Abstract
Background
The field of psychiatry has long sought biomarkers that can objectively diagnose patients, predict treatment response, or identify individuals at risk of illness onset. However, reliable psychiatric biomarkers have yet to emerge. The recent application of machine learning techniques to develop neuroimaging-based biomarkers has yielded promising preliminary results. However, much of the work in this domain has not met best practice standards from the field of machine learning. This is especially true for studies of anxiety, creating uncertainty about the potential for anxiety biomarker development.
Methods
We applied machine learning tools to predict trait anxiety from neuroimaging measurements in humans. Using publicly available data from the Brain Genomics Superstruct Project, we compared a suite of neuroimaging-based machine learning models predicting anxiety within a discovery sample (n=531, 307 female) via k-fold cross-validation, and we tested the final model (a stacked model incorporating region-to-region functional connectivity, amygdala seed-to-voxel connectivity, and volumetric and cortical thickness data) in a held out, unseen test sample (n=348, 209 female).
Results
Though the best model was able to predict anxiety within the discovery sample (cross-validated R2 of .06, permutation test p<.001), the generalization test within the holdout sample failed (R2 of −.04, permutation test p>.05).
Conclusions
In this study, we did not find evidence of a generalizable anxiety biomarker. However, we encourage other researchers to investigate this topic, utilizing large samples and proper methodology, in order to clarify the potential of neuroimaging-based anxiety biomarkers.
Keywords: anxiety, biomarker, machine learning, fMRI, predictive modeling, functional connectivity
Introduction
Biomarkers are objective, reproducible biological measures of medical state (1). Biomarkers can perform an invaluable function by informing treatment plans or indicating the presence, prognosis, or risk level of disease. For example, doctors test for elevated cardiac troponin to assess whether a heart attack has occurred (2) and determine treatment course (3). AIDS is defined by CD4 (T cell) count, (4) and CD4 count is used to gauge opportunistic infection risk (5). The identification of psychiatric biomarkers ready for use in clinical practice has been elusive. Recently, there has been a focus on developing psychiatric biomarkers from neuroimaging data. Thousands of studies have aimed to identify brain-based differences between patients with mental illnesses and mentally healthy patients, and authors often speculate that the neural differences identified could form the basis of a biomarker. However, clinically useful neuroimaging-derived biomarkers have not emerged (6–9).
It has been argued that progress in neuroscience, psychology, and psychiatry could be advanced by placing more emphasis on prediction, rather than explanation alone (8–12). Psychiatric neuroscience has tended to prioritize explanation by favoring theory-driven, tightly controlled studies (often with small samples). Understanding the mechanisms of a psychiatric disorder would clearly advance the ability to develop tests and treatments. However, maximizing prediction of a clinical variable is not usually an explicit goal of these studies. A more direct emphasis on prediction of clinical outcomes may speed psychiatric translation and engender reproducible science.
Traditionally, neuroimaging studies aiming to identify biomarkers tested for significant differences in the means of populations with and without the disorder in a given measure(s) like amygdala-PFC connectivity, or BOLD signal in individual voxels during a task. This work has provided a foundational step in highlighting the areas of the brain where the differences between patients and non-patients are most striking. However, a significant difference in mean between populations does not indicate that an individual could be classified with meaningful accuracy on the basis of that variable in isolation. There may be enough overlap between the populations that one cannot predict illness status with sufficient reliability (12–17).
Recently, there has been interest in using machine learning to develop biomarkers, as machine learning provides many tools that can complement traditional statistical approaches. In machine learning, prediction is typically valued over explanation. In this paper we focus on supervised machine learning models. A model, in this context, is a function that transforms input variables, or “features,” as they are referred to in machine learning, into a prediction of a “target variable,” like patient group or symptom score, by learning the relationship between the features and target variable. Machine learning models are typically multivariate—they leverage the combined effects of many variables to predict group membership, potentially allowing for greater predictive power than any individual predictor (18, 19).
Researchers have applied machine learning tools to neuroimaging measurements in order to differentiate patients and controls, with promising but mixed success (15, 20). We identified 23 papers that have used machine learning to predict anxiety status or traits from neuroimaging data (selection criteria in the Supplement, study characteristics in Table S1) (21–42). Most studies classifying patients versus controls reported accuracies of over 80%, and some over 90%. Below we review methodological considerations important to the interpretation of these studies.
Training and testing a model on different participants, in order to account for overfitting, is fundamental to the practice of machine learning (18, 19). Overfitting is the phenomenon that a model may be able to predict the target variable from the training data (data used to learn the parameters of the model) very well, even perfectly, but fail to perform well on novel examples (test data) that were not used to train the model. It is necessary to apply the model to previously unseen test data to evaluate its performance. This can be done with cross-validation, where the data are iteratively split into training and test sets, with training data used to fit the model and test data used to evaluate it. The 23 studies of anxiety neuromarkers reviewed tended to use cross-validation to assess predictive performance. However, to assess the model’s generalizability, it is crucial, especially if multiple models or variants of the analysis are tested with cross-validation, to additionally test the final model on a completely held out dataset, which only two of the reviewed studies did (25, 40). If the researchers use cross-validation multiple times on the same dataset to assess different types of classifiers, different feature types, or different model hyperparameters (without nested cross-validation), and pick the best result to include or emphasize in the manuscript, the cross-validation accuracy is no longer an unbiased estimate of generalization performance. Stated differently, the researcher risks overfitting via the model selection process (9, 15, 16, 43). There are many examples within the broader field of machine learning-based psychiatric neuroimaging where performance on held out datasets was substantially worse than that obtained by internal cross-validation, suggesting that it is dangerous to assume cross-validation accuracy is an unbiased assessment of how the model will perform on new data (25, 44–46). Using cross-validation alone to assess models is risky because cross-validation accuracy is a quite variable estimator of generalization, particularly with small samples (47). So while cross-validation or some initial validation is a necessary first step for any machine learning model, it is strongly recommended to additionally test the model on a dataset that has been completely held out throughout the analysis process (ideally, an external dataset). However, likely due to the high cost of acquiring data, most of the studies reviewed did not perform a holdout test.
Another limitation of the 23 reviewed studies is small sample size. With the exception of 3 studies that used the Human Connectome Project (HCP) sample (48), none of the studies had an n>181, and most had fewer than 100 participants. The machine learning literature emphasizes the importance of a large sample size. The amount of data available to a model often (but not always) has more influence on success than algorithm choice (49). Data are more easily overfit when the sample size is small, and this includes “procedural” overfitting by testing multiple methods with cross-validation. In neuroimaging datasets, the number of features typically vastly outnumbers the number of subjects, which can also make models more prone to overfitting (10, 43, 47). Due to technical and cultural changes within the field of neuroscience (50, 51), there has been a shift towards collection of large, cross-site neuroimaging datasets and concatenation of existing datasets (e.g., 48, 52–55). This commendable effort will likely be crucial in the emergence of biomarkers. Of course, “large” and “small” are relative terms—machine learning applications in natural language processing or image processing often involve millions (or more!) of samples (56), but by neuroimaging standards, an N approaching or exceeding 1000 is considered large (57).
Three studies that explored trait anxiety prediction utilized the HCP sample (40–42), the largest sample yet studied to answer this question. Two of the three reported the ability to reliably predict anxious personality/neuroticism. Though the HCP studies make unique contributions to the literature, they should not be considered independent evidence because they use the same sample. One study (40) took a particularly compelling approach by performing holdout tests (some successful) with 2 independent datasets.
The present study pursued the development of a neuroimaging-based anxiety biomarker with machine learning tools, utilizing a large sample and testing the proposed model on a completely held out dataset. As reviewed above, this question has only been addressed with one large sample previously. Additionally, nearly all prior reports on this topic have relied solely on cross-validation, and it is unclear whether results will generalize to truly unseen samples. We tested whether trait anxiety could be predicted from neuroimaging measurements with a suite of machine learning algorithms, using a large, publicly available sample. We first considered models of whole-brain, region-to-region resting-state functional connectivity data. Subsequently, we explored the utility of adding grey matter volumetric/thickness measurements and region-to-voxel connectivity data as features. We performed all model comparison within a discovery sample, and tested our final model on a held out dataset. A dimensional approach to studying behavioral systems relevant to psychiatric disorders may be fruitful in linking biology and mental illness (58–60); thus we chose to predict variation in anxiety in a non-clinical sample. We note that our model does not produce true “predictions”, in the sense that the target variable was a measure of current anxiety rather than future anxiety. However, we view this study as an important analytical stepping-stone in the development of pragmatic clinical tools—if challenges arise in predicting current anxiety, the same challenges may be present in predicting future anxiety.
Methods and Materials
Dataset
The data are from the Brain Genomics Superstruct Project (GSP), a large-scale, multi-site brain imaging project (55). The publically released GSP dataset consists of resting-state fMRI and structural MRI scans of 1570 participants. Self-report and behavioral data are available for a subset of participants (n=926). The Supplement details motion- and coverage-related exclusions (47 participants). Data collection and sharing were approved by the Partners Health Care Institutional Review Board and the Harvard University Committee on the Use of Human Subjects in Research.
Data were first split into a discovery sample (n=531 after exclusions), and a final model evaluation sample (referred to as the “holdout” sample, n=348 after exclusions). The holdout neuroimaging data were sequestered (not downloaded) until the final model was tested. The two samples did not differ in age, sex, level of education, estimated IQ, anxiety score, number of runs, motion statistics, site of acquisition, and console software (ps>.32).
Target variable
The target variable was a composite anxiety score derived from several questionnaires administered through an online battery (55). We used a composite score with the rationale that it would be more stable and less idiosyncratic than any individual anxiety-related scale. Four anxiety-related questionnaires were collected for all participants: the Spielberger Trait Anxiety Inventory (61), the NEO-neuroticism scale (62), the Behavioral Inhibition Scale (63), and the Harm Avoidance scale from the Temperament and Character Inventory (64). The composite anxiety score was derived (following 65) by z-scoring each of these 4 scales across participants and taking the mean of these 4 z-scores per participant. In computing the composite anxiety scores for the holdout sample, we performed z-score transformations based on the means and standard deviations of the discovery sample. In the Supplement, we report results for individual scales for the best performing model.
Neuroimaging data collection and preprocessing
Imaging sequences are described in the Supplement. FMRI data were preprocessed with FMRIB Software Library (FSL) 5.0.9 (66) and FreeSurfer 6.0 (http://surfer.nmr.mgh.harvard.edu/) using standard methods for resting-state functional connectivity analysis (see Supplement). Structural data was processed, and region-wise volume and cortical thickness measurements were extracted in FreeSurfer (see Supplement).
Functional connectivity measures
In the model building/selection phase, working with the discovery sample, we evaluated 6 methods for parceling the brain into regions from which to derive connectivity measurements. Table S2 lists the 6 parcellations evaluated. The best performing method in the discovery sample was the FreeSurfer segmentation (67, 68).
To construct region-to-region connectivity features, we extracted the mean BOLD timecourse from each region in the parcellation. We computed the Pearson’s correlation in signal between each pair of regions (transformed with Fisher’s r-to-z). These z-values were used as features in the models.
Some of the stacked models utilized voxelwise connectivity maps. Each voxel’s connectivity to a given seed region was used as a feature. These maps were generated with FSL using FreeSurfer-defined seed regions. See the Supplement for additional details on functional connectivity measures.
Modeling/model selection
Modeling was carried out in python with the scikit-learn package (69). We used R2, calculated on the test data, as an evaluation metric (see Supplement for discussion of R2).
Within the discovery sample, each model was constructed and evaluated with stratified k-fold cross-validation (k=6). For several of the models tested, we tuned a hyperparameter of the model with nested cross-validation (see Supplement for further description of modeling, cross-validation and hyperparameter tuning). Various model classes were evaluated in the discovery sample, including Ridge Regression, Lasso Regression, Partial Least Squares Regression, Principal Components Regression, Random Forest Regression, Support Vector Regression with a linear or polynomial kernel, Relevance Vector Regression, and the “Connectome-based Predictive Modeling” approach (70–72). We also attempted to specifically replicate methods from prior trait anxiety prediction studies (see Supplement). Table S3 lists models tested and hyperparameters tuned.
Several models evaluated in the discovery sample were stacked models. Model stacking is a method of combining predictions from several models (referred to as base models), by building a model where the predictions of base models serve as features. We combined models that made predictions based on different data sources, such as region-to-region connectivity, structural MRI data, and voxelwise connectivity data (see Supplement and Figure S1 for detailed explanation of model stacking.)
To assess the significance of the R2 observed in the best model (the model with the highest R2), we used permutation testing (see Supplement).
To address the possibility that the model could be learning to predict some confound rather than anxiety scores, we performed a control analysis in the discovery sample with the best model, in which we regressed out potential confounds from the features. We also performed an analysis on censored data (see Supplement).
We tested whether the model that had performed best in the discovery sample could predict anxiety in the holdout sample. We retrained this best performing model using the entire discovery sample, and generated predictions of this model for the unseen holdout data. We computed the R2 with these holdout predictions, and assessed significance with a permutation test.
Data and code availability
The imaging data are publically available at (http://neuroinformatics.harvard.edu/gsp/). Some data, including anxiety-related questionnaires, require approval for access (instructions: http://neuroinformatics.harvard.edu/gsp/get). Code used to run the analyses is available upon request.
Results
Sample Characteristics
Across the discovery and holdout samples, participants had a mean age of 21.59 (SD 2.87) ranging from 18–35. Age in the public GSP release is binned by 2 years to protect the privacy of participants, so the mean and SD are not exact. The sample was 59% female. To illustrate the range of anxiety-like phenotypes present in the sample, Figure 1 shows a histogram of scores on the Spielberger Trait Anxiety Index. For reference, this figure illustrates the mean Spielberger Trait Anxiety scores of patient samples with anxiety disorders or PTSD in recent studies. It is apparent from the figure that the GSP participants are not from a clinical sample, but some participants do show levels of anxiety comparable to patient populations.
Figure 1. Sample heterogeneity in trait anxiety.
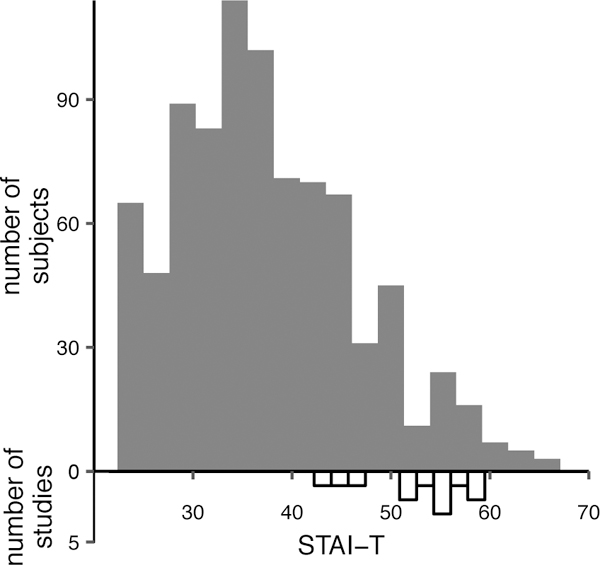
Distribution of Spielberger Trait Anxiety Index (STAI-T) in the complete sample analyzed in this paper (n=879), and in recent clinical studies of anxiety (n=12). Filled bars show the scores in the current sample. Empty bars show mean scores of samples with anxiety disorders or PTSD from recent studies (85–96).
Discovery sample model performance
The model producing the greatest cross-validated R2 (within the discovery sample) was a stacked model that incorporated region-to-region connectivity data, volumetric/cortical thickness data, and voxelwise bilateral amygdala connectivity data, model 1 from Table S3. The regions used as seeds for the region-to-region and amygdala voxelwise connectivity features came from the FreeSurfer segmentation. This model resulted in a cross-validated R2 of .06. Figure 2A shows the relationship between actual anxiety scores and predicted anxiety scores. This level of performance significantly exceeded chance levels (p<.001, as determined by a permutation test, Figure 2B.) Performance of other models tested in the discovery sample is summarized in Table S3. Anxiety could also be predicted from censored connectivity data, and data that had undergone confound regression (see Supplement).
Figure 2. Model performance in the discovery sample.

A. Actual anxiety scores plotted against predicted anxiety scores, in the discovery sample. Model predictions are from the best performing model, model 1 (see Table S3). B. Empirical null distribution of R2 generated in permutation test, in the discovery sample. The dotted black line shows the 95% confidence bound. The solid red line shows the actual R2 of the model using the (unscrambled) data.
Holdout sample test
We tested the best model in the holdout sample after training it with the entire discovery sample. The R2 in the holdout sample was −.04, which failed to achieve significance with a permutation test (Figure 3).
Figure 3. Model performance in the holdout sample.

A. Actual anxiety scores plotted against predicted anxiety scores, in the holdout sample. B. Empirical null distribution of R2 generated in permutation test, in the holdout sample. The dotted black line shows the 95% confidence bound. The solid red line shows the actual R2 of the model using the (unscrambled) data.
One possible explanation for the poor performance in the holdout dataset is that by testing so many models within the discovery sample (see Table S3), we may have overfit the model to the discovery sample through the model selection process. To attempt to understand the reason for the generalization failure, we tested this hypothesis by examining whether a model that was tested early in the model comparison process (with an R2 of .03 in the discovery sample), a ridge regression from region-to-region connectivity data (model 4 from Table S3), outperformed the final model. This ridge model that had been tested early in the model exploration process did not outperform the final model (R2 of −.06 in the holdout sample).
Discussion
In this study, we attempted to predict trait anxiety in a large sample by applying machine learning tools to multimodal neuroimaging data. The best model (determined in the discovery sample) was a stacked model, with three ridge regression base models which used different data sources as features: whole brain region-to-region connectivity data, amygdala seed-to-voxel connectivity data, and grey matter volumetric/thickness data. This model significantly predicted anxiety scores in the discovery sample as assessed by cross-validation, but when tested on a previously unseen holdout sample, it did not successfully predict anxiety scores. Our ability to predict anxiety within the discovery sample is consistent with prior work (21–41). However, when we tested generalizability to a holdout sample, a step most previous studies did not take, the model failed to make accurate out-of-sample predictions. Thus, our findings do not support the hypothesis that anxiety is predictable from neural measurements.
We studied a limited set of brain phenotypes and applied a circumscribed set of approaches. Our study should be considered a proof-of-concept for evaluating relations linking brain functions to behavior rather than decisively addressing the full range of possible associations between neuroimaging measures and anxiety. There are multiple possible explanations for why we were ultimately unable to predict anxiety scores.
One set of possible reasons for our failure to predict anxiety relates to the anxiety phenotype examined. We did not analyze a clinical sample, so perhaps there was not sufficient clinical heterogeneity for the model to learn to make accurate predictions. As shown in Figure 1, there were participants with anxiety scores close to the mean scores of anxiety patients in clinical studies, but the number of participants with scores in this range was relatively small, and perhaps insufficient to train the model. Though the sample used here was large in comparison to those from most fMRI studies, larger samples exist, such as the UK Biobank sample (52), which includes over 10,000 subjects. Prediction might be improved in these larger samples, as the quantity of training data is an important determinant of model performance, and as there might be more subjects in a high anxiety range to inform the model. We encourage other researchers to investigate this question in large open-access samples. However, despite the lack of participants in this high range in the GSP sample, there is still substantial variability, and one would expect that this healthy heterogeneity would be predictable from neural measurements. Additionally, anxiety scores were only assessed once. Multiple assessments might yield a more stable estimate of the phenotype and improve prediction. It has been suggested that the non-biological nature of current diagnostic categories has stymied progress in identifying biological mechanisms of these disorders (6). This argument can be made about continuous variables as well—a measurement may not “carve nature at the joints.” It is likely that our anxiety measure does not reflect a single process; relatedly, two individuals could have the same elevated anxiety score with different underlying brain mechanisms, and this may impair prediction of the score.
Feature-related issues could also have impaired prediction. One limitation to note is that the imaging sequences used lack the spatial and temporal precision of current approaches (data collection began in 2008). It is possible that with more state-of-the-art sequences, prediction would be facilitated. Relatedly, each subject had 6–12 minutes of resting-state data, but recent studies have suggested that substantially improved reliability of connectivity estimates can be obtained with ~15–25 minutes of data (73–75). Others have recently shown that the inclusion of task-based fMRI data can improve connectivity estimates and predictive performance from connectivity data (76, 77). A limitation of the current paper is the unavailability of longer resting-state scans and task-based data in these subjects.
Model selection-related issues could also underlie our failure to predict anxiety in the holdout set. One possibility is that a model exists that could successfully predict anxiety from the measurements obtained, but we did not identify it. However, in the discovery sample, we tested a large range of both linear and non-linear models in combination with different parcellations for extracting connectivity features (see Table S2). Conversely, another concern we had was that we may have tested too many models in the discovery sample, leading to an overfitting of the model selection process to the discovery sample. In other words, a model tested early in the exploration of the discovery sample may have actually outperformed the chosen model when tested on the holdout sample, despite performing worse on the discovery sample. To investigate whether this could be the case, we performed a supplementary test in the holdout sample of a model that had been tested early in the model comparison process. However, this earlier model also failed to accurately predict anxiety scores in the holdout sample. This failure did not support the explanation that we could have obtained better holdout performance had we stopped the model testing in the discovery sample sooner. It does, however, allow for the possibility that anxiety was not predictable from the measurements obtained, but the good performance in the discovery sample was illusory and due to procedural overfitting.
We close by providing suggestions on how to proceed with research on neuroimaging-based psychiatric biomarker development, given our observations in the current study. Previous anxiety biomarker research has tended to use small samples (only one large-scale sample investigated previously (40–42), with mixed results) and evaluate models with cross-validation only. As demonstrated here, it is possible to achieve promising results via internal cross-validation that do not generalize to a held out sample. We therefore recommend that future studies utilize large samples, and test their models on truly unseen holdout data. Heterogeneity in preprocessing and statistical approaches creates problems for interpreting and replicating traditional neuroimaging analyses, but machine learning-based neuroimaging studies arguably suffer from these issues even more. The number of possible models from which to choose is large, methods for assessing generalization vary, and standards of reporting/visualizing feature importance (which also depend on which model is used) are undefined. We therefore recommend further research on methods development that can illuminate best practices (e.g., 78), and that studies attempt to replicate the specific methods of other studies. The continued acquisition of new large samples will also undoubtedly be crucial to biomarker development. One difficulty in this field is that the phenotypes we want to predict may be multidimensional and may not derive from a single biological mechanism (6, 79). We see promise in applying unsupervised machine learning methods to biomarker development, as these methods may circumvent this issue (although see 80). Another class of methods that could help with this issue is multi-output learning, where multiple phenotypes are predicted with the same model (81). This methodology takes advantage of relationships between different target variables (possibly helping to disambiguate cases where subjects have the same anxiety score with different underlying mechanisms) and has been shown to improve upon single-output model predictions (81). Lastly, we note that in our comparisons of models within the discovery sample, a stacked model that combined predictions from multimodal data performed best. Though we interpret this result with caution, as this model did not ultimately successfully predict anxiety, the result is consistent with other neuroimaging biomarker studies suggesting that stacked multimodal models outperform non-stacked models (81, 82).
The potential to develop neuroimaging biomarkers for anxiety is unclear, but some research suggests that success is possible. In this study, we were unable to find evidence of a generalizable anxiety biomarker. Though this research area is proving challenging, some encouraging results have emerged. Outside the field of psychiatry, there have been successful attempts at producing generalizable neuromarkers of psychological states and traits (71, 83, 84). Given the potential of biomarkers to revolutionize psychiatry, it is important to rigorously explore their possible development and application.
Supplementary Material
Acknowledgements
Data were provided by the Brain Genomics Superstruct Project of Harvard University and the Massachusetts General Hospital, (Principal Investigators: Randy Buckner, Joshua Roffman, and Jordan Smoller), with support from the Center for Brain Science Neuroinformatics Research Group, the Athinoula A. Martinos Center for Biomedical Imaging, and the Center for Human Genetic Research. 20 individual investigators at Harvard and MGH generously contributed data to the overall project. This work was supported by R01 DA042855 to EAP. The authors would like to thank Annabel Boeke for suggesting examples of non-psychiatric biomarkers.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors declare no biomedical financial interests or potential conflicts of interest.
References
- 1.Strimbu K, Tavel JA (2010): What are biomarkers? Current Opinion in HIV and AIDS 5:463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jneid H, Anderson JL, Wright RS, Adams CD, Bridges CR, Casey DE, et al. (2012): 2012 ACCF/AHA focused update of the guideline for the management of patients with unstable angina/non–ST-elevation myocardial infarction (updating the 2007 guideline and replacing the 2011 focused update): a report of the American College of Cardiology Foundation/American Heart Association Task Force on Practice Guidelines. Journal of the American College of Cardiology 60:645–681. [DOI] [PubMed] [Google Scholar]
- 3.Morrow DA, Cannon CP, Rifai N, Frey MJ, Vicari R, Lakkis N, et al. (2001): Ability of minor elevations of troponins I and T to predict benefit from an early invasive strategy in patients with unstable angina and non-ST elevation myocardial infarction: results from a randomized trial. Jama 286:2405–2412. [DOI] [PubMed] [Google Scholar]
- 4.Selik RM, Mokotoff ED, Branson B, Owen SM, Whitmore S, Hall HI (2014): Revised surveillance case definition for HIV infection—United States, 2014. Morbidity and Mortality Weekly Report: Recommendations and Reports 63:1–10. [PubMed] [Google Scholar]
- 5.Phair J, Muñoz A, Detels R, Kaslow R, Rinaldo C, Saah A, et al. (1990): The risk of Pneumocystis carinii pneumonia among men infected with human immunodeficiency virus type 1. New England Journal of Medicine 322:161–165. [DOI] [PubMed] [Google Scholar]
- 6.Kapur S, Phillips AG, Insel TR (2012): Why has it taken so long for biological psychiatry to develop clinical tests and what to do about it? Molecular psychiatry 17:1174. [DOI] [PubMed] [Google Scholar]
- 7.Castellanos FX, Di Martino A, Craddock RC, Mehta AD, Milham MP (2013): Clinical applications of the functional connectome. Neuroimage 80:527–540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Paulus MP (2015): Pragmatism instead of mechanism: a call for impactful biological psychiatry. JAMA psychiatry 72:631–632. [DOI] [PubMed] [Google Scholar]
- 9.Gillan CM, Whelan R (2017): What big data can do for treatment in psychiatry. Current Opinion in Behavioral Sciences 18:34–42. [Google Scholar]
- 10.Yarkoni T, Westfall J (2017): Choosing prediction over explanation in psychology: Lessons from machine learning. Perspectives on Psychological Science 12:1100–1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenberg MD, Casey B, Holmes AJ (2018): Prediction complements explanation in understanding the developing brain. Nature communications 9:589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bzdok D, Meyer-Lindenberg A (2018): Machine learning for precision psychiatry: opportunities and challenges. Biological Psychiatry: Cognitive Neuroscience and Neuroimaging 3:223–230. [DOI] [PubMed] [Google Scholar]
- 13.Doyle OM, Mehta MA, Brammer MJ (2015): The role of machine learning in neuroimaging for drug discovery and development. Psychopharmacology 232:4179–4189. [DOI] [PubMed] [Google Scholar]
- 14.Orru G, Pettersson-Yeo W, Marquand AF, Sartori G, Mechelli A (2012): Using support vector machine to identify imaging biomarkers of neurological and psychiatric disease: a critical review. Neuroscience & Biobehavioral Reviews 36:1140–1152. [DOI] [PubMed] [Google Scholar]
- 15.Woo C-W, Chang LJ, Lindquist MA, Wager TD (2017): Building better biomarkers: brain models in translational neuroimaging. Nature neuroscience 20:365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reddan MC, Lindquist MA, Wager TD (2017): Effect size estimation in neuroimaging. JAMA psychiatry 74:207–208. [DOI] [PubMed] [Google Scholar]
- 17.Holmes AJ, Patrick LM (2018): The myth of optimality in clinical neuroscience. Trends in Cognitive Sciences [DOI] [PMC free article] [PubMed]
- 18.Hastie T, Tibshirani R, Friedman J (2009): The elements of statistical learning: Data Mining, Inference, and Prediction Second Edition. New York, NY: Springer Series in Statistics. [Google Scholar]
- 19.Domingos P (2012): A few useful things to know about machine learning. Communications of the ACM 55:78–87. [Google Scholar]
- 20.Wolfers T, Buitelaar JK, Beckmann CF, Franke B, Marquand AF (2015): From estimating activation locality to predicting disorder: a review of pattern recognition for neuroimaging-based psychiatric diagnostics. Neuroscience & Biobehavioral Reviews 57:328–349. [DOI] [PubMed] [Google Scholar]
- 21.Liu F, Xie B, Wang Y, Guo W, Fouche J-P, Long Z, et al. (2015): Characterization of post-traumatic stress disorder using resting-state fMRI with a multi-level parametric classification approach. Brain topography 28:221–237. [DOI] [PubMed] [Google Scholar]
- 22.Frick A, Gingnell M, Marquand AF, Howner K, Fischer H, Kristiansson M, et al. (2014): Classifying social anxiety disorder using multivoxel pattern analyses of brain function and structure. Behavioural brain research 259:330–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang W, Yang X, Lui S, Meng Y, Yao L, Xiao Y, et al. (2015): Diagnostic prediction for social anxiety disorder via multivariate pattern analysis of the regional homogeneity. BioMed research international 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhu H, Qiu C, Meng Y, Yuan M, Zhang Y, Ren Z, et al. (2017): Altered topological properties of brain networks in social anxiety disorder: a resting-state functional MRI study. Scientific Reports 7:43089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantazatos SP, Talati A, Schneier FR, Hirsch J (2014): Reduced anterior temporal and hippocampal functional connectivity during face processing discriminates individuals with social anxiety disorder from healthy controls and panic disorder, and increases following treatment. Neuropsychopharmacology 39:425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hilbert K, Lueken U, Muehlhan M, Beesdo□Baum K (2017): Separating generalized anxiety disorder from major depression using clinical, hormonal, and structural MRI data: A multimodal machine learning study. Brain and behavior 7:e00633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qiao J, Li A, Cao C, Wang Z, Sun J, Xu G (2017): Aberrant Functional Network Connectivity as a Biomarker of Generalized Anxiety Disorder. Frontiers in human neuroscience 11:626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yao Z, Liao M, Hu T, Zhang Z, Zhao Y, Zheng F, et al. (2017): An Effective Method to Identify Adolescent Generalized Anxiety Disorder by Temporal Features of Dynamic Functional Connectivity. Frontiers in human neuroscience 11:492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lueken U, Hilbert K, Wittchen H-U, Reif A, Hahn T (2015): Diagnostic classification of specific phobia subtypes using structural MRI data: a machine-learning approach. Journal of Neural Transmission 122:123–134. [DOI] [PubMed] [Google Scholar]
- 30.Jin C, Jia H, Lanka P, Rangaprakash D, Li L, Liu T, et al. (2017): Dynamic brain connectivity is a better predictor of PTSD than static connectivity. Human brain mapping 38:4479–4496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rangaprakash D, Deshpande G, Daniel TA, Goodman AM, Robinson JL, Salibi N, et al. (2017): Compromised hippocampus□striatum pathway as a potential imaging biomarker of mild□traumatic brain injury and posttraumatic stress disorder. Human brain mapping 38:2843–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rangaprakash D, Dretsch MN, Venkataraman A, Katz JS, Denney TS Jr, Deshpande G (2018): Identifying disease foci from static and dynamic effective connectivity networks: illustration in soldiers with trauma. Human brain mapping 39:264–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Q, Wu Q, Zhu H, He L, Huang H, Zhang J, et al. (2016): Multimodal MRI-based classification of trauma survivors with and without post-traumatic stress disorder. Frontiers in neuroscience 10:292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gong Q, Li L, Du M, Pettersson-Yeo W, Crossley N, Yang X, et al. (2014): Quantitative prediction of individual psychopathology in trauma survivors using resting-state FMRI. Neuropsychopharmacology 39:681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin S, Young CB, Duan X, Chen T, Supekar K, Menon V (2014): Amygdala subregional structure and intrinsic functional connectivity predicts individual differences in anxiety during early childhood. Biological psychiatry 75:892–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hsu W-T, Rosenberg MD, Scheinost D, Constable RT, Chun MM (2018): Resting-state functional connectivity predicts neuroticism and extraversion in novel individuals. Social cognitive and affective neuroscience 13:224–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greening SG, Mitchell DG (2015): A network of amygdala connections predict individual differences in trait anxiety. Human brain mapping 36:4819–4830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nicholson AA, Densmore M, McKinnon MC, Neufeld RW, Frewen PA, Théberge J, et al. (2018): Machine learning multivariate pattern analysis predicts classification of posttraumatic stress disorder and its dissociative subtype: a multimodal neuroimaging approach. Psychological medicine 1–11. [DOI] [PubMed]
- 39.Long J, Huang X, Liao Y, Hu X, Hu J, Lui S, et al. (2014): Prediction of post-earthquake depressive and anxiety symptoms: a longitudinal resting-state fMRI study. Scientific reports 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takagi Y, Sakai Y, Abe Y, Nishida S, Harrison BJ, Martínez-Zalacaín I, et al. (2018): A common brain network among state, trait, and pathological anxiety from whole-brain functional connectivity. NeuroImage 172:506–516. [DOI] [PubMed] [Google Scholar]
- 41.Nostro AD, Müller VI, Varikuti DP, Pläschke RN, Hoffstaedter F, Langner R, et al. (2018): Predicting personality from network-based resting-state functional connectivity. Brain Structure and Function 223:2699–2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dubois J, Galdi P, Han Y, Paul LK, Adolphs R (2018): Resting-state functional brain connectivity best predicts the personality dimension of openness to experience. Personality Neuroscience 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Whelan R, Garavan H (2014): When optimism hurts: inflated predictions in psychiatric neuroimaging. Biological psychiatry 75:746–748. [DOI] [PubMed] [Google Scholar]
- 44.Schnack HG, Nieuwenhuis M, van Haren NE, Abramovic L, Scheewe TW, Brouwer RM, et al. (2014): Can structural MRI aid in clinical classification? A machine learning study in two independent samples of patients with schizophrenia, bipolar disorder and healthy subjects. Neuroimage 84:299–306. [DOI] [PubMed] [Google Scholar]
- 45.Yahata N, Morimoto J, Hashimoto R, Lisi G, Shibata K, Kawakubo Y, et al. (2016): A small number of abnormal brain connections predicts adult autism spectrum disorder. Nature communications 7:11254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabuncu MR, Konukoglu E, Initiative AsDN (2015): Clinical prediction from structural brain MRI scans: a large-scale empirical study. Neuroinformatics 13:31–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Varoquaux G (2017): Cross-validation failure: small sample sizes lead to large error bars. Neuroimage [DOI] [PubMed]
- 48.Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, et al. (2013): The WU-Minn human connectome project: an overview. Neuroimage 80:62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banko M, Brill E (2001): Scaling to very very large corpora for natural language disambiguation. Proceedings of the 39th annual meeting on association for computational linguistics: Association for Computational Linguistics, pp 26–33. [Google Scholar]
- 50.Holmes AJ, Yeo BT (2015): From phenotypic chaos to neurobiological order. Nature neuroscience 18:1532. [DOI] [PubMed] [Google Scholar]
- 51.Bzdok D, Yeo BT (2017): Inference in the age of big data: Future perspectives on neuroscience. Neuroimage 155:549–564. [DOI] [PubMed] [Google Scholar]
- 52.Alfaro-Almagro F, Jenkinson M, Bangerter NK, Andersson JL, Griffanti L, Douaud G, et al. (2018): Image processing and Quality Control for the first 10,000 brain imaging datasets from UK Biobank. Neuroimage 166:400–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, et al. (2014): The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain imaging and behavior 8:153–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nooner KB, Colcombe S, Tobe R, Mennes M, Benedict M, Moreno A, et al. (2012): The NKI-Rockland sample: a model for accelerating the pace of discovery science in psychiatry. Frontiers in neuroscience 6:152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Holmes AJ, Hollinshead MO, O’Keefe TM, Petrov VI, Fariello GR, Wald LL, et al. (2015): Brain Genomics Superstruct Project initial data release with structural, functional, and behavioral measures. Scientific data 2:150031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Halevy A, Norvig P, Pereira F (2009): The unreasonable effectiveness of data. IEEE Intelligent Systems 24:8–12. [Google Scholar]
- 57.Smith SM, Nichols TE (2018): Statistical challenges in “big data” human neuroimaging. Neuron 97:263–268. [DOI] [PubMed] [Google Scholar]
- 58.Insel T, Cuthbert B, Garvey M, Heinssen R, Pine DS, Quinn K, et al. (2010): Research domain criteria (RDoC): toward a new classification framework for research on mental disorders. Am Psychiatric Assoc [DOI] [PubMed]
- 59.Allardyce J, Suppes T, Van Os J (2007): Dimensions and the psychosis phenotype. International journal of methods in psychiatric research 16:S34–S40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Andrews G, Brugha T, Thase ME, Duffy FF, Rucci P, Slade T (2007): Dimensionality and the category of major depressive episode. International Journal of Methods in Psychiatric Research 16:S41–S51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Spielberger CD (1970): STAI manual for the state-trait anxiety inventory. Self-Evaluation Questionnaire 1–24.
- 62.Costa PT, McCrae RR (1992): Normal personality assessment in clinical practice: The NEO Personality Inventory. Psychological assessment 4:5. [Google Scholar]
- 63.Carver CS, White TL (1994): Behavioral inhibition, behavioral activation, and affective responses to impending reward and punishment: the BIS/BAS scales. Journal of personality and social psychology 67:319. [Google Scholar]
- 64.Cloninger CR (1987): A systematic method for clinical description and classification of personality variants: A proposal. Archives of general psychiatry 44:573–588. [DOI] [PubMed] [Google Scholar]
- 65.Holmes AJ, Lee PH, Hollinshead MO, Bakst L, Roffman JL, Smoller JW, et al. (2012): Individual differences in amygdala-medial prefrontal anatomy link negative affect, impaired social functioning, and polygenic depression risk. Journal of Neuroscience 32:18087–18100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TE, Johansen-Berg H, et al. (2004): Advances in functional and structural MR image analysis and implementation as FSL. Neuroimage 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- 67.Fischl B, Van Der Kouwe A, Destrieux C, Halgren E, Ségonne F, Salat DH, et al. (2004): Automatically parcellating the human cerebral cortex. Cerebral cortex 14:11–22. [DOI] [PubMed] [Google Scholar]
- 68.Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, et al. (2002): Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron 33:341–355. [DOI] [PubMed] [Google Scholar]
- 69.Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, et al. (2011): Scikit-learn: Machine learning in Python. Journal of machine learning research 12:2825–2830. [Google Scholar]
- 70.Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, et al. (2015): Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nature neuroscience 18:1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosenberg MD, Finn ES, Scheinost D, Papademetris X, Shen X, Constable RT, et al. (2016): A neuromarker of sustained attention from whole-brain functional connectivity. Nature neuroscience 19:165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rosenberg M, Finn E, Scheinost D, Constable R, Chun M (2017): Characterizing attention with predictive network models. Trends in cognitive sciences 21:290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson JS, Ferguson MA, Lopez-Larson M, Yurgelun-Todd D (2011): Reproducibility of single-subject functional connectivity measurements. American journal of neuroradiology 32:548–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Laumann TO, Gordon EM, Adeyemo B, Snyder AZ, Joo SJ, Chen M-Y, et al. (2015): Functional system and areal organization of a highly sampled individual human brain. Neuron 87:657–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hacker CD, Laumann TO, Szrama NP, Baldassarre A, Snyder AZ, Leuthardt EC, et al. (2013): Resting state network estimation in individual subjects. Neuroimage 82:616–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Elliott ML, Knodt AR, Cooke M, Kim MJ, Melzer TR, Keenan R, et al. (2019): General Functional Connectivity: shared features of resting-state and task fMRI drive reliable and heritable individual differences in functional brain networks. NeuroImage 189:516–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Greene AS, Gao S, Scheinost D, Constable RT (2018): Task-induced brain state manipulation improves prediction of individual traits. Nature communications 9:2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dadi K, Rahim M, Abraham A, Chyzhyk D, Milham M, Thirion B, et al. (2018): Benchmarking functional connectome-based predictive models for resting-state fMRI [DOI] [PubMed]
- 79.Drysdale AT, Grosenick L, Downar J, Dunlop K, Mansouri F, Meng Y, et al. (2017): Resting-state connectivity biomarkers define neurophysiological subtypes of depression. Nature medicine 23:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Dinga R, Schmaal L, Penninx B, van Tol MJ, Veltman DJ, van Velzen L, et al. (2019): Evaluating the evidence for biotypes of depression: Methodological replication and extension of. NeuroImage: Clinical 101796. [DOI] [PMC free article] [PubMed]
- 81.Rahim M, Thirion B, Bzdok D, Buvat I, Varoquaux G (2017): Joint prediction of multiple scores captures better individual traits from brain images. NeuroImage 158:145–154. [DOI] [PubMed] [Google Scholar]
- 82.Liem F, Varoquaux G, Kynast J, Beyer F, Masouleh SK, Huntenburg JM, et al. (2017): Predicting brain-age from multimodal imaging data captures cognitive impairment. Neuroimage 148:179–188. [DOI] [PubMed] [Google Scholar]
- 83.Beaty RE, Kenett YN, Christensen AP, Rosenberg MD, Benedek M, Chen Q, et al. (2018): Robust prediction of individual creative ability from brain functional connectivity. Proceedings of the National Academy of Sciences 201713532. [DOI] [PMC free article] [PubMed]
- 84.Chang LJ, Gianaros PJ, Manuck SB, Krishnan A, Wager TD (2015): A sensitive and specific neural signature for picture-induced negative affect. PLoS biology 13:e1002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wiemer J, Schulz SM, Reicherts P, Glotzbach-Schoon E, Andreatta M, Pauli P (2014): Brain activity associated with illusory correlations in animal phobia. Social cognitive and affective neuroscience 10:969–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Barrera TL, Cully JA, Amspoker AB, Wilson NL, Kraus-Schuman C, Wagener PD, et al. (2015): Cognitive–behavioral therapy for late-life anxiety: Similarities and differences between Veteran and community participants. Journal of anxiety disorders 33:72–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pletti C, Dalmaso M, Sarlo M, Galfano G (2015): Gaze cuing of attention in snake phobic women: the influence of facial expression. Frontiers in psychology 6:454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Makovac E, Mancini M, Fagioli S, Watson DR, Meeten F, Rae CL, et al. (2018): Network abnormalities in generalized anxiety pervade beyond the amygdala-pre-frontal cortex circuit: Insights from graph theory. Psychiatry Research: Neuroimaging 281:107–116. [DOI] [PubMed] [Google Scholar]
- 89.Kim D-H, Lee J-H (2016): A Preliminary study on the Biased Attention and Interpretation in the Recognition of Face-body Compound of the Individuals with Social Anxiety. Frontiers in psychology 7:414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Naegeli C, Zeffiro T, Piccirelli M, Jaillard A, Weilenmann A, Hassanpour K, et al. (2018): Locus coeruleus activity mediates hyperresponsiveness in posttraumatic stress disorder. Biological psychiatry 83:254–262. [DOI] [PubMed] [Google Scholar]
- 91.Masdrakis VG, Legaki E-M, Vaidakis N, Ploumpidis D, Soldatos CR, Papageorgiou C, et al. (2015): Baseline heartbeat perception accuracy and short-term outcome of brief cognitive-behaviour therapy for panic disorder with agoraphobia. Behavioural and cognitive psychotherapy 43:426–435. [DOI] [PubMed] [Google Scholar]
- 92.Raboni MR, Alonso FF, Tufik S, Suchecki D (2014): Improvement of mood and sleep alterations in posttraumatic stress disorder patients by eye movement desensitization and reprocessing. Frontiers in behavioral neuroscience 8:209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Prats E, Domínguez E, Pailhez G, Bulbena A, Fullana MA (2014): Effectiveness of cognitive-behavioral group therapy for panic disorder in a specialized unit. Actas Esp Psiquiatr 42:176–184. [PubMed] [Google Scholar]
- 94.Newman MG, Lafreniere LS, Jacobson NC (2018): Relaxation-induced anxiety: Effects of peak and trajectories of change on treatment outcome for generalized anxiety disorder. Psychotherapy Research 28:616–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Jergović M, Bendelja K, Savić Mlakar A, Vojvoda V, Aberle N, Jovanovic T, et al. (2015): Circulating levels of hormones, lipids, and immune mediators in post-traumatic stress disorder–a 3-month follow-up study. Frontiers in psychiatry 6:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Keller-Ross ML, Schlinder-Delap B, Doyel R, Larson G, Hunter SK (2014): Muscle fatigability and control of force in men with posttraumatic stress disorder. Medicine & Science in Sports & Exercise 46:1302–1313. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The imaging data are publically available at (http://neuroinformatics.harvard.edu/gsp/). Some data, including anxiety-related questionnaires, require approval for access (instructions: http://neuroinformatics.harvard.edu/gsp/get). Code used to run the analyses is available upon request.


