Abstract
OBJECTIVE
Maternal gestational diabetes mellitus (GDM) has been associated with adverse outcomes in the offspring. Growing evidence suggests that the epigenome may play a role, but most previous studies have been small and adjusted for few covariates. The current study meta-analyzed the association between maternal GDM and cord blood DNA methylation in the Pregnancy and Childhood Epigenetics (PACE) consortium.
RESEARCH DESIGN AND METHODS
Seven pregnancy cohorts (3,677 mother-newborn pairs [317 with GDM]) contributed results from epigenome-wide association studies, using DNA methylation data acquired by the Infinium HumanMethylation450 BeadChip array. Associations between GDM and DNA methylation were examined using robust linear regression, with adjustment for potential confounders. Fixed-effects meta-analyses were performed using METAL. Differentially methylated regions (DMRs) were identified by taking the intersection of results obtained using two regional approaches: comb-p and DMRcate.
RESULTS
Two DMRs were identified by both comb-p and DMRcate. Both regions were hypomethylated in newborns exposed to GDM in utero compared with control subjects. One DMR (chr 1: 248100345–248100614) was located in the OR2L13 promoter, and the other (chr 10: 135341870–135342620) was located in the gene body of CYP2E1. Individual CpG analyses did not reveal any differentially methylated loci based on a false discovery rate–adjusted P value threshold of 0.05.
CONCLUSIONS
Maternal GDM was associated with lower cord blood methylation levels within two regions, including the promoter of OR2L13, a gene associated with autism spectrum disorder, and the gene body of CYP2E1, which is upregulated in type 1 and type 2 diabetes. Future studies are needed to understand whether these associations are causal and possible health consequences.
Introduction
Gestational diabetes mellitus (GDM) is one of the most common pregnancy complications, with prevalence estimates ranging from 2% to 25% depending on the screening and diagnostic criteria used and the population examined (1,2). In addition to the adverse pregnancy and delivery outcomes associated with GDM, which can include preeclampsia, macrosomia, and shoulder dystocia (3), women diagnosed with GDM are four times more likely to have children who develop metabolic syndrome later in life and twice as likely to have children who become overweight or obese (4). There is also evidence that maternal GDM during pregnancy alters fetal growth trajectories (5) and adversely affects neurodevelopment (6,7). Thus, understanding the molecular changes related to prenatal exposure to GDM could have widespread implications for children’s health.
One potential mechanism underlying such a diverse array of GDM-associated outcomes is epigenetic dysregulation. In support of this, a growing number of studies have observed associations between GDM and cord blood DNA methylation patterns (8–17). However, the majority of studies have been small (e.g., <100 participants or <30 GDM cases), adjusted for few if any covariates, and used lenient or no adjustment for multiple testing (8–10,12–15,17), which may have contributed to a lack of replication of results across studies.
There has therefore been a call for research on GDM and offspring DNA methylation within larger studies (18). The current study conducted a meta-analysis of results from epigenome-wide association studies (EWAS) of GDM and cord blood DNA methylation patterns from seven cohorts participating in the Pregnancy and Childhood Epigenetics (PACE) consortium (19). Additionally, we conducted a look-up in our meta-analysis results for CpGs that were previously identified as differentially methylated in prior publications.
Research Design and Methods
Participating Cohorts
All cohorts in the PACE consortium (19) were invited to participate in the current meta-analysis. Seven cohorts, representing eight countries, participated, contributing a total of 317 GDM case and 3,360 control subjects (Table 1 and Supplementary Table 1). These cohorts are the Avon Longitudinal Study of Parents and Children (ALSPAC), the Genome-Wide Population-Based Association Study of Extremely Overweight Young Adults (GOYA), the Healthy Start Study, Proyecto Infancia y Medio Ambiente (INMA), the Prediction and Prevention of Preeclampsia and Intrauterine Growth Restriction (PREDO) study, Project Viva, and a pooled analysis of three cohorts: the Rhea Study (RHEA), the ENVIRonmental influence ON early AGEing (ENVIRONAGE) study, and the Piccolipiù study (RHEA/ENVIRONAGE/Piccolipiù). Cohort details are described in the Supplementary Data. Each cohort received ethics approval and informed consent from participants prior to data collection, and the current meta-analysis was approved by the Health Sciences Institutional Review Board of the University of Southern California.
Table 1.
Characteristics of participating cohorts*
| Cohort | Location | Participant enrollment years | GDM screening approach | GDM classification criteria and source of information | GDM case subjects (n) | Control subjects (n) |
|---|---|---|---|---|---|---|
| ALSPAC | U.K. | 1991–1992 | Selective | Physician-diagnosed GDM (medical records) | 22 | 867 |
| GOYA | Denmark | 1996–2002 | Selective | Physician-diagnosed GDM based on a one-step 75-g OGTT (medical records) + self-report | 28 | 404 |
| Healthy Start Study | U.S. | 2009–2014 | Universal | Physician-diagnosed GDM based on a two-step 100-g OGTT, Carpenter-Coustan criteria (medical records) | 32 | 534 |
| INMA | Spain | 2004–2007 | Selective | Two-step 100-g OGTT, Carpenter-Coustan criteria (medical records) | 12 | 144 |
| PREDO | Finland | 2006–2010 | Universal | One-step 75-g OGTT, IADPSG criteria (medical records) | 180 | 600 |
| RHEA/ENVIRONAGE/Piccolipiù (pooled) | Greece/Belgium/Italy | 2007–2008/2010–ongoing/2011–2015 | Universal/universal/universal | Two-step 100-g OGTT, Carpenter-Coustan criteria (medical records)/two-step 100-g OGTT, Carpenter-Coustan criteria (medical records)/self-report | 20 | 352 |
| Project Viva | U.S. | 1999–2002 | Universal | Two-step 100-g OGTT, Carpenter-Coustan criteria (medical records) | 23 | 459 |
Additional details on GDM classification and other characteristics of each cohort are included in Supplementary Data.
GDM
Participants diagnosed with type 1 or type 2 diabetes prior to the index pregnancy were excluded from analyses. The criteria used to classify GDM cases are summarized by cohort in Table 1 and are also described in more detail in the Supplementary Data. For all cohorts except Piccolipiù, GDM was primarily classified based on information that was abstracted from medical records. Due to a lack of international consensus, the criteria used to classify GDM differ by country and have changed over time. In the U.S. and some European countries, GDM is often diagnosed using a two-step approach, which entails universal screening with a 50-g glucose challenge test, followed by a 100-g 3-h oral glucose tolerance test (OGTT) for those who test positive (20). In contrast, some European countries have adopted the International Association of Diabetes and Pregnancy Study Groups (IADPSG) guidelines (21), which recommend a one-step approach, in which a 75-g 2-h OGTT is performed for all women at 24–28 weeks’ gestation. Furthermore, some countries use a selective approach and only administer GDM diagnostic tests to women with traditional risk factors. GDM cases from the Healthy Start Study, Project Viva, RHEA, and ENVIRONAGE were classified based on the two-step approach, using the Carpenter-Coustan criteria (22). GDM cases from PREDO were classified based on the IADPSG one-step approach. Piccolipiù identified GDM cases based on self-reported questionnaire data collected at delivery, and all but one case was confirmed using medical record data (IADPSG one-step approach [21]). GDM cases from INMA were diagnosed using a selective screening approach, where women at high risk for GDM were administered a glucose challenge test, followed by a diagnostic OGTT, using the Carpenter-Coustan criteria (22). GDM cases from ALSPAC and GOYA were diagnosed based on the practices at the time in the U.K. and Denmark, respectively, in which diagnostic tests were only performed for women 1) at high risk for GDM based on established risk factors or 2) with glycosuria (23,24). Given anticipated underreporting of GDM in the medical records, information from telephone interviews was also used to classify GDM cases in GOYA.
Methylation Measurements
Cord blood DNA was bisulfite converted using a EZ-96 DNA Methylation Kit (Zymo Research Corporation, Irvine). Each cohort measured DNA methylation using the Infinium HumanMethylation450 BeadChip array (Illumina, San Diego, CA), either at Illumina or in cohort-specific laboratories, and each cohort conducted its own quality control and normalization of data, as described in Supplementary Data. Since the PACE consortium has observed that extreme outliers (greater than three times the interquartile range) can have a large impact on results, they were removed prior to analyses. For all analyses, normalized, untransformed β values were evaluated as outcomes.
Cohort-Specific Statistical Analyses
Cohorts ran independent EWAS models according to the same analysis plan, using robust linear regression, as this method controls for possible heteroscedasticity and potential outliers. Only singleton pregnancies were included in analyses. GDM was modeled as the exposure of interest, and the cord blood DNA methylation level at each CpG was modeled as the outcome. Regression models were adjusted for hypothesized confounders, which included newborn’s sex, maternal age, maternal education level, maternal BMI (prepregnancy or early pregnancy), maternal smoking status during pregnancy (ever vs. never), and maternal genetic ancestry (if available) or maternal race/ethnicity. Cohort-specific details for covariate assessment are described in Supplementary Data. First, we adjusted only for this baseline set of covariates (results are presented in Supplementary Data), such that results could be compared with previous studies, which have generally not accounted for cord blood cell heterogeneity. However, our final model was additionally adjusted for cord blood cell fractions, including B cells, CD8+ T cells, CD4+ T cells, granulocytes, natural killer cells, monocytes, and nucleated red blood cells, which were estimated using a cord blood reference panel (25). We also examined results from two of the larger participating cohorts (PREDO and Project Viva) after additional adjustment for parity. Since results were very similar (Supplementary Tables 2 and 3), parity was not included in the final model.
Meta-analyses
METAL (26) was used to conduct inverse variance–weighted fixed-effects meta-analyses, using results from the cohort-specific analyses. Control probes, probes mapping to the X and Y chromosomes, and probes that have been shown to cross-hybridize or that target polymorphic CpGs or contain single nucleotide polymorphisms (SNPs) at the single base pair (bp) extension (27) were excluded. A total of 380,878 CpGs were therefore included in the meta-analyses. Probes were annotated to hg19 using the IlluminaHumanMethylation450kanno.ilmn12.hg19 R package (28). After meta-analyses were complete, a second analyst ran shadow meta-analyses to rule out potential human error. CpGs were considered differentially methylated if the false discovery rate (FDR)–adjusted P value (PFDR) was <0.05.
Potential heterogeneity between studies was assessed using the Cochran Q statistic and I2. Additionally, leave-one-out meta-analyses (i.e., comparison of results after the sequential removal of one cohort and a meta-analysis of the remaining six cohorts) were conducted to evaluate the influence of each individual cohort on the results.
Differentially methylated regions (DMRs) were identified from meta-analysis results by taking the intersection of DMRs identified using two different software programs: comb-p (29) and DMRcate (30). comb-p identifies regions enriched for low P values, uses the Stouffer-Liptak method to correct for autocorrelation, and adjusts for multiple testing using the Sidak correction (29). DMRcate calculates two smoothed estimates for each chromosome (one weighted by F statistics and one not) and uses a Satterthwaite approximation to compare these estimates; it then adjusts for multiple testing using the FDR method (30). These approaches were selected because they can be applied to meta-analysis results. Windows of 500 and 1,000 bp were compared for each approach. For comb-p, a P value threshold of 1 × 10−3 was used to specify the start of each region, and a distance of 200 bp was selected for extending the region. For DMRcate, the default settings were used, as recommended (30), and FDR thresholds of 0.05 and 0.01 were compared.
Sensitivity Analyses
Since the seven participating cohorts represent different geographic regions and differ in the timing of participant recruitment and the criteria used to classify GDM cases, we ran a series of sensitivity meta-analyses. We compared meta-analysis results after restricting to 1) cohorts with GDM cases identified by selective versus universal screening, 2) cohorts with GDM cases identified using a one-step 75-g versus a two-step 100-g OGTT, 3) European versus U.S. cohorts, and 4) cohorts that recruited participants prior to 2004 versus after 2004.
Look-up Analyses
In an effort to replicate previous findings, a look-up of CpGs previously identified as differentially methylated by GDM status was conducted within results from the meta-analyses (both with and without adjustment for estimated cell proportions). Relevant studies were identified in PubMed using the following search terms: Gestational diabetes AND DNA methylation. We focused on studies that 1) were not included in the current meta-analyses, 2) included >10 GDM cases, 3) measured DNA methylation in cord blood using the Infinium HumanMethylation450K, MethylationEPIC, or HumanMethylation27 BeadChip array, 4) adjusted for multiple testing using any method, and 5) provided effect estimates and P values for individual CpGs. Two studies met these criteria (9,12). These studies collectively reported a total of 110 differentially methylated CpGs, none of which were common. Additionally, nine CpGs within two genes (MEST and NR3C1) that were identified as differentially methylated by GDM status in both cord blood and placenta in a previous candidate gene study (11), which are represented on the 450K array, were evaluated. Of these 119 CpGs, 32 were cross-reactive or polymorphic or the CpG probe contained a SNP at the single bp extension (27). These 32 CpGs were therefore excluded, leaving a total of 87 CpGs for the look-up analyses.
Results
Study Characteristics
Characteristics of participating studies are shown in Table 1 and Supplementary Table 1. The number (%) of GDM cases per study ranged from 12 (7.7%) for INMA to 180 (23.1%) for PREDO. The majority of participants were of European ancestry, and approximately half of the newborns were male (N = 1,900 [51.7%]).
Meta-analyses for the Individual CpG Results
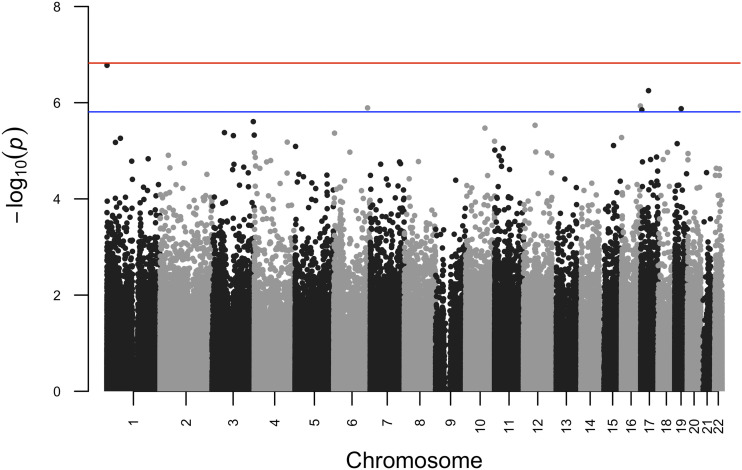
Probe numbers and the level of inflation (λ) for individual cohort results are shown in Supplementary Table 4. The λ for the meta-analyses was 1.15. Meta-analysis results are summarized in a Manhattan plot (Fig. 1). No CpGs were identified as differentially methylated by GDM status based on a PFDR < 0.05, but six were identified based on a PFDR < 0.10 (Table 2). While the directions of effect were generally consistent for the Healthy Start Study, INMA, PREDO, Project Viva, and the pooled analysis of RHEA/ENVIRONAGE/Piccolipiù, they often differed for ALSPAC or GOYA (Table 2). For five of the CpGs, there was not strong evidence of heterogeneity (I2 < 10.0, Pheterogeneity > 0.36), but for one CpG (cg11723077), there was evidence of moderate heterogeneity (I2 = 38.7, P = 0.13). However, effect estimates were similar across the leave-one-out meta-analyses (results shown in Supplementary Fig. 1 and Supplementary Table 5).
Figure 1.
Manhattan plot summarizing results for meta-analyses of the associations between maternal GDM and cord blood DNA methylation. Meta-analyses were run using METAL on results from robust linear regression models, which adjusted for newborn’s sex, maternal age, maternal education, maternal BMI (prepregnancy or in early pregnancy), maternal smoking status during pregnancy (ever vs. never), maternal genetic ancestry (if available) or maternal race/ethnicity, and estimated cord blood cell fractions. Blue and red lines indicate log10(P values) that are equivalent to a PFDR of 0.10 and a PFDR of 0.05, respectively.
Table 2.
CpGs with a PFDR < 0.10 in the meta-analysis of maternal GDM exposure*
| CpG | Genomic position | % methylation difference (95% CI)† | Direction by cohort‡ | Raw P value | FDR-corrected P value | Pheterogeneity§ | I2 | Relation to CpG island‖ | Target gene¶ |
|---|---|---|---|---|---|---|---|---|---|
| cg00812770 | chr 1:1073510 | 0.8 (0.5, 1.1) | + − + + + + + | 1.7 × 10−7 | 0.06 | 0.55 | 0.0 | South shore | LINC01342 |
| cg11723077 | chr 6: 158508188 | −1.0 (−1.4, −0.6) | − + − − − − − | 1.3 × 10−6 | 0.09 | 0.13 | 38.7 | South shore | SYNJ2 |
| cg22791932 | chr 16: 88537374 | 0.8 (0.4, 1.1) | + + + − + + + | 1.2 × 10−6 | 0.09 | 0.74 | 0.0 | Island | ZFPM1 |
| cg17588003 | chr 17: 5138696 | −1.4 (−2.0, −0.8) | + − − − − − − | 1.4 × 10−6 | 0.09 | 0.67 | 0.0 | Open sea | C17orf87 |
| cg11187204 | chr 17: 36480526 | −1.6 (−2.1, −1.0) | + + − − − − − | 5.2 × 10−8 | 0.09 | 0.57 | 0.0 | Open sea | NA |
| cg10139436 | chr 19: 30219558 | −0.4 (−0.5, −0.2) | + + − − − − − | 1.3 × 10−6 | 0.09 | 0.36 | 9.7 | South shelf | NA |
NA, not applicable.
*Results are from inverse variance–weighted fixed-effects meta-analyses, conducted using METAL. Each cohort independently ran robust linear regression models, with adjustment for newborn’s sex, maternal age (in years), maternal BMI (early pregnancy or prepregnancy), maternal smoking status during pregnancy, maternal education, maternal genetic ancestry (if available) or maternal race/ethnicity, and estimated proportions of B cells, CD8+ T cells, CD4+ T cells, granulocytes, natural killer cells, monocytes, and nucleated red blood cells in cord blood.
†% difference in newborn DNA methylation and 95% CI in comparison of the GDM case group with the control group.
‡Direction of association between GDM and methylation at the locus of interest by cohort, ordered as follows: ALSPAC, GOYA, the Healthy Start Study, INMA, PREDO, RHEA/ENVIRONAGE/Piccolipiù, Project Viva.
§The heterogeneity P value and I2 were calculated by METAL using Cochran Q test for heterogeneity.
‖Relation to CpG island from the UCSC Genome Browser, annotated using the Infinium HumanMethylation450K Manifest File (28).
¶Target gene name(s) from the UCSC Genome Browser, annotated using the Infinium HumanMethylation450K Manifest File (28).
Look-up Analysis Results
The full look-up analysis results are presented in Supplementary Tables 6 and 7 within the Supplementary Data. Of the 87 CpGs examined, 4 were differentially methylated (uncorrected P < 0.05) in the same direction in the meta-analysis that accounted for cell heterogeneity (Supplementary Table 7). These four CpGs (cg01203331, cg03345925, cg08471713, and cg20507276) were annotated to a total of seven genes: NOP56, SNORD56, SNORD57, SNORD86, ZC3H3, MEOX1, and OR2L13, respectively. However, based on the 87 tests conducted, FDR-corrected P values exceeded 0.05 for all of the CpGs evaluated.
DMRs Identified from the Meta-analysis Results
Using individual CpG results from the meta-analyses, comb-p identified five regions that were differentially methylated by GDM status (Supplementary Table 8). comb-p results were the same when either a 500 or 1,000 bp window was used. DMRcate identified two DMRs when using an FDR threshold of 0.10. One DMR was identified when using the 500 bp window (chr 1: 248100407–248100614) and the other when using the 1,000 bp window (chr 10: 135341870–135342620) (Supplementary Table 9). Both of these DMRs overlapped two DMRs that had also been identified by comb-p (Table 3). One was located in the promoter region of OR2L13 and was also annotated to pseudogene CLK3P2. The second overlapped a CpG island in the gene body of CYP2E1. Percent methylation levels in both regions were lower in the GDM case, compared with control, group, and effect estimates were generally consistent for the individual CpGs contained within each region (Supplementary Fig. 2). DMRcate did not identify any DMRs when using an FDR threshold of 0.05.
Table 3.
DMRs identified by both comb-p and DMRcate*
| DMR | CpGs comprising the DMR | Direction of association | Nearby gene(s) | Regulatory feature group/gene group/relation to island |
|---|---|---|---|---|
| comb-p (500 and 1,000 bp window) | ||||
| chr 1: 248100345–248100614 | cg00785941, cg03748376, cg04028570, cg08260406, cg08944170, cg20434529, cg20507276 | − | OR2L13, CLK3P2 | Promoter-associated/first exon: 5′UTR or TSS200/island or north shore |
| chr 10: 135342218–135342413 | cg10862468, cg25330361 | − | CYP2E1 | NA/body/island |
| DMRcate (500 bp window)chr 1: 248100407–248100614 | cg00785941, cg03748376, cg04028570, cg08260406, cg08944170, cg20507276 | − | OR2L13 | Promoter-associated/first exon/5′UTR/island |
| DMRcate (1,000 bp window) | cg00321709, cg10862468, cg19469447, cg23400446, cg24530264, cg25330361 | − | CYP2E1 | Unclassified/body/island |
| chr 10: 135341870–135342620 |
NA, not applicable. UTR, untranslated region.
DMRs were identified from meta-analysis results for individual CpGs, which used results from robust linear regression models that were adjusted for newborn sex, maternal age (in years), maternal BMI (prepregnancy or early pregnancy), maternal education, maternal smoking status during pregnancy (ever vs. never), maternal genetic ancestry (if available) or maternal race/ethnicity, and estimated cord blood cell fractions.
Sensitivity Analysis Results
Results were generally similar for the six CpGs, with a PFDR < 0.10, and also for CpGs within the two DMRs identified by comb-p and DMRcate, after restricting to cohorts with GDM cases identified by a one-step 75-g OGTT versus a two-step 100-g OGTT or using selective versus universal screening. They were also generally similar for U.S. versus European cohorts and for cohorts that recruited participants prior to versus after 2004 (Supplementary Figs. 3–5).
Conclusions
While previous studies have investigated associations between maternal GDM and newborn DNA methylation (8–16), the majority have been small, used lenient or no adjustment for multiple testing, did not consider regional methylation differences, and adjusted for a limited number of covariates. In particular, few studies have adjusted for cell heterogeneity, an important source of variability in DNA methylation (31). Results have been inconsistent between these previous studies, raising questions of robustness and reproducibility. The current study therefore conducted meta-analyses of EWAS results from seven cohorts (3,677 mother-newborn pairs [317 with GDM]) participating in the PACE consortium (19), which examined associations between GDM and cord blood DNA methylation, after adjustment for a larger number of potential confounders. We evaluated methylation differences at both the regional and individual CpG level.
Using two dimension reduction approaches (comb-p [29] and DMRcate [30]), we identified two regions that are differentially methylated by GDM status. One of the DMRs identified by the meta-analysis (chr 1: 248100276–248100614) is located in the promoter region of OR2L13, a gene that codes for an olfactory receptor (9). Methylation levels in this region were lower in cord blood from GDM-exposed, compared with -unexposed, newborns. This finding is consistent with a previous study by Quilter et al. (9), which observed lower cord blood methylation levels at a CpG located in this DMR (cg20507276) among GDM-exposed newborns. This same CpG has also been identified as differentially methylated in both blood and buccal cells from autism spectrum disorder (ASD) case versus control subjects (32). While the mechanism by which OR2L13 may contribute to ASD is currently unknown, olfactory dysfunction has been associated with more severe social impairments among individuals with ASD (32). Since children exposed to maternal GDM in utero have a higher risk of developing ASD (6), future investigation into the potential mediating role of OR2L13 in GDM-associated ASD is merited. Methylation levels in the second DMR (chr 10: 135341933–135342560) were also lower in the GDM case, compared with control, group. This DMR is located in a CpG island within the gene body of CYP2E1, which codes for an enzyme that is highly expressed in the liver and metabolizes ethanol, numerous drugs, and certain protoxicants (33). Although, to our knowledge, the CpGs within this DMR have not previously been associated with in utero exposure to GDM, increased CYP2E expression has been observed in peripheral blood from individuals with type 1 and type 2 diabetes (33).
In contrast with the DMR results, we did not identify any individual differentially methylated CpGs when we used a conservative PFDR threshold of 0.05. When we used a more lenient PFDR threshold of 0.10, six individual CpGs (cg00812770, cg11723077, cg22791932, cg17588003, cg11187204, and cg10139436) were identified as differentially methylated by GDM status, none of which had been identified in the previous studies that we reviewed. Three of these CpGs (cg11723077, cg22791932, and cg17588003) were annotated to genes—SYNJ2, ZFPM1, and C17orf87, respectively—and a fourth CpG (cg00812770) was located in a long intergenic noncoding RNA (LINC01342). The remaining two CpGs were not annotated to any genes, and the potential consequences of altered methylation at these loci are currently unclear.
The 13 CpGs comprising the two DMRs identified by both comb-p and DMRcate were not identified as differentially methylated in individual CpG meta-analyses, likely due to the greater statistical power of the DMR approaches. Additionally, the six CpGs identified as differentially methylated based on a PFDR < 0.10 in the individual CpG analyses were not identified by either comb-p or DMRcate. It is possible that these six CpGs are false positives, since they did not reach statistical significance after application of a more conservative threshold of PFDR < 0.05. However, two of these CpGs (cg11187204 and cg10139436) also resided in intergenic regions that are either CpG poor or sparsely represented on the 450K array, which would have precluded their identification using regional approaches.
In our examination of 87 CpGs that have previously been associated with GDM status (9,11,12), only 4 were found to be differentially methylated in the same direction in the current meta-analysis, based on an uncorrected P < 0.05. Since these previous studies were similarly conducted in predominately European populations, differences in race or ethnicity are likely not driving these discrepancies. However, some of the prior findings may be false positives due to small sample sizes, insufficient control for multiple testing, or a lack of adjustment for important confounding factors, such as maternal BMI. Other potential explanations for the lack of replication include differences in exclusion criteria and the fact that these previous studies stratified by fetal sex (9) or GDM treatment type (11,12), which was not feasible for the current meta-analysis.
Importantly, the seven cohorts participating in the current meta-analysis represent eight countries and multiple time periods. Since the criteria used to classify GDM differ by country and have changed across time, the severity of disease among GDM case subjects, and the proportion of control subjects with undiagnosed GDM or hyperglycemia, may have varied between cohorts. Nevertheless, we did not observe evidence of heterogeneity for the majority of meta-analysis results. Furthermore, results were generally similar across a series of sensitivity analyses, which stratified cohorts based on geographic location, time, and the criteria used for GDM classification. It is therefore possible that there may be a linear relationship between maternal glucose levels and cord blood DNA methylation. However, while there is some evidence for this (34), additional studies are needed to determine whether maternal glucose is the main mechanism through which GDM alters DNA methylation and, if so, whether there is a clear threshold below which maternal glucose does not alter cord blood methylation.
The current study had many notable strengths. By meta-analyzing results from multiple cohorts, we were able to increase the statistical power of the study and adjust for a large number of potential confounders, including estimated cell fractions. We also used stringent adjustments for multiple testing to reduce the chance of identifying false positives. Another strength of the study was the evaluation of DMRs (using two different approaches) in addition to individual CpGs.
However, our meta-analyses also had limitations. First, there may have been an overall underestimation of GDM cases, since GDM in several cohorts was diagnosed based on a selective approach. This may have resulted in some participants being misclassified as control subjects, which would have biased results toward the null. Another important consideration is that regression models were adjusted for maternal BMI because it is a risk factor for GDM (35) and may impact cord blood DNA methylation (36). However, this may have also biased results toward the null, since GDM in several cohorts was diagnosed selectively based on traditional risk factors, including obesity. Importantly, women with GDM may have utilized different strategies to manage their disease. However, this information was not available for all cohorts, and the number of GDM case subjects adhering to particular management strategies or treatments was very small for most cohorts, so these differing subsets of GDM case subjects could not be evaluated separately. We also could not evaluate potential differences by fetal sex due to the small number of GDM case subjects per cohort. Additionally, since the gestational age at OGTT was not available for all participants, we were unable to adjust for this covariate. Another potential limitation was our focus on cord blood DNA methylation, which may not reflect methylation patterns in other tissues. However, cord blood DNA methylation has been associated with several outcomes that have been associated with in utero exposure to GDM, such as early childhood weight and adiposity (37) and ASD (38). While we excluded CpGs that overlapped SNPs and also CpG probes with SNPs at the single bp extension (27), we cannot rule out the possibility that some of the differentially methylated CpGs and regions identified in this meta-analysis may be driven by genetic, rather than epigenetic, differences between GDM case and control subjects, which merits future investigation. Finally, since the majority of individuals in the seven participating cohorts were of European ancestry, results from the current meta-analysis may not be generalizable to other populations.
Conclusion
In a meta-analysis of integrated EWAS results from seven pregnancy cohorts, comprising data from 3,677 mother-newborn pairs, GDM was associated with lower cord blood methylation levels within the promoter region of OR2L13 and the gene body of CYP2E1. Given that reduced methylation in the OR2L13 promoter has previously been associated with both GDM status and ASD, its potential role in mediating this relationship should be evaluated in future studies. Additionally, since CYP2E1 is upregulated in peripheral blood from individuals with type 1 and type 2 diabetes, the impact of reduced methylation within this gene among GDM-exposed newborns on subsequent health merits future investigation. Finally, the inability to replicate many results from previous studies of GDM exposure and cord blood DNA methylation highlights the importance of conducting EWAS meta-analyses using data from multiple cohorts.
Supplementary Material
Article Information
Acknowledgments. Acknowledgments for each study can be found in the Supplementary Data.
Funding. Funding for authors and cohorts can be found in the Supplementary Data.
Duality of Interest. D.L.D. has received personal fees from Novartis. D.A.L. has received support from Roche Diagnostics and Medtronic for work unrelated to that presented here. No other potential conflicts of interest relevant to this article were reported.
Author Contributions. C.G.H. conducted the meta-analysis and wrote the first draft of the manuscript. C.G.H., B.C., R.F., J.J., T.K., S.L., H.E.M., L.A.S., S.R.-S., A.P.S., P.Y., C.L.-A., A.B., E.B.B., V.L.C., D.C., D.D., D.L.D., A.G., Z.H., E.K., J.M.T.L., D.A.L., A.L., T.S.N., E.A.N., E.O., C.P., M.P., K.R., C.L.R., G.C.S., T.I.A.S., J.S., M.V., W.Z., M.-F.H., and C.V.B. provided feedback on the manuscript. C.G.H., C.L.-A., M.-F.H., and C.V.B. drafted the meta-analysis plan. C.G.H. and C.V.B. conceptualized the study. B.C., R.F., J.J., T.K., S.L., H.E.M., L.A.S., S.R.-S., A.P.S., and P.Y. contributed to cohort-specific statistical analyses. R.F. conducted the shadow meta-analyses. A.B., E.B.B., V.L.C., D.C., D.D., D.L.D., A.G., Z.H., E.K., J.M.T.L., D.A.L., A.L., T.S.N., E.A.N., E.O., C.P., M.P., K.R., C.L.R., G.C.S., T.I.A.S., J.S., M.V., W.Z., and M.-F.H. oversaw cohort-specific analyses. M.-F.H. and C.V.B. supervised the meta-analysis. C.G.H. is the guarantor of this work and, as such, had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Prior Presentation. Parts of this study were presented in abstract form at the 2019 Developmental Origins of Health and Disease World Congress, 20–23 October 2019, Melbourne, Australia.
Footnotes
This article contains Supplementary Data online at http://care.diabetesjournals.org/lookup/suppl/doi:10.2337/dc19-0524/-/DC1.
M.-F.H. and C.V.B. contributed equally to this work.
Where authors are identified as personnel of the International Agency for Research on Cancer/World Health Organization, the authors alone are responsible for the views expressed in this article, and they do not necessarily represent the decisions, policy, or views of the International Agency for Research on Cancer/World Health Organization.
References
- 1.DeSisto CL, Kim SY, Sharma AJ. Prevalence estimates of gestational diabetes mellitus in the United States, Pregnancy Risk Assessment Monitoring System (PRAMS), 2007-2010. Prev Chronic Dis 2014;11:E104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sacks DA, Hadden DR, Maresh M, et al.; HAPO Study Cooperative Research Group . Frequency of gestational diabetes mellitus at collaborating centers based on IADPSG consensus panel-recommended criteria: the Hyperglycemia and Adverse Pregnancy Outcome (HAPO) Study. Diabetes Care 2012;35:526–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mack LR, Tomich PG. Gestational diabetes: diagnosis, classification, and clinical care. Obstet Gynecol Clin North Am 2017;44:207–217 [DOI] [PubMed] [Google Scholar]
- 4.Clausen TD, Mathiesen ER, Hansen T, et al. . Overweight and the metabolic syndrome in adult offspring of women with diet-treated gestational diabetes mellitus or type 1 diabetes. J Clin Endocrinol Metab 2009;94:2464–2470 [DOI] [PubMed] [Google Scholar]
- 5.Brand JS, West J, Tuffnell D, et al. . Gestational diabetes and ultrasound-assessed fetal growth in South Asian and White European women: findings from a prospective pregnancy cohort. BMC Med 2018;16:203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wan H, Zhang C, Li H, Luan S, Liu C. Association of maternal diabetes with autism spectrum disorders in offspring: a systemic review and meta-analysis. Medicine (Baltimore) 2018;97:e9438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fraser A, Almqvist C, Larsson H, Långström N, Lawlor DA. Maternal diabetes in pregnancy and offspring cognitive ability: sibling study with 723,775 men from 579,857 families. Diabetologia 2014;57:102–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finer S, Mathews C, Lowe R, et al. . Maternal gestational diabetes is associated with genome-wide DNA methylation variation in placenta and cord blood of exposed offspring. Hum Mol Genet 2015;24:3021–3029 [DOI] [PubMed] [Google Scholar]
- 9.Quilter CR, Cooper WN, Cliffe KM, et al. . Impact on offspring methylation patterns of maternal gestational diabetes mellitus and intrauterine growth restraint suggest common genes and pathways linked to subsequent type 2 diabetes risk. FASEB J 2014;28:4868–4879 [DOI] [PubMed] [Google Scholar]
- 10.Ruchat S-M, Houde A-A, Voisin G, et al. . Gestational diabetes mellitus epigenetically affects genes predominantly involved in metabolic diseases. Epigenetics 2013;8:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.El Hajj N, Pliushch G, Schneider E, et al. . Metabolic programming of MEST DNA methylation by intrauterine exposure to gestational diabetes mellitus. Diabetes 2013;62:1320–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haertle L, El Hajj N, Dittrich M, et al. . Epigenetic signatures of gestational diabetes mellitus on cord blood methylation. Clin Epigenetics 2017;9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang J, Lee CN, Li HY, Hsu KH, Lin SY. Genome-wide DNA methylation variation in maternal and cord blood of gestational diabetes population. Diabetes Res Clin Pract 2017;132:127–136 [DOI] [PubMed] [Google Scholar]
- 14.Yang IV, Zhang W, Davidson EJ, Fingerlin TE, Kechris K, Dabelea D. Epigenetic marks of in utero exposure to gestational diabetes and childhood adiposity outcomes: the EPOCH study. Diabet Med 2018;35:612–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Weng X, Liu F, Zhang H, et al. . Genome-wide DNA methylation profiling in infants born to gestational diabetes mellitus. Diabetes Res Clin Pract 2018;142:10–18 [DOI] [PubMed] [Google Scholar]
- 16.Chen D, Zhang A, Fang M, et al. . Increased methylation at differentially methylated region of GNAS in infants born to gestational diabetes. BMC Med Genet 2014;15:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kang J, Lee CN, Li HY, Hsu KH, Wang SH, Lin SY. Association of interleukin-10 methylation levels with gestational diabetes in a Taiwanese population. Front Genet 2018;9:222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bouchard L Epigenetics and fetal metabolic programming: a call for integrated research on larger cohorts. Diabetes 2013;62:1026–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Felix JF, Joubert BR, Baccarelli AA, et al. . Cohort profile: Pregnancy And Childhood Epigenetics (PACE) Consortium. Int J Epidemiol 2018;47:22–23u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Moyer VA; U.S. Preventive Services Task Force . Screening for gestational diabetes mellitus: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:414–420 [DOI] [PubMed] [Google Scholar]
- 21.Metzger BE, Gabbe SG, Persson B, et al.; International Association of Diabetes and Pregnancy Study Groups Consensus Panel . International Association of Diabetes and Pregnancy Study Groups recommendations on the diagnosis and classification of hyperglycemia in pregnancy. Diabetes Care 2010;33:676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carpenter MW, Coustan DR. Criteria for screening tests for gestational diabetes. Am J Obstet Gynecol 1982;144:768–773 [DOI] [PubMed] [Google Scholar]
- 23.Fraser A, Nelson SM, Macdonald-Wallis C, Lawlor DA. Associations of existing diabetes, gestational diabetes, and glycosuria with offspring IQ and educational attainment: the Avon Longitudinal Study of Parents and Children. Exp Diabetes Res 2012;2012:963735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Olsen SF, Houshmand-Oeregaard A, Granström C, et al. . Diagnosing gestational diabetes mellitus in the Danish National Birth Cohort. Acta Obstet Gynecol Scand 2017;96:563–569 [DOI] [PubMed] [Google Scholar]
- 25.Andrews SV, Bakulski KM. FlowSorted. CordBlood. 450k User’s Guide Methylation Dataset on Sorted Cord Blood Cells, 2019
- 26.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics 2010;26:2190–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen YA, Lemire M, Choufani S, et al. . Discovery of cross-reactive probes and polymorphic CpGs in the Illumina Infinium HumanMethylation450 microarray. Epigenetics 2013;8:203–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hansen KD IlluminaHumanMethylation450kanno.ilmn12.hg19: Annotation for Illumina's 450k Methylation Arrays. R Package Version 0.6.0, 2016
- 29.Pedersen BS, Schwartz DA, Yang IV, Kechris KJ. Comb-p: software for combining, analyzing, grouping and correcting spatially correlated P-values. Bioinformatics 2012;28:2986–2988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peters T, Peters MT. biocViews DifferentialMethylation G, GenomeAnnotation D, OneChannel T, MultipleComparison Q. Package ‘DMRcate’. 2016
- 31.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol 2014;15:R31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dall’Aglio L, Muka T, Cecil CAM, et al. . The role of epigenetic modifications in neurodevelopmental disorders: a systematic review. Neurosci Biobehav Rev 2018;94:17–30 [DOI] [PubMed] [Google Scholar]
- 33.Wang Z, Hall SD, Maya JF, Li L, Asghar A, Gorski JC. Diabetes mellitus increases the in vivo activity of cytochrome P450 2E1 in humans. Br J Clin Pharmacol 2003;55:77–85 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houde AA, Ruchat SM, Allard C, et al. . LRP1B, BRD2 and CACNA1D: new candidate genes in fetal metabolic programming of newborns exposed to maternal hyperglycemia. Epigenomics 2015;7:1111–1122 [DOI] [PubMed] [Google Scholar]
- 35.Ramos GA, Caughey AB. The interrelationship between ethnicity and obesity on obstetric outcomes. Am J Obstet Gynecol 2005;193:1089–1093 [DOI] [PubMed] [Google Scholar]
- 36.Sharp GC, Salas LA, Monnereau C, et al. . Maternal BMI at the start of pregnancy and offspring epigenome-wide DNA methylation: findings from the pregnancy and childhood epigenetics (PACE) consortium. Hum Mol Genet 2017;26:4067–4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kresovich JK, Zheng Y, Cardenas A, et al. . Cord blood DNA methylation and adiposity measures in early and mid-childhood. Clin Epigenetics 2017;9:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andrews SV, Ellis SE, Bakulski KM, et al. . Cross-tissue integration of genetic and epigenetic data offers insight into autism spectrum disorder. Nat Commun 2017;8:1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.