Abstract
The cellular response to genotoxic DNA double strand breaks (DSBs) uses a multitude of post-translational modifications to localise, modulate and ultimately clear DNA repair factors in a timely and accurate manner. Ubiquitination is well established as vital to the DSB response, with a carefully co-ordinated pathway of histone ubiquitination events being a central component of DSB signalling. Other ubiquitin-like modifiers (Ubl) including SUMO and NEDD8 have since been identified as playing important roles in DSB repair. In the last five years ∼20 additional Ub/Ubl proteases have been implicated in the DSB response. The number of proteases identified highlights the complexity of the Ub/Ubl signal present at DSBs. Ub/Ubl proteases regulate turnover, activity and protein–protein interactions of DSB repair factors both catalytically and non-catalytically. This not only ensures efficient repair of breaks but has a role in channelling repair into the correct DSB repair sub-pathways. Ultimately Ub/Ubl proteases have essential roles in maintaining genomic stability. Given that deficiencies in many Ub/Ubl proteases promotes sensitivity to DNA damaging chemotherapies, they could be attractive targets for cancer treatment.
Keywords: DNA synthesis and repair, double strand break, DUB, SENP, sumoylation, ubiquitin
Introduction
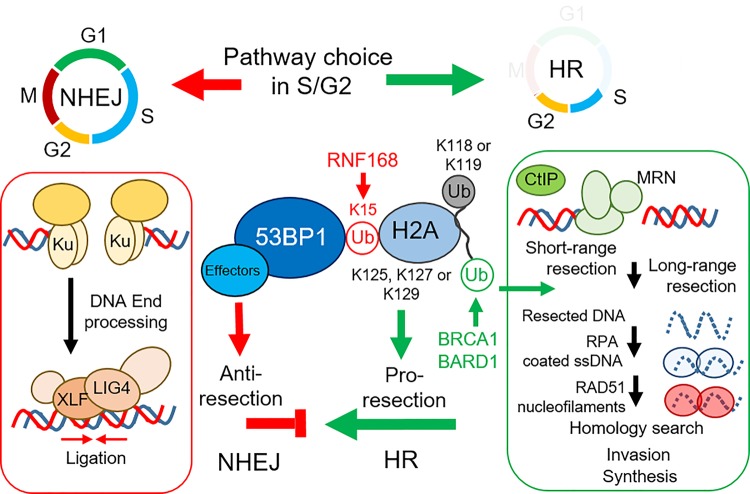
DNA double stranded breaks are highly toxic lesions that occur when both strands of DNA are broken. The breaks can be formed from endogenous processes such as DNA replication, or exogenous sources such ionising radiation. Two major pathways repair double strand breaks (DSBs) (Figure 1). Non-Homologous End Joining (NHEJ) occurs throughout the cell cycle and uses small regions of homology present in DNA overhangs followed by direct re-ligation for repair. If the overhangs are not compatible loss of nucleotides can lead to mutations. The second pathway Homologous Recombination (HR) is restricted to late S/G2 phases of the cell cycle as it relies on homologous DNA sequences, provided by sister chromatids, for homology directed repair. HR repair is generally error free [1]. Ub plays essential roles in orchestrating both repair pathways. A central function of Ub in DSB repair is maintaining the balance between NHEJ and HR in S/G2 cells. RNF168 mono-ubiquitination on K15 of H2A or H2AX promotes the recruitment of the pro-NHEJ factor 53BP1, while BRCA1–BARD1 mediated mono-ubiquitination of H2A K125/K127 or K129 promotes displacement of 53BP1 and subsequent DNA end-resection, the essential precursor step for HR [2]. Maintaining the correct balance is critical, as too much NHEJ can be mutagenic, but un-restrained DNA end-resection is also toxic. To co-ordinate DSB repair Ub and the Ubl SUMO and NEDD8 play multiple roles, they promote the recruitment and retention of repair factors at damaged chromatin, alter protein–protein interactions and enable clearance and termination of repair signalling [3,4]. As all of these processes require exquisite temporal and spatial tuning, a plethora of Ub/Ubl proteases are deployed by DNA damage signalling to ensure efficient and accurate DSB repair [5]. The profound consequences that result from the mis-regulation of these proteases offers fascinating insight into the intricacies of DSB repair.
Figure 1. NHEJ occurs at all stages of the cell cycle.
Ku70/80 heterodimers bind and stabilise the ends of the DSB. Other factors are also recruited by the Ku dimer. DNA end processing chemically modifies the bases to allow re-ligation. Ligase 4 (LIG4), XLF and other repair factors ligate the broken DNA ends. HR is restricted to S/G2, initial DNA 3′ end resection is initiated by the MRN complex and CtIP. Further resection promoted by the BRCA1–BARD1 heterodimer, EXO1 and DNA2 promote long range resection. The ssDNA generated by end resection is coated in the RPA heterotrimeric complex, which is then displaced by RAD51 nucleofilaments with the aid of the BRCA1–PALB2–BRCA2 complex. After RAD51 filament formation, homology search followed by invasion of the sister chromatid and synthesis of new DNA occurs. As the biology of these later processes in the context of Ub/Ubl modifications is poorly understood they are not illustrated here. As NHEJ can also occur in S/G2 pathway choice between HR and NHEJ occurs. The pro-NHEJ factor 53BP1 is recruited by K15 Ub modified H2A/H2AX while modification of K125/K127 or K129 by mono-Ub aids in nucleosome remodelling and displacement of 53BP1, this favours DNA end-resection and HR. Thus maintaining the balance of Ub signals dictates repair pathway choice.
Ku70/80 and the MRN complex
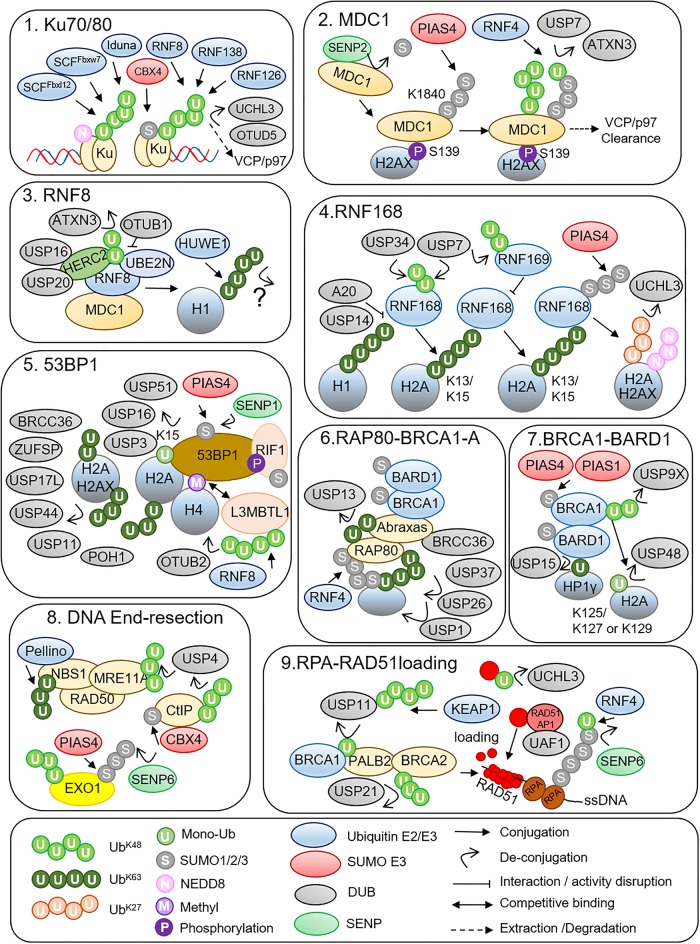
DSBs are rapidly detected by two complexes, the Ku70/Ku80 heterodimer which promotes NHEJ and MRN (MRE11A/RAD50/NBS1) which, with the exonuclease CtIP promotes HR [6,7]. Multiple Ub-E3 ligases ubiquitinate Ku heterodimers which ultimately results in extraction by the VCP/p97 complex [8,9]. The importance of this regulation is highlighted by the toxic effects of trapped Ku heterodimers which not only prevent NHEJ repair termination but also block the early steps in HR repair [9]. Ubiquitination of Ku heterodimers is countered by at least two DUBs (De-ubiquitinating proteases). UCHL3 de-ubiquitinates and stabilises Ku80 which is required for Ku80 retention at DSBs [10]. OTUD5 also stabilises Ku80, loss of OTUD5 reduces NHEJ repair efficiency [11] (Figure 2).
Figure 2. The DNA bound Ku70/80 heterodimer is ubiquinated and SUMOylated by multiple E3 ligases.
Ultimately this signals for Ku70/80 dimer removal from chromatin by VCP/p97. MDC1 is constitutively protected from excessive SUMOylation through its interaction with SENP2. This interaction is disrupted by ATM signalling which allows the DSB located MDC1 to be hyperSUMOylated by PIAS4 which also localises to DSBs. The SUMOylated MDC1 is then recognised by the SUMO directed Ub ligase RNF4 which ubiquitinates MDC1 and promotes extraction from chromatin by VCP. The RNF4 step can be countered by ATXN3, slowing MDC1 turnover at DSBs. RNF8 is recruited by MDC1 where it forms K63-Ub linkages on H1.2. HUWE1 also contributes to the generation of Ub chains on H1.2. OTUB1 inhibits RNF8 activity by disrupting Ub recognition by the E2 UBE2N. Conversely, HERC2 stimulates RNF8–UBE2N activity in a SUMOylation dependent manner. HERC2 also interacts with two DUBs, USP16 and USP20 both of which are involves in DSB repair signalling. ATXN3 counters RNF8 auto-ubiquitination. RNF168 interacts with RNF8 generated H1.2K63-Ub and amplifies this Ub signal through recognition of its own Ub product — K15 mono-ubiquitinated H2A or H2AX. Two DUBs, (A20 and USP14) interfere with RNF168's initial recruitment to H1.2. Two more DUBs (USP7 and USP34) are needed to protect RNF168 from auto-degradation. The paralog RNF169 competes with RNF168 for H2A/H2AXK15-Ub binding. 53BP1 recognises the K15-Ub modified H2A or H2AX generated by RNF168. Additionally, 53BP1 is recruited by H4K20me2, a modification that is competitively bound by L3MBTL1. Ubiquitination and extraction from chromatin of L3MBTL1 is required for efficient 53BP1 spreading along chromatin. The turnover of L3MBTL1 is antagonised by OTUB2. Multiple DUBs regulate the spread of 53BP1 via regulating the amplitude of Ub/UbK63 at DSBs, some of these DUBs directly counter the H2A/H2AXK15-Ub modification while others may act on other uncharacterised K63-Ub modified proteins. RAP80 and the BRCA1-A complex are recruited to DSBs in a K63-Ub dependent fashion, although RAP80 also recognises SUMO modifications (most likely generated by RNF4), suggesting that the RAP80–BRCA1-A complex may recognise a different signal to 53BP1. BRCA1 recruitment into the BRCA1-A complex is thought to divert its activity from promoting DNA end-resection. At least two DUBs (USP26 and USP37) specifically counter the Ub signal that promotes RAP80–BRCA1 accrual. USP13 trims Ub from the RAP80 UIM domain to allow efficient Ub recognition. BRCA1–BARD1 also recruit to DSBs independently of K63-Ub signalling. Ubiquitination inhibits the BARD1- HP1γ interaction needed for efficient accumulation of BRCA1–BARD1 at DSBs. This disruptive ubiquitination event is countered by USP15. The ubiquitination and turnover of BRCA1 is countered by USP9X. BRCA1–BARD1 also generates mono-Ub on the far C terminus of H2A (K125, K127 or K129). This signal promotes DNA end-resection and aids in the displacement of 53BP1 from DSBs. USP48 restricts the spread of H2AK125/127/129-Ub preventing excessive DNA end-resection. The MRN complex and CtIP, both of which are essential for the early steps in DNA end-resection are ubiquitinated, USP4 antagonises these modifications. SUMOylation promotes degradation of the DNA exonuclease EXO1, to control DNA end-resection which is countered by SENP6. The loading of the RPA hetero-trimer onto single stranded DNA (generated through DNA end-resection) and its exchange with RAD51 are essential steps in HR. The RPA70 subunit is SUMOylated and degraded by RNF4, this is countered by SENP6. Loading of RAD51 onto ssDNA is promoted by BRCA2 in the BRCA1–PALB2–BRCA2 complex. This complex can only form due to the activity of USP11 which removes an inhibitory Ub modification that is disruptive to BRCA1–PALB2 interactions. USP11 levels are cell cycle regulated by KEAP1. USP1–UAF1 non-catalytically aids in the loading of RAD51, UCHL3 de-ubiquitinates RAD51 promoting its ability to load onto DNA, while USP21 protects the unstable BRCA2 protein from degradation.
The MRN complex performs several activities, including DNA unwinding, promotion of DNA end resection and recruitment of the master DSB kinase ATM to the DSB [6]. ATM phosphorylates multiple DSB repair proteins, including histone H2AX at Ser139 (γH2AX) [12]. Pellino generates K63-Ub chains on NBS1 promoting ATM activation and HR repair, suggesting that Ub signalling also regulates MRN activity [13]. Indeed, USP4 is recruited to DSBs through interaction with NBS1 where it also regulates CtIP ubiquitination [14].
MDC1 (Mediator of Damage Checkpoint 1)
MDC1 interacts with γH2AX via its BRCT repeats [15], and has multiple roles in DSB signalling, it aids in the retention of ATM which further propagates the γH2AX signal along damaged chromatin and it recruits RNF8 [16,17]. While MDC1 recruitment is phosphorylation dependent, turnover at DSBs in G1 requires SUMOylation and ubiquitination. Following recruitment to DSBs MDC1 is SUMOylated by PIAS4 [18–22]. MDC1 SUMOylation promotes recognition by RNF4, an Ub-E3 that interacts with SUMOylated substrates [23]. This in turn promotes VCP/p97 dependent extraction of MDC1 from DSBs [20–22,24]. The basal SUMOylation state of MDC1 is maintained by the SUMO protease SENP2 which dissociates from MDC1 shortly after DSB induction. This allows un-restrained SUMOylation of MDC1 by PIAS4 on DSBs which ultimately triggers MDC1 removal by VCP. In the absence of SENP2 the constitutively hyperSUMOylated MDC1 undergoes premature PIAS4-RNF4-VCP dependent clearance from DSBs resulting in ablated RNF8 accrual and K63-Ub signalling [24]. The RNF4 dependent clearance of MDC1 is antagonised by ATXN3, a DUB that is rapidly recruited to DSBs by SUMO [25]. USP7 also interacts with and stabilises the MDC1–MRN complex [26].
RNF8
RNF8 is recruited to DSBs via its FHA domain that recognises phosphorylated MDC1 [27], where along with its partner E2 (UBE2N) K63-Ub chains are conjugated on histone H1 [28]. HUWE1 primes the ubiquitination of H1 prior to extension by RNF8 [29]. Several DUB dependent modes of RNF8 regulation have been identified. OTUB1 blocks the generation of K63-Ub through a non-enzymatic activity that involves disruption of the RNF8–UBE2N catalytic cycle [30,31]. The RNF8–UBE2N complex is also regulated by HERC2 which stimulates the generation of K63-Ub at DSBs by stabilising or promoting the interaction of RNF8 with UBE2N [32]. This activity is promoted by SUMOylation of HERC2 through PIAS4 [33]. HERC2 interacts with USP20, a DUB that is recruited to DSBs and is involved in checkpoint signalling in response to replication stress and HR repair [34–37]. HERC2 also interacts with and stabilises USP16 which is involved in H2A de-ubiquitination in DSB repair [38]. RNF8 auto-ubiquitination is countered by ATXN3, which is required to balance VCP/p97 dependent extraction from damaged chromatin [39].
RNF168
H1K63-Ub is recognised by a MIU domain of RNF168, which conjugates mono-ubiquitin to H2A or H2AX at K15 [28,40]. This serves as a docking site for the resection antagonist/pro-NHEJ factor 53BP1 [41,42]. A second MIU domain in RNF168 also recognises its own Ub product, allowing spreading and amplification of H2A/H2AX-UbK15 [28]. RNF168 is de-stabilised following DSB induction and at least two DUBs, USP34 and USP7 are involved in countering RNF168 turnover. Loss of either DUBs results in reduced RNF168 and less K63-Ub signalling at DSBs [43,44]. RNF168 accumulation at damaged chromatin is also regulated by DUBs. A20 interacts with RNF168 upon DSB induction and disrupts recruitment to H1K63-Ub non-catalytically [45]. USP14 also recruits to DSBs where it interacts with RNF168 via its MIU1 domain and impairs RNF168 Ub signalling [46].
USP3, USP51 and USP16 de-ubiquitinate H2A/H2AXK15-Ub with varying degrees of specificity [47–49]. Loss of these DUBs causes excessive spreading of H2A/H2AXK15-Ub and its downstream reader 53BP1. In addition to K63-Ub, RNF168 generates K27-Ub conjugates, which are required for the recruitment of 53BP1 and RAP80–BRCA1 [50]. Although the biology of K27-Ub in DSB repair is not well understood, it has since been shown that RNF168 generated K27-Ub conjugates can be antagonised by UCHL3 [51].
H2A/H2AXK15-Ub is also recognised by the RNF168 paralog RNF169 which competes with 53BP1. RNF169 therefore limits the recruitment of the pro-NHEJ/anti-resection factor 53BP1 and promotes HR repair [41,52–55]. Like RNF168, RNF169 is also stabilised by USP7 [56].
Several DUBs were characterised for their roles in DSB signalling prior to the identification of H2A/H2AXK15-Ub generated by RNF168 and therefore it is not clear if they only target this modification, other sites on H2A/H2AX or non-histone K63-Ub modified proteins. USP44, USP17L2/DUB3, USP11, ZUFSP and POH1 loss promotes the excessive spreading of Ub at DSBs and concomitant excessive accumulation of 53BP1 [57–65].
H2a-K118/K119
H2A is predominantly mono-ubiquitinated at K118/K119 by BMI1:RING1B E3 ligase. H2AK118/K119-Ub has important, but not fully understood the role in DSB repair [66]. BAP1 is the dominant H2AK118/K119-Ub DUB [67] and is rapidly and transiently recruited to DSBs [37,68,69]. BAP1 loss specifically reduces HR but not NHEJ [68,69].
53BP1
53BP1 is a major adapter protein that recruits to DSBs via histone phosphorylation [70], methylation [71] and ubiquitination [42]. While 53BP1 has no enzymatic activity it does recruit a number of other proteins that antagonise DNA end-resection and subsequent HR pathways mediated by BRCA1. The major effectors of 53BP1 include PTIP, RIF1 and the Rev7-Shieldin-CST complexes [72].
The Tudor domain of 53BP1 recognises H4K20me2, which is also recognised with higher avidity by L3MBTL1 and JMJD2A [73,74]. To allow maximal 53BP1 recruitment these factors need to be displaced from chromatin. L3MBTL1 is K48 ubiquitinated by RNF8 and extracted by VCP/p97 from chromatin [73]. This ubiquitination is antagonised by OTUB2, a DUB that is recruited to DSBs and slows L3MBTL1 Ub dependent removal from chromatin. This limits excessive 53BP1 spreading [37,75]. SUMOylation at DSBs is also important for 53BP1 recruitment, and 53BP1 is itself SUMOylated by PIAS4 in G1 [18,76]. SUMOylation of 53BP1 is antagonised by SENP1, as displacement of SENP1 from nuclear pores reduces 53BP1 SUMOylation and disrupts NHEJ repair [77]. The 53BP1 interacting partner RIF1 is also PIAS4 SUMOylated with similar kinetics to 53BP1 [76].
RAP80 and the BRCA1-A complex
RAP80 is a reader of K63-Ub polymers at DSBs and serves as a docking module for the BRCA1-A complex [78–81]. In addition to K63-Ub, RAP80 interacts with SUMO chains, likely through its recognition of mixed K63-Ub-SUMO linkages for which it has an eighty times higher binding preference over K63-Ub chains. RNF4 is the likely source of these linkages due to its localisation at DSBs and ability to generate mixed K63-Ub-SUMO conjugates [82,83].
BRCC36 (BRCA1/2 Containing Complex 36) was the first DUB characterised in DSB repair [84] and resides within the BRCA1-A complex. BRCC36 de-ubiquitinates H2A/H2AX and RAP80. Loss of BRCC36 results in extensive spreading of K63-Ub at DSBs, suggesting its role is to limit the amplitude of K63-Ub at DSBs [85,86]. BRCA1-A components, including BRCC36 are antagonists of HR repair. The BRCA1-A complex has been proposed to divert BRCA1–BARD1 Ub ligase activity to dampen DNA end-resection. Loss of BRCA1-A components causes a hyper-resection phenotype [87,88]. USP26 and USP37 are both recruited to DSBs and act on conjugates generated by RNF168. However, these DUBs act on the RAP80 pathway rather than the 53BP1 pathway. USP26/USP37 depletion increases size but not number of BRCA1 foci. This excessive spreading of BRCA1 can be countered by depletion of RAP80, suggesting the BRCA1 is being recruited into enlarged BRCA1-A complexes. In cells depleted of USP26/USP37, BRCA1 interacts less efficiently with PALB2, which bridges interaction with BRCA2. As BRCA2 is required for RAD51 loading this is a likely cause of failed HR in these cells [57,58]. The K63-Ub dependent interaction by RAP80 is disrupted by ubiquitination of lysines in close proximity to its UIM domain. USP13 counters this modification to enable recruitment of RAP80 and the BRCA1-A complex to DSBs [89]. BRCA1 recruitment in S/G2 is also regulated by USP1–UAF1, which antagonises K63-Ub chains that serve as recruiters for BRCA1. Unlike several other DUBs that antagonise K63-Ub, over-expression of USP1 only affected BRCA1 recruitment and not 53BP1. USP1 is locally inactivated by K11-Ub modification generated by APCCdh1/Ube2S which is also localised to DSBs. The APCCdh1 dependent inactivation of USP1 thus removes the restraint on K63-Ub generation in a cell cycle dependent manner [90].
BRCA1–BARD1
BARD1 aids in the recruitment of its heterodimeric partner BRCA1 to DSBs through a number of different interacting partners, including HP1γ [91]. The BARD1 BRCT repeats are modified by K63-Ub which disrupts interaction with HP1γ and therefore localisation of BRCA1–BARD1 to DSBs. This inhibitory ubiquitination is removed by USP15, a cytoplasmic DUB that accumulates at DSBs through interaction with BARD1 [37,92]. The constitutive turnover of BRCA1 is countered by USP9X, loss of which causes reduction in BRCA1 protein levels and reduced HR repair [93]. The BRCA1–BARD1 heterodimer constitutes an additional source of H2A ubiquitination in the DSB repair response, being responsible for mono-ubiquitination of the extreme C terminus of H2A at K125/127/129 [94]. This ubiquitination promotes chromatin alterations which facilitate long range DNA end resection [95]. This modification is regulated by USP48 which specifically cleaves mono-ubiquitinated H2AK125/127/129Ub. USP48 controls the extent of H2AK125/127/129 ubiquitination to prevent hyper-resection. In the absence of USP48, DNA is over-resected and the cells utilise the single strand annealing (SSA) sub-pathway in addition to HR. SSA is mutagenic due to the use of homologous repeats for bridging DSB ends, resulting in sections of DNA being deleted [48].
CtIP
CtIP activity is essential for the early commitment to HR and is extensively regulated by both Ub and SUMO E3 ligases [96,97]. Ubiquitination and SUMOylation are important for regulating CtIP steady state, promoting recruitment to DSBs and limiting excessive resection. USP4 recruits to DSBs and interacts with both the MRN complex and CtIP [14,98]. In its inactive ubiquitinated form USP4 cannot interact with these factors but due to auto-de-ubiquitination these modifications are removed, enabling interaction with MRN-CtIP. It is not fully understood how USP4 regulates CtIP activity [14,98].
EXO1
EXO1 is degraded after DSB induction as a means to limit hyper-resection [99]. EXO1 is SUMOylated by PIAS4 in response to DSB induction, which promotes its degradation independently of RNF4. SENP6 antagonises EXO1 SUMOylation promoting stabilisation [100]. UCHL5 is also required for BLM/EXO1 dependent end resection through de-ubiquitinating the INO80 subunit NFRKB, although the function of this subunit in regulating end resection is not currently understood [37].
RPA complex
USP1 both directly and indirectly — through its partner UAF1 affects HR repair [101–103]. UAF1 is critical to the HR response through its association with RAD51AP1–RAD51–DNA complex which is required for efficient RAD51 loading and unloading [101–103]. The RAD51AP1 interaction is mediated through a SUMO-like domain, interestingly SUMO has previously been shown to be important for RAD51 foci formation [104]. The RPA70 subunit is SUMOylated in response to DSB induction due to the release of the constitutive interaction with SENP6. The SUMOylation of RPA70 improves interaction with RAD51 which is required for the exchange of RPA subunits for RAD51 during HR [105].
BRCA2 and RAD51
Assembly of the BRCA1–PALB2–BRCA2 complex is restricted to S/G2 phases of the cell cycle. The Cullin3 adapter KEAP1 is responsible for preventing the formation of this complex outside of S/G2 by conjugating Ub on the interface between BRCA1 and PALB2. This disruptive ubiquitination is countered by USP11 [106,107]. USP11 turnover is regulated by KEAP1 in a cell cycle dependent manner, with USP11 protein levels at their lowest in G1. Thus cell cycle regulated turnover of USP11 allows a Ub dependent modification event to disrupt the formation of the BRCA1–PALB2–BRCA2 complex. This prevents RAD51 loading from occurring outside of S/G2 phases. Cells deficient of USP11 are defective in RAD51 loading due to failure in the formation of this complex, resulting in loss of HR [107,108]. USP21 interacts with, and stabilises the BRCA2–RAD51 complex by antagonising degradative BRCA2 ubiquitination. Down-regulation of USP21 results in failure of RAD51 loading and HR repair [109]. UCHL3 interacts with RAD51 and de-ubiquitinates residues that disrupt the RAD51–BRCA2 interaction. In the absence of UCHL3, RAD51 interaction with BRCA2 is disrupted, leading to reduced RAD51 foci formation and HR repair efficiency [110].
Chromatin relaxation
USP8 forms a complex with the early DDR factor BRIT1. Under basal conditions BRIT1 is K63-Ub modified, which is antagonised by USP8. Low levels of BRIT1 K63-Ub modification are needed for recruitment to γH2AX which promotes the further recruitment of chromatin remodellers. The subsequent relaxation of chromatin then allows accumulation of downstream repair factors [111]. SENP7 maintains steady state SUMOylation levels of the chromatin component KAP1. ATM dependent phosphorylation of KAP1 is required to disrupt the SUMO dependent interaction with the CHD3 subunit of the NuRD complex. This disruption allows dispersion of the NuRDCHD3 complex from DSBs and subsequent chromatin relaxation [112]. In the absence of SENP7 the hyperSUMOylated KAP1 retains interaction with CHD3 resulting in failure of chromatin relaxation and HR repair [113]. The NuRD complex also recruits USP11 to DSBs promoting both histone de-ubiquitination and de-acetylation which enforces proper chromatin remodelling post DSB induction [65].
Maintaining the free pool of Ub/Ubl for effective DSB response
Disruption of proteasome function results in attenuation of Ub driven DSB repair which can be overcome by suppling excess free Ub [62,114]. In addition to the proteasome, Ub/Ubl proteases are required for maintaining modifier availability, either by processing immature precursors or by recycling modifiers from substrates. Only six SUMO proteases are responsible for the bulk of SUMO maturation and recycling. Loss of SENP6 provokes a HR defect due to failure in RPA70 deSUMOylation and RAD51 filament formation [105], however, SENP6 is the major polySUMO2/3 protease, depletion of which causes substantial alterations in SUMO2/3 homeostasis in cells. These pleiotropic effects contribute to the HR defect, as re-supply of SUMO2 restores HR in SENP6 depleted cells [113]. Depletion of SENP2 also causes SUMO starvation resulting in HR failure that can be rescued with additional free SUMO [24].
NEDDylation and DSB repair
NEDD8 conjugates are enriched at DSBs [115]. NEDD8 likely plays multiple roles in DSB resolution, but a role in H4 NEDDylation by RNF111, required for RNF168 accrual has been proposed [115]. To add further complexity RNF168 has also been proposed to NEDDylate H2A acting as a competitor for Ub signalling [116]. Additionally RNF111 NEDDylates CtIP and thus regulates DNA end-resection [117]. NEDDylation is reversed by just two known active proteases — the JAMM type DUB CSN5 within the COP9 signalosome and SENP8 [118]. Given the role of NEDDylation in the DSB repair response it is unsurprising that the COP9 signalosome and its DUB component CSN5 are recruited to DSBs in a NEDD8 dependent fashion. Disruption of this complex reduces HR [37,119].
Ub/Ubl proteases and cancer
Imbalances in DSB associated Ub/Ubl protease activity can be caused by mutations or expression alterations. For example, inactivating mutations in BAP1 are common in mesothelioma, renal clear cell carcinomas and uveal melanomas. Cell lines carrying these mutations are particularly sensitive to ionising radiation and PARP inhibitors due to the important roles BAP1 plays in HR repair [68,69,120]. The amplification of the long arm of chromosome 3q is common in epithelial cancers, including lung squamous cell carcinoma, oesophageal, cervical and ovarian cancer. The 3q amplification encompasses the RNF168, USP13 and SENP2 genes. Over-expression of each of these individually promotes resistance to irradiation through altered Ub/SUMO signalling and DSB repair kinetics [24,89,121]. Several other DUBs are amplified in cancers including UCHL3 in breast cancer [110] and USP21 in hepatocellular carcinoma [109]. Dependency on Ub/Ubl proteases for cancer survival could make these useful targets in patient stratification. Indeed, inhibition of specific DUBs is currently being investigated as a means to enhance sensitivity to chemo/radiotherapies [122]. USP1 inhibitors have been proven effective in BRCA1 mutant tumours as USP1 is required for replication fork stability in the absence of functional BRCA1 [123]. The USP13/USP10 inhibitor Spautin-1 improves the anti-cancer activity of PARP inhibitors in a ovarian cancer model in mice [89]. USP7 inhibitors are also effective at sensitising therapy resistant CLL cells to HR directed therapies [124].
Perspectives
The sheer number and lack of redundancy of DSB associated Ub/Ubl proteases highlights the complexity of Ub/Ubl signalling in genomic stability (summarised in Table 1). In many cases, the reduction or inactivation of a single Ub/Ubl protease is sufficient to entirely block DSB repair.
The RNF8–RNF168–K63-Ub signalling node generates easily detectable DSB associated foci that can be visualised by a number of Ub specific antibodies such as FK2 and K63-Ub. As these modifications are read by 53BP1 which also forms readily detectable foci much of the initial research in the field focused on DUBs that regulate this step, indeed ∼8 DUBs have so far been identified that regulate 53BP1 dependent foci spreading [2]. However in more recent years Ub/Ubl proteases that regulate the earliest steps of DSB repair, the ordered clearance of repair factors and the later steps of RAD51 loading have been identified, suggesting Ub/Ubl modifiers are involved in multiple steps of DSB repair. Even greater nuance in Ub/Ubl modifier roles in DSB repair has been highlighted by a number of DUBs that remove disruptive Ub conjugates that impair protein–protein interactions needed for DSB repair.
- Further layers of complexity arise from the multiple Ub chain types now implicated in DSB repair. Additionally SUMOylation is unlikely to act separately from ubiquitination as co-modification and mixed chains are important signalling elements of the DSB response [125]. Therefore the diversity of chains types present at DSBs is likely many times greater than currently appreciated. SUMOylation is essential for the recruitment, activity and clearance of several DSB repair factors but we know relatively little concerning the activity of deSUMOylases in the DSB response, indeed there appears to be little redundancy between SENP enzymes as depletion of each causes specific DSB repair defects [24,105,113]. Finally, in both NHEJ and HR repair pathways there are multiple steps that are regulated by Ub/Ubls but the roles for their respective proteases await discovery.
Table 1. Summary table of the different roles played by Ub/Ubl proteases in the DSB response.
Function Ub/Ubl Protease Ku dimer retention UCHL3, OTUD5 MDC1 retention SENP2, ATXN3, USP7 RNF8 stabilisation ATXN3 RNF8–UBE2N catalysis antagonist OTUB1 RNF168 stabilisation USP34, USP7 RNF168 accumulation antagonist A20, USP14 H2A/H2AXK13Ub spread antagonist USP3, USP51, USP16 H2A-K118/119Ub antagonist BAP1 K63-Ub/53BP1 spread antagonist USP44, DUB3, USP11, ZUFSP, POH1, BRCC36, USP26, USP37 53BP1 spread (methyl dependent) antagonist OTUB2 RAP80–BRCA1-A complex regulators BRCC36, USP26, USP37, USP13, USP1-UAF1 BRCA1–BARD1 accumulation USP15 BRCA1 stabilisation USP9X H2A-K125/K127/K129 antagonist USP48 CtIP-MRN regulators USP4 EXO1 stabilisation SENP6, UCHL5 RPA–RAD51 interaction USP1-UAF1, SENP6 BRCA2 stabilisation USP21 RAD51 loading USP11, UCHL3 Chromatin remodellers USP8, SENP7, USP11 Free SUMO pool regulators SENP2, SENP6 Note that many proteases play multiple roles in DSB signalling e.g. USP11.
Abbreviations
- DSBs
double strand breaks
- HR
homologous recombination
- NHEJ
non-homologous end joining
- SSA
single strand annealing
Competing Interests
The Author declares that there are no competing interests associated with this manuscript.
References
- 1.Scully R., Panday A., Elango R. and Willis N.A. (2019) DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 10.1038/s41580-019-0152-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Uckelmann M. and Sixma T.K. (2017) Histone ubiquitination in the DNA damage response. DNA Repair 56, 92–101 10.1016/j.dnarep.2017.06.011 [DOI] [PubMed] [Google Scholar]
- 3.Garvin A.J. and Morris J.R. (2017) SUMO, a small, but powerful, regulator of double-strand break repair. Philos. Trans. R. Soc. Lond. B Biol. Sci. 372, 20160281 10.1098/rstb.2016.0281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brown J.S., Lukashchuk N., Sczaniecka-Clift M., Britton S., le Sage C., Calsou P. et al. (2015) Neddylation promotes ubiquitylation and release of Ku from DNA-damage sites. Cell Rep. 11, 704–714 10.1016/j.celrep.2015.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nishi R. (2017) Balancing act: to be, or not to be ubiquitylated. Mutat. Res. 803, 43–50 10.1016/j.mrfmmm.2017.07.006 [DOI] [PubMed] [Google Scholar]
- 6.Syed A. and Tainer J.A. (2018) The MRE11-RAD50-NBS1 complex conducts the orchestration of damage signaling and outcomes to stress in DNA replication and repair. Annu. Rev. Biochem. 87, 263–294 10.1146/annurev-biochem-062917-012415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shibata A., Jeggo P. and Lobrich M. (2018) The pendulum of the Ku-Ku clock. DNA Repair 71, 164–171 10.1016/j.dnarep.2018.08.020 [DOI] [PubMed] [Google Scholar]
- 8.Kragelund B.B., Weterings E., Hartmann-Petersen R. and Keijzers G. (2016) The Ku70/80 ring in non-homologous end-joining: easy to slip on, hard to remove. Front. Biosci. 21, 514–527 10.2741/4406 [DOI] [PubMed] [Google Scholar]
- 9.van den Boom J., Wolf M., Weimann L., Schulze N., Li F.H., Kaschani F., et al. (2016) VCP/p97 extracts sterically trapped Ku70/80 rings from DNA in double-strand break repair. Mol. Cell 64, 189–198 10.1016/j.molcel.2016.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nishi R., Wijnhoven P.W.G., Kimura Y., Matsui M., Konietzny R., Wu Q. et al. (2018) The deubiquitylating enzyme UCHL3 regulates Ku80 retention at sites of DNA damage. Sci. Rep. 8, 17891 10.1038/s41598-018-36235-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Vivo A., Sanchez A., Yegres J., Kim J., Emly S. and Kee Y. (2019) The OTUD5-UBR5 complex regulates FACT-mediated transcription at damaged chromatin. Nucleic Acids Res. 47, 729–746 10.1093/nar/gky1219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S. and Bonner W.M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- 13.Ha G.H., Ji J.H., Chae S., Park J., Kim S., Lee J.K., et al. (2019) Pellino1 regulates reversible ATM activation via NBS1 ubiquitination at DNA double-strand breaks. Nat. Commun. 10, 1577 10.1038/s41467-019-09641-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu H.L., Zhang H.X., Wang X.H., Tian Q.S., Hu Z.H., Peng C.M., et al. (2015) The deubiquitylating enzyme USP4 cooperates with CtIP in DNA double-strand break end resection. Cell Rep. 13, 93–107 10.1016/j.celrep.2015.08.056 [DOI] [PubMed] [Google Scholar]
- 15.Stucki M., Clapperton J.A., Mohammad D., Yaffe M.B., Smerdon S.J. and Jackson S.P. (2005) MDC1 directly binds phosphorylated histone H2AX to regulate cellular responses to DNA double-strand breaks. Cell 123, 1213–1226 10.1016/j.cell.2005.09.038 [DOI] [PubMed] [Google Scholar]
- 16.Savic V., Yin B., Maas N.L., Bredemeyer A.L., Carpenter A.C., Helmink B.A. et al. (2009) Formation of dynamic gamma-H2AX domains along broken DNA strands is distinctly regulated by ATM and MDC1 and dependent upon H2AX densities in chromatin. Mol. Cell 34, 298–310 10.1016/j.molcel.2009.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huen M.S.Y., Grant R., Manke I., Minn K., Yu X.C., Yaffe M.B. et al. (2007) RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131, 901–914 10.1016/j.cell.2007.09.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galanty Y., Belotserkovskaya R., Coates J., Polo S., Miller K.M. and Jackson S.P. (2009) Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 10.1038/nature08657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morris J.R., Boutell C., Keppler M., Densham R., Weekes D., Alamshah A., et al. (2009) The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 462, 886–890 10.1038/nature08593 [DOI] [PubMed] [Google Scholar]
- 20.Galanty Y., Belotserkovskaya R., Coates J. and Jackson S.P. (2012) RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev. 26, 1179–1195 10.1101/gad.188284.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yin Y.L., Seifert A., Chua J.S., Maure J.F., Golebiowski F. and Hay R.T. (2012) SUMO-targeted ubiquitin E3 ligase RNF4 is required for the response of human cells to DNA damage. Genes Dev. 26, 1196–1208 10.1101/gad.189274.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo K.T., Zhang H.X., Wang L.W., Yuan J. and Lou Z.K. (2012) Sumoylation of MDC1 is important for proper DNA damage response. EMBO J. 31, 3008–3019 10.1038/emboj.2012.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun H., Leverson J.D. and Hunter T. (2007) Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 26, 4102–4112 10.1038/sj.emboj.7601839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garvin A.J., Walker A.K., Densham R.M., Chauhan A.S., Stone H.R., Mackay H.L., et al. (2019) The deSUMOylase SENP2 coordinates homologous recombination and nonhomologous end joining by independent mechanisms. Genes Dev. 33, 333–347 10.1101/gad.321125.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pfeiffer A., Luijsterburg M.S., Acs K., Wiegant W.W., Helfricht A., Herzog L.K., et al. (2017) Ataxin-3 consolidates the MDC1-dependent DNA double-strand break response by counteracting the SUMO-targeted ubiquitin ligase RNF4. EMBO J. 36, 1066–1083 10.15252/embj.201695151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Su D.X., Ma S., Shan L., Wang Y., Wang Y.J., Cao C., et al. (2018) Ubiquitin-specific protease 7 sustains DNA damage response and promotes cervical carcinogenesis. J. Clin. Invest. 128, 4280–4296 10.1172/JCI120518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kolas N.K., Chapman J.R., Nakada S., Ylanko J., Chahwan R., Sweeney F.D., et al. (2007) Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318, 1637–1640 10.1126/science.1150034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thorslund T., Ripplinger A., Hoffmann S., Wild T., Uckelmann M., Villumsen B., et al. (2015) Histone H1 couples initiation and amplification of ubiquitin signalling after DNA damage. Nature 527, 389–393 10.1038/nature15401 [DOI] [PubMed] [Google Scholar]
- 29.Mandemaker I.K., van Cuijk L., Janssens R.C., Lans H., Bezstarosti K., Hoeijmakers J.H. et al. (2017) DNA damage-induced histone H1 ubiquitylation is mediated by HUWE1 and stimulates the RNF8-RNF168 pathway. Sci. Rep. 7, 15353 10.1038/s41598-017-15194-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakada S., Tai I., Panier S., Al-Hakim A., Iemura S., Juang Y.C., et al. (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466, 941–946 10.1038/nature09297 [DOI] [PubMed] [Google Scholar]
- 31.Wiener R., Zhang X.B., Wang T. and Wolberger C. (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483, 618–622 10.1038/nature10911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bekker-Jensen S., Danielsen J.R., Fugger K., Gromova I., Nerstedt A., Bartek J. et al. (2010) HERC2 coordinates ubiquitin-dependent assembly of DNA repair factors on damaged chromosomes. Nat. Cell Biol. 12, 80–86 10.1038/ncb2008 [DOI] [PubMed] [Google Scholar]
- 33.Danielsen J.R., Povlsen L.K., Villumsen B.H., Streicher W., Nilsson J., Wikstrom M. et al. (2012) DNA damage-inducible SUMOylation of HERC2 promotes RNF8 binding via a novel SUMO-binding Zinc finger. J. Cell Biol. 197, 179–187 10.1083/jcb.201106152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shanmugam I., Abbas M., Ayoub F., Mirabal S., Bsaili M., Caulder E.K. et al. (2014) Ubiquitin-specific peptidase 20 regulates Rad17 stability, checkpoint kinase 1 phosphorylation and DNA repair by homologous recombination. J. Biol. Chem. 289, 22739–22748 10.1074/jbc.M114.550459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yuan J., Luo K.T., Deng M., Li Y.H., Yin P., Gao B.W. et al. (2014) HERC2-USP20 axis regulates DNA damage checkpoint through Claspin. Nucleic Acids Res. 42, 13110–13121 10.1093/nar/gku1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhu M., Zhao H.C., Liao J. and Xu X.Z. (2014) HERC2/USP20 coordinates CHK1 activation by modulating CLASPIN stability. Nucleic Acids Res. 42, 13074–13081 10.1093/nar/gku978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nishi R., Wijnhoven P., le Sage C., Tjeertes J., Galanty Y., Forment J.V. et al. (2014) Systematic characterization of deubiquitylating enzymes for roles in maintaining genome integrity. Nat. Cell Biol. 16, 1016–1026 10.1038/ncb3028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z., Yang H.R. and Wang H.B. (2014) The histone H2A deubiquitinase USP16 interacts with HERC2 and fine-tunes cellular response to DNA damage. J. Biol. Chem. 289, 32883–32894 10.1074/jbc.M114.599605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh A.N., Oehler J., Torrecilla I., Kilgas S., Li S.D., Vaz B., et al. ) The p97-Ataxin 3 complex regulates homeostasis of the DNA damage response E3 ubiquitin ligase RNF8. EMBO J. 38, e102361 10.15252/embj.2019102361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattiroli F., Vissers J.H.A., van Dijk W.J., Ikpa P., Citterio E., Vermeulen W. et al. (2012) RNF168 ubiquitinates K13-15 on H2A/H2AX to drive DNA damage signaling. Cell 150, 1182–1195 10.1016/j.cell.2012.08.005 [DOI] [PubMed] [Google Scholar]
- 41.Panier S., Ichijima Y., Fradet-Turcotte A., Leung C.C.Y., Kaustov L., Arrowsmith C.H. et al. (2012) Tandem protein interaction modules organize the ubiquitin-dependent response to DNA double-strand breaks. Mol. Cell 47, 383–395 10.1016/j.molcel.2012.05.045 [DOI] [PubMed] [Google Scholar]
- 42.Fradet-Turcotte A., Canny M.D., Escribano-Diaz C., Orthwein A., Leung C.C.Y., Huang H., et al. (2013) 53BP1 is a reader of the DNA-damage-induced H2A Lys 15 ubiquitin mark. Nature 499, 50–54 10.1038/nature12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu Q.Z., Sharma N., He J.S., Wani G. and Wani A.A. (2015) USP7 deubiquitinase promotes ubiquitin-dependent DNA damage signaling by stabilizing RNF168*. Cell Cycle 14, 1413–1425 10.1080/15384101.2015.1007785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sy S.M., Jiang J., O W.S., Deng Y.Q. and Huen M.S.Y. (2013) The ubiquitin specific protease USP34 promotes ubiquitin signaling at DNA double-strand breaks. Nucleic Acids Res. 41, 8572–8580 10.1093/nar/gkt622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang C.Z., Zang W.C., Tang Z.F., Ji Y.P., Xu R.D., Yang Y.F., et al. (2018) A20/TNFAIP3 regulates the DNA damage response and mediates tumor cell resistance to DNA-damaging therapy. Cancer Res. 78, 1069–1082 10.1158/0008-5472.CAN-17-2143 [DOI] [PubMed] [Google Scholar]
- 46.Sharma A., Alswillah T., Singh K., Chatterjee P., Willard B., Venere M. et al. (2018) USP14 regulates DNA damage repair by targeting RNF168-dependent ubiquitination. Autophagy 14, 1976–1990 10.1080/15548627.2018.1496877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma N., Zhu Q.Z., Wani G., He J.S., Wang Q.E. and Wani A.A. (2014) USP3 counteracts RNF168 via deubiquitinating H2A and gamma H2AX at lysine 13 and 15. Cell Cycle 13, 106–114 10.4161/cc.26814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uckelmann M., Densham R.M., Baas R., Winterwerp H.H.K., Fish A., Sixma T.K. et al. (2018) USP48 restrains resection by site-specific cleavage of the BRCA1 ubiquitin mark from H2A. Nat. Commun. 9, 229 10.1038/s41467-017-02653-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang Z.Q., Zhang H.L., Liu J., Cheruiyot A., Lee J.H., Ordog T. et al. (2016) USP51 deubiquitylates H2AK13,15ub and regulates DNA damage response. Genes Dev. 30, 946–959 10.1101/gad.271841.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M.G., Aebersold R. et al. (2015) RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 10, 226–238 10.1016/j.celrep.2014.12.021 [DOI] [PubMed] [Google Scholar]
- 51.Zhang X.F., Smits A.H., van Tilburg G.B.A., Jansen P., Makowski M.M., Ovaa H. et al. (2017) An interaction landscape of ubiquitin signaling. Mol. Cell 65, 941–955.e8 10.1016/j.molcel.2017.01.004 [DOI] [PubMed] [Google Scholar]
- 52.Poulsen M., Lukas C., Lukas J., Bekker-Jensen S. and Mailand N. (2012) Human RNF169 is a negative regulator of the ubiquitin-dependent response to DNA double-strand breaks. J. Cell Biol. 197, 189–199 10.1083/jcb.201109100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chen J., Feng W.J., Jiang J., Deng Y.Q. and Huen M.S.Y. (2012) Ring finger protein RNF169 antagonizes the ubiquitin-dependent signaling cascade at sites of DNA damage. J. Biol. Chem. 287, 27715–27722 10.1074/jbc.M112.373530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitevski-LeBlanc J., Fradet-Turcotte A., Kukic P., Wilson M.D., Portella G., Yuwen T., et al. (2017) The RNF168 paralog RNF169 defines a new class of ubiquitylated histone reader involved in the response to DNA damage. eLife 6, e23872 10.7554/eLife.23872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hu Q., Botuyan M.V., Cui G.F., Zhao D.B.A. and Mer G. (2017) Mechanisms of ubiquitin-nucleosome recognition and regulation of 53BP1 chromatin recruitment by RNF168/169 and RAD18. Mol. Cell 66, 473–487.e9 10.1016/j.molcel.2017.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.An L.W., Jiang Y.Y., Ng H.H.W., Man E.P.S., Chen J., Khoo U.S. et al. (2017) Dual-utility NLS drives RNF169-dependent DNA damage responses. Proc. Natl Acad. Sci. U.S.A. 114, E2872–E2881 10.1073/pnas.1616602114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mosbech A., Lukas C., Bekker-Jensen S. and Mailand N. (2013) The deubiquitylating enzyme USP44 counteracts the DNA double-strand break response mediated by the RNF8 and RNF168 ubiquitin ligases. J. Biol. Chem. 288, 16579–16587 10.1074/jbc.M113.459917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Typas D., Luijsterburg M.S., Wiegant W.W., Diakatou M., Helfricht A., Thijssen P.E. et al. (2016) The de-ubiquitylating enzymes USP26 and USP37 regulate homologous recombination by counteracting RAP80 (vol 43, pg 6919, 2015). Nucleic Acids Res. 44, 2976–2976 10.1093/nar/gkv1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Delgado-Diaz M.R., In Y.M., Berg A., Freire R. and Smits V.A.J. (2017) Dub3 controls DNA damage signalling by direct deubiquitination of H2AX (vol 8, pg 884, 2014). Mol. Oncol. 11, 1112 10.1002/1878-0261.12097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwasna D., Rehman S.A.A., Natarajan J., Matthews S., Madden R., De Cesare V., et al. (2018) Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol. Cell 70, 150–164.e6 10.1016/j.molcel.2018.02.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Haahr P., Borgermann N., Guo X.H., Typas D., Achuthankutty D., Hoffmann S. et al. (2018) ZUFSP deubiquitylates K63-linked polyubiquitin chains to promote genome stability. Mol. Cell 70, 165–174.e6 10.1016/j.molcel.2018.02.024 [DOI] [PubMed] [Google Scholar]
- 62.Butler L.R., Densham R.M., Jia J.Y., Garvin A.J., Stone H.R., Shah V. et al. (2012) The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 31, 3918–3934 10.1038/emboj.2012.232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kakarougkas A., Ismail A., Katsuki Y., Freire R., Shibata A. and Jeggo P.A. (2013) Co-operation of BRCA1 and POH1 relieves the barriers posed by 53BP1 and RAP80 to resection. Nucleic Acids Res. 41, 10298–10311 10.1093/nar/gkt802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu M., Liu K., Mao Z.B., Luo J.Y., Gu W. and Zhao W.H. (2016) USP11 is a negative regulator to gamma H2AX ubiquitylation by RNF8/RNF168. J. Biol. Chem. 291, 959–967 10.1074/jbc.M114.624478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ting X., Xia L., Yang J.G., He L., Si W.Z., Shang Y.F. et al. (2019) USP11 acts as a histone deubiquitinase functioning in chromatin reorganization during DNA repair. Nucleic Acids Res. 47, 9721–9740 10.1093/nar/gkz726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ismail I.H., Andrin C., McDonald D. and Hendzel M.J. (2010) BMI1-mediated histone ubiquitylation promotes DNA double-strand break repair. J. Cell Biol. 191, 45–60 10.1083/jcb.201003034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sahtoe D.D., van Dijk W.J., Ekkebus R., Ovaa H. and Sixma T.K. (2016) BAP1/ASXL1 recruitment and activation for H2A deubiquitination. Nat. Commun. 7, 10292 10.1038/ncomms10292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ismail I.H., Davidson R., Gagne J.P., Xu Z.Z., Poirier G.G. and Hendzel M.J. (2014) Germline mutations in BAP1 impair its function in DNA double-strand break repair. Cancer Res. 74, 4282–4294 10.1158/0008-5472.CAN-13-3109 [DOI] [PubMed] [Google Scholar]
- 69.Yu H., Pak H., Hammond-Martel I., Ghram M., Rodrigue A., Daou S., et al. (2014) Tumor suppressor and deubiquitinase BAP1 promotes DNA double-strand break repair. Proc. Natl Acad. Sci. U.S.A. 111, 285–290 10.1073/pnas.1309085110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kleiner R.E., Verma P., Molloy K.R., Chait B.T. and Kapoor T.M. (2015) Chemical proteomics reveals a gamma H2AX-53BP1 interaction in the DNA damage response. Nat. Chem. Biol. 11, 807–814 10.1038/nchembio.1908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Botuyan M.V., Lee J., Ward I.M., Kim J.E., Thompson J.R., Chen J.J. et al. (2006) Structural basis for the methylation state-specific recognition of histone H4-K20 by 53BP1 and Crb2 in DNA repair. Cell 127, 1361–1373 10.1016/j.cell.2006.10.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Setiaputra D. and Durocher D. (2019) Shieldin - the protector of DNA ends. EMBO Rep. 20, e47560 10.15252/embr.201847560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Acs K., Luijsterburg M.S., Ackermann L., Salomons F.A., Hoppe T. and Dantuma N.P. (2011) The AAA-ATPase VCP/p97 promotes 53BP1 recruitment by removing L3MBTL1 from DNA double-strand breaks. Nat. Struct. Mol. Biol. 18, 1345–1350 10.1038/nsmb.2188 [DOI] [PubMed] [Google Scholar]
- 74.Mallette F.A., Mattiroli F., Cui G.F., Young L.C., Hendzel M.J., Mer G. et al. (2012) RNF8-and RNF168-dependent degradation of KDM4A/JMJD2A triggers 53BP1 recruitment to DNA damage sites. EMBO J. 31, 1865–1878 10.1038/emboj.2012.47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kato K., Nakajima K., Ui A., Muto-Terao Y., Ogiwara H. and Nakada S. (2014) Fine-tuning of DNA damage-dependent ubiquitination by OTUB2 supports the DNA repair pathway choice. Mol. Cell 53, 617–630 10.1016/j.molcel.2014.01.030 [DOI] [PubMed] [Google Scholar]
- 76.Kumar R. and Fang C.C. (2017) Dynamics of RIF1 SUMOylation is regulated by PIAS4 in the maintenance of genomic stability. Sci. Rep. 7, 17367 10.1038/s41598-017-16934-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Duheron V., Nilles N., Pecenko S., Martinelli V. and Fahrenkrog B. (2017) Localisation of Nup153 and SENP1 to nuclear pore complexes is required for 53BP1-mediated DNA double-strand break repair. J. Cell Sci. 130, 2306–2316 10.1242/jcs.198390 [DOI] [PubMed] [Google Scholar]
- 78.Wang B., Matsuoka S., Ballif B.A., Zhang D., Smogorzewska A., Gygi S.P. et al. (2007) Abraxas and RAP80 form a BRCA1 protein complex required for the DNA damage response. Science 316, 1194–1198 10.1126/science.1139476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Sobhian B., Shao G.Z., Lilli D.R., Culhane A.C., Moreau L.A., Xia B. et al. (2007) RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316, 1198–1202 10.1126/science.1139516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim H., Chen J.J. and Yu X.H. (2007) Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316, 1202–1205 10.1126/science.1139621 [DOI] [PubMed] [Google Scholar]
- 81.Yan J., Kim Y.S., Yang X.P., Li L.P., Liao G., Xia F. et al. (2007) The ubiquitin-interacting motif-containing protein RAP80 interacts with BRCA1 and functions in DNA damage repair response. Cancer Res. 67, 6647–6656 10.1158/0008-5472.CAN-07-0924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Guzzo C.M., Berndsen C.E., Zhu J.M., Gupta V., Datta A., Greenberg R.A. et al. (2012) DNA damage RNF4-dependent hybrid SUMO-ubiquitin chains are signals for RAP80 and thereby mediate the recruitment of BRCA1 to sites of DNA damage. Sci. Signal. 5, ra88 10.1126/scisignal.2003485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hu X., Paul A. and Wang B. (2012) Rap80 protein recruitment to DNA double-strand breaks requires binding to both small ubiquitin-like modifier (SUMO) and ubiquitin conjugates. J. Biol. Chem. 287, 25510–25519 10.1074/jbc.M112.374116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Dong Y.S., Hakimi M.A., Chen X.W., Kumaraswamy E., Cooch N.S., Godwin A.K. et al. (2003) Regulation of BRCC, a holoenzyme complex containing BRCA1 and BRCA2, by a signalosome-like subunit and its role in DNA repair. Mol. Cell 12, 1087–1099 10.1016/S1097-2765(03)00424-6 [DOI] [PubMed] [Google Scholar]
- 85.Shao G., Lilli D.R., Patterson-Fortin J., Coleman K.A., Morrissey D.E. and Greenberg R.A. (2009) The Rap80-BRCC36 de-ubiquitinating enzyme complex antagonizes RNF8-Ubc13-dependent ubiquitination events at DNA double strand breaks. Proc. Natl Acad. Sci. U.S.A. 106, 3166–3171 10.1073/pnas.0807485106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Patterson-Fortin J., Shao G., Bretscher H., Messick T.E. and Greenberg R.A. (2010) Differential regulation of JAMM domain deubiquitinating enzyme activity within the RAP80 complex. J. Biol. Chem. 285, 30971–30981 10.1074/jbc.M110.135319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Coleman K.A. and Greenberg R.A. (2011) The BRCA1-RAP80 complex regulates DNA repair mechanism utilization by restricting end resection. J. Biol. Chem. 286, 13669–13680 10.1074/jbc.M110.213728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ng H.M., Wei L.Z., Lan L. and Huen M.S.Y. (2016) The Lys(63)-deubiquitylating enzyme BRCC36 limits DNA break processing and repair. J. Biol. Chem. 291, 16197–16207 10.1074/jbc.M116.731927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Li Y.H., Luo K.T., Yin Y.J., Wu C.M., Deng M., Li L. et al. (2017) USP13 regulates the RAP80–BRCA1 complex dependent DNA damage response. Nat. Commun. 8, 15752 10.1038/ncomms15752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ha K., Ma C.X., Lin H., Tang L.C., Lian Z.S., Zhao F., et al. (2017) The anaphase promoting complex impacts repair choice by protecting ubiquitin signalling at DNA damage sites. Nat. Commun. 8, 15751 10.1038/ncomms15751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Wu W.W., Nishikawa H., Fukudal T., Vittal V., Asano M., Miyoshi Y. et al. (2015) Interaction of BARD1 and HP1 is required for BRCA1 retention at sites of DNA damage. Cancer Res. 75, 1311–1321 10.1158/0008-5472.CAN-14-2796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Peng Y.H., Liao Q.C., Tan W., Peng C.M., Hu Z.H., Chen Y.L., et al. (2019) The deubiquitylating enzyme USP15 regulates homologous recombination repair and cancer cell response to PARP inhibitors. Nat. Commun. 10, 1224 10.1038/s41467-019-09232-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lu Q., Zhang F.L., Lu D.Y., Shao Z.M. and Li D.Q. (2019) USP9X stabilizes BRCA1 and confers resistance to DNA-damaging agents in human cancer cells. Cancer Med. 8, 6730–6740 10.1002/cam4.2528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kalb R., Mallery D.L., Larkin C., Huang J.T.J. and Hiom K. (2014) BRCA1 is a histone-H2A-specific ubiquitin ligase. Cell Rep. 8, 999–1005 10.1016/j.celrep.2014.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Densham R.M., Garvin A.J., Stone H.R., Strachan J., Baldock R.A., Daza-Martin M., et al. (2016) Human BRCA1-BARD1 ubiquitin ligase activity counteracts chromatin barriers to DNA resection. Nat. Struct. Mol. Biol. 23, 647–655 10.1038/nsmb.3236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Himmels S.F. and Sartori A.A. (2016) Controlling DNA-end resection: an emerging task for ubiquitin and SUMO. Front. Genet. 7, 152 10.3389/fgene.2016.00152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Soria-Bretones I., Cepeda-Garcia C., Checa-Rodriguez C., Heyer V., Reina-San-Martin B., Soutoglou E. et al. (2017) DNA end resection requires constitutive sumoylation of CtIP by CBX4. Nat. Commun. 8, 113 10.1038/s41467-017-00183-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wijnhoven P., Konietzny R., Blackford A.N., Travers J., Kessler B.M., Nishi R. et al. (2015) USP4 auto-deubiquitylation promotes homologous recombination. Mol. Cell 60, 362–373 10.1016/j.molcel.2015.09.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Tomimatsu N., Mukherjee B., Harris J.L., Boffo F.L., Hardebeck M.C., Potts P.R. et al. (2017) DNA-damage-induced degradation of EXO1 exonuclease limits DNA end resection to ensure accurate DNA repair. J. Biol. Chem. 292, 10779–10790 10.1074/jbc.M116.772475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bologna S., Altmannova V., Valtorta E., Koenig C., Liberali P., Gentili C., et al. (2015) Sumoylation regulates EXO1 stability and processing of DNA damage. Cell Cycle 14, 2439–2450 10.1080/15384101.2015.1060381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cukras S., Lee E., Palumbo E., Benavidez P., Moldovan G.L. and Kee Y. (2016) The USP1-UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair. Cell Cycle 15, 2636–2646 10.1080/15384101.2016.1209613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Liang F.S., Longerich S., Miller A.S., Tang C., Buzovetsky O., Xiong Y. et al. (2016) Promotion of RAD51-mediated homologous DNA pairing by the RAD51AP1-UAF1 complex. Cell Rep. 15, 2118–2126 10.1016/j.celrep.2016.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Murai J., Yang K.L., Dejsuphong D., Hirota K., Takeda S. and D'Andrea A.D. (2011) The USP1/UAF1 complex promotes double-strand break repair through homologous recombination. Mol. Cell. Biol. 31, 2462–2469 10.1128/MCB.05058-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shima H., Suzuki H., Sun J.Y., Kono K., Shi L., Kinomura A., et al. (2013) Activation of the SUMO modification system is required for the accumulation of RAD51 at sites of DNA damage. J. Cell Sci. 126, 5284–5292 10.1242/jcs.133744 [DOI] [PubMed] [Google Scholar]
- 105.Dou H., Huang C., Singh M., Carpenter P.B. and Yeh E.T.H. (2010) Regulation of DNA repair through deSUMOylation and SUMOylation of replication protein A complex. Mol. Cell 39, 333–345 10.1016/j.molcel.2010.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Schoenfeld A.R., Apgar S., Dolios G., Wang R. and Aaronson S.A. (2004) BRCA2 is ubiquitinated in vivo and interacts with USP11, a deubiquitinating enzyme that exhibits prosurvival function in the cellular response to DNA damage. Mol. Cell. Biol. 24, 7444–7455 10.1128/MCB.24.17.7444-7455.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Orthwein A., Noordermeer S.M., Wilson M.D., Landry S., Enchev R.I., Sherker A., et al. (2015) A mechanism for the suppression of homologous recombination in G1 cells. Nature 528, 422–426 10.1038/nature16142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wiltshire T.D., Lovejoy C.A., Wang T., Xia F., O'Connor M.J. and Cortez D. (2010) Sensitivity to poly(ADP-ribose) polymerase (PARP) inhibition identifies ubiquitin-specific peptidase 11 (USP11) as a regulator of DNA double-strand break repair. J. Biol. Chem. 285, 14565–14571 10.1074/jbc.M110.104745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Liu J.P., Kruswick A., Dang H., Tran A.D., Kwon S.M., Wang X.W. et al. (2017) Ubiquitin-specific protease 21 stabilizes BRCA2 to control DNA repair and tumor growth. Nat. Commun. 8, 137 10.1038/s41467-017-00206-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Luo K.T., Li L., Li Y.H., Wu C.M., Yin Y.J., Chen Y.P. et al. (2016) A phosphorylation-deubiquitination cascade regulates the BRCA2-RAD51 axis in homologous recombination. Genes Dev. 30, 2581–2595 10.1101/gad.289439.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ge C.M., Che L.X., Ren J.Y., Pandita R.K., Lu J., Li K.Y. et al. (2015) BRUCE regulates DNA double-strand break response by promoting USP8 deubiquitination of BRIT1. Proc. Natl Acad. Sci. U.S.A. 112, E1210–E1219 10.1073/pnas.1418335112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Goodarzi A.A., Kurka T. and Jeggo P.A. (2011) KAP-1 phosphorylation regulates CHD3 nucleosome remodeling during the DNA double-strand break response. Nat. Struct. Mol. Biol. 18, 831–U112 10.1038/nsmb.2077 [DOI] [PubMed] [Google Scholar]
- 113.Garvin A.J., Densham R., Blair-Reid S.A., Pratt K.M., Stone H.R., Weekes D. et al. (2013) The deSUMOylase SENP7 promotes chromatin relaxation for homologous recombination DNA repair. EMBO Rep. 14, 975–983 10.1038/embor.2013.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dantuma N.P., Groothuis T.A.M., Salomons F.A. and Neefjes J. (2006) A dynamic ubiquitin equilibrium couples proteasomal activity to chromatin remodeling. J. Cell Biol. 173, 19–26 10.1083/jcb.200510071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ma T., Chen Y.B., Zhang F., Yang C.Y., Wang S.M. and Yu X.C. (2013) RNF111-Dependent neddylation activates DNA damage-induced ubiquitination. Mol. Cell 49, 897–907 10.1016/j.molcel.2013.01.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Li T.T., Guan J.H., Huang Z.J., Hu X. and Zheng X.F. (2014) RNF168-mediated h2a neddylation antagonizes ubiquitylation of H2A and regulates DNA damage repair. J. Cell Sci. 127, 2238–2248 10.1242/jcs.138891 [DOI] [PubMed] [Google Scholar]
- 117.Jimeno S., Fernandez-Avila M.J., Cruz-Garcia A., Cepeda-Garcia C., Gomez-Cabello D. and Huertas P. (2015) Neddylation inhibits CtIP-mediated resection and regulates DNA double strand break repair pathway choice. Nucleic Acids Res. 43, 987–999 10.1093/nar/gku1384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Brown J.S. and Jackson S.P. (2015) Ubiquitylation, neddylation and the DNA damage response. Open Biol. 5, 150018 10.1098/rsob.150018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Meir M., Galanty Y., Kashani L., Blank M., Khosravi R., Fernandez-Avila M.J., et al. (2015) The COP9 signalosome is vital for timely repair of DNA double-strand breaks. Nucleic Acids Res. 43, 4517–4530 10.1093/nar/gkv270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pena-LlopiS S., Vega-Rubin-De-Celis S., Liao A., Leng N., Pavia-Jimenez A., Wang S., et al. (2012) BAP1 loss defines a new class of renal cell carcinoma. Nat. Genet. 44, 751–759 10.1038/ng.2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Chroma K., Mistrik M., Moudry P., Gursky J., Liptay M., Strauss R., et al. (2017) Tumors overexpressing RNF168 show altered DNA repair and responses to genotoxic treatments, genomic instability and resistance to proteotoxic stress. Oncogene 36, 2405–2422 10.1038/onc.2016.392 [DOI] [PubMed] [Google Scholar]
- 122.Harrigan J.A., Jacq X., Martin N.M. and Jackson S.P. (2018) Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–77 10.1038/nrd.2017.152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Lim K.S., Li H., Roberts E.A., Gaudiano E.F., Clairmont C., Sambel L.A., et al. (2018) USP1 is required for replication fork protection in BRCA1-deficient tumors. Mol. Cell 72, 925–941.e4 10.1016/j.molcel.2018.10.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Agathanggelou A., Smith E., Davies N.J., Kwok M., Zlatanou A., Oldreive C.E., et al. (2017) USP7 inhibition alters homologous recombination repair and targets CLL cells independently of ATM/p53 functional status. Blood 130, 156–166 10.1182/blood-2016-12-758219 [DOI] [PubMed] [Google Scholar]
- 125.Morris J.R. and Garvin A.J. (2017) SUMO in the DNA double-stranded break response: similarities, differences, and cooperation with ubiquitin. J. Mol. Biol. 429, 3376–3387 10.1016/j.jmb.2017.05.012 [DOI] [PubMed] [Google Scholar]