Summary
Centromeres are rapidly evolving across eukaryotes, despite performing a conserved function to ensure high fidelity chromosome segregation. CENP-A chromatin is a hallmark of a functional centromere in most organisms. Due to its critical role in kinetochore architecture, the loss of CENP-A is tolerated in only a few organisms, many of which possess holocentric chromosomes. Here, we characterize the consequence of the loss of CENP-A in the fungal kingdom. Mucor circinelloides, an opportunistic human pathogen, lacks CENP-A along with the evolutionarily conserved CENP-C, but assembles a monocentric chromosome with a localized kinetochore complex throughout the cell cycle. Mis12 and Dsn1, two conserved kinetochore proteins were found to colocalize to a short region, one in each of nine large scaffolds, comprised of an ~200-bp AT-rich sequence followed by a centromere-specific conserved motif that echoes the structure of budding yeast point centromeres. Resembling fungal regional centromeres, these core centromere regions are embedded in large genomic expanses devoid of genes yet marked by Grem-LINE1s, a novel retrotransposable element silenced by the Dicer-dependent RNAi pathway. Our results suggest that these hybrid features of point and regional centromeres arose from the absence of CENP-A, thus defining novel mosaic centromeres in this early-diverging fungus.
eTOC Blurb
Mucor circinelloides lacks CENP-A and displays monocentric chromosomes with defined mosaic centromeres. Navarro-Mendoza et al. describe these mosaic centromeres as a hybrid of point and regional centromeres with a conserved, short DNA motif flanked by large stretches of Grem-LINE1 retrotransposons; a unique organization never described in fungi.
Introduction
Accurate chromosome segregation is crucial to maintain genome integrity during cell division. The timely attachment of microtubules to centromere DNA is essential to achieve proper chromosome segregation. This is accomplished by a specialized multilayered protein complex, the kinetochore which links microtubules to centromere DNA. This protein bridge is divided into two layers – the inner and outer kinetochore. The fundamental inner kinetochore protein is the histone H3 variant CENP-A. It binds directly to centromere DNA and lays the foundation to recruit other essential proteins of the kinetochore complex, playing a fundamental role in centromere structure and function, and hence, precise chromosome segregation [1,2]. CENP-A is also found at all identified neocentromeres [3] and at the active centromeres of dicentric chromosomes [4], acting as an epigenetic determinant of centromeric identity.
Despite its conserved function, the centromere is one of the most rapidly evolving regions of the genome [5]. This so-called “centromere paradox” has led to centromeres of diverse sizes and content. The first centromeres identified in Saccharomyces cerevisiae were found to be point centromeres - small regions of ~120 bp defined by specific DNA sequences [6,7]. In contrast to point centromeres described in only a few budding yeasts of the phylum Ascomycota, most other fungi and metazoans have regional centromeres that are larger, ranging from a few kilobases to several megabases [8]. Regional centromeres are often interspersed with repetitive sequences and are mostly defined by epigenetic factors rather than DNA sequence per se [9]. While the centromeres described thus far are confined to one region of the DNA, there are organisms in which their entire chromosomes are loaded with kinetochores and display a parallel separation of the two chromatids. Such chromosomes possess holocentromeres and have been found in several clades of insects [10] and nematodes like Caenorhabditis elegans [11]. Despite this remarkable variation in the types of centromeres, most organisms have their centromere loci defined by the binding of CENP-A. However, there are a few exceptions, including some insect lineages and kinetoplastids, which have independently lost CENP-A. The kinetoplastid Trypanosoma brucei has evolved a unique set of proteins to perform the role of the kinetochore complex [12], whereas all insects that have recurrently lost CENP-A have transitioned from monocentricity to holocentricity [10].
Neither holocentricity nor lack of CENP-A has ever been reported in the fungal kingdom until a recent study showed that this protein could be absent in the Mucoromycotina subphylum [13], which belongs to the early diverging fungi. Kinetochore structure and centromere function are well-studied in species of the Dikarya, whereas centromere identity as well as the kinetochore architecture remain largely unexplored in basal fungi because these are challenging to manipulate genetically. Mucor circinelloides, a fungus of the subphylum Mucoromycotina, is an exception and molecular tools are available to modify its genome [14]. M. circinelloides, as with other Mucorales, causes an infectious disease known as mucormycosis, which have been associated with high mortality rates due to rhino-orbital-cerebral, pulmonary, or cutaneous infections [15]. Despite its medical significance and research efforts to identify new antifungal targets, the genome biology and cell cycle of this fungus remain poorly understood. Using evolutionarily conserved kinetochore proteins as tools, we investigated the dynamics of the kinetochore complex during nuclear division and unveiled the unique characteristics of the centromeres of M. circinelloides. Studying distinct and divergent factors involved in the cell cycle of this organism will open avenues for the development of species-specific antifungal drugs.
Results
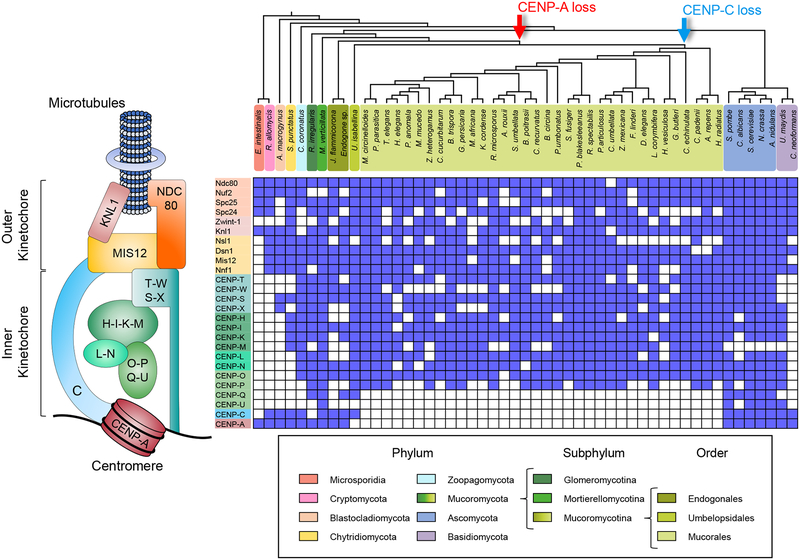
Last Mucoralean common ancestor lost CENP-A and CENP-C, and the descendant Mucoromycotina lack centromere-specific histone variants
A recent study of kinetochore evolution in eukaryotes reported the striking absence of two well conserved inner kinetochore proteins, CENP-A and CENP-C, in two species of the Mucoromycotina [13]. To ascertain the presence or loss of well-studied kinetochore proteins across the entire subphylum, we analyzed the curated proteomes of 75 fungal species, including 55 species of Mucoromycotina and 20 species from other fungal clades (Table S1). The kinetochore protein sequences from Mus musculus, Ustilago maydis, S. cerevisiae, and Schizosaccharomyces pombe served as queries to conduct iterative Blast and Hmmer searches against these proteomes. These sequences were confirmed as putative orthologs by the presence of conserved Pfam domains or by obtaining a matching first hit in a reciprocal Blastp search against the initial four species: mouse, U. maydis, budding, and fission yeasts (Table S1, protein sequences at Mendeley data DOI: 10.17632/wpyyb58h8v.1).
To confirm the presence or absence of centromere-specific histone H3 CENP-A in each fungal species, 289 histone H3 homologs were analyzed, identifying 27 putative CENP-A proteins (Figure S1, Table S1, protein sequences at Mendeley data DOI: 10.17632/wpyyb58h8v.1). These proteins share common features with well-known CENP-A proteins [16], such as an N-terminal tail that varies significantly in length and sequence compared to the canonical H3 counterparts (Figure S1B), the absence of a conserved glutamine residue in the first α-helix (69Q) and a phenylalanine residue (84F) (Figure S1C), a longer and divergent first loop (Figure S1C), and a non-conserved C-terminal tail (Figure S1D). In addition, the histone fold domain (HFD) sequences of the putative CENP-A proteins have diverged rapidly from the conserved canonical H3 sequences [16] (Figure S1E). Nine additional H3 homologs also share several CENP-A common features but were regarded as rare histones because they lack these conserved substitutions. The analysis revealed that CENP-A was absent in most of the Mucoromycotina clade, specifically in the orders Umbelopsidales and Mucorales; nevertheless, we identified putative CENP-A proteins in all Endogonalean species (Figure 1). Similarly, CENP-C is present in the Endogonales and Umbelopsidales but not in the Mucorales (Figure 1). These findings suggest that the last Mucoromycotina common ancestor possessed CENP-A, which was lost after the Endogonales diverged from the rest of the clade. In the absence of CENP-A, CENP-C was presumably lost soon after. In addition, CENP-Q is absent in all Mucoralean species, and CENP-U is lost in the whole subphylum. These proteins have been recurrently lost across the fungal kingdom, as evidenced by their absence in species of all phyla of early-diverging fungi and most species in Basidiomycota (Figure 1). The absence of orthologs that were present in closely-related species should be taken with caution because this could be a result of the incomplete state of their genome assemblies and annotation.
Figure 1. Distribution of the kinetochore complex across the Mucoromycotina subphylum.
Concise kinetochore schematic showing the most conserved protein in eukaryotes is shown on the left corresponding to the kinetochore proteins analyzed in matrix on the right. The matrix displays the presence or absence of 26 kinetochore across a cladogram of 51 fungal lineages (top) to show the relationships among them. fungal phylum is represented and color-coded, differentiating the Mucoromycota into Glomeromycotina, Mortierellomycotina, and Mucoromycotina subphyla to provide a emphasis on the latter. Arrows at divergence events in the cladogram mark the hypothetical loss of proteins CENP-A (red) and CENP-C (blue) in these clades. See also Figure S1, Tables S1 and S4.
Despite these relevant losses, the remaining kinetochore proteins were well conserved among most species of Mucoromycotina, especially in M. circinelloides. Therefore, we hypothesized that another histone protein could have taken the role of CENP-A as a part of centromeric nucleosome for kinetochore assembly. We searched the M. circinelloides genome sequence for histone protein-coding genes and examined their sequences seeking distinctive features of histone variants. Genes encoding mammalian histone variants have intronic sequences and polyadenylation [poly(A)] signals, in contrast to canonical histones H3.1 and H3.2 [17]. However, fungal canonical histones may have intronic sequences and poly(A) signals [18]. Thus, the analysis of possible variants focused on the presence of substitutions in relevant amino acid residues targeted by post-translational modifications [19] rather than intronic and poly(A) sequences. The M. circinelloides genome contains four histone H3- and three histone H4-coding genes, named histone H three (hht1–4) and histone H four (hhf1–3), respectively (Figure S2A), and all of them feature a putative poly(A) signal (5’-AATAAA-3’). Three of the four histone H3-coding genes are intronless and their protein sequence identical, except the intron-containing gene hht4 (Figure S2A). Also, the amino acid residues at positions 32, 43, and 97 in the Hht4 protein sequence differ compared to the conserved H3 sequence (Figure S2B). Similarly, histone H4-coding genes hhf2 and hhf3 lack introns and encode identical proteins, whereas hhf1 has intronic sequences (Figure S2A) and its product has a longer N-terminal tail (Figure S2C).
To test if the minor differences observed in Hht4 and Hhf1 could have resulted in a centromere-specific binding protein, the cellular localization was analyzed and compared to their canonical histone counterpart Hhf3. We constructed alleles that contained each histone-coding gene fused in-frame at the C-terminus with the enhanced green fluorescent protein (eGFP) flanked by each of the histone gene native promoter and terminator sequences, and a selectable marker (Figure S3). Homokaryotic strains expressing fluorescent histones Hht4, Hhf1, or Hhf3 were obtained by transforming a wild-type strain with their respective eGFP-tagged alleles. Integration by homologous recombination at the corresponding native loci was confirmed by PCR (Figure S3 and Table S2).
M. circinelloides is a saprophytic fungus with defined sexual and asexual lifecycles [20]. The asexual reproductive cycle is the preferred culture method to maintain this organism under laboratory conditions, due to fast developing mycelia and sporangiospores that contain asexual spores [14]. Asexual spores from these strains were observed with a confocal microscope under two stages of germination: ungerminated and 4-h pre-germinated spores, that had started swelling and forming germ tubes. In both conditions, all fluorescent histone fusions were localized encompassing the entire nucleus instead of forming kinetochore-like nuclear clusters (Figure S2D). Most species of the Mucoromycotina are coenocytic fungi and have multinucleated spores, as does the M. circinelloides wild-type strain used in this study which has large spores with a high number of nuclei [21]. We established that nuclear division in M. circinelloides is asynchronous by observing the nuclear distribution of Hht4-eGFP protein in pre-germinated spores during a time-lapse imaging session (Video S1). Altogether, these findings indicate that either none of these histone variants functions like CENP-A in kinetochore assembly or Mucor centromeres are holocentric in nature. We could test these two possibilities by studying other kinetochore proteins that are evolutionarily conserved and easily identifiable in M. circinelloides.
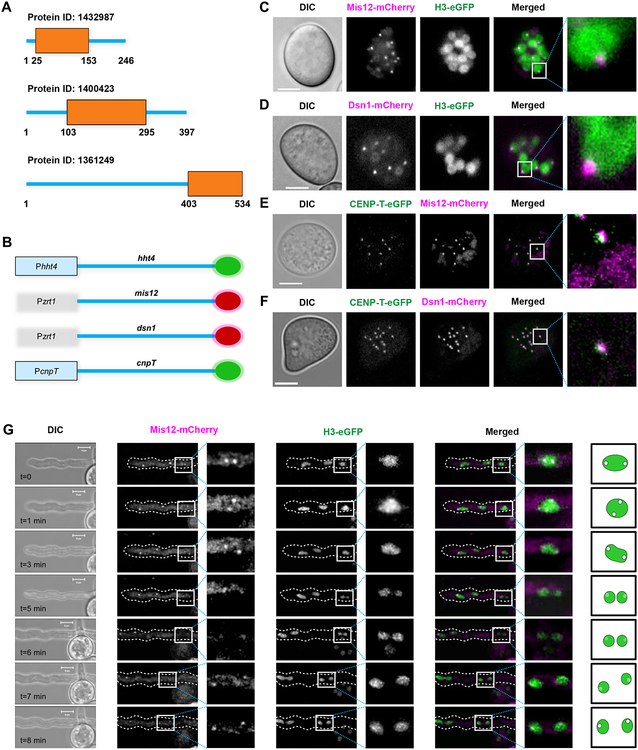
Mis12 and CENP-T complexes show discrete nuclear localization
Analyzing the nuclear localization and assembly dynamics of the kinetochore complex of M. circinelloides could offer insights into its architecture. In the absence of CENP-A or another similar histone H3 variant, and CENP-C, that could anchor the kinetochore formation, we focused our attention on the evolutionarily conserved and obvious homologs of inner kinetochore proteins that are expected to be closer to the centromere DNA in this organism (Figure 2A). We designed fluorescent kinetochore fusion proteins of Mis12, Dsn1, and CENP-T because these are well-conserved in almost all species of the Mucoromycotina. First, we constructed alleles to tag the Mis12 and Dsn1 proteins with the red fluorescent protein mCherry, both at their C-termini and N-termini, aiming to integrate the alleles in their endogenous loci following a similar strategy as for the epitope tagging of the histone genes described above. Homologous recombination in the desired transformants was confirmed by PCR, and six colonies from the four tagged alleles were checked for fluorescent localization. Unfortunately, fluorescent signals were not detected in any of the four different tagged strains possibly due to low levels of expression.
Figure 2. Inner and outer kinetochore proteins colocalize in M. circinelloides nuclei in the absence of CENP-A and CENP-C.
(A) Graphical representation of Mis12, Dsn1, and CENP-T sequence features showing individual Pfam domains PF05859, PF08202, PF15511 respectively identified in M. circinelloides. (B) Schematic to represent C-terminal tagging of histone H3 and kinetochore proteins with mCherry (red circles), eGFP (green circles). (C-F) Confocal microscope imaging showing the cellular localization of well-conserved mCherry-tagged outer kinetochore proteins Mis12 (C, E) and Dsn1 (D, F), eGFP-tagged histone H3 (C, D), and eGFP-tagged inner kinetochore CENP-T (E, F) in pre-germinated spores of M. circinelloides strains expressing fluorescent fusion proteins. (G) Time-lapsed confocal image displaying the cellular localization of mCherry-tagged Mis12 and eGFP-tagged histone H3 in a germinative tube sprouting from a spore. A discontinuous white perimeter outlines the germinative tube in the fluorescence images, and a white square indicates a nuclear division event. Cartoon for the zoomed image represents the localization and signal intensity of Mis12-mCherry in the nucleus. A calibrated scale (white bar) is provided for size comparison (5 μm) and mCherry fluorescent signal is colored as magenta in the merged images in C-G. See also Figures S2 and S3, Table S2, and Video S1.
As an alternative approach, the Mis12 and Dsn1 proteins were tagged with mCherry at each end, N-terminal and C-terminal, separately, and overexpressed from a strong promoter (Pzrt1) in M. circinelloides (Figures 2B and S3). The alleles were designed to target integration into the carRP locus, a gene encoding an enzyme involved in carotenogenesis. Thus, the mutant strains obtained should lack colored-carotenes and display an albino phenotype [14]. After confirming the integration of the alleles in the carRP locus by PCR (Figures S3E and S3F, Table S2), one strain harboring each construct was used as a parental strain to integrate the allele for H3-eGFP expression at its own locus as previously described, allowing monitoring colocalization of the kinetochore proteins within the nucleus. Once the double integration was confirmed, two independent strains of each type were selected for fluorescent localization.
Mis12 and Dsn1 displayed a clear localization signal forming small fluorescent puncta within the nucleus marked by histone H3 (Figures 2C and 2D, respectively). The microscopy screening confirmed that the expression of both N-terminal and C-terminal tagged proteins was similar. The Mis12 and Dsn1 localization patterns revealed a single cluster of kinetochores as observed in many fungal species. This result strongly suggests that M. circinelloides has monocentric chromosomes with a localized kinetochore even in the absence of CENP-A.
Next, another evolutionarily conserved inner kinetochore protein CENP-T, which is known to interact with DNA, was tagged with eGFP at the C-terminus following the same procedure used for the histone tagging and integrated by homologous recombination at the endogenous locus in strains expressing the Mis12-mCherry and Dsn1-mCherry fusion proteins. Integration of the alleles at the desired loci was confirmed by PCR in both parental backgrounds (Figure S3D), and the localization of the kinetochore proteins was examined by analyzing their fluorescent signal in pre-germinated spores. CENP-T colocalized with Mis12 and Dsn1 in each nucleus (Figures 2E and 2F, respectively), indicating that all three proteins assemble together in the kinetochore ensemble. Time-lapse imaging during spore germination of the double-tagged strains allowed the analysis of nuclear division and kinetochore structure during the cell cycle in live cells. The Mis12 and Dsn1 proteins clustered at the nuclear periphery during division (Figure 2G), a typical feature of fungal kinetochore proteins, suggesting all these three proteins are constitutively present at the kinetochore during the cell cycle.
M. circinelloides exhibits small mosaic centromeres associated with RNAi-suppressed L1-like retrotransposons
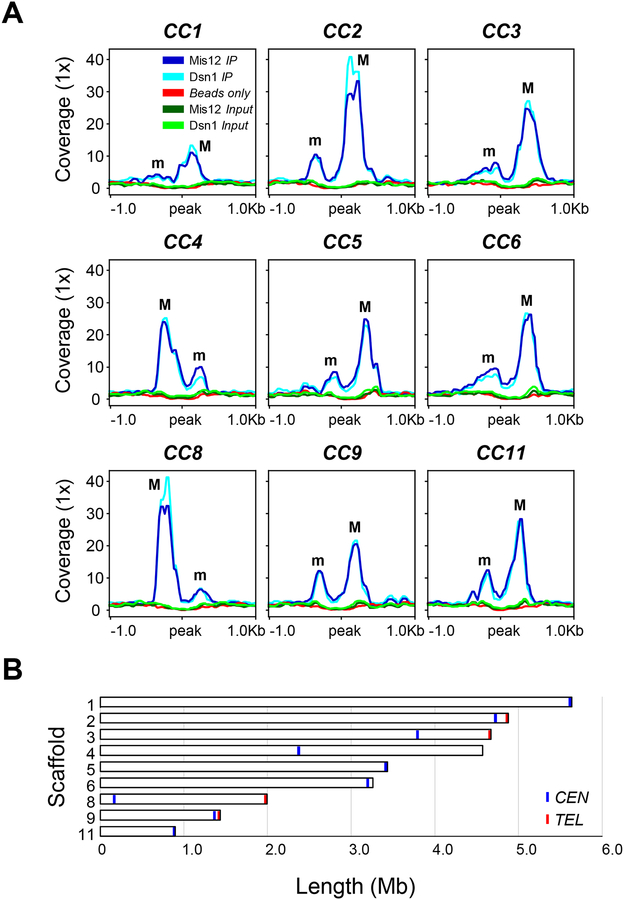
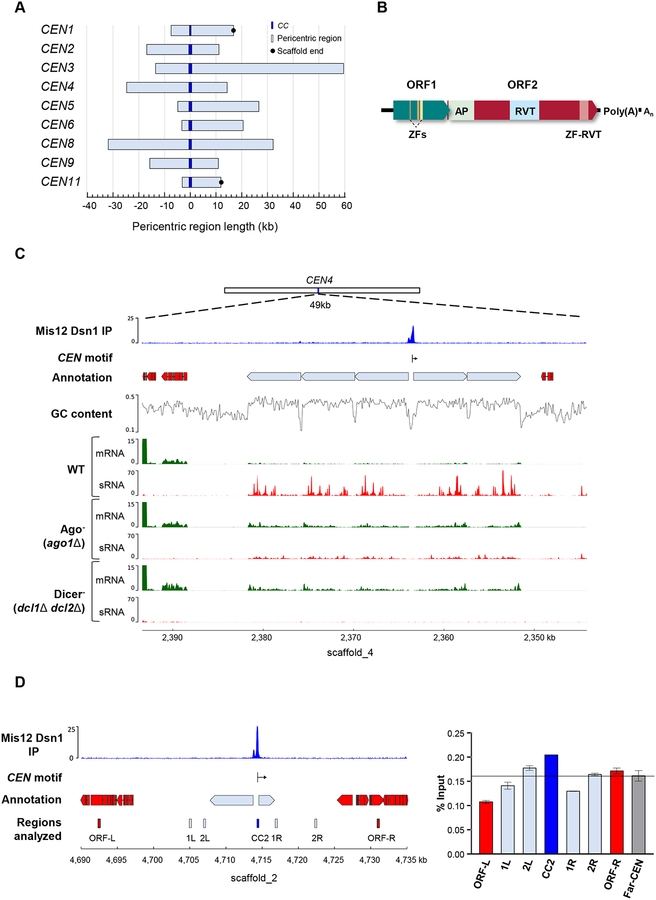
The centromere regions of early-diverging fungi have never been described, possibly because of their genetic intractability. The generation of kinetochore-tagged strains of M. circinelloides offered the opportunity to identify the nature of centromeres in an organism that lost CENP-A and CENP-C. To map M. circinelloides centromeres, chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) was performed with Mis12- and Dsn1-mCherry proteins. Illumina sequencing was conducted in two independently immunoprecipitated (IP DNA) samples for each kinetochore protein, one for Mis12-mCherry and the other for Dsn1-mCherry, using RFP-Trap MA beads (Chromotek) for mCherry immunoprecipitation. Input control samples consisting of total sheared DNA (Input DNA) were also sequenced for each tagged strain, as well as mock binding control (beads only DNA) samples with MA beads without antibody. Raw sequencing data were processed, and reads were aligned to a newly generated PacBio genome assembly of Mucor circinelloides f. lusitanicus MU402 strain (available at JGI http://genome.jgi.doe.gov/Muccir1_3/Muccir1_3.home.html, hereafter the M. circinelloides genome), the parental strain of the kinetochore protein-tagged strains. Out of the whole genome assembly of 24 scaffolds, which has an average GC content of 42.19%, 11 major scaffolds encompass more than 90% of the genome.
Both Mis12 and Dsn1 IP DNA sequence reads were compared to the corresponding control Input DNA reads to identify statistically significant kinetochore protein-enriched genomic loci in both IP DNA samples (FDR ≤ 5×10−5, fold enrichment ≥ 1.6; Data S1). The result of this analysis was visualized in a genome browser to ensure that the enrichment was robust and absent in either the Input or beads only DNA controls (Figure S4). By this approach, we detected nine significantly enriched peaks that overlapped in both Mis12 and Dsn1 IP DNA samples, and thus we defined kinetochore protein-bound regions as core centromeres (CCs) (Figure 3A) that were designated by the corresponding scaffold number. Each of these core CEN sequences was located in a different major scaffold of the genome assembly (Figure 3B), indicating that each chromosome was assembled into an average of one to three scaffolds. The identification of five repeated regions matching the telomeric sequence (5’-TTAGGG-3’) at the 3’-end of scaffolds 2, 3, 8, 9, and 10, suggested that CC2 and CC9 display a subtelomeric localization (Figure 3B). Because the length of the overlapping peaks was minimally different in the two kinetochore IP DNA samples, DNA sequence enriched with at least one of the kinetochore proteins was assumed to be the length of kinetochore binding region on each chromosome. In addition, contiguous enriched regions were added to each CC, defining the core centromere regions (Data S1).
Figure 3. M. circinelloides displays nine centromeres.
(A) Putative kinetochore-binding regions showing color-coded enrichment of immunoprecipitated DNA (IP DNA) from Mis12 and Dsn1 mCherry-tagged strains compared to their corresponding input (Input DNA) and binding controls (Beads only). 1 kb flanking sequences from the center of the enrichment peak are shown. Major (M) and minor (m) kinetochore-binding regions are indicated. Each kinetochore-protein data was obtained from a pool of duplicated IP DNA samples. (B) Scaled ideogram of the nine scaffolds (white) of M. circinelloides genome assembly that contain a kinetochore-binding region (blue), showing the telomeric repeats (red). See also Figure S4 and Data S1.
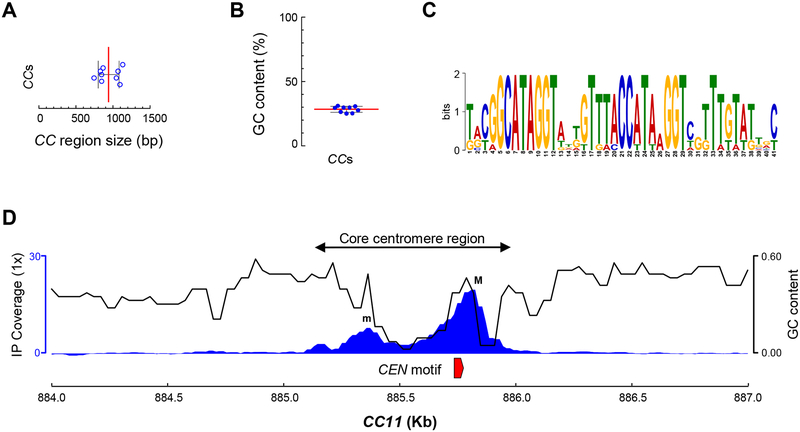
The average length of core centromere regions of M. circinelloides is 941 bp (Figure 4A). These unusually short regions are consistently AT-rich (71% average) (Figure 4B), resembling point centromeres of S. cerevisiae and its related budding yeast species. Further analysis revealed a 41-bp DNA sequence motif conserved in all nine kinetochore protein-bound core centromere regions (Figure 4C). An extensive search of the presence of this motif across the whole genome of M. circinelloides revealed that the 41-bp sequence is centromere-specific, occurs as a single-copy element at each core centromere, and is absent in the rest of the genome. Moreover, a closer inspection of the core centromere regions revealed a conserved pattern. Each core centromere is composed of a minor peak enriched of kinetochore proteins spaced by a highly AT-rich stretch spanning the major binding peak of kinetochore proteins that starts with the 41-bp conserved motif (Figure 4D).
Figure 4. The centromeres of M. circinelloides are short, AT rich and harbor a conserved DNA motif.
(A) Size distribution of the nine core centromere (CC) regions. (B) GC content across the nine core centromere (CC) sequences. The median (red line) and standard deviation (black lines) are shown in A and B. (C) 41 bp centromere-specific DNA motif conserved in all nine CC regions and absent in the remainder of the genome. A score of 2 bits in the logo indicates that the nucleotide is present at that position in all nine CC regions. (D) A genomic view of CEN11 illustrating all nine core centromere regions. Kinetochore-binding region enrichment (IP coverage) as the average of both immunoprecipitation signals minus Input and beads only controls (left axis, blue), GC content across the region (right axis, black), and the centromere-specific DNA motif described in e (red) are shown. See also Figure S4 and Data S1.
Surrounding the core centromere regions, both upstream and downstream, are stretches of genomic sequences completely devoid of genes. These pericentric regions are considerably larger than the core and vary in length, ranging from 15 to 75 kb (Figure 5A). These pericentric regions contain a variable number of large repeats oriented away from the core centromere regions (Figure S5). These repeats cluster in nine groups according to their similarity (≥ 95% identity and ≥ 70% coverage across their entire sequence). Clusters 1, 2 and 3 contain all of the full-length elements, which were termed autonomous elements, while the remaining clusters are composed of incomplete repeats or remnants (Figure S6A). A pairwise comparison of all of the repeats revealed a high similarity in their aligned sequences (≥ 90% identity), though the truncated sequences of remnants showed higher differences accounting for the gaps in the alignment.
Figure 5. M. circinelloides core centromeres are present in large ORF-free pericentric regions having retrotransposable elements regulated by the Dicer-dependent RNAi pathway and bind H3 nucleosomes.
(A) Genomic sequences lacking annotated genes (light blue) that flank the CC regions (dark blue) served as the reference points to calculate the length both upstream and downstream. Black circles at either end of the regions indicate an abrupt, non-telomeric end of scaffold. (B) Schematic of the Grem-LINE1 interspersed in the pericentromeric regions. Open Reading Frames (ORF) and protein domains predicted from their coding sequences are shown as colored boxes [Zf, zinc finger (PF00098 and PF16588); AP, AP endonuclease (PTHR22748); RVT, reverse transcriptase (PF00078); and zf-RVT, zinc-binding in reverse transcriptase (PF13966)]. (C) CEN4 is exemplified by low GC content, lack of genes and transcripts, which are shown in different tracks displaying the kinetochore-binding region enrichment (IP, an average of both IP DNA signals minus Input and beads only controls), annotation of genes (red blocks) and transposable elements (light blue blocks), CEN-specific DNA motif position (vertical line) and direction (arrow), GC content, and transcriptomic data of mRNA (green) and sRNAs (red) in M. circinelloides wild-type, ago1, and double dcl1 dcl2 deletion mutant strains after 48 h of growth in rich media. (D) Genomic location of CEN2 showing the regions studied for histone H3 occupancy as labelled and colored rectangles. ChIP assays were performed using polyclonal antibodies against histone H3. Primers were designed for CC2 region, pericentric regions (1L, 2L, 1R, 2R), flanking ORFs (ORF-L, ORF-R) and a far-CEN control ORF ~2 Mb away from CEN2 (Far-CEN). IP samples were analyzed by real-time PCR using these primers. The y-axis denotes the qPCR value as a percentage of the total chromatin input with standard error mean (SEM), from each region tested. The experiment was repeated three times with similar results. See also Figures S5 and S6, Tables S2 and S3, and Data S1.
Each autonomous repeat comprises two sequential open reading frames (ORFs) with an overlapping codon. The first ORF encodes a product containing several zinc finger domains, and the second codes for a protein with AP endonuclease and reverse transcriptase domains (Figure 5B, Table S3). Based on their high similarity, their ORF domain architecture, the presence of a poly(A) signal at their 3’-end, and the absence of long terminal repeats (LTR) at either end, they were classified as a single non-LTR retrotransposon. Phylogenetic profiling revealed that these retrotransposons belong to the LINE1 clade (Figure S6B) and are closely related to other LINE1-like retrotransposons of Mucoromycotina species, so they were termed Genomic Retrotransposable Element of Mucoromycotina (Grem) LINE1-like. A search of the M. circinelloides genome revealed that the Grem-LINE1s are found specifically in the pericentromeric regions; or at the non-telomeric ends of three small scaffolds, oriented away from the end (Table S3). Four major scaffolds end abruptly in a putative centromere region, suggesting that they may be linked to these minor scaffolds that contain the Grem-LINE1 elements at one end. Broadening the search across 55 genomes of species of Mucoromycotina identified the Grem-LINE1 elements in most Mucorales and Umbelopsidales (≥ 30% identity and ≥ 50% coverage), but they could not be found in any of the Endogonalean genomes, indicating an inverse correlation between the presence of Grem-LINE1s and CENP-A (Figure 1 and Table S4).
RNAi silences expression and prevents transposition of retrotransposable elements in several fungal species featuring functional RNAi [22,23]. M. circinelloides possess an elaborate RNAi mechanism that protects the genome against invading elements, that could prevent the movement of retrotransposable elements. The two Dicer enzymes, particularly Dcl2, and one of the three Argonaute proteins (Ago1), play a pivotal role in this protective RNAi-related pathway [24]. To analyze the transcriptional landscape of the pericentric regions, previously generated and publicly available transcriptomic raw data for mRNA and small RNAs were reprocessed and aligned to the M. circinelloides genome assembly. The pericentric regions were almost devoid of transcription in the wild-type strain, although the retrotransposons were modestly expressed. Indeed, a high number of sRNAs corresponding to the retrotransposons were detected (Figures 5C and S5), indicating an inverse correlation between mRNA and sRNA levels. This inverse correlation was confirmed by analyzing the transcriptomic profile of mutant strains lacking essential components of the RNAi machinery, Ago1 or Dcl1/Dcl2. In these RNAi-deficient mutants, the pattern was reversed; high mRNA levels from the retrotransposons were observed, while the production of sRNAs originating from those regions was considerably lower than in the wild-type strain. These findings suggest that the retrotransposons contained in the pericentric regions of M. circinelloides are silenced by the Dicer-dependent RNAi pathway.
In most organisms, deposition of CENP-A at the centromere-specific nucleosomes is correlated to depletion of histone H3 [25–27]. Because CENP-A is absent in M. circinelloides, we quantified the presence of histone H3 at the centromere-specific nucleosomes by performing ChIP with anti-histone H3 antibodies and a qPCR analysis. The occupancy of histone H3 was examined by qPCR in four regions across the scaffold_2: the spanning kinetochore protein-binding region (core centromere, CC2), the ORF-free pericentromeric region, the centromere-flanking ORFs and the control region far-CEN ORF ~2 Mb away from the centromere (Figure 5D). There are no significant differences in histone H3 occupancy across the analyzed regions, indicating that H3 is not depleted at either the kinetochore protein binding region or the pericentric region compared to the flanking ORFs and the control far-CEN ORF (Figure 5D).
Discussion
Mucoralean species are the causative agents of mucormycosis, an emerging fungal infection with extremely poor clinical outcomes, probably as a consequence of their innate resistance to all current antifungal drugs [28,29] and their ability to evade host innate immunity [30,31]. In spite of their clinical relevance, limited information is available on essential biological processes in these fungal pathogens, especially nuclear division and cell cycle. In this study, we identified major kinetochore components in M. circinelloides, and utilized them to identify and characterize nine chromosomal loci as centromeres.
In most eukaryotes, the centrochromatin is composed of nucleosomes containing the histone H3 variant CENP-A as a hallmark of centromere identity. The most striking observation of our study was the absence of this centromere determining protein in all Mucoralean species. Absence of CENP-A was accompanied by loss of CENP-C, while most other kinetochore protein orthologs were present in these species. Loss of a previously-thought indispensable protein like CENP-A raised questions on the evolutionary bases of centromere and kinetochore structure. Absence of CENP-A had been previously observed in certain insect orders including butterflies and moths, bugs and lice, earwigs, and dragonflies which in all known cases led to a transition to holocentricity, suggesting that the presence of a centromere-specific protein is dispensable for segregation of a chromosome with localized kinetochore[10].
Our study is the first to demonstrate loss of core kinetochore proteins in the fungal kingdom, reinforcing that the absence of CENP-A can be tolerated in eukaryotic organisms. Surprisingly, punctate localization patterns of other conserved kinetochore proteins including Mis12, Dsn1, and CENP-T throughout the cell cycle indicate that these organisms still retain a monocentric arrangement of a canonical kinetochore structure, as opposed to the holocentric insects that lack CENP-A. Kinetochore proteins in M. circinelloides were observed to be localized at the periphery of each nucleus in the asexual spores as well as mycelia. Three types of nuclear divisions have been studied in multinucleated fungi – synchronous, asynchronous, and parasynchronous [32]. In all ascomycetes except S. pombe and Zymoseptoria tritici, kinetochore proteins are centromere-localized and clustered throughout the cell cycle [33,34]. In the basidiomycete C. neoformans, kinetochore proteins transition between clustering and declustering states in different stages of the cell cycle [35].
We have observed asynchronous nuclear divisions during germination of asexual spores of M. circinelloides, a multinucleated and coenocytic fungus. The kinetochore was found to be consistently present as a cluster in all stages of mitosis, implying that the basic kinetochore structure in this organism is constitutively assembled throughout the cell cycle. Overall, these findings hint that the role of CENP-A is being carried out by either a distinct protein that has a newly evolved function or by one of the known kinetochore proteins like CENP-T, which also has a histone-fold DNA binding domain. We also tried to find whether one of the histone H3 variants plays a role of centromeric histone, but none of them displayed kinetochore-like localization, indicating that they are probably not involved in replacing CENP-A function in chromosome segregation mediated by the kinetochore machinery. Usually, histone H3 is depleted at the centromeres as compared to other non-centromeric loci in the genome. We observed the presence of H3 at the relatively short kinetochore protein-binding region at a level that is similar to the pericentric region or gene bodies. Thus, we propose that centromeres in this organism contain histone H3 at the centromere core. Further studies will provide the exact composition of centromere-specific nucleosomes in this basal fungus.
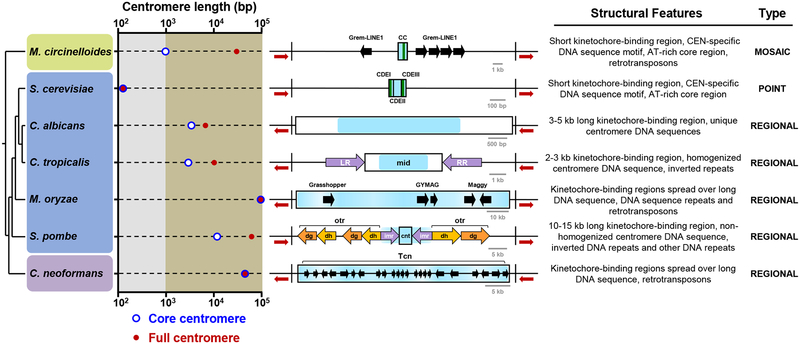
A ChIP-based analysis using Dsn1 and Mis12 revealed the first glimpse of the centromere structure of this Mucoromycotina species. The centromeres in this organism possess features of both point and regional centromeres, resulting in a unique mosaic; thus, we named them mosaic centromeres (Figure 6). One of their distinctive features is a small kinetochore protein binding domain (~750–1150 bp) defined by the presence of an AT-rich sequence stretch (~70–80% AT, ~100 bp), which is followed by a conserved centromere-specific 41-bp sequence motif. This motif is only found in M. circinelloides centromeres and marks the start of the major kinetochore-binding region. Overall, this pattern resembles the distribution of conserved elements in S. cerevisiae point centromeres, in which two conserved Centromere Determining Elements (CDEI and CDEIII) are separated by an AT-rich non conserved DNA sequence stretch (CDEII) [7]. The kinetochore of S. cerevisiae binds CDEI and CDEIII in a sequence-dependent manner through Cbf1 [36] and the CBF3 complex [37,38], respectively. The presence of a conserved DNA motif in the centromeres of M. circinelloides suggests that it could be a binding site for an unknown centromere-specific protein in this species.
Figure 6. M. circinelloides possesses a mosaic of point and regional centromeres.
Length plot of the core centromere (kinetochore-bound region, blue circles) and the full centromere (defined as the region required for centromere function, red dots) of representative fungi belonging to the Mucoromycota (green panel), Ascomycota (blue panel) and Basidiomycota (purple panel). The size axis is divided in short (≤1 kb, gray area) and long regions (>1 kb, brown area). The centromeres of M. circinelloides (CEN5), S. cerevisiae (CEN3 [7]), Candida albicans (CEN7 [39]), C. tropicalis (Scnt 3 [40]), Magnaporthe oryzae Guy11 (CEN2 [49]), S. pombe (CEN2 [52]), and C. neoformans (CEN10 [23]) are used for comparison and classified as point, regional and mosaic centromeres according to their structural features. In organisms where functionality of the centromere regions could not be experimentally determined, the ORF-free region spanning the kinetochore binding region is considered as the full centromere. Line diagrams represent the structural features of each centromere (drawn to scale). The ORF-free region at the centromere is defined by black vertical boundaries and flanking ORFs are shown as red arrows. Core centromeres (rectangles); kinetochore-bound regions (blue area); centromere-specific DNA motifs next to an AT-rich core (green lines); Grem-LINE1, Grasshopper, GYMAG, Maggy, Tcn retrotransposons (black arrows); inverted repeats (purple arrows); and other DNA repeats (light and dark orange arrows) are shown. Repeats/retrotransposons sizes are not drawn to scale. CC, core centromere; CDE, centromere DNA elements; LR, left repeat; RR, right repeat; otr, outer repeats containing dg/dh elements; imr, innermost repeats; cnt, central core.
Other features of M. circinelloides mosaic centromeres are reminiscent of fungal regional centromeres like the ones described in several Candida spp. [39,40]. The pericentric regions of M. circinelloides are large (~15–75 kb), gene-free, and transcriptionally silenced sequences that are interspersed by Grem-LINE1s, which are repeats of a LINE1-like non-LTR retrotransposable element. Several retroelements have been identified in the centromeres and neocentromeres of highly diverged eukaryotes: non-LTR LINE1-like elements in mammals [41–46] and Jockey elements in the fruit fly [47], and LTR retrotransposons in plants [48] and fungi [23,49]. Based on these findings, a model has been proposed that CENP-A is recruited by genomic sequences rich in repeats and transposable elements and thus, gives rise to the centromeres [50]. The Grem-LINE1s are arranged in a systematic pattern, oriented away from the core centromeric regions and thus forming a large inverted repeat in most of the centromeres. Similar organization and specific orientation of inverted repeats also appears flanking the core centromeres of S. pombe [51,52] and Candida tropicalis [40], performing an important role in centromere function [40,53]. M. circinelloides is the first organism to display an enrichment in retrotransposable elements in their centromeric regions in the absence of CENP-A, suggesting that these non-LTR elements determine the centromere identity. Thus, we hypothesize that identifying Grem-LINE1 clusters in other species of Mucoromycotina could help mapping the location of the putative pericentromeric regions of the corresponding species.
RNAi machinery is essential to control the mobility of transposable elements, but also to maintain centromere structure and stability [23]. Fungi that have lost essential components of the RNAi machinery have shorter pericentric regions and fewer transposable elements than closely related RNAi-proficient species. M. circinelloides has a functional and complex RNAi system with Dicer-dependent and -independent pathways [24]. Our results demonstrate that the expression of the centromeric Grem-LINE1s is suppressed by the Dicer-dependent RNAi pathway, which may have resulted in accumulation of these elements in the centromeres [23].
Overall this study has uncovered for the first time the centromeres of an early-diverging fungus, which are characterized by a mosaic of features from both point and regional centromeres found in the fungal kingdom (Figure 6). Several studies on rapidly-evolving centromeres suggest that point centromeres may be a recently evolved state as compared to regional centromeres [9]. Therefore, we hypothesize that the last mucoralean common ancestor had regional centromeres with pericentric regions that harbored retrotransposable elements contained by an active RNAi machinery. The loss of CENP-A triggered an evolutionary rearrangement from typical regional centromeres to the smaller kinetochore-binding regions that defines the mosaic centromeres of M. circinelloides.
STAR Methods
Lead contact and materials availability
All the reagents generated by this study are available for sharing. Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Victoriano Garre (vgarre@um.es).
Experimental model and subject details
Fungal strains and culture conditions
All the strains used and generated in this work derive from M. circinelloides f. lusitanicus CBS277.49. The double auxotroph MU402 [14] (Ura−, Leu−) or the MU636 (Ura+, Leu−) strain was used for transformation with cassettes containing the selectable marker for uracil pyrG or for leucine leuA, respectively. Single tagged strains in kinetochore proteins MU840, MU842, MU844, and MU846 were used as parental strains for transformation with the Hht4-eGFP cassette to obtain double-tagged strains. All the generated strains in this work are listed in the Key Resources Table (KRT).
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| RFP-Trap_MA antibody | Chromotek | Cat#rtma; RRID: AB_2631363 |
| Anti-Histone H3 antibody | Abcam | Cat#ab1791; RRID: AB_302613 |
| Bacterial and Virus Strains | ||
| Escherichia coli Stellar™ Competent Cells HST08 strain | Takara | Cat#636763 |
| Biological Samples | ||
| Chemicals, Peptides, and Recombinant Proteins | ||
| Formaldehyde Solution | Sigma Aldrich | Cat#F8775 |
| Glycine | Sigma Aldrich | Cat#G8898 |
| Protease Inhibitor Cocktail | Sigma Aldrich | Cat#P8215 |
| HEPES | Sigma Aldrich | Cat#H3375 |
| NaCl | Sigma Aldrich | Cat#S7653 |
| Triton X-100 | Sigma Aldrich | Cat#X100 |
| DOC | Sigma Aldrich | Cat#D6750 |
| EDTA | Sigma Aldrich | Cat#EDS |
| Tris | Sigma Aldrich | Cat#T1503 |
| SDS | Sigma Aldrich | Cat#L3771 |
| LiCl | Sigma Aldrich | Cat#L4408 |
| NP40 | Sigma Aldrich | Cat#NP40 |
| RNase A | Sigma Aldrich | Cat#R6513 |
| Proteinase K | Sigma Aldrich | Cat#1.07393 |
| Phenol | Sigma Aldrich | Cat#P1037 |
| Chloroform | Sigma Aldrich | Cat#C2432 |
| Isoamyl alcohol | Sigma Aldrich | Cat#309435 |
| Glycogen | Thermo Fisher | Cat#R0561 |
| Na-Acetate | Sigma Aldrich | Cat#S2889 |
| Ethanol | Sigma Aldrich | Cat#51976 |
| Critical Commercial Assays | ||
| In-Fusion HD cloning | Takara | Cat#638909 |
| TruSeq ChIP Library Prep Kit | Illumina | Cat#IP-202-1012 |
| SensiFAST SYBR® No ROX Kit | Bioline | Cat#BIO-98020 |
| Deposited Data | ||
| ChIP-seq raw data and processed files | This study | GEO: GSE132687 |
| M. circinelloides f. lusitanicus MU402 raw sequences and assembled genome | Joint Genome Institute | https://genome.jgi.doe.gov/portal/Muccir1_3/Muccir1_3.download.html |
| R7B wild-type strain mRNA | [54] | NCBI SRA: SRR1611144 |
| ago1 deletion mutant strain mRNA | [54] | NCBI SRA: SRR1611151 |
| double dcl1 dcl2 deletion mutant strain mRNA | [54] | NCBI SRA: SRR1611171 |
| R7B wild-type strain sRNA | [55] | NCBI SRA: SRR039123 |
| ago1 deletion mutant strain sRNA | [56] | NCBI SRA: SRR836082 |
| double dcl1 dcl2 deletion mutant strain sRNA | [55] | NCBI SRA: SRR039128 |
| Kinetochore ortholog sequences in Mucoromycotina and other fungi | This study | https://doi.org/10.17632/wpyyb58h8v.1 |
| Experimental Models: Organisms/Strains | ||
| Mucor circinelloides: MU402: leuA−, pyrG- | [14] | N/A |
| Mucor circinelloides: MU636: leuA−, pyrG+ | This study | N/A |
| Mucor circinelloides: MU837: hht4-eGFP::leuA, pyrG+ | This study | N/A |
| Mucor circinelloides: MU838: hht4-eGPF::leuA, pyrG+ | This study | N/A |
| Mucor circinelloides: MU839: hht4-eGPF::leuA, pyrG+ | This study | N/A |
| Mucor circinelloides: MU840: carRP::mCherry-mis12-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU841: carRP::mCherry-mis12-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU842: carRP::mis12-mCherry-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU843: carRP::mis12-mCherry-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU844: carRP::mCherry-dsn1-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU845: carRP::mCherry-dsn1-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU846: carRP::dsn1-mCherry-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU847: carRP::dsn1-mCherry-pyrG, leuA− | This study | N/A |
| Mucor circinelloides: MU851: carRP::mCherry-mis12-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU852: carRP::mCherry-mis12-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU853: carRP::mis12-mCherry-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU854: carRP::mis12-mCherry-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU855: carRP::mCherry-dsn1-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU856: carRP::mCherry-dsn1-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU857: carRP::dsn1-mCherry-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU858: carRP::dsn1-mCherry-pyrG, hht4-eGFP::leuA | This study | N/A |
| Mucor circinelloides: MU657: cnpT::cnpT-eGFP-leuA, carRP::dsn 1-mCherry-pyrG | This study | N/A |
| Mucor circinelloides: MU658: cnpT::cnpT-eGFP-leuA, carRP::dsn 1-mCherry-pyrG | This study | N/A |
| Mucor circinelloides: MU659: cnpT::cnpT-eGFP-leuA, carRP::mis12-mCherry-pyrG | This study | N/A |
| Mucor circinelloides: MU660: cnpT::cnpT-eGFP-leuA, carRP::mis12-mCherry-pyrG | This study | N/A |
| Mucor circinelloides: MU661: hhf1-eGFP::leuA, pyrG− | This study | N/A |
| Mucor circinelloides: MU662: hhf3-eGFP::leuA, pyrG− | This study | N/A |
| Mucor circinelloides: MU663: hhf3-eGFP::leuA, pyrG− | This study | N/A |
| Mucor circinelloides: MU411: dcl1::pyrG, dcl2::leuA | This study | N/A |
| Mucor circinelloides: MU413: ago1::pyrG, leuA- | This study | N/A |
| Oligonucleotides | ||
| Oligonucleotides | This study | See Table S2 |
| Recombinant DNA | ||
| pBluescript II SK+ | Agilent | Cat#212205 |
| pMAT1477 | [14] | N/A |
| pMAT1915 | This study | N/A |
| pMAT1917 | This study | N/A |
| pMAT1919 | This study | N/A |
| pMAT1916 | This study | N/A |
| pMAT1918 | This study | N/A |
| Software and Algorithms | ||
| Zeiss Zen v2.5 | Zeiss | https://www.zeiss.com/microscopy/us/products/microscope-software/zen.html#introduction |
| BLAST+ v2.7.1 | [57] | ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/2.7.1/ |
| HMMER v3.2.1 | European Bioinformatics Institute | http://www.hmmer.org |
| Pfam-A v32.0 | [58] | ftp://ftp.ebi.ac.uk/pub/databases/Pfam/releases/Pfam32.0/ |
| iTOL v4 | [59] | https://itol.embl.de/ |
| MUSCLE v3.8.1551 | [60] | https://www.drive5.com/muscle/muscle_src_3.8.1551.tar.gz |
| JTT substitution model | [61] | |
| MEGA v10.0.5 | [62] | https://www.megasoftware.net |
| FASTQC v0.11.8 | Babraham Institute | https://www.bioinformatics.babraham.ac.uk/projects/fastqc/fastqc_v0.11.8.zip |
| Trim Galore! V0.6.2 | Babraham Institute | https://github.com/FelixKrueger/TrimGalore/releases/tag/0.6.2 |
| BWAv0.7.8 | [63] | https://github.com/lh3/bwa/releases/tag/0.7.8 |
| STAR v2.7.1 | [64] | https://github.com/alexdobin/STAR/releases/tag/2.7.1a |
| deepTools v3.2.1 | [65] | https://github.com/deeptools/deepTools/releases/tag/3.2.1 |
| MACS2 v2.1.1 | [66] | https://github.com/taoliu/MACS/releases/tag/2015.4.20 |
| IGV v2.4.1 | [67] | https://software.broadinstitute.org/software/igv/download |
| MEME v5.0.5 | [68] | http://meme-suite.org/meme-software/5.0.5/meme-5.0.5.tar.gz |
| bedTools v2.28 | [69] | https://github.com/arq5x/bedtools2/releases/tag/v2.28.0 |
| Gviz v1.29.0 | [70] | http://bioconductor.riken.jp/packages/devel/bioc/src/contrib/Gviz_1.29.0.tar.gz |
| RepeatScout v1.0.5 | [71] | https://bix.ucsd.edu/repeatscout/RepeatScout-1.0.5.tar.gz |
| RepeatMasker v4.0.9 | Institute for Systems Biology | http://www.repeatmasker.org/RepeatMasker-open-4-0-9-p2.tar.gz |
| MAFFT v7.407 | [72] | https://mafft.cbrc.jp/alignment/software/mafft-7.429-with-extensions-src.tgz |
| CD-HIT v4.8.1 | [73] | https://github.com/weizhongli/cdhit/releases/tag/V4.8.1 |
| Emboss v6.6.0 | [74] | ftp://emboss.open-bio.org/pub/EMBOSS/MYEMBOSS-6.6.0.tar.gz |
| Repbase v24.04 | [75] | https://www.girinst.org/server/RepBase/protected/RepBase24.04.fasta.tar.gz |
| GraphPad Prism v6.01 | GraphPad Software | https://www.graphpad.com/ |
| Other | ||
| Binding Control Magnetic Agarose | Chromotek | Cat#bmab-20 |
| Protein A-Sepharose® 4B, Fast Flow from Staphylococcus aureus | Sigma Aldrich | Cat#P9424 |
After transformation, to select uracil prototrophy M. circinelloides spores were plated in minimal medium with Casamino Acids (MMC) [14], and for leucine prototrophy spores were plated in medium yeast nitrogen base (YNB) [14], both adjusted at pH 3.2 for transformation and colony isolation. M. circinelloides spores were cultured in rich medium yeast-peptone-glucose (YPG)[14] for optimal growth and sporulation at pH 4.5. The microscopy and ChIP assays were performed with pre-germinated spores growing in YPG pH 4.5 for 3 hours at 250 rpm and 26°C.
Method details
Construction of the fluorescent strains
Alleles containing fluorescent fusion protein coding-sequences were designed to either integrate them in their corresponding endogenous locus, or in the carRP locus. The Hht4-eGFP, Hhf1-eGFP, Hhf4-eGFP and CENP-T-eGFP alleles contained the ORF of each gene fused in-frame at their C-termini with the eGFP sequence, followed by the leuA gene as a selectable marker. The alleles were obtained by overlapping PCR using 5’-modified primers harboring restriction sites for easy cloning. Briefly, each allele was obtained by fusing the 1-kb fragment upstream the stop codon of each gene, the 0.7-kb eGFP fragment (ending in a stop codon), the 3.4-kb leuA fragment, and the 1-kb sequence downstream the stop codon of each tagged gene. All the linear alleles were digested with appropriate restriction enzymes, ligated into the plasmid Bluescript SK (+), and cloned into Escherichia coli.
The Mis12-mCherry and Dsn1-mCherry constructions were designed to allow the integration and ectopic overexpression of the fused proteins in the carRP locus. First, a universal mCherry-expression vector was constructed, containing the strong promoter Pzrt1, the ORF of the mCherry coding-gene including the stop codon, and the pyrG gene as a selectable marker (named pMAT1915). To do that, a fragment containing the 1-kb downstream and upstream sequences from the carRP gene flanking the Pzrt1 promoter was amplified with an inverse PCR using the plasmid pMAT1477 [14] as a template. Then, the mCherry and pyrG fragments were added by In-Fusion cloning (Takara) using primers with appropriate overlapping 5’-tails (Table S2). Once the universal pMAT1915 vector was constructed, it was used for the C-terminal tagging of mis12 or dsn1, fusing their ORFs without the stop codon upstream and in-frame with the mCherry ORF by In-Fusion cloning. The resulting plasmids were pMAT1917 and pMAT1919, respectively for mis12 and dsn1. For the N-terminal tagging, the plasmid pMAT1915 was inverse-amplified using primers that exclude the stop codon of the mCherry fragment, and the mis12 and dsn1 ORFs, including their stop codons, were cloned in-frame. The resulting plasmids were pMAT1916 and pMAT1918 for mCherry-Mis12 and mCherry-Dsn1 expression, respectively.
Stellar™ competent cells (Takara) were used for transformation by heat-shock following the supplier procedures. The correct construction of all the generated plasmids was confirmed by restriction analysis and Sanger sequencing.
For transformation, the alleles were released from the vector backbone by restriction digestion and used to transform M. circinelloides protoplasts by electroporation [14]. Spores from individual colonies were collected and plated in selective media to ensure homokaryosis. After at least five vegetative cycles, the integration in the target locus was confirmed by PCR using primers that amplified the whole locus (Table S2), producing PCR-fragments that allowed discrimination between the mutant and the wild-type locus (Figures S3). Genomic DNA purification was performed following the procedure previously published [54].
Microscopy imaging and acquisition
Freshly isolated M. circinelloides spores were washed twice with 1X phosphate buffered saline (PBS). The spores were resuspended in PBS and 10 l suspension was placed on a slide, covered with a coverslip and sealed with clear nail polish. The images were acquired at room temperature using laser scanning inverted confocal microscope LSM 880-Airyscan (Zeiss, Plan Apochromat 63×, NA oil 1.4) equipped with highly sensitive photodetectors. The filters used were GFP/FITC 488, mCherry 561 for excitation and GFP/FITC 500/550 band pass, mCherry 565/650 long pass for emission. Z- stack images were taken at every 0.5 m and processed using Zeiss Zen software. All the images were displayed after the maximum intensity projection of images using Zeiss Zen software.
Live cell imaging
Freshly isolated M. circinelloides spores were incubated in YPG growth medium at 26°C for 2 h. Then, the spores to be imaged were pelleted at 3000 xg and washed twice with 1X phosphate buffered saline (PBS). The spores were resuspended in PBS and 10 l suspension was placed on a slide containing thin 2% agarose patch containing 2% dextrose and covered with a coverslip. Live cell imaging was performed at 30°C in a temperature-controlled chamber of an inverted confocal microscope (Zeiss, LSM-880) using a Plan Apochromat 100X NA oil 1.4 objective and GaAsp photodetectors. Images were collected at 60-s intervals with 0.5 μm Z‐steps using GFP/FITC 488, mCherry 561 for excitation. GFP/FITC 500/550 band pass or GFP 495/500 and mCherry 565/650 or mCherry 580‐750 band pass for emission. All the images were displayed after the maximum intensity projection of images at each time using Zeiss Zen software.
Ortholog search
Seventy five fungal proteomes were retrieved from the Joint Genome Institute (JGI) Mycocosm genome portal [55] (Table S1). BLAST+ [56] v2.7.1 individual protein-protein BLASTp, iterative BLASTp (PSI-BLAST) and iterative HMMER v3.2.1 (http://www.hmmer.org) jackhammer searches (E-value ≤ 1×10−5) were conducted against these proteomes using M. musculus, U. maydis, S. cerevisiae, and S. pombe protein sequences (Table S1) as queries. In addition, an hmmsearch was launched using the Pfam-A [57] HMM models for each kinetochore protein [Pfam gathering threshold (GA) as cut-off value]. All the putative orthologs found were used to perform a reciprocal BLASTp against M. musculus, U. maydis, S. cerevisiae, and S. pombe protein sequences. Also, Pfam-A protein domains were searched in all the possible orthologs using HMMER hmmscan. Protein sequences that either lacked appropriate Pfam domains or failed to produce a hit in a reciprocal blastp search were discarded, resulting in a matrix of putative orthologs for each given species (Table S1). The cladogram showing the relationship among species was drawn using the tree data from JGI Mycocosm [55] and the interactive Tree of Life[58] (iTOL v4) tool. To determine CENP-A presence or absence in each given species, all 289 protein sequences of histone H3 putative orthologs (Table S1) were aligned using MUSCLE[59] v3.8.1551, and the phylogenetic relationship of their HFD was inferred by the neighbor-joining method employing the Jones, Taylor, and Thornton (JTT) substitution model [60], with a bootstrap procedure of 1,000 iterations (MEGA[61] v10.0.5).
Chromatin immunoprecipitation and sequencing
The ChIP protocol for M. circinelloides was adapted from previous studies in other fungal models [23]. Briefly, spores from the strains MU842 (expressing Mi12-mCherry) and MU846 (expressing Dsn1-mCherry) were used for independent IP experiments. 107 spores/mL of these strains were germinated in 100 ml of liquid YPG pH 4.5 medium, shaking at 250 rpm at 26°C for 3 hours. Then, cells were fixed by adding formaldehyde at a final concentration of 1% for 20 minutes. The fixation was quenched by adding glycine to a final concentration of 135 mM. Cells were pelleted and ground with liquid nitrogen, and 150 mg of ground pellet was resuspended in ChIP lysis Buffer (50 mM HEPES pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% DOC, and 1 mM EDTA), with a protease inhibitor cocktail. The sample was sonicated using a Bioruptor (Diogenode) with pulses of 30s ON/OFF for 50 cycles to obtain a chromatin fragmentation between 100 and 300 bp.
The lysates were clarified by centrifugation at 12,000 xg for 10 min at 4°C. At this step, 100 L were frozen at −20°C for Input DNA control. The samples were then divided into 450 L for the antibody immunoprecipitation and 450 L for the mock control. For IP 25 L of Red Fluorescent Protein (RFP)-Trap Magnetic Agarose (MA) beads (Chromotek) were added and 25 L of MA beads (Chromotek) to the binding control. Samples were incubated at 4°C overnight. After incubation, the beads were washed twice with 1 mL of the low salt wash buffer (20 mM Tris pH 8.0, 150 mM NaCl, 0.1% SDS, 1 % Triton X-100, and 2 mM EDTA), twice with 1 mL of the high salt wash buffer (20 mM Tris pH 8.0, 500 mM NaCl, 0.1% SDS, 1% Triton X-100 and 2 mM EDTA), once with the LiCl wash buffer (10 mM Tris pH 8.0, 1 mM EDTA, 0.25 M LiCl, 1% NP40, and 1% DOC), and finally once with TE (10mM Tris pH 8.0, and 1 mM EDTA). To elute the DNA, 250 L of TES (50 mM Tris pH 8.0, 10 mM EDTA, 1% SDS) was added to each sample and incubated for 10 min at 65°C, twice. The immunoprecipitated samples and the Input controls were de-crosslinked by incubating at 65°C overnight. The samples were treated with 199 g of RNase A (Sigma Aldrich) and 190 g of Proteinase K (Sigma Aldrich) for 2 hours at 50°C. For DNA purification, 1 volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added, the aqueous phase was recovered, and then 1 volume of chloroform:isoamyl alcohol (24:1) was added. After centrifugation, the aqueous phase was recovered, and DNA was precipitated with 20 g of glycogen (Thermo Fisher), 1/10 volume of Na-Acetate 3 M (pH 5.2), and 1 volume of ethanol 100%. Pulled DNA from duplicated samples were precipitated together for sequencing. Samples incubated at −20°C overnight were centrifuged for 10 min at 12,000 xg and the pellets were washed with ethanol 70%. Each sample was resuspended in 30 L of MilliQ water. The quality of the DNA was analyzed by QuBit (Thermo Fisher) and sent to Novogene, which prepared the libraries using TruSeq ChIP Library Prep Kit and sequenced them with Illumina Hiseq 2500 High-Output v4 single-end reads of 50 bp.
Histone H3-ChIP assays were performed in the same way with the following changes. The protoplasts were obtained as described before [14] and then sonicated as above. Native ChIP was performed [23] using anti-H3 antibody (Abcam ab1791). The DNA pellet was dissolved in 25 l of MilliQ water. All samples (Input, IP with or without antibodies) were used for PCR. The Input and IP DNA was subsequently used for qPCR using the primers listed in Table S2 and SensiFAST™ SYBR® No ROX Kit. Histone H3 enrichment was determined by the percentage input method using the formula: 100 × 2adjusted ct input-adjusted ct IP. The adjusted Ct value is the dilution factor (log2 of dilution factor) subtracted from the Ct value of the Input or IP DNA samples. Three technical replicates were taken for qPCR analysis and standard error of mean was calculated.
Sequencing data analysis
Raw reads from Chromatin Immunoprecipitation and RNA sequencing (ChIP- and RNA-seq) were quality-checked with FASTQC v0.11.8 and adapters removed with Trim Galore! v0.6.2 (http://www.bioinformatics.babraham.ac.uk/projects/). Reads were aligned to the Mucor circinelloides f. lusitanicus MU402 reference genome Muccir1_3 (referred to as the M. circinelloides genome throughout the text, available at JGI http://genome.jgi.doe.gov/Muccir1_3/Muccir1_3.home.html), using the Burrows-Wheeler Aligner [62] (BWA v0.7.8) algorithm for ChIP- and sRNA-seq reads and STAR [63] v2.7.1a for mRNA-seq data. The number of overlapping aligned reads per 25-bp bin was used as a measure of coverage, obtaining normalized bigWig coverage files with deepTools [64] v3.2.1 bamCoverage function. For RNA-seq data, coverage was normalized to bins per million mapped reads (BPM). For ChIP-seq data, reads were normalized per genomic content (1× normalization). ChIP enrichment peaks were identified by Model-based Analysis of ChIP-seq [65] (MACS2 v2.1.1) callpeak function and sorted by fold-enrichment and FDR values (Data S1). Enriched peaks (fold-enrichment ≥ 1.6, FDR ≤ 5×10−5) were manually curated by analyzing the IP coverage with the Integrative Genomics Viewer [66] (IGV v2.4.1) genome browser, ensuring that peaks were present in both Mis12 and Dsn1 IP samples, and absent in either the input and binding controls. The resulting centromeric sequences were analyzed with Multiple Em for Motif Elicitation [67] (MEME v5.0.5) to discover conserved DNA motifs on both strands. GC content every 25-bp bin was calculated with bedTools [68] v2.28. Normalized coverage, GC content and genetic elements annotation files were rendered with either Gviz [69] v1.29.0 for whole-chromosome visualization or deepTools pyGenomeTracks module for close-in genomic views.
Transposable element analysis
The M. circinelloides genome assembly of strain MU402 was searched for repeats using RepeatScout [70] v1.0.5 and RepeatMasker (http://www.repeatmasker.org) v4.0.9. The repeats located in the pericentromeric regions were aligned using MAFFT [71] v7.407 and clustered with CD-HIT [72] v4.8.1, generating groups of sequences that shared ≥ 95% identity and ≥ 70% coverage and calculating their p-distance (nucleotide changes per site). A BLASTn search against M. circinelloides genome assembly was conducted using a representative repeated sequence from each cluster to locate similar sequences across the entire genome, obtaining hits with ≥ 80% identity and ≥ 20% coverage (Table S3). The pericentromeric repeats were examined with getorf from Emboss [73] v6.6.0, obtaining all ORFs ≥ 600 nt. Protein domains in these ORFs translated sequences were predicted by hmmsearch using the Pfam-A v32.0 database. The reverse transcriptase (RVT) domains found in the repeated sequences were aligned with MUSCLE to create a consensus sequence, and this consensus sequence was aligned with 281 RVT domains of well-known non-LTR retrotransposons from Repbase [74] 24.04. A neighbor-joining phylogenetic tree was inferred from this alignment using the JTT substitution model and a bootstrap procedure of 1000 iterations (MEGA v10.0.5). To assess the presence of the retrotransposable elements in other species of Mucoromycotina, a tBLASTn search was launched against the 55 available genomes at JGI (Table S4) using the translated sequence of both ORFs combined, recovering hits with ≥ 50% coverage and E-value ≤ 10−5.
Quantification and statistical analysis
Statistical details of experiments are detailed in the Results, Figures and Figure legends, including the number of biological and technical replicates, the definition of center, dispersion and precision measures (mean, SEM and SD). Statistical analyses were performed with GraphPad Prism 6.
Data and code availability
ChIP-seq raw data and processed files are deposited at the Gene Expression Omnibus (GEO) repository under GSE132687 accession number and are publicly available. PacBio raw data and assembly of the genome of M. circinelloides f. lusitanicus MU402 are publicly available at the JGI website (http://genome.jgi.doe.gov/Muccir1_3/Muccir1_3.download.html) and subjected to the JGI Data Usage Policy. Both mRNA and sRNA-seq data were already available at the Sequence Read Archive (SRA) and can be accessed with the following run accession numbers: SRR1611144 (R7B wild-type strain mRNA) [75], SRR1611151 (ago1 deletion mutant strain mRNA) [75], SRR1611171 (double dcl1 dcl2 deletion mutant strain mRNA) [75], SRR039123 (R7B wild-type strain sRNA) [76], SRR836082 (ago1 deletion mutant strain sRNA) [77], SRR039128 (double dcl1 dcl2 deletion mutant strain sRNA) [76]. The protein sequences of the kinetochore orthologs identified in this study are accessible at Mendeley data (DOI: 10.17632/wpyyb58h8v.1), as well as all the main and supplementary material.
Supplementary Material
Orthologs identified in each species analyzed, showing their correspondent ID in the JGI platform, Pfam domains (name, ID, coordinates within the translated sequence, e-value and score).
(A) Genomic coordinates of the core centromere (CC) defined by the both Mis12 and Dsn1 kinetochore proteins binding regions; and of the entire centromere, including the upstream and downstream pericentric regions. (B, C) MACS2 called peaks for Mis12 IP/Input (B) and Dsn1 IP/Input (C). Rows colored red are above established threshold (FDR ≤ 5×10−7, fold enrichment ≥ 1.6).
Protein domains [name and ID (Pfam or Panther), start and end in the translated product, as well as predicted e-value, score, and bias] for the ORFs [numbered (#) and showing start and end in the DNA sequence] found in each Grem-LINE1 (showing genomic location and length).
Genomic location of Grem-LINE1s across species of Mucoromycotina showing the percentage of identity compared to a representative of M. circinelloides Grem-LINE1s.
A pre-germinated spore of M. circinelloides showing the distribution of histone H3-eGFP fusion protein encompassing the nuclei. Red arrows mark single events of nuclear division.
Highlights.
Mucorales have lost CENP-A and CENP-C, two essential components of the kinetochore
Mucor shows a monocentric arrangement of its kinetochore throughout the cell cycle
Its nine centromeres are a mosaic of point and regional centromeres
Grem-LINE1 retrotransposons surrounding the centromeres are being silenced by RNAi
Acknowledgments
This work was supported by the Ministerio de Economía y Competitividad, Spain (grant number BFU2015-65501-P, cofinanced by FEDER, and RYC-2014-15844) and Ministerio de Ciencia, Innovación y Universidades, Spain (grant number PGC2018-097452-B-I00, cofinanced by FEDER). CPA and MINM were supported by predoctoral fellowships from the Ministerio de Educación, Cultura y Deporte, Spain (grants number FPU14/01983 and FPU14/01832 respectively). We acknowledge financial support from Tata Innovation Fellowship, Department of Biotechnology, Government of India (BT/HRD/35/01/03/2017), DBT-BUILDER programme support at JNCASR (BT/INF22/SP27679/2018) and intramural support from JNCASR to KS. SP is supported by DST-WOS-A postdoctoral fellowship (SR/WOS-A/LS-207/2018). Supported in part by NIH/NIAID R37 grant AI39115-21 and R01 grant AI50113-15 to JH, and by the CIFAR program Fungal Kingdom: Threats & Opportunities, for which JH serves as Co-Director and Fellow. We thank B. Suma at the confocal imaging facility of the Molecular Biology and Genetics Unit, JNCASR for assistance in imaging. Work conducted by the US Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, was supported by the Office of Science of the US Department of Energy under Contract No. DE-AC02-05CH11231. We thank Alexander Idnurm (University of Melbourne, Australia), Luis M. Corrochano (University of Seville, Spain), Timothy Y. James (University of Michigan, USA), Mathew E. Smith (University of Florida, USA), Joseph W. Spatafora (Oregon State University, USA), Jason E. Stajich (University of California-Riverside, USA), and Gregory Bonito (Michigan State University, USA) for sharing fungal genome sequences to analyze the distribution of kinetochore proteins. We also thank Thomas D. Petes (Duke University School of Medicine, USA) and Beth A. Sullivan (Duke University School of Medicine, USA) for critical reading of the manuscript and the members of KS lab, VG lab, and JH lab for valuable discussions during bi-weekly Skype meetings.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests
The authors declare no competing interests.
References
- 1.McKinley KL, and Cheeseman IM (2016). The molecular basis for centromere identity and function. Nat. Rev. Mol. Cell Biol. 17, 16–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cheeseman IM (2014). The kinetochore. Cold Spring Harb. Perspect. Biol 6, a015826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marshall OJ, Chueh AC, Wong LH, and Choo KHA (2008). Neocentromeres: new insights into centromere structure, disease development, and karyotype evolution. Am. J. Hum. Genet 82, 261–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Earnshaw WC, and Migeon BR (1985). Three related centromere proteins are absent from the inactive centromere of a stable isodicentric chromosome. Chromosoma 92, 290–6. [DOI] [PubMed] [Google Scholar]
- 5.Henikoff S (2001). The centromere paradox: stable inheritance with rapidly evolving DNA. Science 293, 1098–1102. [DOI] [PubMed] [Google Scholar]
- 6.Clarke L, and Carbon J (1980). Isolation of a yeast centromere and construction of functional small circular chromosomes. Nature 287, 504–9. [DOI] [PubMed] [Google Scholar]
- 7.Clarke L, and Carbon J (1985). The structure and function of yeast centromeres. Annu. Rev. Genet 19, 29–55. [DOI] [PubMed] [Google Scholar]
- 8.Friedman S, and Freitag M (2017). Centrochromatin of fungi In Centromeres and kinetochores. Progress in Molecular and Subcellular Biology, Black B, ed. (Springer, Cham: ), pp. 85–109. [DOI] [PubMed] [Google Scholar]
- 9.Malik HS, and Henikoff S (2009). Major evolutionary transitions in centromere complexity. Cell 138, 1067–1082. [DOI] [PubMed] [Google Scholar]
- 10.Drinnenberg IA, DeYoung D, Henikoff S, and Malik HS (2014). Recurrent loss of CenH3 is associated with independent transitions to holocentricity in insects. eLife 3, 717–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steiner FA, and Henikoff S (2014). Holocentromeres are dispersed point centromeres localized at transcription factor hotspots. eLife 3, e02025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akiyoshi B, and Gull K (2014). Discovery of unconventional kinetochores in kinetoplastids. Cell 156, 1247–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Hooff JJ, Tromer E, van Wijk LM, Snel B, and Kops GJ (2017). Evolutionary dynamics of the kinetochore network in eukaryotes as revealed by comparative genomics. EMBO Rep. 18, e201744102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nicolás FE, Navarro-Mendoza MI, Pérez-Arques C, López-García S, Navarro E, Torres-Martínez S, and Garre V (2018). Molecular tools for carotenogenesis analysis in the mucoral Mucor circinelloides In Microbial Carotenoids. Methods in Molecular Biology, Barreiro C and Barredo JL, eds. (Humana Press, New York, NY: ). [DOI] [PubMed] [Google Scholar]
- 15.Prakash H, and Chakrabarti A (2019). Global epidemiology of mucormycosis. J. Fungi 5, E26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik HS, and Henikoff S (2003). Phylogenomics of the nucleosome. Nat. Struct. Biol 10, 882–891. [DOI] [PubMed] [Google Scholar]
- 17.Osley M (1991). The regulation of histone synthesis in the cell cycle. Annu. Rev. Biochem 60, 827–861. [DOI] [PubMed] [Google Scholar]
- 18.Yun CS, and Nishida H (2011). Distribution of introns in fungal histone genes. PLoS One 6, 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luense LJ, Wang X, Schon SB, Weller AH, Lin Shiao E, Bryant JM, Bartolomei MS, Coutifaris C, Garcia BA, and Berger SL (2016). Comprehensive analysis of histone post-translational modifications in mouse and human male germ cells. Epigenetics Chromatin 9, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schipper MAA (1973). A study on variability in Mucor hyemalis and related species. Stud. Mycol 4, 1–40. [Google Scholar]
- 21.Li CH, Cervantes M, Springer DJ, Boekhout T, Ruiz-Vazquez RM, Torres-Martinez SR, Heitman J, and Lee SC (2011). Sporangiospore size dimorphism is linked to virulence of Mucor circinelloides. PLoS Pathog. 7, e1002086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith KM, Phatale PA, Sullivan CM, Pomraning KR, and Freitag M (2011). Heterochromatin is required for normal distribution of Neurospora crassa CenH3. Mol. Cell. Biol 31, 2528–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yadav V, Sun S, Billmyre RB, Thimmappa BC, Shea T, Lintner R, Bakkeren G, Cuomo CA, Heitman J, and Sanyal K (2018). RNAi is a critical determinant of centromere evolution in closely related fungi. Proc. Natl. Acad. Sci 115, 3108–3113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Torres-Martínez S, and Ruiz-Vázquez RM (2016). RNAi pathways in Mucor: A tale of proteins, small RNAs and functional diversity. Fungal Genet. Biol 90, 44–52. [DOI] [PubMed] [Google Scholar]
- 25.Mizuguchi G, Xiao H, Wisniewski J, Smith MM, and Wu C (2007). Nonhistone Scm3 and histones CenH3-H4 assemble the core of centromere-specific nucleosomes. Cell 129, 1153–1164. [DOI] [PubMed] [Google Scholar]
- 26.Thakur J, Talbert PB, and Henikoff S (2015). Inner kinetochore protein interactions with regional centromeres of fission yeast. Genetics 201, 543–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kapoor S, Zhu L, Froyd C, Liu T, and Rusche LN (2015). Regional centromeres in the yeast Candida lusitaniae lack pericentromeric heterochromatin. Proc. Natl. Acad. Sci 112, 12139–12144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Calo S, Shertz-Wall C, Lee SC, Bastidas RJ, Nicolás FE, Granek JA, Mieczkowski P, Torres-Martínez S, Ruiz-Vázquez RM, Cardenas ME, et al. (2014). Antifungal drug resistance evoked via RNAi-dependent epimutations. Nature 513, 555–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caramalho R, Tyndall JDA, Monk BC, Larentis T, Lass-Flörl C, and Lackner M (2017). Intrinsic short-tailed azole resistance in mucormycetes is due to an evolutionary conserved aminoacid substitution of the lanosterol 14α-demethylase. Sci. Rep 7, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andrianaki AM, Kyrmizi I, Thanopoulou K, Baldin C, Drakos E, Soliman SSM, Shetty AC, McCracken C, Akoumianaki T, Stylianou K, et al. (2018). Iron restriction inside macrophages regulates pulmonary host defense against Rhizopus species. Nat. Commun 9, 3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pérez-Arques C, Navarro-Mendoza MI, Murcia L, Lax C, Martínez-García P, Heitman J, Nicolás FE, and Garre V (2019). Mucor circinelloides thrives inside the phagosome through an Atf-mediated germination pathway. mBio 10, 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gladfelter AS, Hungerbuehler AK, and Philippsen P (2006). Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 172, 347–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu X, McLeod I, Anderson S, Yates JR, and He X (2005). Molecular analysis of kinetochore architecture in fission yeast. EMBO J. 24, 2919–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schotanus K, Soyer JL, Connolly LR, Grandaubert J, Happel P, Smith KM, Freitag M, and Stukenbrock EH (2015). Histone modifications rather than the novel regional centromeres of Zymoseptoria tritici distinguish core and accessory chromosomes. Epigenetics Chromatin 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kozubowski L, Yadav V, Chatterjee G, Sridhar S, Yamaguchi M, Kawamoto S, Bose I, Heitman J, and Sanyal K (2013). Ordered kinetochore assembly in the human-pathogenic basidiomycetous yeast Cryptococcus neoformans. mBio 4, e00614–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hemmerich P, Stoyan T, Wieland G, Koch M, Lechner J, and Diekmann S (2000). Interaction of yeast kinetochore proteins with centromere-protein/transcription factor Cbf1. Proc. Natl. Acad. Sci 97, 12583–12588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sorger PK, Doheny KF, Hieter P, Kopski KM, Huffaker TC, and Hyman AA (1995). Two genes required for the binding of an essential Saccharomyces cerevisiae kinetochore complex to DNA. Proc. Natl. Acad. Sci 92, 12026–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pietrasanta LI, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, and Hansma H (1999). Probing the Saccharomyces cerevisiae centromeric DNA (CEN DNA)-binding factor 3 (CBF3) kinetochore complex by using atomic force microscopy. Proc. Natl. Acad. Sci 96, 3757–3762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sanyal K, Baum M, and Carbon J (2004). Centromeric DNA sequences in the pathogenic yeast Candida albicans are all different and unique. Proc. Natl. Acad. Sci 101, 11374–11379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chatterjee G, Sankaranarayanan SR, Guin K, Thattikota Y, Padmanabhan S, Siddharthan R, and Sanyal K (2016). Repeat-associated fission yeast-like regional centromeres in the ascomycetous budding yeast Candida tropicalis. PLoS Genet. 12, e1005839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miga KH, Newton Y, Jain M, Altemose N, Willard HF, and Kent WJ (2014). Centromere reference models for human chromosomes X and Y satellite arrays. Genome Res. 24, 697–707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chueh AC, Northrop EL, Brettingham-Moore KH, Choo KHA, and Wong LH (2009). LINE retrotransposon RNA is an essential structural and functional epigenetic component of a core neocentromeric chromatin. PLoS Genet. 5, e1000354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Carbone L, Harris RA, Mootnick AR, Milosavljevic A, Martin DIK, Rocchi M, Capozzi O, Archidiacono N, Konkel MK, Walker JA, et al. (2012). Centromere remodeling in Hoolock leuconedys (Hylobatidae) by a new transposable element unique to the gibbons. Genome Biol. Evol 4, 648–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.de Sotero-Caio CG, Cabral-de-Mello DC, Calixto M. da S., Valente GT, Martins C, Loreto V, de Souza MJ, and Santos N (2017). Centromeric enrichment of LINE-1 retrotransposons and its significance for the chromosome evolution of Phyllostomid bats. Chromosom. Res 25, 313–325. [DOI] [PubMed] [Google Scholar]
- 45.Longo MS, Carone DM, Green ED, O’Neill MJ, and O’Neill RJ (2009). Distinct retroelement classes define evolutionary breakpoints demarcating sites of evolutionary novelty. BMC Genomics 10, 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O’Neill RJW, O’Neill MJ, and Graves JAM (1998). Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature 393, 68–72. [DOI] [PubMed] [Google Scholar]
- 47.Chang C-H, Chavan A, Palladino J, Wei X, Martins NMC, Santinello B, Chen C-C, Erceg J, Beliveau BJ, Wu C-T, et al. (2019). Islands of retroelements are major components of Drosophila centromeres. PLoS Biol. 17, e3000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Presting GG (2018). Centromeric retrotransposons and centromere function. Curr. Opin. Genet. Dev 49, 79–84. [DOI] [PubMed] [Google Scholar]
- 49.Yadav V, Yang F, Reza MH, Liu S, Valent B, Sanyal K, and Naqvi NI (2019). Cellular dynamics and genomic identity of centromeres in cereal blast fungus. mBio 10, e01581–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Klein SJ, and O’Neill RJ (2018). Transposable elements: genome innovation, chromosome diversity, and centromere conflict. Chromosom. Res 26, 5–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Steiner NC, Hahnenberger KM, and Clarke L (1993). Centromeres of the fission yeast Schizosaccharomyces pombe are highly variable genetic loci. Mol. Cell. Biol 13, 4578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pidoux AL, and Allshire RC (2004). Kinetochore and heterochromatin domains of the fission yeast centromere. Chromosom. Res 12, 521–534. [DOI] [PubMed] [Google Scholar]
- 53.Baum M, Ngan VK, and Clarke L (1994). The centromeric K-type repeat and the central core are together sufficient to establish a functional Schizosaccharomyces pombe centromere. Mol. Biol. Cell 5, 747–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Navarro-Mendoza MI, Pérez-Arques C, Murcia L, Martínez-García P, Lax C, Sanchis M, Capilla J, Nicolás FE, and Garre V (2018). Components of a new gene family of ferroxidases involved in virulence are functionally specialized in fungal dimorphism. Sci. Rep 8, 7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grigoriev IV, Nikitin R, Haridas S, Kuo A, Ohm R, Otillar R, Riley R, Salamov A, Zhao X, Korzeniewski F, et al. (2014). MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 42, D699–D704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, and Madden TL (2009). BLAST+: architecture and applications. BMC Bioinformatics 10, 421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47, D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Letunic I, and Bork P (2019). Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 47, W256–W259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Edgar RC (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jones DT, Taylor WR, and Thornton JM (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci 8, 275–82. [DOI] [PubMed] [Google Scholar]
- 61.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, and Kumar S (2011). MEGA5: Molecular Evolutionary Genetics Analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol 28, 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Li H, and Durbin R (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dobin A, and Gingeras TR (2015). Mapping RNA-seq Reads with STAR In Current Protocols in Bioinformatics (Hoboken, NJ, USA: John Wiley & Sons, Inc.), pp. 11.14.1–11.14.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, and Manke T (2016). deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 44, W160–W165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, et al. (2008). Model-based Analysis of ChIP-Seq (MACS). Genome Biol. 9, R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, and Mesirov JP (2011). Integrative genomics viewer. Nat. Biotechnol 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, and Noble WS (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, and Hall IM (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hahne F, and Ivanek R (2016). Visualizing genomic data using Gviz and Bioconductor In Statistical genomics, Mathé E and Davis S, eds. (Humana Press, New York, NY: ), pp. 335–351. [DOI] [PubMed] [Google Scholar]
- 70.Price AL, Jones NC, and Pevzner PA (2005). De novo identification of repeat families in large genomes. Bioinformatics 21, i351–i358. [DOI] [PubMed] [Google Scholar]
- 71.Katoh K, and Standley DM (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fu L, Niu B, Zhu Z, Wu S, and Li W (2012). CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rice P, Longden I, and Bleasby A (2000). EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16, 276–7. [DOI] [PubMed] [Google Scholar]
- 74.Bao W, Kojima KK, and Kohany O (2015). Repbase update, a database of repetitive elements in eukaryotic genomes. Mob. DNA 6, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nicolás FE, Vila A, Moxon S, Cascales MD, Torres-Martínez S, Ruiz-Vázquez RM, and Garre V (2015). The RNAi machinery controls distinct responses to environmental signals in the basal fungus Mucor circinelloides. BMC Genomics 16, 237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nicolas FE, Moxon S, de Haro JP, Calo S, Grigoriev IV, Torres-Martínez S, Moulton V, Ruiz-Vázquez RM, and Dalmay T (2010). Endogenous short RNAs generated by Dicer 2 and RNA-dependent RNA polymerase 1 regulate mRNAs in the basal fungus Mucor circinelloides. Nucleic Acids Res. 38, 5535–5541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Cervantes M, Vila A, Nicolás FE, Moxon S, de Haro JP, Dalmay T, Torres-Martínez S, and Ruiz-Vázquez RM (2013). A single argonaute gene participates in exogenous and endogenous RNAi and controls cellular functions in the basal fungus Mucor circinelloides. PLoS One 8, e69283. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Orthologs identified in each species analyzed, showing their correspondent ID in the JGI platform, Pfam domains (name, ID, coordinates within the translated sequence, e-value and score).
(A) Genomic coordinates of the core centromere (CC) defined by the both Mis12 and Dsn1 kinetochore proteins binding regions; and of the entire centromere, including the upstream and downstream pericentric regions. (B, C) MACS2 called peaks for Mis12 IP/Input (B) and Dsn1 IP/Input (C). Rows colored red are above established threshold (FDR ≤ 5×10−7, fold enrichment ≥ 1.6).
Protein domains [name and ID (Pfam or Panther), start and end in the translated product, as well as predicted e-value, score, and bias] for the ORFs [numbered (#) and showing start and end in the DNA sequence] found in each Grem-LINE1 (showing genomic location and length).
Genomic location of Grem-LINE1s across species of Mucoromycotina showing the percentage of identity compared to a representative of M. circinelloides Grem-LINE1s.
A pre-germinated spore of M. circinelloides showing the distribution of histone H3-eGFP fusion protein encompassing the nuclei. Red arrows mark single events of nuclear division.
Data Availability Statement
ChIP-seq raw data and processed files are deposited at the Gene Expression Omnibus (GEO) repository under GSE132687 accession number and are publicly available. PacBio raw data and assembly of the genome of M. circinelloides f. lusitanicus MU402 are publicly available at the JGI website (http://genome.jgi.doe.gov/Muccir1_3/Muccir1_3.download.html) and subjected to the JGI Data Usage Policy. Both mRNA and sRNA-seq data were already available at the Sequence Read Archive (SRA) and can be accessed with the following run accession numbers: SRR1611144 (R7B wild-type strain mRNA) [75], SRR1611151 (ago1 deletion mutant strain mRNA) [75], SRR1611171 (double dcl1 dcl2 deletion mutant strain mRNA) [75], SRR039123 (R7B wild-type strain sRNA) [76], SRR836082 (ago1 deletion mutant strain sRNA) [77], SRR039128 (double dcl1 dcl2 deletion mutant strain sRNA) [76]. The protein sequences of the kinetochore orthologs identified in this study are accessible at Mendeley data (DOI: 10.17632/wpyyb58h8v.1), as well as all the main and supplementary material.