines had reduced RAPTOR and MCL1 levels, with loss of RAPTOR correlating with evidence of reduced mTOR activity. Thus the p.Ala696Val variant resulted in both defective TGFβ and mTOR signalling, and was associated with the two most severely affected subjects. USP9X has also other substrates that are encoded by genes whose LOF are associated with NDDs (CTNNB1, ITCH, NUAK1, PEX5, SMAD4, SMURF1, DCX, MIB1, SOX2, HERC2, NONO, RPGRIP1L, PRICKLE 1, PRICKLE 2, MTORC1; (1)). Importantly, USP9X functions upstream of all these substrates by maintaining their stability (and hence function) via deubiquitylation, and therefore, any loss of interaction between USP9X has potential to cause the neurodevelopmental pathology associated with that substrate.
USP9X sits at the “hub” of a protein interactome network enriched with NDD genes. It is also known that USP9X regulates processes relevant to NDDs through this NDD network, including neurogenesis, migration, neurite growth and synaptogenesis. Resolving the molecular, cellular and developmental pathologies underpinning USP9X variants is likely to converge on pathologies of NDDs of diverse genetic origins and potentially offer a point for intervention.
Supplementary Material
KEY RESOURCES TABLE
| Resource Type | Specific Reagent or Resource | Source or Reference | Identifiers | Additional Information |
|---|---|---|---|---|
| Add additional rows as needed for each resource type | Include species and sex when applicable. | Include name of manufacturer, company, repository, individual, or research lab. Include PMID or DOI for references; use “this paper” if new. | Include catalog numbers, stock numbers, database IDs or accession numbers, and/or RRIDs. RRIDs are highly encouraged; search for RRIDs at https://scicrunch.org/resources. | Include any additional information or notes if necessary. |
| Antibody | Rabbit anti-USP9X A301–351A | Bethyl Laboratories | A301–351A, RRID:AB_938084 | |
| Bacterial or Viral Strain | ||||
| Biological Sample | ||||
| Cell Line | Human Dermal Fibrobalsts | This Paper | N/A | |
| Chemical Compound or Drug | ||||
| Commercial Assay Or Kit | Dual-Luciferase® Reporter Assay System | Promega | E1910 | |
| Deposited Data; Public Database | ||||
| Genetic Reagent | ||||
| Organism/Strain | Emx1-Cre mice (C57Bl6) | Shigeyoshi Itohara; PMID: 10963597 | N/A | |
| Organism/Strain | Usp9x-LoxP/LoxP mice (129SvJ/C57Bl6) | Stephen Wood; PMID: 23861879 | N/A | |
| Peptide, Recombinant Protein | Human Recombinant TGFβ | R&D Systems | 240-B | |
| Recombinant DNA | ||||
| Sequence-Based Reagent | ||||
| Software; Algorithm | ANNOVAR | Hakonarson, H.; PMID: 20601685 | N/A | |
| Software; Algorithm | ||||
| Transfected Construct | pGL3-CAGA-Luc | Gauthier, JM; PMID: 9606191 | N/A | |
| Other | Proteomics Service | Australian Proteome Analysis Facility (APAF) | N/A |
ACKNOWLEDGEMENTS
We are grateful for the support and contributions of all families involved in this study. We are thankful for the funding received from Creola Pora with the help of her friends and colleagues. This work was supported by SFARI Explorer Grant 527556 to M. Piper, S. Wood and L. Jolly. L. Jolly is supported by Australian Research Council ARC DE160100620. J. Gecz is supported by grants from the National Health and Medical Research Council of Australia: Program Grant (628952) and Research Fellowship (1041920). P. Penzes is supported by National Institute of Health grant R01MH107182. USA 26 was evaluated through the Duke Genome Sequencing Clinic, supported by the Duke University Health system, and partially funded by UCB Celltech. I. Cutcutache, M. Page and M. Armstrong are employees of UCB. C. López-Otín and O. Santiago-Fernández were supported by Programa EDP SOLIDARIA 2016 (Fundación EDP). D. Koboldt, D. Bartholomew and T. Mihalic Mosher were supported by The Research Institute, Nationwide Children’s Hospital. T. M. Pierson was funded by the Cedars-Sinai Diana and Steve Marienhoff Fashion Industries Guild Endowed Fellowship in Pediatric Neuromuscular Diseases and the Undiagnosed Diseases Program. The DDD study presents independent research commissioned by the Health Innovation Challenge Fund [grant number HICF-1009–003], a parallel funding partnership between Wellcome and the Department of Health, and the Wellcome Sanger Institute [grant number WT098051]. The views expressed in this publication are those of the author(s) and not necessarily those of Wellcome or the Department of Health. The study has UK Research Ethics Committee approval (10/H0305/83, granted by the Cambridge South REC, and GEN/284/12 granted by the Republic of Ireland REC). The research team acknowledges the support of the National Institute for Health Research, through the Comprehensive Clinical Research Network. This study makes use of DECIPHER (http://decipher.sanger.ac.uk), which is funded by the Wellcome. Authors from the Undiagnosed Diseases Network include Loren Pena, Vandana Shashi, Kelly Schoch and Jennifer A. Sullivan. We acknowledge the contributions of other collaborating members: Maria T. Acosta, David R. Adams, Aaron Aday, Mercedes E. Alejandro, Patrick Allard, Euan A. Ashley, Mahshid S. Azamian, Carlos A. Bacino, Guney Bademci, Eva Baker, Ashok Balasubramanyam, Dustin Baldridge, Deborah Barbouth, Gabriel F. Batzli, Alan H. Beggs, Hugo J. Bellen, Jonathan A. Bernstein, Gerard T. Berry, Anna Bican, David P. Bick, Camille L. Birch, Stephanie Bivona, Carsten Bonnenmann, Devon Bonner, Braden E. Boone, Bret L. Bostwick, Lauren C. Briere, Elly Brokamp, Donna M. Brown, Matthew Brush, Elizabeth A. Burke, Lindsay C. Burrage, Manish J. Butte, Olveen Carrasquillo, Ta Chen Peter Chang, Hsiao-Tuan Chao, Gary D. Clark, Terra R. Coakley, Laurel A. Cobban, Joy D. Cogan, F. Sessions Cole, Heather A. Colley, Cynthia M. Cooper, Heidi Cope, William J. Craigen, Precilla D’Souza, Surendra Dasari, Mariska Davids, Jean M. Davidson, Jyoti G. Dayal, Esteban C. Dell’Angelica, Shweta U. Dhar, Naghmeh Dorrani, Daniel C. Dorset, Emilie D. Douine, David D. Draper, Annika M. Dries, Laura Duncan, David J. Eckstein, Lisa T. Emrick, Christine M. Eng, Gregory M. Enns, Cecilia Esteves, Tyra Estwick, Liliana Fernandez, Carlos Ferreira, Elizabeth L. Fieg, Paul G. Fisher, Brent L. Fogel, Irman Forghani, Noah D. Friedman, William A. Gahl, Rena A. Godfrey, Alica M. Goldman, David B. Goldstein, Jean-Philippe F. Gourdine, Alana Grajewski, Catherine A. Groden, Andrea L. Gropman, Melissa Haendel, Rizwan Hamid, Neil A. Hanchard, Frances High, Ingrid A. Holm, Jason Hom, Alden Huang, Yong Huang, Rosario Isasi, Fariha Jamal, Yong-hui Jiang, Jean M. Johnston, Angela L. Jones, Lefkothea Karaviti, Emily G. Kelley, David M. Koeller, Isaac S. Kohane, Jennefer N. Kohler, Deborah Krakow, Donna M. Krasnewich, Susan Korrick, Mary Koziura, Joel B. Krier, Jennifer E. Kyle, Seema R. Lalani, Byron Lam, Brendan C. Lanpher, Ian R. Lanza, C. Christopher Lau, Jozef Lazar, Kimberly LeBlanc, Brendan H. Lee, Hane Lee, Roy Levitt, Shawn E. Levy, Richard A. Lewis, Sharyn A. Lincoln, Pengfei Liu, Xue Zhong Liu, Sandra K. Loo, Joseph Loscalzo, Richard L. Maas, Ellen F. Macnamara, Calum A. MacRae, Valerie V. Maduro, Marta M. Majcherska, May Christine V. Malicdan, Laura A. Mamounas, Teri A. Manolio, Thomas C. Markello, Ronit Marom, Martin G. Martin, Julian A. Martínez-Agosto, Shruti Marwaha, Thomas May, Jacob McCauley, Allyn McConkie-Rosell, Colleen E. McCormack, Alexa T. McCray, Jason D. Merker, Thomas O. Metz, Matthew Might, Eva Morava-Kozicz, Paolo M. Moretti, Marie Morimoto, John J. Mulvihill, David R. Murdock, Avi Nath, Stan F. Nelson, J. Scott Newberry, John H. Newman, Sarah K. Nicholas, Donna Novacic, Devin Oglesbee, James P. Orengo, Stephen Pak, J. Carl Pallais, Christina GS. Palmer, Jeanette C. Papp, Neil H. Parker, John A. Phillips III, Jennifer E. Posey, John H. Postlethwait, Lorraine Potocki, Barbara N. Pusey, Genecee Renteri, Chloe M. Reuter, Lynette Rives, Amy K. Robertson, Lance H. Rodan, Jill A. Rosenfeld, Robb K. Rowley, Ralph Sacco, Jacinda B. Sampson, Susan L. Samson, Mario Saporta, Judy Schaechter, Timothy Schedl, Daryl A. Scott, Lisa Shakachite, Prashant Sharma, Kathleen Shields, Jimann Shin, Rebecca Signer, Catherine H. Sillari, Edwin K. Silverman, Janet S. Sinsheimer, Kevin S. Smith, Lilianna Solnica-Krezel, Rebecca C. Spillmann, Joan M. Stoler, Nicholas Stong, David A. Sweetser, Cecelia P. Tamburro, Queenie K.-G. Tan, Mustafa Tekin, Fred Telischi, Willa Thorson, Cynthia J. Tifft, Camilo Toro, Alyssa A. Tran, Tiina K. Urv, Tiphanie P. Vogel, Daryl M. Waggott, Colleen E. Wahl, Nicole M. Walley, Chris A. Walsh, Melissa Walker, Jennifer Wambach, Jijun Wan, Lee-kai Wang, Michael F. Wangler, Patricia A. Ward, Katrina M. Waters, Bobbie-Jo M. Webb-Robertson, Daniel Wegner, Monte Westerfield, Matthew T. Wheeler, Anastasia L. Wise, Lynne A. Wolfe, Jeremy D. Woods, Elizabeth A. Worthey, Shinya Yamamoto, John Yang, Amanda J. Yoon, Guoyun Yu, Diane B. Zastrow, Chunli Zhao and Stephan Zuchner. The principle investigator of the Undiagnosed Disease Network is William Gahl (gahlw@mail.nih.gov).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DISCLOSURES
J. A. Rosenfeld: The Department of Molecular and Human Genetics at Baylor College of Medicine receives revenue from clinical genetic testing conducted at Baylor Genetics Laboratories. L. A. Pérez-Jurado is scientific advisor and founding partner of qGenomics Laboratory. Carlos López-Otín is a scientific advisor and shareholder of DreamGenics Ltd. All other authors report no biomedical financial interests or potential conflicts of interest.
REFERENCES
- 1.Murtaza M, Jolly LA, Gecz J, Wood SA (2015): La FAM fatale: USP9X in development and disease. Cell Mol Life Sci. 72:2075–2089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pantaleon M, Kanai-Azuma M, Mattick JS, Kaibuchi K, Kaye PL, Wood SA (2001): FAM deubiquitylating enzyme is essential for preimplantation mouse embryo development. Mech Dev. 109:151–160. [DOI] [PubMed] [Google Scholar]
- 3.Reijnders MR, Zachariadis V, Latour B, Jolly L, Mancini GM, Pfundt R, et al. (2016): De Novo Loss-of-Function Mutations in USP9X Cause a Female-Specific Recognizable Syndrome with Developmental Delay and Congenital Malformations. Am J Hum Genet. 98:373–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Homan CC, Kumar R, Nguyen LS, Haan E, Raymond FL, Abidi F, et al. (2014): Mutations in USP9X are associated with X-linked intellectual disability and disrupt neuronal cell migration and growth. Am J Hum Genet. 94:470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paemka L, Mahajan VB, Ehaideb SN, Skeie JM, Tan MC, Wu S, et al. (2015): Seizures are regulated by ubiquitin-specific peptidase 9 X-linked (USP9X), a de-ubiquitinase. PLoS Genet. 11:e1005022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal P, Chen YT, Schilling B, Gibson BW, Hughes RE (2012): Ubiquitin-specific peptidase 9, X-linked (USP9X) modulates activity of mammalian target of rapamycin (mTOR). J Biol Chem. 287:21164–21175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bridges CR, Tan MC, Premarathne S, Nanayakkara D, Bellette B, Zencak D, et al. (2017): USP9X deubiquitylating enzyme maintains RAPTOR protein levels, mTORC1 signalling and proliferation in neural progenitors. Sci Rep. 7:391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen X, Zhang B, Fischer JA (2002): A specific protein substrate for a deubiquitinating enzyme: Liquid facets is the substrate of Fat facets. Genes Dev. 16:289–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dupont S, Mamidi A, Cordenonsi M, Montagner M, Zacchigna L, Adorno M, et al. (2009): FAM/USP9x, a deubiquitinating enzyme essential for TGFbeta signaling, controls Smad4 monoubiquitination. Cell. 136:123–135. [DOI] [PubMed] [Google Scholar]
- 10.Mouchantaf R, Azakir BA, McPherson PS, Millard SM, Wood SA, Angers A (2006): The ubiquitin ligase itch is auto-ubiquitylated in vivo and in vitro but is protected from degradation by interacting with the deubiquitylating enzyme FAM/USP9X. J Biol Chem. 281:38738–38747. [DOI] [PubMed] [Google Scholar]
- 11.Overstreet E, Fitch E, Fischer JA (2004): Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development. 131:5355–5366. [DOI] [PubMed] [Google Scholar]
- 12.Premarathne S, Murtaza M, Matigian N, Jolly LA, Wood SA (2017): Loss of Usp9x disrupts cell adhesion, and components of the Wnt and Notch signaling pathways in neural progenitors. Sci Rep. 7:8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Taya S, Yamamoto T, Kanai-Azuma M, Wood SA, Kaibuchi K (1999): The deubiquitinating enzyme Fam interacts with and stabilizes beta-catenin. Genes Cells. 4:757–767. [DOI] [PubMed] [Google Scholar]
- 14.Tseng LC, Zhang C, Cheng CM, Xu H, Hsu CH, Jiang YJ (2014): New classes of mind bomb-interacting proteins identified from yeast two-hybrid screens. PLoS One. 9:e93394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, Avello M, Schirle M, McWhinnie E, Feng Y, Bric-Furlong E, et al. (2013): Deubiquitinase FAM/USP9X interacts with the E3 ubiquitin ligase SMURF1 protein and protects it from ligase activity-dependent self-degradation. J Biol Chem. 288:2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jolly LA, Taylor V, Wood SA (2009): USP9X enhances the polarity and self-renewal of embryonic stem cell-derived neural progenitors. Mol Biol Cell. 20:2015–2029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Oishi S, Premarathne S, Harvey TJ, Iyer S, Dixon C, Alexander S, et al. (2016): Usp9x-deficiency disrupts the morphological development of the postnatal hippocampal dentate gyrus. Sci Rep. 6:25783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stegeman S, Jolly LA, Premarathne S, Gecz J, Richards LJ, Mackay-Sim A, et al. (2013): Loss of Usp9x disrupts cortical architecture, hippocampal development and TGFbeta-mediated axonogenesis. PLoS One. 8:e68287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liang CC, Park AY, Guan JL (2007): In vitro scratch assay: a convenient and inexpensive method for analysis of cell migration in vitro. Nat Protoc. 2:329–333. [DOI] [PubMed] [Google Scholar]
- 20.Jolly LA, Nguyen LS, Domingo D, Sun Y, Barry S, Hancarova M, et al. (2015): HCFC1 loss-of-function mutations disrupt neuronal and neural progenitor cells of the developing brain. Hum Mol Genet. 24:3335–3347. [DOI] [PubMed] [Google Scholar]
- 21.Harris L, Dixon C, Cato K, Heng YH, Kurniawan ND, Ullmann JF, et al. (2013): Heterozygosity for nuclear factor one x affects hippocampal-dependent behaviour in mice. PLoS One. 8:e65478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers DC, Fisher EM, Brown SD, Peters J, Hunter AJ, Martin JE (1997): Behavioral and functional analysis of mouse phenotype: SHIRPA, a proposed protocol for comprehensive phenotype assessment. Mamm Genome. 8:711–713. [DOI] [PubMed] [Google Scholar]
- 23.Stuchlik A, Petrasek T, Prokopova I, Holubova K, Hatalova H, Vales K, et al. (2013): Place avoidance tasks as tools in the behavioral neuroscience of learning and memory. Physiol Res. 62 Suppl 1:S1–S19. [DOI] [PubMed] [Google Scholar]
- 24.Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, et al. (2009): DECIPHER: Database of Chromosomal Imbalance and Phenotype in Humans Using Ensembl Resources. Am J Hum Genet. 84:524–533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. (2015): Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 17:405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Q, Dong A, Walker JR, Bountra C, Arrowsmith CH, Edwards AM, et al. (2018): Crystal structure of a peptidase. http://www.rcsb.org/structure/5WCH.
- 27.Wang K, Li M, Hakonarson H (2010): ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38:e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M (2019): CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47:D886–D894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pejaver V, Urresti J, Lugo-Martinez J, Pagel KA, Lin GN, Nam H, et al. (2019): MutPred2: inferring the molecular and phenotypic impact of amino acid variants. bioRxiv. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ebisawa T, Fukuchi M, Murakami G, Chiba T, Tanaka K, Imamura T, et al. (2001): Smurf1 interacts with transforming growth factor-beta type I receptor through Smad7 and induces receptor degradation. J Biol Chem. 276:12477–12480. [DOI] [PubMed] [Google Scholar]
- 31.Wiese KE, Nusse R, van Amerongen R (2018): Wnt signalling: conquering complexity. Development. 145. [DOI] [PubMed] [Google Scholar]
- 32.Crino PB (2016): The mTOR signalling cascade: paving new roads to cure neurological disease. Nat Rev Neurol. 12:379–392. [DOI] [PubMed] [Google Scholar]
- 33.Vucic D, Dixit VM, Wertz IE (2011): Ubiquitylation in apoptosis: a post-translational modification at the edge of life and death. Nat Rev Mol Cell Biol. 12:439–452. [DOI] [PubMed] [Google Scholar]
- 34.Schiller M, Javelaud D, Mauviel A (2004): TGF-beta-induced SMAD signaling and gene regulation: consequences for extracellular matrix remodeling and wound healing. J Dermatol Sci. 35:83–92. [DOI] [PubMed] [Google Scholar]
- 35.Berletch JB, Ma W, Yang F, Shendure J, Noble WS, Disteche CM, et al. (2015): Escape from X inactivation varies in mouse tissues. PLoS Genet. 11:e1005079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kashima R, Hata A (2018): The role of TGF-beta superfamily signaling in neurological disorders. Acta Biochim Biophys Sin (Shanghai). 50:106–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bialas AR, Stevens B (2013): TGF-beta signaling regulates neuronal C1q expression and developmental synaptic refinement. Nat Neurosci. 16:1773–1782. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Gomes FC, Sousa Vde O, Romao L (2005): Emerging roles for TGF-beta1 in nervous system development. Int J Dev Neurosci. 23:413–424. [DOI] [PubMed] [Google Scholar]
- 39.Caraci F, Gulisano W, Guida CA, Impellizzeri AA, Drago F, Puzzo D, et al. (2015): A key role for TGF-beta1 in hippocampal synaptic plasticity and memory. Sci Rep. 5:11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fukushima T, Liu RY, Byrne JH (2007): Transforming growth factor-beta2 modulates synaptic efficacy and plasticity and induces phosphorylation of CREB in hippocampal neurons. Hippocampus. 17:5–9. [DOI] [PubMed] [Google Scholar]
- 41.Snijders Blok L, Madsen E, Juusola J, Gilissen C, Baralle D, Reijnders MR, et al. (2015): Mutations in DDX3X Are a Common Cause of Unexplained Intellectual Disability with Gender-Specific Effects on Wnt Signaling. Am J Hum Genet. 97:343–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoubridge C, Harvey RJ, Dudding-Byth T (2019): IQSEC2 mutation update and review of the female-specific phenotype spectrum including intellectual disability and epilepsy. Hum Mutat. 40:5–24. [DOI] [PubMed] [Google Scholar]
- 43.Fieremans N, Van Esch H, de Ravel T, Van Driessche J, Belet S, Bauters M, et al. (2015): Microdeletion of the escape genes KDM5C and IQSEC2 in a girl with severe intellectual disability and autistic features. Eur J Med Genet. 58:324–327. [DOI] [PubMed] [Google Scholar]
- 44.Jansen S, Kleefstra T, Willemsen MH, de Vries P, Pfundt R, Hehir-Kwa JY, et al. (2016): De novo loss-of-function mutations in X-linked SMC1A cause severe ID and therapy-resistant epilepsy in females: expanding the phenotypic spectrum. Clin Genet. 90:413–419. [DOI] [PubMed] [Google Scholar]
- 45.Epi KC, Epilepsy Phenome/Genome P, Allen AS, Berkovic SF, Cossette P, Delanty N, et al. (2013): De novo mutations in epileptic encephalopathies. Nature. 501:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sakakibara N, Morisada N, Nozu K, Nagatani K, Ohta T, Shimizu J, et al. (2019): Clinical spectrum of male patients with OFD1 mutations. J Hum Genet. 64:3–9. [DOI] [PubMed] [Google Scholar]
- 47.Bostrom C, Yau SY, Majaess N, Vetrici M, Gil-Mohapel J, Christie BR (2016): Hippocampal dysfunction and cognitive impairment in Fragile-X Syndrome. Neurosci Biobehav Rev. 68:563–574. [DOI] [PubMed] [Google Scholar]
- 48.Penzes P, Buonanno A, Passafaro M, Sala C, Sweet RA (2013): Developmental vulnerability of synapses and circuits associated with neuropsychiatric disorders. J Neurochem. 126:165–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nadel L (2003): Down’s syndrome: a genetic disorder in biobehavioral perspective. Genes Brain Behav. 2:156–166. [DOI] [PubMed] [Google Scholar]
- 50.Deboer T, Wu Z, Lee A, Simon TJ (2007): Hippocampal volume reduction in children with chromosome 22q11.2 deletion syndrome is associated with cognitive impairment. Behav Brain Funct. 3:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.