Abstract
The ataxia telangiectasia-mutated and Rad3-related (ATR) serine/threonine kinase plays a central role in the repair of replication-associated DNA damage, the maintenance of S and G2/M-phase genomic stability, and the promotion of faithful mitotic chromosomal segregation. A number of stimuli activate ATR, including persistent single-stranded DNA at stalled replication folks, R loop formation, hypoxia, ultraviolet light, and oxidative stress, leading to ATR-mediated protein phosphorylation. Recently, hydrogen sulfide (H2S), an endogenous gasotransmitter, has been found to regulate multiple cellular processes through complex redox reactions under similar cell stress environments. Three enzymes synthesize H2S: cystathionine-β-synthase, cystathionine γ-lyase, and 3-mercaptopyruvate sulfurtransferase. Since H2S can under some conditions cause DNA damage, we hypothesized that ATR activity may regulate cellular H2S concentrations and H2S-syntheszing enzymes. Here we show that human colorectal cancer cells carrying biallelic knock-in hypomorphic ATR mutations have lower cellular H2S concentrations than do syngeneic ATR wild-type cells, and all three H2S-synthesizing enzymes show lower protein expression in the ATR hypomorphic mutant cells. Additionally, ATR serine 428 phosphorylation is altered by H2S donor and H2S synthesis enzyme inhibition, while the oxidative-stress induced phosphorylation of the ATR-regulated protein CHK1 on serine 345 is increased by H2S synthesis enzyme inhibition. Lastly, inhibition of H2S production potentiated oxidative stress-induced double-stranded DNA breaks in the ATR hypomorphic mutant compared to ATR wild-type cells. Our findings demonstrate that the ATR kinase regulates and is regulated by H2S.
Keywords: Hydrogen sulfide, H2S, ATR, cystathionine-β-synthase, cystathionine γ-lyase, 3-mercaptopyruvate sulfurtransferase, CHK1, nicotinamide phosphoribosyltransferase
1. Introduction
The ataxia telangiectasia-mutated and Rad3-related (ATR) serine/threonine kinase plays a central role in maintaining genomic stability [1–3]. Located at 3q23, ATR consists of a 2,644-amino acid residue phosphatidylinositol 3-kinase-related family member with overlapping sequence and functional homologies to the DNA-dependent and ataxia telangiectasia-mutated protein kinases [4]. Together, these proteins are central in coordinating the DNA damage response (DDR), which functions to recognize DNA damage and initiate intracellular pathways that repair genomic damage [5]. The ATR consensus phosphorylation site occurs at serine or threonine residues followed by glutamine residues (SQ/TQ), with kinase activation correlating with ATR serine 435 (ATR-pS435) and threonine 1989 (ATR-pT1989) phosphorylations [6–8]. Specifically ATR-pS435 is required for ATR-XPA complex formation, which promotes nucleotide excision repair at sites of photodamaged DNA [9,10]. ATR also regulates the small ubiquitin like modifier (SUMO) system, particularly the sumoylation of proteins that protect cells from replication stress and fork breakage [11].
ATR was first identified as being essential for embryonic development with ATR-deficient mouse embryos showing inviability, accompanied by shattered chromosomes [12,13]. Individuals with hypomorphic ATR mutations have Seckel syndrome type 1, characterized by primordial dwarfism, avian faces, accelerated aging, micrognathia, microcephaly, growth retardation, intellectual disability, and defects in the DDR [14]. Complete ablation of ATR function results in rapid cell death [12–14].
ATR maintains genomic stability by safeguarding replication S-phase fork integrity, regulating cell cycle progression, initiating cell cycle checkpoints following genotoxic insults, and by associating with centromeres where it promotes faithful chromosomal segregation at mitosis [2,4,7–10,15,16]. Specifically, ATR recognizes single-stranded DNA (ssDNA) coated by Replication Protein A, which commonly occurs following DNA damage or at stalled DNA replication folks [15,16]. In combination with other proteins (including ATRIP, TopBP1, and the 911 complex), ATR phosphorylates multiple protein substrates, including the checkpoint kinase 1 (CHK1), initiating cellular DNA damage responses [15–20]. ATR is also activated by hypoxia, cellular mechanical, and oxidative stressors [21–23].
Recent studies have shown that hydrogen sulfide (H2S) is an important cellular gasotransmitter, functioning in neuromodulation, cytoprotection, oxygen sensing, angiogenesis, and vascular tone regulation [24,25]. H2S is synthesized by three enzymes: cystathionine-β-synthase (CBS), cystathionine γ-lyase (CSE), and 3-mercaptopyruvate sulfurtransferase (3-MST) [24,25]. Presently there is no data demonstrating a role for H2S in the DDR or ATR activities. However, under certain conditions H2S can directly induce DNA damage, suggesting that it could activate the DDR [5,26,27]. Additionally, H2S can either promote or suppress cell cycle progression, likely due to lower endogenous H2S concentrations promoting cell proliferation and higher H2S concentrations inhibiting it [28–30]. Since ATR responds to DNA damage, we hypothesized that it may play a role in regulating cellular H2S concentrations and levels of the H2S-synthesizing enzymes [11]. Here we examined the role of the ATR kinase in H2S regulation.
2. Materials and methods
2.1. Materials
Monobromobimane (MBB), Tris (2-carboxyethyl)phosphine hydrochloride (TCEP), sulfosalicylic acid (SSA), 1-fluoro-2,4-dinitrobenzene (DNFB), TPP® tissue culture dishes, NU6027, penicillin/streptomycin, and t-BOOH were purchased from Sigma (St. Louis, MO). Fetal bovine serum, Dulbecco’s modified Eagle’s medium (DMEM) (standard liquid media) were from Invitrogen (Rockville, MD). β-cyano-L-alanine and diallyl trisulfide were purchased from the Cayman Chemical Company (Ann Arbor, MI). Antibodies used were anti-ATR-pS435 (Cell Signaling Technology Inc, Danvers, MA, catalog number 2853s), anti-total-ATR (Invitrogen, Waltham, MA USA, catalog number PA1–450), anti-CHK1-pS345 (Invitrogen, catalog number PA5–34625), anti-total-CHK1(Abcam, Cambridge, MA, catalog number ab47574), anti-3-MST (Santa Cruz Biotechnology, Santa Cruz, CA, catalog number sc-135993), anti-CSE (Santa Cruz Biotechnology, catalog number sc-101924), anti-CBS (Santa Cruz Biotechnology, catalog number sc-67154), and anti-Visfatin (Nampt, Bethyl Laboratories, Montgomery TX, catalog number A300–779A), rabbit anti-GADPH (Sigma, catalog number G9545), and anti-beta-actin (abcam, catalog number ab8227). Secondary antibodies were goat anti-rabbit IgG (catalog numbers ab6721 & ab2040, Abcam), goat anti-mouse IgG (catalog number ab205719, Abcam), and goat anti-mouse IgG (catalog number sc358920, Santa Cruz).
2.2. Cells
The human colon cancer cell line DLD1 cell line containing wild-type ATR (ATR cells) or the ATR-Seckel knock-in hypomorphic mutation (ATR-H cells) were a kind gift from Dr. Fred Bunz of the Department of Radiation Oncology and Molecular Radiation Sciences, The Johns Hopkins University School of Medicine [31]. The cells were cultured in DMEM with 5% FBS, 1% penicillin/streptomycin. Only low-passage cells were used to avoid the possibility that high passage ATR-H cells might lose the hypomorphic mutation and hence be altered [31].
2.3. Colony forming-efficiency assay
Colony forming-efficiency (CEFA) experiments were performed as previously described [32]. In brief, exponentially growing cells were plated for 12 hours at 2,000 cells/100 mm tissue culture dish in 10 ml appropriate media. The cells were pretreated for 2 hours with either an H2S inhibitor (β-cyano-L-alanine, 1 mM) or donor (diallyl trisulfide, 20 uM), washed 3X with 1X phospate buffered saline, the media replaced, and the cells were immediately treated with for 15 minutes with 50, 100, and 200 μM t-butyl hydroperoxide (t-BOOH). Following treatment, the cells were washed 3X with phosphate-buffered saline, the media replaced and the cells cultured for 11 days. The resulting colonies were fixed and stained by water: methanol addition (1:1) containing crystal violet (1 g/L). “Colonies” consisted of cell clusters containing greater than 50 cells when counted by dissecting microscopy.
2.4. H2S Measurements
Bioavailable sulfide levels were measured as previously reported [33,34]. Levels of free sulfide (H2S) in ATR and ATR-H cells were measured by high performance liquid chromatography (HPLC) after derivatization with excess MBB as stable products sulphide-dibimane (SDB). Briefly, ATR and ATR-H cells were homogenized in Tris·HCl buffer [100 mM Tris·HCl (pH 9.5) and 0.1 mM diethylenetriaminepentaacetic acid (DTPA)]. Cell lysates were derivatized with MBB and then measured by Shimadzu Prominence 20A equipment with RF-10AXL (excitation wavelength: 390 mm and emission wavelength: 475 mm) and an Eclipse XDB-C18 column (4.6 × 250 mm, 5 μm). Typical retention times of SDB were 16.5 minutes. H2S levels were calculated according to the standard SDB [34].
2.5. Western blotting
To prepare whole cell lysates from ATR and ATR-H cells, the cells were grown in 6-well plates, the medium was removed, and 300 μl of SDS sample buffer (62.5 mM Tris-HCl (pH 6.8), 2% w/v SDS, 10% glycerol, 50 mM dithiothreitol, 0.1% w/v bromphenol blue) was added to each well. Following lysis in SDS sample buffer, lysates were harvested with cell scrapers and collected in Eppendorf tubes. The lysates were boiled, centrifuged, and frozen at −20°C until gel electrophoresis was performed. The protein concentrations were measured by Bio-rad DC protein assay. Twenty μgs of total protein supernatant extract was mixed with 2X SDS loading buffer according to protein concentration. Lysates separated by SDS-PAGE were transferred to polyvinylidene difluoride membranes, and membranes were blocked in 5% nonfat dry milk before the addition of primary antibodies. Densitometry was performed with ImageJ software version 1.45s. All western blots were performed at least in triplicate.
2.6. Quantitative Real-Time Polymerase Chain Reaction
Total cellular RNA was isolated from ATR and ATR-H cultured cells using Trizol according to the manufacturer’s instruction. One μg of RNA was reverse transcribed using iScript cDNA synthesis kit (Bio-rad, 1708891). Quantitative real-time polymerase chain reaction was performed using SYBR Green Master Mix (Bio-rad, 1708882), and gene expression was quantified using the 2−ΔΔCT method. All genes of interest were normalized to the housekeeping gene GAPDH. The primers used in polymerase chain reaction reactions are listed in Table 1.
Table 1.
Forward and reverse primers used to quantify CBS, CSE, and 3-MST mRNA levels in ATR and ATR-H cell lines. GAPDH primers were used for control normalizations.
| Gene | Species | Forward | Reverse |
|---|---|---|---|
| GAPDH | Human | ACAGTCAGCCGCATCTTC | CGCCCAATACGACCAAATC |
| CSE | Human | GCCTTTGCTTCAGGTTTAGC | CCTTCTGGGTGGGGTTTGT |
| CBS | Human | AGGATGAACACAGGCAAT | AAAAACCCAAACACGCAAAC |
| 3-MST | Human | ACCGTGAACATCCCCTTC | TTCTTCTCCTGGAACAGATG |
2.7. Ultraviolet ATR and ATR-H Cell Treatment
ATR and ATR-H cells were treated with ultraviolet (UV) light by removing the media from the tissue culture dishes and exposing the cells to UV light using a CL-1000 UV Crosslinker (UVP/Analytik Jena, Atkinson, NH, USA) at 15,000 μJ/cm2 (150 × 100 μJ/cm2). Following UV treatment with media was replaced and the cells were incubated for 45 minutes and harvested. Untreated cells were subjected to the same procedures without the UV exposure.
2.8. Chromosomal preparation and analysis
One 100-mm tissue culture plate/treatment of logarithmically growing ATR and ATR-H cells at 50% confluence were treated with or without, 1 mM H2S inhibitor β-cyano-L-alanine for two hours, followed by a 15-minute treatment with 10 μM t-BOOH. Following this treatment, the cells were washed 3X with phosphate-buffered saline, the media replaced, and the cells cultured for 1 hour. Colcemid (100 ng/ml) was then added for 4 hours and the cells harvested by washing 2X in 1 × PBS, followed by trypsinization and transference to a 15 ml tube. Two-ml of DMEM with 5% FBS was added/tube and the cells were pelleted 5 minutes at 500 × g. The cells were re-suspended in 5 ml 0.075 mM KCl and incubated 15 minutes at 37°C. About 200 μl of fresh methanol: glacial acetic acid at 3:1 (Carnoy’s fixative) was added, the cells gently vortexed, and pelleted for 5 minutes at 500 × g. The supernatant was removed and 5 ml of Carnoy’s fixative added with gentle vortexing. The cells were pelleted for 5 minutes at 500 × g, the supernatant removed, and 5 ml of Carnoy’s fixative was added with gentle vortexing. Chromosomal preparations were made by pelleting the preparations for 5 minutes at 500 × g, removing the supernatant, dropping on slides, drying for 30 minutes at 90°C, Giemsa staining, washing, and cover-slipping. Each data point represents 5,000 individual chromosomal observations preformed under oil immersion microscopy done in triplicate.
2.9. Statistical analysis
The significance for all the experiments in this paper were calculated by using prism software version 5.02 (GraphPad Inc., San Diego, CA). The P values are given in each figure.
3. Results
3.1. Compared to ATR wild-type cells, syngeneic ATR-H cells are preferentially sensitive to oxidative stress following H2S modulators in the CEFA
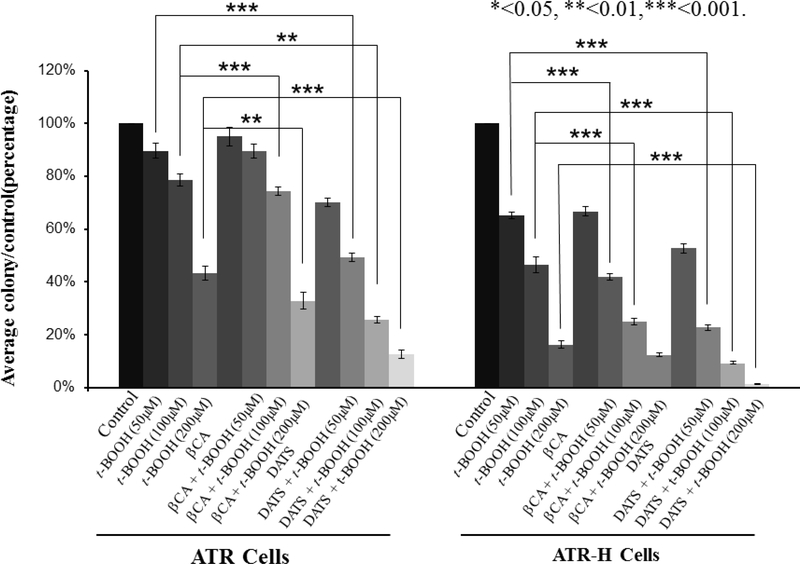
To initiate these studies, colony efficiency formation in ATR and ATR-H cells was examined with either pharmacologic H2S inhibition or H2S supplementation, followed by a 15-minute treatment with 50, 100, or 200 μM t-BOOH, culturing for 11 days, followed by fixation and analysis. As shown in Figure 1, the ATR-H cells exhibited greater sensitivity to increasing t-BOOH exposures compared to ATR cells. Pretreatment with the H2S inhibitor (2 hours with 1 mM β-cyano-L-alanine) followed by the same t-BOOH exposures, slightly increased the sensitivity of the ATR cells to different t-BOOH concentrations. Under the same conditions, the ATR-H cells were significantly more sensitive to the same t-BOOH exposures compared to the syngeneic ATR wild-type cells, (Fig. 1). For both cell types, colony formation suppression was t-BOOH dose-dependent and interestingly, β-cyano-L-alanine by itself suppressed ATR-H cell colony formation. H2S donor pretreatment (2 hours with 20 μM diallyl trisulfide) significantly decreased ATR and ATR-H cell colony formation, with the ATR-H cells significantly more sensitive to the H2S donor (Fig.1). Based on the above data, we conclude that the ATR hypomorphic mutation confers increased cellular sensitivity to pharmacologic perturbations in H2S metabolism, both with and without exogenous oxidative stress.
Figure 1.
The effects of t-BOOH and H2S inhibitor and donor treatments in the colony-forming efficiency assay with ATR and ATR-H cells. Twelve hours after plating in appropriate media, exponentially growing ATR and ATR-H cells were treated for two hours with either an H2S inhibitor (1 mM β-cyano-L-alanine, βCA) or an H2S donor (20 μM diallyl trisulfide, DATS), and subjected to 15 minutes 50, 100, or 200 μM t-BOOH oxidative stress. After 11 days, the cells were fixed, stained, and the colonies counted. Data indicates survival as a percentage of untreated cells.
3.2. Cellular H2S concentrations are lower in the hypomorphic ATR-H mutants compared to wild-type ATR cells
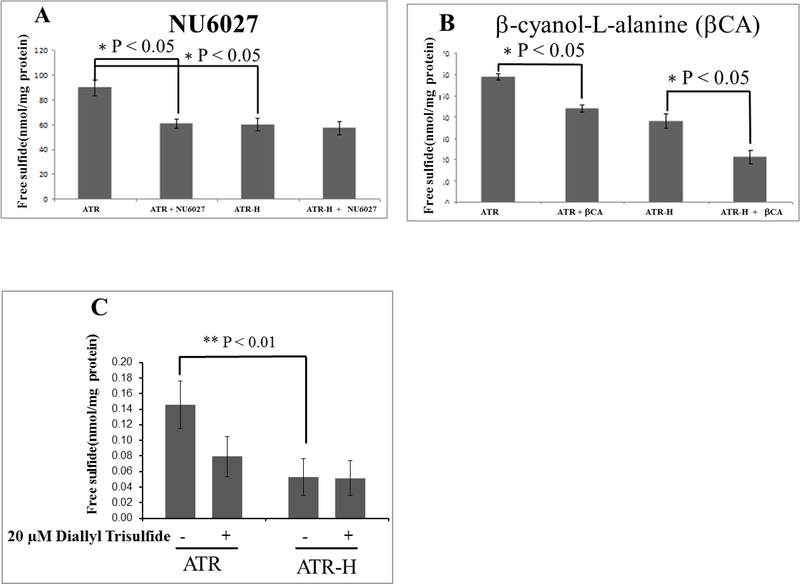
We used HPLC to analyze cellular H2S levels following MBB derivatization [33]. As shown in Figure 2A, free H2S was significantly lower in the ATR-H cells compared to the ATR cells. Additionally, a 2 hour treatment with 12 μM NU6027, an ATR inhibitor, significantly decreased the cellular free H2S concentrations in the ATR, but not the ATR-H cells (Fig 2A). In addition, as shown in Figure 2B, a 2 hour treatment with 1 mM H2S synthesis inhibitor β-cyano-L-alanine significantly lowered free H2S concentrations in both cell types, demonstrating that this H2S inhibitor worked in our assays. Last, ATR and ATR-H cells were treated 2 hours with 20 μM diallyl trisulfide. As shown in Figure 2C, diallyl trisulfide treatment suppressed the cellular free H2S concentrations in the ATR, but not the ATR-H cells. Thus, cellular H2S levels are diminished by two different perturbations of ATR activity, the hypomorphic Seckel syndrome ATR mutation and ATR protein inhibition by NU6027 (Fig. 2A). Additionally, exogenous H2S suppressed cellular H2S levels in the wild-type cells, but not the hypomorphic mutants, indicating possible differences in cellular H2S processing due to the hypomorphic mutant ATR protein.
Figure 2.
Free cellular H2S concentrations in the ATR wild-type cells compared to the ATR-H mutant cells. Figure 2A, H2S concentrations were compared in ATR and ATR-H, with and without the ATR kinase inhibitor NU6027. The cells were treated for two hours with 12 μM NU6027 in standard media and harvested (2A). Figure 2B, H2S concentrations were compared in ATR and ATR-H, with and without the CBS and CSE inhibitor β-cyanol-L-alanine. The cells were treated with 1 mM β-cyanol-L-alanine for two hours in standard media and harvested (2B). Figure 2C, H2S concentrations were compared in ATR and ATR-H, with and without 20μM diallyl trisulfide for two hours in standard media and harvested (2C). “βCA” = 1 mM β-cyanol-L-alanine. Free H2S is in nmol/mg protein.
3.3. CBS, CSE, and 3-MST protein expression is lower in the hypomorphic ATR-H mutants compared to wild-type ATR cells
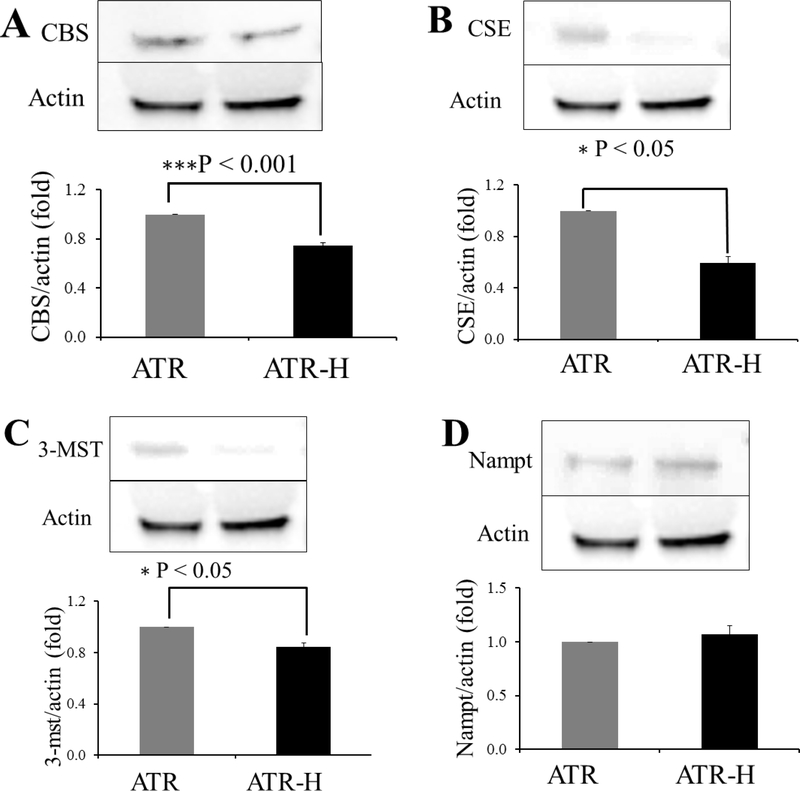
The lower H2S concentrations in the ATR-H cells raised the possibility that CBS, CSE, and 3-MST proteins levels might be altered in these cells compared to the wild-type ATR cells. We employed western blotting to examine possible differences in protein expression of these enzymes in ATR and ATR-H cells. We also examined the expression of nicotinamide phosphoribosyltransferase (Nampt), as some reports indicate that Nampt, CBS, and CSE may be co-regulated [35–37]. As shown in Figures 3A–3C, CBS, CSE, and 3-MST protein levels were all significantly lower in the ATR-H cells compared to the wild-type ATR cells. Importantly, Nampt levels were not significantly different between the two cell types, demonstrating that the hypomorphic ATR mutation does not affect Nampt protein expression (Fig. 3D). Based on this, we conclude that wild-type ATR protein activity plays a role in the maintenance of the H2S-synthesizng protein levels, but not that of Nampt.
Figure 3.
Representative western blots for CBS (3A), CSE (3B), 3-MST (3C), and Nampt (3D) comparing ATR and ATR -H cells protein expression with β-actin as a control protein. All western blots were performed at least in triplicate.
3.4. CBS, CSE, and 3-MST mRNA are not significantly different in the ATR-H cells compared to ATR cells
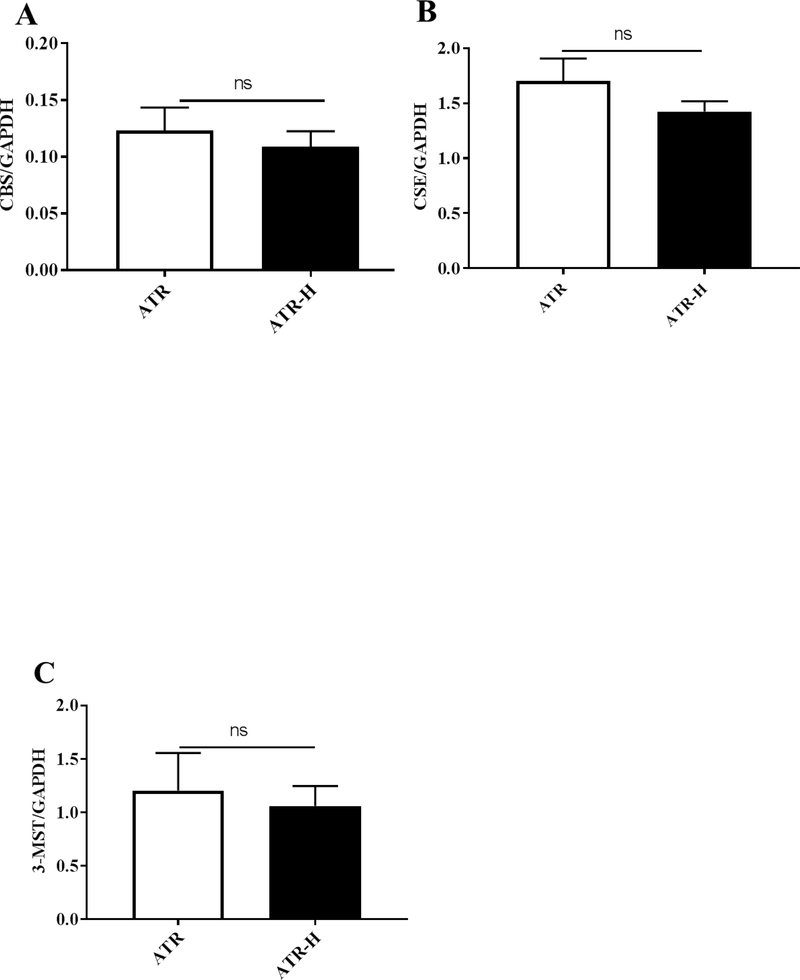
Since CBS, CSE, and 3-MST protein levels were lower in the ATR-H cells compared to the ATR cells, we examined mRNA levels of each of these genes in the two cell types using GADPH mRNA as a control. As shown in Figures 4A–4C, the levels of each mRNA where reduced in the ATR-H cells compared to the ATR cells, but not significantly lower. Based on these data we conclude that the CBS, CSE, and 3-MST protein levels are lower in the ATR-H cell line due to either a lower rate of protein translation or decreased protein stabilities in the ATR-H cell line.
Figure 4.
Quantitative real-time polymerase chain reaction analyses of CBS, CSE, and 3-MST mRNA levels in the ATR and ATR-H cell lines. Statistical analyses revealed no significant differences between CBS, CSE, and 3-MST mRNA levels between the two cell lines. The primers used in the quantitative real-time polymerase chain reactions are given in Table 1. GADPH was used as a control.
3.5. H2S inhibitor or donor exposures modulate ATR protein serine 435 phosphorylation in the ATR wild-type cells
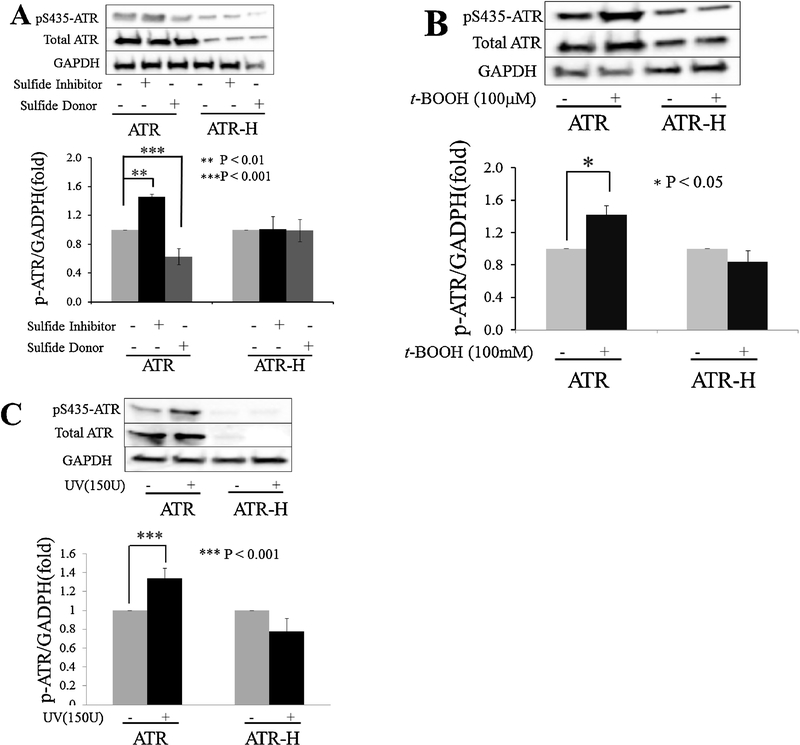
The ATR kinase is partially regulated by phosphorylations on serine 435 and threonine 1989, with these phosphorylations correlating with kinase activation and with ATR-pS435 also promoting nucleotide excision repair at sites of photodamaged DNA [6–10]. Since the above data supports a role for the ATR kinase regulating H2S, we hypothesized that H2S may in turn regulate ATR activity. To test this, we employed an anti-ATR-pS435 antibody and performed western blots on ATR and ATR-H cells following a 2 hour treatment with either 1 mM β-cyano-L-alanine (an H2S inhibitor), or 20 μM diallyl trisulfide (an H2S donor). As shown in Figure 5A, β-cyano-L-alanine increased ATR-pS435 in the wild-type cells, while diallyl trisulfide inhibited this phosphorylation. In the ATR-H cells, ATR-pS435 levels were low and not affected by any treatment, indicating that the hypomorphic ATR mutation lacks a normal phosphorylation pattern on this amino acid moiety. To examine the possibility that ATR-pS435 was modulated by the H2S inhibitor and donor at a low level, in Figure 5A the ratio of ATR-H-pS435/GAPDH protein levels was designated as one unit in the western blot graph and this measurement was compared to the ATR-H-pS435/GAPDH levels in the ATR-H cells treated with the H2S inhibitor or donor. No changes in ATR-H-pS435 were detected in the ATR-H cells (Fig. 5A). The ATR-pS435/GAPDH ratio was also used to analyze the ATR cells treated with the H2S inhibitor and donor, t-BOOH, and UV (Figs. 5A–5C).
Figure 5.
ATR protein phosphorylation on serine-435 with an H2S donor and inhibitor, or t-BOOH treatments. ATR and ATR-H cells were treated with 1mM β-cyanol-L-alanine or 20 μM diallyl trisulfide for two hours and harvested (5A). In Figure 5B, ATR and ATR-H cells were treated with 100 μM t-BOOH for 15 minutes, incubated in standard media for 45 minutes, and harvested. In Figure 5C, ATR and ATR-H cells were treated with 15,000 μJ/cm2 UV light and the cells were harvested 45 minutes later.
Since the H2S inhibitor β-cyano-L-alanine increased ATR-pS435 by ~50% compared to untreated cell phosphorylation levels, we treated ATR and ATR-H cells with 100 μM t-BOOH for 15 minutes, incubated the cells in media for 45 more minutes, and examined ATR-pS435 levels. As shown in Figure 5B, this treatment induced ATR-pS435 levels ~50% in ATR wild type cells, indicating that with moderate oxidative stress, ATR-pS435 levels change roughly as much as does H2S synthesis inhibition. The ATR-H cells again showed minimal ATR-pS435 that was not increased by t-BOOH exposure (Fig. 5B). As in Figure 4A, an untreated ATR-H-pS435/GAPDH ratio designated as one unit in the western blot graph was compared to the other ATR-H cell treatments (Fig. 5B).
ATR-pS435 regulates nucleotide excision repair at sites of DNA photodamage and is increased following UV exposure [6–10]. We next treated ATR and ATR-H cells with 15,000 μJ/cm2 UV light. As shown in Figure 5C, ATR cell UV treatment resulted in increased ATR-pS435 levels. No serine 435 phosphorylation increase was seen in the UV treated ATR-H cells (Fig. 5C). As in Figures 5A and 5B, the untreated ATR-H-pS435/GAPDH ratio was designated as one unit in the western blot graph and compared to the UV treated ATR-H cells. In Figures 5A–5C, whole ATR protein levels were also examined. When the ATR-pS435/whole ATR and ATR-H-pS435/whole ATR protein ratios were examined and analyzed in the same manner as the ATR-pS435/actin and ATR-H-pS435/actin ratios, the changes in ATR serine 435 phosphorylation levels were the same following H2S donor and inhibitor, t-BOOH, and UV treatments, while those in the ATR-H cells did not change.
Based on the above data, we conclude that both H2S inhibitor or donor modulate ATR serine 435 phosphorylation in the wild type, but not the ATR-H Seckel hypomorphic mutant cells. Additionally, since UV light also induces this phosphorylation, H2S may play an important role in regulating nucleotide excision repair by the ATR protein.
3.6. Induction of CHK1 serine 345 phosphorylation by t-BOOH exposure is modulated by H2S inhibitor pretreatment
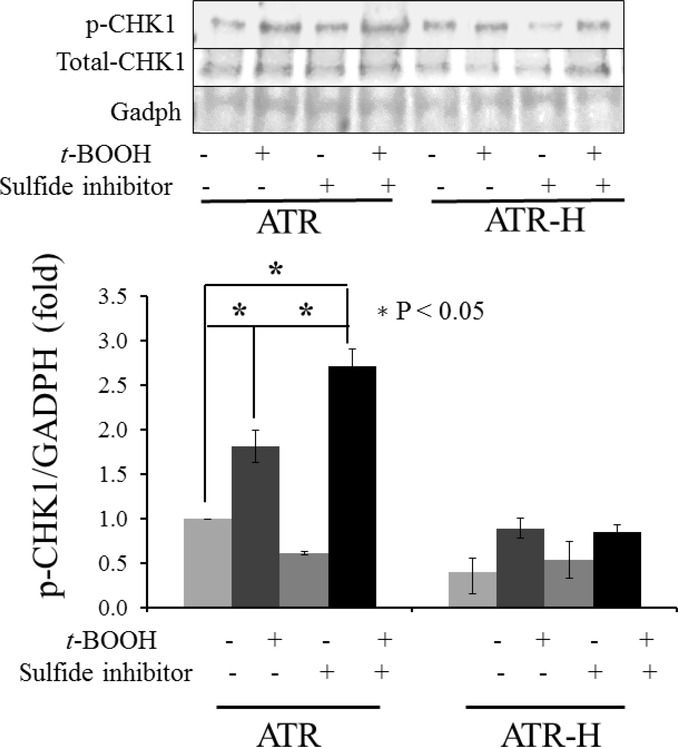
The ATR kinase specifically phosphorylates CHK1 on serine 345, an event required for checkpoint-mediated cell cycle arrest and for faithful chromosomal segregation during mitosis [2,38]. Based on the data presented above, we hypothesized that H2S synthesis inhibition by β-cyano-L-alanine would augment CHK1 serine-345 phosphorylation. We examined CHK1 serine-345 phosphorylation in ATR and ATR-H cells, with and without a 2 hour pretreatment with 1 mM β-cyano-L-alanine followed by a low 10 μM t-BOOH exposure for 15 minutes, followed by cell harvest at 45 minutes. As shown in Figure 6, pretreatment of ATR cells with 1 mM β-cyano-L-alanine significantly increased CHK1 serine-345 phosphorylation following a 15-minute 10 μM t-BOOH exposure, compared to the same t-BOOH treatment without the β-cyano-L-alanine pretreatment. Thus, modulation of intracellular H2S can also alter oxidative stress-induced ATR kinase activity. The hypomorphic ATR-H mutants failed to show significant CHK1 serine-345 phosphorylation level changes with any treatment (Fig. 6). H2S donor pretreatment did not significantly alter CHK1 serine-345 phosphorylation levels in the ATR wild-type cells following t-BOOH treatment. Since H2S is rapidly converted into other portions of the cellular sulfur pool, which can exert both pro-oxidant and anti-oxidant effects, these results are being further analyzed in our lab [24,25]. In this experiment, we also included a whole CHK1 protein control. CHK1 protein levels did not show significant variation between the ATR and ATR-H cells. For this reason, we designated the untreated ATR cell CHK1 phospho-serine-345/GADPH ratio as one unit on the western blot and compared this ratio to all other treatments of the ATR and ATR-H cells. When the untreated ATR cell CHK1 phospho-serine-345/whole CHK1 protein ratio was used, the changes in the CHK1 phosphorylation were the same as with the GAPDH control. Thus, whole CHK1 or GAPDH proteins were thus equally useful as western blot loading controls. Based on these observations, pharmacologic suppression of cellular H2S potentiates ATR kinase activity following oxidant exposure. This potentiation is not seen in the ATR-H hypomorphic mutant cells.
Figure 6.
CHK1 serine 345 phosphorylation in ATR and ATR-H cells with H2S synthesis inhibition followed by t-BOOH treatment was examined. ATR and ATR-H cells were pretreated with 1mM β-cyanol-L-alanine for two hours, then 15 minutes with 10μM t-BOOH, and harvested following a 45-minute incubation in standard media.
3.7. H2S synthesis inhibition preferentially increases dsDNA breaks in ATR-H cells by itself and when combined with t-BOOH treatment
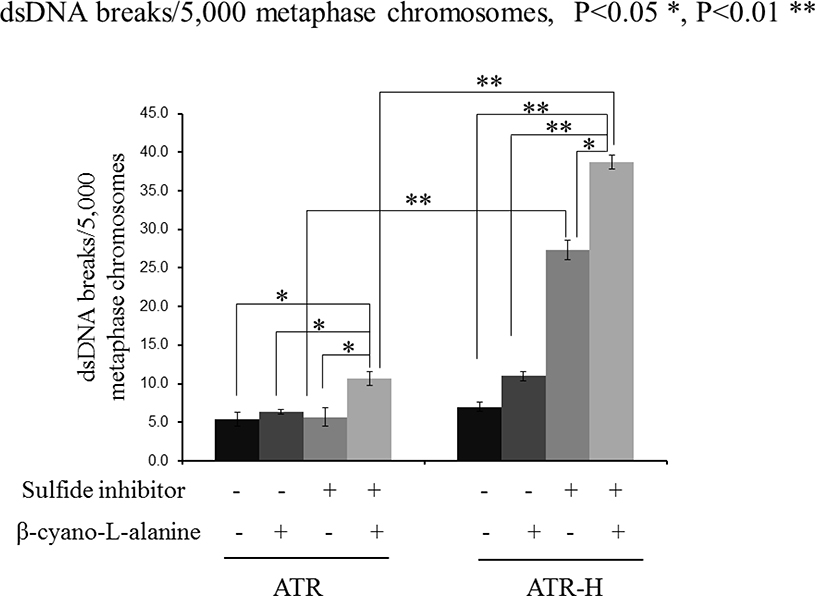
During S-phase ATR prevents stalled replication forks from degenerating in dsDNA breaks [1–3]. Since H2S regulates several ATR functions and the ATR-H cells show reduced H2S metabolism and increased oxidant sensitivity (see above), we hypothesized that H2S inhibition would preferentially effect genomic stability in ATR-H cells following oxidative stress. To do this we examined dsDNA breaks by oil immersion microscopy in metaphase colcemid-blocked, Giemsa stained, metaphase chromosomal preparations pretreated with or without an H2S inhibitor (1 mM β-cyano-L-alanine), t-BOOH, or both combined. We chose this method of dsDNA quantification, as enzymatic methods to measure DNA damage often depend on correctly functioning DNA repair enzymes, which are at least partially dysregulated in the ATR-H cells, making the assay results meaningless [39]. As shown in Figure 7, 10 μM t-BOOH treatment alone significantly increased dsDNA break formation in ATR-H cells, but not the ATR cells. However, a two-hour pretreatment with the H2S inhibitor followed by t-BOOH treatment significantly increased dsDNA breaks in both cells types. Interestingly, H2S inhibition alone significantly increased dsDNA breaks in ATR-H cells, but not in the ATR wild type cells (Fig. 7). Additionally the number of breaks was much higher in the ATR-H cells compared to the ATR cells. Since an H2S donor did not alter CHK1 serine-345 phosphorylation, it was not analyzed by dsDNA break studies. These findings indicate that upon being stressed by pharmacologic H2S inhibition alone or pharmacologic H2S inhibition combined with oxidative stress, the ATR-H cells show increased genomic instability. These data suggest that H2S likely plays a role in the maintenance of genomic stability by the ATR protein kinase.
Figure 7.
dsDNA break formation in ATR and ATR-H cells following H2S synthesis inhibition. ATR and ATR-H cells were pretreated with 1mM β-cyanol-L-alanine for two hours, then 15 minutes with t-BOOH, cultured one hour in standard media, treated with colcemid for four hours, and harvested. dsDNA breaks in Giemsa stained, Colcemid-treated cells, were counted under oil immersion microscopy. The t-BOOH concentration was 10μM.
Discussion
Here we show for the first time that the hypomorphic Seckel syndrome type 1 ATR mutation causes lower cellular H2S concentrations compared to syngeneic ATR kinase wild type cells. ATR cell treatment with the pharmacologic ATR inhibitor NU6027 also lowered cellular H2S concentrations in the ATR, but not ATR-H cells. Additionally, treating ATR cells with the H2S donor diallyl trisulfide suppressed ATR cell H2S levels, but not in the ATR-H cells. Last, the CBS and CSE inhibitor β-cyano-L-alanine lowered H2S levels in both cell types. Taken together, this data indicates that the ATR kinase regulates cellular H2S concentrations. It also demonstrates that there are two loci of H2S concentration control: 1) the activities of the CBS and CSE enzymes and 2) the functioning of the ATR kinase. The observation that diallyl trisulfide suppressed H2S concentrations in the ATR, but not ATR-H cells, suggests that ATR may have a role in responding to excess H2S levels. However, as the H2S levels are already low in the ATR-H cells, the lack of suppression may be due to the ATR-H cells not biochemically requiring further H2S level suppression. Further work is needed to examine this question, particularly to examine if ATR kinase inhibition by NU6027 affects CBS, CSE, and 3-MST protein stabilities.
The ATR-H cells were more sensitive to the toxic effects of t-BOOH in the CEFA compared to wild-type cells, demonstrating that the hypomorphic ATR mutation causes deficits in cellular anti-oxidant responses. Additionally, H2S inhibitor treatment combined with t-BOOH preferentially lowered ATR-H cell colony formation and interestingly, the H2S inhibitor by itself, suppressed colony formation in the ATR-H, but not the ATR cells. The H2S donor exerted toxic effects on both cell types, with preferential toxicity observed in the ATR-H cells. Thus, different patterns between the two cells lines were seen. In the ATR cells, the H2S inhibitor only significantly potentiated t-BOOH toxicity at the two higher doses, while in the ATR-H cells 200 μM t-BOOH and H2S inhibitor pretreatment with 200 μM t-BOOH were also not significantly different. These findings are likely due to 50 μM t-BOOH being too small a dose to significantly effect the ATR cells, even with the inhibitor pretreatment. The lack of a significant difference the ATR-H cells treated with 200 μM t-BOOH, with and without the H2S inhibitor, may be due to this higher t-BOOH dose being highly toxic to the hypomorphic mutant cells, making the biochemical contribution of the H2S inhibitor insignificant. However, taken together these results indicate that the perturbations in H2S metabolism seen in the ATR-H cells makes them more vulnerable to the toxic effects of t-BOOH when combined with pharmacologic alterations in cellular H2S metabolism.
These data implied that the function of ATR in the maintenance of genomic stability might be compromised in the ATR-H mutant cells. Direct measurements of dsDNA breaks by oil immersion microscopy revealed that the hypomorphic ATR-H mutant cells where significantly more vulnerable to dsDNA break formation than wild type cells following H2S inhibitor pretreatment, followed both with and without subsequent 10 μM t-BOOH treatment. Interestingly, H2S inhibitor treatment alone significantly induced dsDNA breaks in the ATR-H cells, but not the wild-type cells. As expected, there was a positive correlation between dsDNA breaks in the ATR-H cells with H2S synthesis inhibition, both with and without t-BOOH treatment. Lower dsDNA breaks were seen in the ATR cells, likely due to the very low 10 μM t-BOOH dose employed. This data suggests that when cellular H2S levels fall to very low levels, the ability to maintain genomic integrity is compromised, and even exacerbated in the ATR-H cell line. Our data also implies that exogenous H2S may induce DNA damage. This relationship is being further studied in the lab.
The ATR-H cells also had statistically significant lower CBS, CSE, and 3-MST protein expression levels compared to wild type cells, while Nampt levels were the same. Although here we did not examine the enzymatic activities of these proteins, possibly the lower ATR-H H2S levels are due to less protein present to synthesize H2S. Examination of CBS, CSE, and 3-MST mRNA levels between the two cell lines did not show significant differences in mRNA expression levels for these gene products. Based on this it is likely that the levels of the H2S-synthesizing protein are lower in the ATR-H cells due to lower protein stabilities or decreased mRNA translation.
We next examined the effects of H2S donors and synthesis inhibitors on ATR phosphorylation. The H2S donor suppressed ATR-pS435 in the ATR cells, while the H2S synthesis inhibitor increased it, demonstrating that cellular H2S concentrations modulate this phosphorylation. These changes were not seen in the ATR-H cells. Control experiments demonstrated that t-BOOH and UV exposure also induced this phosphorylation in ATR, but not ATR-H cells. Based on these observations H2S bioavailability plays a role in regulating ATR kinase activation and may modulate its role in nucleotide excision repair. Additionally, since this phosphorylation correlates with ATR kinase activation and is suppressed by increased H2S levels and increased by low H2S levels, our results suggest that increased ATR-pS435 may function sensing and responding to changes in cellular H2S concentrations [6–10]. Last, the ATR-mediated serine 345 CHK1 phosphorylation induced by a low concentration of t-BOOH treatment was potentiated by pretreatment with an H2S inhibitor, further confirming ATR kinase activity itself, is modulated by cellular H2S concentrations. Thus, ATR-initiated signal transduction is linked to cellular H2S bioavailability and metabolism.
ATR regulates intra-S-phase and G2/M-phase checkpoints via CHK1 phosphorylation, stabilizes stalled replication forks, preventing dsDNA break formation, promotes homologous recombination, regulates replication origin firing at specialized start sites, and promotes faithful chromosomal segregation during mitosis [2,9–23,40]. The later function likely depends on the CHK1 serine 345 phosphorylation [2]. The data presented here implies that H2S plays a role in regulating these ATR kinase-dependent functions via regulating this phosphorylation. Additionally, ATR inhibitors are being examined in preclinical and clinical studies as single agents, or paired with radiotherapy, as novel cancer therapies [40]. Increased H2S cellular concentrations have been shown to promote cancer cell growth in several different cancer types, although very high concentrations of H2S exert cytotoxic effects [41]. Our data suggests that ATR inhibitors could exert anti-cancer effects, in part, as H2S synthesis inhibitors. Further research on this is needed. Last, since an H2S donor attenuated the ATR serine 435 phosphorylation and this phosphorylation is necessary for ATR activation, and ATR activity is necessary for cell survival, high H2S levels may contribute to cell death via inhibition of ATR kinase function and activation [6–8,12–14].
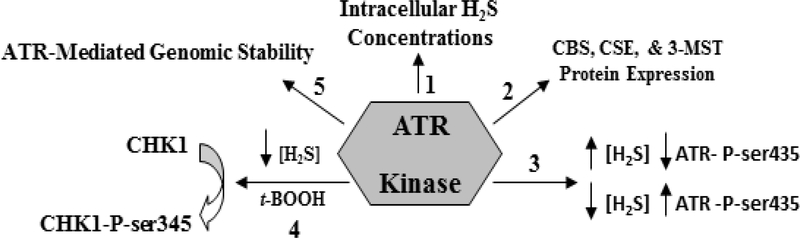
Here we present for the first time that a major constituent of the DDR, the ATR kinase, regulates and is regulated by intracellular H2S concentrations. These findings represent a deeper understanding of H2S regulation of cell survival and are summarized in Figure 8. Additionally, many ATR and CHK1-dependent cellular functions, such as nucleotide excision repair, cellular checkpoint initiation, and chromosomal segregation during mitosis, may also be regulated by intracellular H2S concentrations, which requires further investigation.
Figure 8.
A summary of the findings presented in this paper. The ATR kinase regulates intracellular H2S concentrations (arrow 1) and the levels of the three H2S-synthesizing enzymes (arrow 2). Increased in intracellular H2S concentrations decreased ATR serine 435 phosphorylation, while decreased in intracellular H2S concentrations increased this phosphorylation (arrow 3). Attenuation of intracellular H2S synthesis also potentiates CHK1 serine 345 phosphorylation following ATR cell exposure to low t-BOOH concentration, an event not seen in ATR-H cells (arrow 4). Lastly, low cellular H2S concentrations in the hypomorphic ATR-H cells increase genomic instability by itself, and when combined with a low dose of t-BOOH. Taken together, our data suggests that the ATR kinase regulates and is in turn regulated by H2S.
Highlights.
Here we show for the first time that the ATR kinase regulates intracellular hydrogen sulfide concentrations as a pharmacologic ATR kinase inhibitor lowers cellular hydrogen sulfide levels and hydrogen sulfide levels are also lower syngeneic cells that carry a knock-in hypomorphic ATR Seckel mutant ATR protein. Additionally, ATR serine 435 phosphorylation is suppressed by an exogenous hydrogen sulfide donor and increased by a hydrogen sulfide synthesis inhibitor, indicating that ATR may be in turn regulated by hydrogen sulfide concentrations. We also show that ATR kinase activity is modulated by hydrogen sulfide, as treatment of ATR wild type cells with a hydrogen sulfide synthesis inhibitor potentiates CHK1 protein phosphorylation by the ATR kinase following exposure to low-levels of oxidative stress. Lastly, we present data indicating that hydrogen sulfide may influence genomic stability in conjunction with ATR kinase activity. We believe these are novel findings and show a new function for the ATR kinase.
Acknowledgements
This work was supported by NIH Grants HL113303 to C.G.K. and GM121307 to C.G.K and R.E.S.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Yazinski SA, Zou L. Functions, Regulation, and Therapeutic Implications of the ATR Checkpoint Pathway. Annu Rev Genet. 50 (2016) 155–173. [DOI] [PubMed] [Google Scholar]
- [2].Kabeche L, Nguyen HD, Buisson R, Zou L. A mitosis-specific and R loop-driven ATR pathway promotes faithful chromosome segregation. Science. 359 (2018) 108–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Buisson R, L Boisvert J, H Benes C, Zou L. Distinct but Concerted Roles of ATR, DNA-PK, and Chk1 in Countering Replication Stress during S Phase. Mol Cell. 59 (2015) 1011–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Durocher D, Jackson JP, DNA-PK ATM and ATR as sensors of DNA damage: variations on a theme? Curr Opin Cell Biol. 13 (2001) 225–231. [DOI] [PubMed] [Google Scholar]
- [5].Hanawalt PC, Historical perspective on the DNA damage response. DNA Repair. 36 (2015) 2–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Kim ST, Substrate specificities and identification of putative substrates of ATM kinase family members. J. Biol. Chem 274 (1999) 37538–37543. [DOI] [PubMed] [Google Scholar]
- [7].Daub H, Olsen JV, Bairlein M, Gnad F, Oppermann FS, Korner R, Greff Z, Keri G, Stemmann O, Mann M, Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol Cell. 31(2008) 438–448. [DOI] [PubMed] [Google Scholar]
- [8].Dephoure N, Zhou C, Villen J, Beausoleil SA, Bakalarski CE, Elledge SJ, P Gygi S, A quantitative atlas of mitotic phosphorylation. Proc Natl Acad Sci U S A. 105 (2008) 10762–10767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Jarrett SG, M Horrell E, Christian PA, Vanover JC, Boulanger MC, Zou Y, D’O JA, PKA-Mediated Phosphorylation of ATR Promotes Recruitment of XPA to UVInduced DNA Damage. Mol Cell. 54 (2014) 999–1011. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- [10].Jarrett SG, M Wolf Horrell E, Boulanger MC, D’Orazio JA, Defining the Contribution of MC1R Physiological Ligands to ATR Phosphorylation at Ser435, a Predictor of DNA Repair in Melanocytes. J Invest Dermatol. 135 (2015) 3086–3095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Munk S, Sigurðsson JO, Xiao Z, Batth TS, Franciosa G, von Stechow L, Lopez-A.J. Contreras, A.C.O. Vertegaal, J.V. Olsen, Proteomics Reveals Global Regulation of Protein SUMOylation by ATM and ATR Kinases during Replication Stress. Cell Rep. 21 (2017) 546–558. [DOI] [PubMed] [Google Scholar]
- [12].Brown EJ, Baltimore D, ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 14 (2000) 397–402. [PMC free article] [PubMed] [Google Scholar]
- [13].de Klein A, Muijtjens M, van Os R, Verhoeven Y, Smit B, Carr AM, Lehmann AR, Hoeijmakers JH, Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 10 (2000) 479–482. [DOI] [PubMed] [Google Scholar]
- [14].O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA, A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat. Genet 33 (2003) 497–501. [DOI] [PubMed] [Google Scholar]
- [15].Awasthi P, Foiani M, Kumar A, ATM and ATR signaling at a glance. J. Cell Sci 128 (2015) 4255–4262. [DOI] [PubMed] [Google Scholar]
- [16].Cimprich KA, Cortez D, ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol 9 (2008) 616–627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Lucca C, Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene. 23 (2004) 1206–1213. [DOI] [PubMed] [Google Scholar]
- [18].Kumagai A, TopBP1 activates the ATR-ATRIP complex. Cell. 124 (2006) 943–955. [DOI] [PubMed] [Google Scholar]
- [19].Matsuoka S, ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 316 (2007) 1160–1166. [DOI] [PubMed] [Google Scholar]
- [20].Hammond EM, Dorie MJ, Giaccia AJ, ATR/ATM targets are phosphorylated by ATR in response to hypoxia and ATM in response to reoxygenation. J. Biol. Chem. 278 (2003) 12207–12213. [DOI] [PubMed] [Google Scholar]
- [21].Jeremy W, Yogin P, Lentz BL, Yan YS, APE2 is required for ATR-Chk1 checkpoint activation in response to oxidative stress. Proc. Natl. Acad. Sci. U S A 110 (2013) 10592–10597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Kulkarni A, Das KC, Differential roles of ATR and ATM in p53, Chk1, and histone H2AX phosphorylation in response to hyperoxia: ATR-dependent ATM activation. Am. J. Physiol. Lung Cell. Mol. Physiol 294 (2008) L998–L1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hurley PJ, Wilsker D, Bunz F, Human cancer cells require ATR for cell cycle progression following exposure to ionizing radiation. Oncogene 26 (2007) 2535–2542. [DOI] [PubMed] [Google Scholar]
- [24].Kolluru GK, Shen X, Yuan S, Kevil CG, Gasotransmitter heterocellular signaling. Antioxid. Redox Signal. 26 (2017) 936–960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kimura H, Production and physiological effects of hydrogen sulfide. Antioxid. Redox Signal. 20 (2014) 783–793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Attene-Ramos MS, Wagner ED, Gaskins HR, Plewa MJ, Hydrogen sulfide induces direct radical-associated DNA damage. Mol Cancer Res. 5 (2007) 455–459. [DOI] [PubMed] [Google Scholar]
- [27].Attene-Ramos MS, Nava GM, Muellner MG, Wagner ED, Plewa MJ, Gaskins HR, DNA damage and toxicogenomic analyses of hydrogen sulfide in human intestinal epithelial FHs 74 int cells. Environ Mol Mutagen. 51 (2010) 304–314. [DOI] [PubMed] [Google Scholar]
- [28].Deplancke B, Gaskins HR, Hydrogen sulfide induces serum-independent cell cycle entry in nontransformed rat intestinal epithelial cells. FASEB J. 17 (2003) 1310–1312. [DOI] [PubMed] [Google Scholar]
- [29].Pei Y, Wu B, Cao Q, Wu L, Yang G, Hydrogen sulfide mediates the anti-survival effect of sulforaphane on human prostate cancer cells. Toxicol Appl Pharmacol.257 (2011) 420–428. [DOI] [PubMed] [Google Scholar]
- [30].Hellmich MR, Szabo C, Hydrogen Sulfide and Cancer. Handb Exp Pharmacol. 230 (2015) 233–241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hurley PJ, Wilsker D, Bunz F, Human cancer cells require ATR for cell cycle progression following exposure to ionizing radiation. Oncogene. 2007 26 (2007) 2535–2542. [DOI] [PubMed] [Google Scholar]
- [32].Shackelford RE, Fu Y, Manuszak RP, Brooks TC, P Sequeira A, Wang S, Lowery-Nordberg M, Chen A, Iron chelators reduce chromosomal breaks in ataxia-telangiectasia cells. DNA Repair. 5 (2006) 1327–1336. [DOI] [PubMed] [Google Scholar]
- [33].Leskova A, Pardue S, Glawe JD, G Kevil C, Shen X, Role of thiosulfate in hydrogen sulfide-dependent redox signaling in endothelial cells. Am J Physiol Heart Circ Physiol. 313 (2017) H256–H264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Shen X, Peter EA, Bir S, Wang R, Kevil CG. Analytical measurement of discrete hydrogen sulfide pools in biological specimens. Free Radic Biol Med. 15 (2012):2276–2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sanokawa-Akakura R, Ostrakhovitch EA, Akakura S, Goodwin S, Tabibzadeh S. A H2S-Nampt dependent energetic circuit is critical to survival and cytoprotection from damage in cancer cells. PLoS One. 9 (2014) e108537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Shackelford R, Abdulsattar J, Wei E, Cotelingam J, Coppola D, Herrera G, Increased Nicotinamide Phosphoribosyltransferase and Cystathionine β-Synthase in Renal Oncocytomas, Renal Transitional Cell Carcinoma and Renal Clear Cell Carcinoma. Anticancer Research 37 (2017) 3423–3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Patel S, Ansari J, Meram A, Abdulsattar J, Cotelingam J, Coppola D D, Ghali G G, Shackelford R, Increased nicotinamide phosphoribosyltransferase and Cystathionine-Beta-Synthase in oral cavity squamous cell carcinomas. International Journal of Clinical and Experimental Medicine. 10 (2017) 702–707. [Google Scholar]
- [38].Capasso H, Palermo C, Wan S, Rao H, John UP, O’Connell MJ, Walworth NC, Phosphorylation activates Chk1 and is required for checkpoint-mediated cell cycle arrest. J Cell Sci. 115 (2002) 4555–4564. [DOI] [PubMed] [Google Scholar]
- [39].Burma S, Chen BP, Murphy M, Kurimasa A, Chen DJ. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem. 276 (2001) 42462–42467. [DOI] [PubMed] [Google Scholar]
- [40].Qiu Z, L Oleinick N, Zhang J. ATR/CHK1 inhibitors and cancer therapy. Radiother Oncol. 126(2018) 450–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wu D, Si W, Wang M, Lv S, Ji A, Li Y. Hydrogen sulfide in cancer: Friend or foe? Nitric Oxide. 50 (2015) 38–45. [DOI] [PubMed] [Google Scholar]