Abstract
To function properly, mitochondria utilize products of 37 mitochondrial and >1,000 nuclear genes, which should be compatible with each other. Discordance between mitochondrial and nuclear genetic ancestry could contribute to phenotypic variation in admixed populations. Here we explored potential mito-nuclear incompatibility in six admixed human populations from the Americas: African Americans, African Caribbeans, Colombians, Mexicans, Peruvians, and Puerto Ricans. By comparing nuclear vs. mitochondrial ancestry in these populations, we first show that mtDNA copy number decreases with increasing discordance between nuclear and mitochondrial DNA ancestry. The direction of this effect is consistent across mtDNA haplogroups of different geographic origins. This observation suggests suboptimal regulation of mtDNA replication when its components are encoded by nuclear and mtDNA genes with different ancestry. Second, while most populations analyzed exhibit no such trend, in African Americans and Puerto Ricans, we find a significant enrichment of ancestry at nuclear-encoded mitochondrial genes towards the source populations contributing the most prevalent mtDNA haplogroups (African and Native American, respectively). This possibly reflects compensatory effects of selection in recovering mito-nuclear interactions optimized in the source populations. Our results provide evidence of mito-nuclear interactions in human admixed populations and we discuss their implications for human health and disease.
Mitochondria participate in vital functions of eukaryotic cells, such as generation of ATP via oxidative phosphorylation (OXPHOS), regulation of calcium uptake, apoptosis, and metabolism of essential nutrients. Even though mitochondria harbor their own genome (mitochondrial DNA, or mtDNA) with 37 genes, >1,000 of genes involved in mitochondrial function are encoded by the nuclear genome. Thus, mitochondrial functions rely on fine-tuned mito-nuclear interactions, resulting in mito-nuclear coevolution over time. Mito-nuclear coevolution has been demonstrated in non-human organisms, in which it manifests as mito-nuclear incompatibilities arising in inter-specific and inter-population hybrids. Whether these incompatibilities exist, and contribute to phenotypic variation, in humans is presently unknown. In this study, we explore mito-nuclear incompatibility in recently admixed human populations.
Evidence of mito-nuclear coevolution comes primarily from laboratory crosses of model organisms, such as fruit flies 1-7, wasps 8-10, seed beetles 11, marine copepods 12-15, and yeast 16-18. Inter-population hybrids in these organisms frequently exhibit reduced viability and fecundity 5,6,12-14, which can be explained by altered expression of OXPHOS genes 2,19, reduced OXPHOS activity, decreased ATP production 3, altered mtDNA copy number 13,19, and elevated oxidative damage 14. Fitness can often be restored if the hybrids are backcrossed with the maternal line but not with the paternal line 13, suggesting that their reduced fitness was caused by differences in ancestry between mitochondrial and nuclear genomes, hereafter called ‘mito-nuclear DNA discordance’. The phenotypic consequences of mito-nuclear discordance are known as mito-nuclear incompatibility. Mito-nuclear incompatibility has also been observed in naturally occurring populations 20-22. For example, two populations of the eastern yellow robin carrying different mtDNA haplotypes exhibit high differentiation at nuclear-encoded mitochondrial genes, suggesting that mito-nuclear incompatibility may have maintained the divergence between the two populations in spite of continued gene flow 21. In another example, the admixture fraction at nuclear-encoded mitochondrial loci in hybrids of two killifish populations, who also have divergent mtDNA haplogroups, appears to be correlated with OXPHOS activity 20.
Currently, we know very little about the extent of mito-nuclear incompatibility and its contribution to phenotypic variation in humans. It is established that altered interactions between mtDNA- and nuclear-encoded factors can modulate disease phenotypes for cardiomyopathy, predisposition to type 2 diabetes, and possibly hearing loss and Huntington’s disease 23,24. However, no elevated mito-nuclear linkage disequilibrium (LD) was found across a set of 51 human populations from the Human Genome Diversity Project 25-27. Mito-nuclear LD is expected if selection had favored certain allelic combinations between mtDNA and nuclear-encoded mitochondrial genes across diverging human populations 25. A recent study has also shown that nuclear-encoded mitochondrial genes are significantly underrepresented in Neanderthal, but not Denisovan, introgressed regions in the human genome 28. Such a depletion is expected because there is no evidence of introgression of Neanderthal and Denisovan mtDNA into modern humans 29,30. Therefore, if admixture between Neanderthals/Denisovans and humans results in mito-nuclear incompatibility, selection should favor human-specific alleles at nuclear-encoded mitochondrial genes.
In this manuscript, we explored signatures of mito-nuclear incompatibility and coevolution in six admixed human populations from the Americas studied as part of the 1,000 Genomes Project 31,32: (1) African Americans from the Southwest (ASW); (2) African Caribbeans from Barbados (ACB); (3) Colombians from Medellin, Colombia (CLM); (4) Mexicans from Los Angeles (MXL); (5) Peruvians from Lima, Peru (PEL); and (6) Puerto Ricans from Puerto Rico (PUR) (see Methods and Table 1). These populations are characterized by admixture among Africans, Europeans, and Native Americans. Our hypothesis is that increasing discordance between nuclear and mitochondrial ancestry in admixed individuals will lead to an increase in the degree of mito-nuclear incompatibility (Fig. 1A). For instance, incompatibility might arise between mtDNA origins of replication and nuclear-encoded mtDNA replication machinery 33,34 and therefore, might lead to a decrease in mtDNA replication efficiency. If our hypothesis is correct, then mtDNA copy number should decrease with increasing degree of mito-nuclear DNA discordance in admixed individuals (Fig. 1A). To test this hypothesis, we determined the mtDNA copy number based on the ratio between the depth of reads aligning to the mtDNA and the nuclear genome from publicly available sequence alignments of DNA extracted from lymphoblastoid cell lines (LCLs; see Methods). To compute the degree of mito-nuclear DNA discordance, we first determined mtDNA haplogroups and assigned their origins to African, European, or Native American ancestry (see Methods and Figs. 2C and 2D). Next, nuclear ancestry proportions were estimated using unsupervised clustering with ADMIXTURE 35 from SNP genotype data, which included individuals representing ancestry from three source populations: (1) Europeans (represented by Utah residents of Northern and Western European ancestry – CEU), (2) Africans (represented by Yoruba from Nigeria – YRI), and (3) Native Americans (represented by the Aymara, Nahuan, Quechua, and Maya; see Methods for more details and Figs. 2A and 2B). The degree of mito-nuclear discordance was computed as the proportion of nuclear ancestry of a different origin than the mtDNA. For example, for individuals carrying the African mtDNA haplogroup (L), mito-nuclear discordance is one minus the African ancestry fraction in the nuclear genome.
Table 1. The number of individuals from each population used in the study.
NA - data not available or not used in this study.
| Ancestry group |
Population | Number of individuals with sequence alignments |
Number of individuals with genotype data |
Data Source |
|---|---|---|---|---|
| Admixed | African Americans from the Southwest (ASW) | 60 | 61 | 1,000 Genomes Project Data32 |
| African Caribbean from Barbados (ACB) | 96 | 96 | 1,000 Genomes Project Data32 | |
| Mexicans from Los Angeles (MXL) | 67 | 64 | 1,000 Genomes Project Data32 | |
| Peruvians (PEL) | 86 | 85 | 1,000 Genomes Project Data32 | |
| Puerto Ricans (PUR) | 105 | 104 | 1,000 Genomes Project Data32 | |
| Colombians (CLM) | 94 | 94 | 1,000 Genomes Project Data32 | |
| African | Yoruba from Nigeria (YRI) | 109 | 108 | 1,000 Genomes Project Data32 |
| European | Utah residents of Northern and Western European ancestry (CEU) | 99 | 99 | 1,000 Genomes Project Data32 |
| Native American | Aymara | NA | 25 | Mao et al. 69 |
| Nahuan | NA | 14 | Mao et al.69 | |
| Quechua | NA | 24 | Mao et al.69 | |
| Maya | NA | 25 | Mao et al.69 | |
| Total | 717 | 799 |
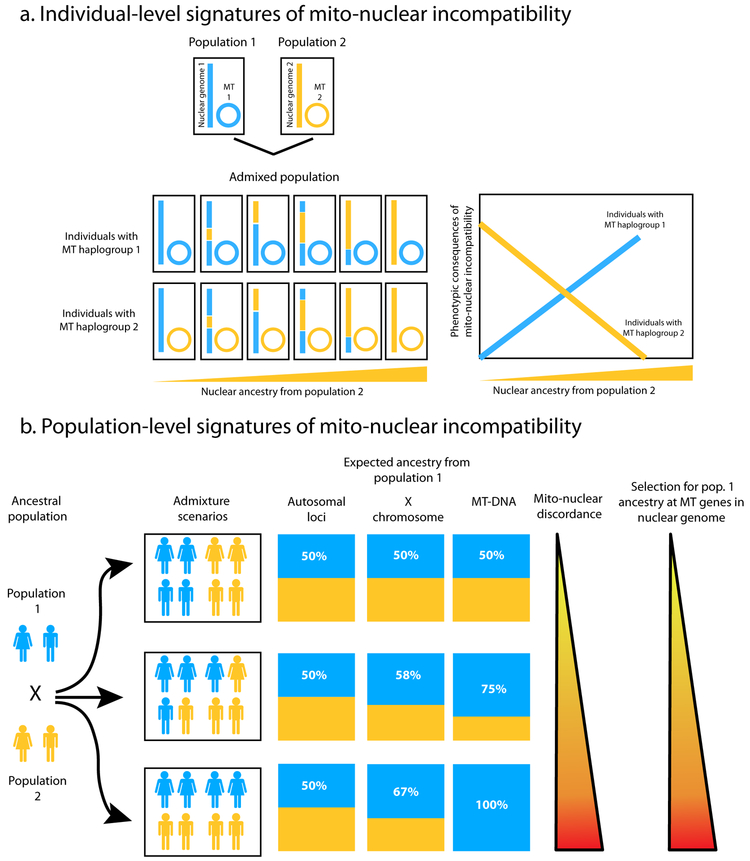
Figure 1. Expected signatures of mito-nuclear incompatibility at the level of (a) individuals or (b) populations.
(a) The nuclear genome of admixed individuals might contain varying levels of ancestry from multiple ancestral populations, whereas mtDNA derives its ancestry from only one population. Increasing discordance (concordance) between mitochondrial and nuclear ancestry in individuals can lead to increasing (decreasing) levels of mito--nuclear incompatibility, which can have phenotypic consequences. (b) Sex--biased admixture can lead to varying levels of ancestry proportions on different chromosome types based on their modes of inheritance. In extreme cases, the mtDNA ancestry can be entirely from one population even though the nuclear ancestry is highly admixed. This discordance in ancestry between the nuclear and mitochondrial genomes might result in selective pressure for ‘matching’ ancestry at nuclear-encoded mitochondrial genes.
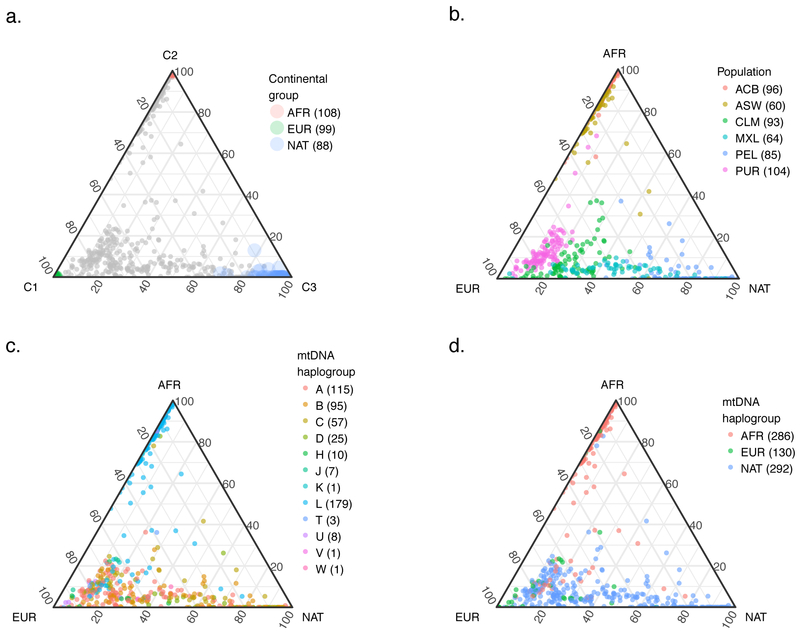
Figure 2. Ternary plots showing the distribution of African, European, and Native American ancestry in the samples analyzed (samples sizes are shown in parentheses).
(a) ADMIXTURE components 1, 2, and 3 correspond to European (EUR), African (AFR), and Native American (NAT) ancestry, respectively. The grey points are the admixed samples from the 1,000 Genomes dataset and the colored points are samples serving as proxies for source populations. (b) The ancestry structure of admixed populations. (c) Distribution of mtDNA haplogroups among admixed individuals. (d) mtDNA haplogroups grouped by region where they are thought to have been most commonly found prior to admixture. ACB: African Caribbeans from Barbados; ASW: African Americans from South Western USA; CLM: Colombians from Colombia; MXL: Mexicans from Los Angeles, USA; PEL: Peruvians from Lima, Peru; PUR: Puerto Ricans from Puerto Rico.
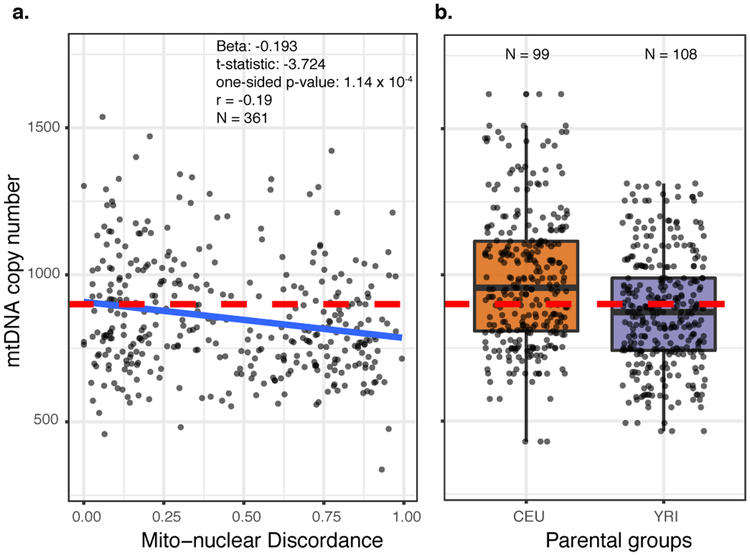
We found a significant negative correlation between mtDNA copy number and mito-nuclear DNA discordance in admixed individuals (Fig. 3A; Beta = −0.193, one-sided P-value = 1.14 × 10−04), suggesting that mtDNA copy number declines as nuclear ancestry becomes increasingly dissimilar to mtDNA ancestry. The y-intercept, i.e. when mito-nuclear DNA discordance is zero, is similar to the median mtDNA copy number in the individuals from source populations, Europeans (represented by CEU) and Africans (represented by YRI), who are not admixed (Fig. 3B). The negative correlation between mtDNA copy number and mito-nuclear DNA discordance in admixed individuals is consistent across mtDNA haplogroups from three different geographic origins – Native American (Beta = −1.06; P = 1.53 × 10−05), African (Beta = −0.63; P = 0.026), and European (Beta = −0.30; P = 0.370) – even though it is not statistically significant in every case (Fig. S1). Moreover, copy number for mtDNA of one geographic origin decreases with increasing nuclear ancestry from each of the other two geographic origins (panels outside of the top-left to bottom-right diagonal in Fig. S1). For instance, the mtDNA copy number decreases with increase in both African and European ancestry in individuals with Native American mtDNA haplogroups. Overall, our results show that mtDNA copy number is negatively correlated with mito-nuclear DNA discordance, consistent with mito-nuclear incompatibility in admixed individuals (Fig. 1A). The lack of statistical power, especially in individuals carrying European haplogroups, is likely due to the sex-biased nature of admixture in Americans.
Figure 3. mtDNA copy number in (a) admixed and (b) non-admixed populations.
(a) mtDNA copy number is negatively correlated with the discordance between mitochondrial and nuclear DNA ancestry. Standardized beta coefficient, t-statistic, one-sided P-value, and correlation coefficient are shown. The discordance score is one minus the ancestry proportion from the same source population as for the mtDNA haplogroup. The red dashed line is the median mtDNA copy number calculated across the individuals from European (CEU) and African (YRI) source populations, plotted separately in (b).
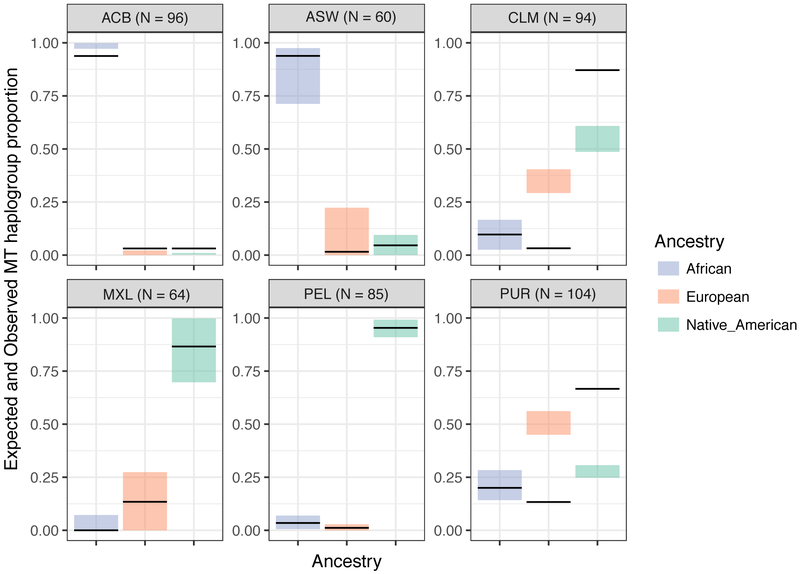
It is established that European gene flow in the Americas was male-biased 36-39, and we replicate this observation in the studied admixed populations. We show that African mtDNA haplogroups are more frequent in the African Caribbeans (frequency = 0.94) and African Americans (0.93), and Native American mtDNA haplogroups are more frequent in Colombians (0.87), Mexicans (0.87), Peruvians (0.95), and Puerto Ricans (0.67), consistent with predominantly non-European female contribution to admixture (Fig. 4). We validate this by showing that the observed mtDNA haplogroup frequencies conform with expectations based on the proportion of females from each of the three source populations derived by comparing the ancestry fractions on the autosome and X-chromosome (see Fig. 4 and Methods). Two exceptions to this result are Colombians and Puerto Ricans, in whom the frequency of Native American mtDNA haplogroups is much higher than expected (Fig. 4; African Caribbeans also exhibit a departure from expectation, but it is small in magnitude). This is an interesting observation that cannot be explained simply by post-admixture drift (see Methods) and therefore, requires further exploration. Nevertheless, it is clear that the mtDNA haplogroups in American populations are predominantly non-European.
Figure 4. Expected vs. observed frequency of mtDNA haplogroups from each of the three source populations.
The colored boxes show the 95% bootstrap interval of the expected mtDNA haplogroup frequency based on the proportion of ancestral females estimated from the ancestry proportions on the X- chromosome and autosomes (Fig. 5). The black horizontal lines show the observed frequency of mtDNA haplogroup. While in most cases the observed mtDNA haplogroup frequency falls within range of expectations, in some cases (in CLM and PUR), the observed frequencies greatly deviate from expectation. ACB: African Caribbeans from Barbados; ASW: African Americans from South Western USA; CLM: Colombians from Colombia; MXL: Mexicans from Los Angeles, USA; PEL: Peruvians from Lima, Peru; PUR: Puerto Ricans from Puerto Rico.
We leveraged this finding to test whether selection has acted against mito-nuclear incompatibilities arising in admixed populations in the Americas. Our reasoning is that, if the mtDNA haplogroups in these populations are predominantly non-European, co-adapted mito-nuclear combinations might be disrupted in individuals carrying European alleles at nuclear-encoded mitochondrial genes, resulting in a reduction in fitness. Hence, post-admixture selection would favor, and lead to an enrichment of, non-European ancestry at these genes compared to the rest of the genome (Fig. 1B). To evaluate this prediction, we analyzed local, i.e. SNP-specific, ancestry inferred from the genotypes of the studied admixed individuals by the 1,000 Genomes Admixture Group 40. Next, we computed the deviations in African, Native American, and European ancestry at each SNP by subtracting them from the mean global nuclear ancestry proportion separately for each admixed group (see Methods). For example, if the frequency of African and European ancestry at a SNP is 0.3 and 0.7, respectively, in a population where both source populations contributed equally (i.e. mean admixture fraction is 0.5), the deviations are −0.2 and 0.2, respectively. The expected deviation in ancestry across neutral loci is zero and its variance due to drift is a function of the time since admixture and effective population size 41,42. For nuclear-encoded mitochondrial genes, we expect local ancestry to deviate, on average, from neutral expectations in favor of the source population contributing the highest proportion of mtDNA (Figs. 1B). To assess whether the data support this expectation, we downloaded a list of nuclear genes from MitoCarta 2.0 43,44 and split them into mitochondrial (N = 960) vs. non-mitochondrial (N = 17,456; Table S5). The mitochondrial genes encode proteins with experimental evidence of mitochondrial localization, whereas the non-mitochondrial genes have no such evidence 43. We next followed a published approach 25 and further split the 960 mitochondrial genes into two subsets –002D 167 high-confidence, or ‘High-mt’, genes (known genes encoding proteins that are part of the mtDNA replication and transcription machinery, and of ribosomal and OXPHOS complexes) and remaining 793 ‘Low-mt’ genes (Table S5). We calculated gene-specific ancestry deviation by averaging across all SNPs in 5-Mb windows flanking the midpoint of the coding sequence. Subsequently, we tested whether the mean ancestry at mitochondrial genes (High-mt or Low-mt) deviates significantly from that at non-mitochondrial genes, which we used as a proxy for the amount of drift experienced by neutrally evolving loci. We used this approach, as opposed to a genome-wide scan because our ability to detect such a signature at individual loci is limited due to: (1) the large number of genes involved in mitochondrial function (>1,000) 45, (2) the low amount of genetic differentiation among human populations (Fst ≈ 0.1) 46, (3) only a relatively recent admixture history in the populations analyzed 36,37,39, and (4) the large number of tests involved in an agnostic genome-wide search. We used a bootstrap approach to generate a distribution of the mean deviation in ancestry for each functional gene category (for 167 High-mt, 793 Low-mt, and 17,456 non-mitochondrial genes, see Methods for more details). We show using simulations (see Methods) that our approach has adequate statistical power to detect selection with the given sample sizes at these time scales (Fig. S12).
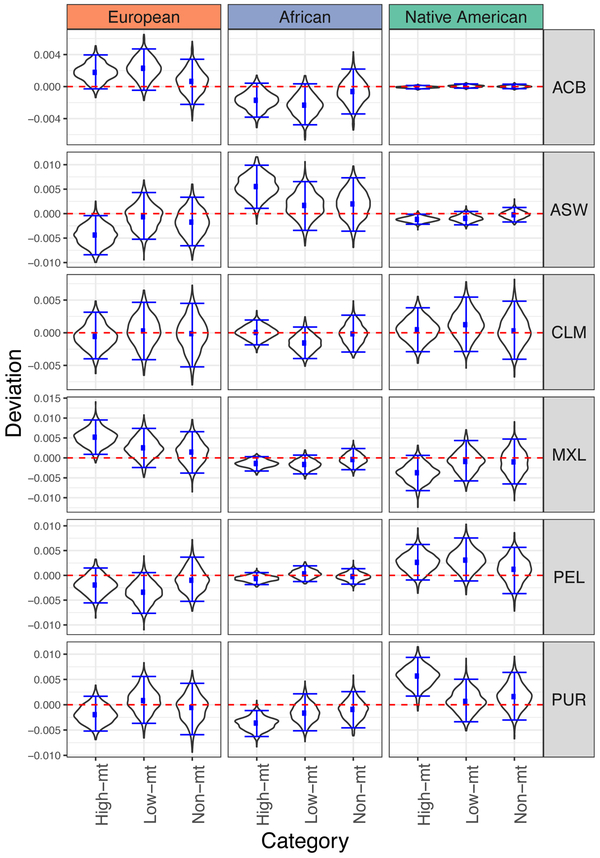
We found a significant enrichment in Native American ancestry at High-mt genes in Puerto Ricans, and a significant enrichment in African ancestry at High-mt genes in African Americans (Fig. 5). Because Native American mtDNA haplogroups are more frequent in Puerto Ricans, and African mtDNA haplogroups are more frequent in African Americans (Fig. 4), these results are consistent with our prediction (Fig. 1B). However, in three out of six admixed populations analyzed, the mean ancestry of nuclear-encoded mitochondrial genes does not significantly deviate from expectation (the 95% bootstrapped confidence interval spans the zero line; Fig. 5), consistent with no evidence of selection for local ancestry at such genes. Surprisingly, we also observe a significant enrichment in European ancestry at High-mt genes in Mexicans (Fig. 5), which contradicts our hypothesis. This lack of a consistent pattern of enrichment across admixed populations paints a complex picture of selection acting on nuclear-encoded mitochondrial genes, requiring further exploration.
Figure 5. Systematic deviations in local ancestry for different functional categories of genes.
The y--axis shows local ancestry deviation and the x--axis lists the functional categories. ‘High-mt’ (167 genes) are nuclear genes that encode important subunits of mitochondrial replication, transcription and the OXPHOS complexes. ‘Low-mt’ (793 genes) are nuclear genes that were inferred to have mitochondrial function by Mitocarta 2.0 43, but are not part of the ‘High-mt’ gene set. ‘Non-mt’ (17,456 genes) are genes that do not have known or inferred mitochondrial function based on Mitocarta 2.0 43. A block bootstrap approach (see Methods) was used to generate the distributions. Briefly, we sampled 167 windows of 5 Mb spanning the genes, with replacement, within each category and calculated the mean local ancestry deviation across these windows. This was repeated for 1,000 bootstraps to generate the distributions. The horizontal bars indicate the empirical 95% confidence interval of the mean ancestry deviation. ACB: African Caribbeans from Barbados; ASW: African Americans from South Western USA; CLM: Colombians from Colombia; MXL: Mexicans from Los Angeles, USA; PEL: Peruvians from Lima, Peru; PUR: Puerto Ricans from Puerto Rico.
Discussion
We report that mtDNA copy number decreases as the nuclear and mtDNA become increasingly dissimilar in ancestry among human admixed individuals. Because mito-nuclear compatibility is important for regulation of mtDNA copy number, its reduction due to mito-nuclear DNA discordance might reflect the incompatibility between the origins of mtDNA replication and nuclear-encoded proteins involved in mtDNA replication 47. Variation across populations in mtDNA binding affinity of proteins involved in mtDNA replication, such as polymerase-γ (POLG), mtDNA helicase (TWINKLE), and mitochondrial single strand binding proteins (mtSSB) 34. Future functional experiments should validate effects of mtDNA haplogroup and nuclear DNA combinations on mtDNA copy number directly. This can be performed, for instance, in cybrids – cellular hybrid lines carrying the same nuclear genetic background but different mtDNA haplotypes 48. Our results and the suggested future experiments are important because there is a growing body of evidence implicating mtDNA copy number as a biomarker for different diseases including diabetes 49, Parkinson’s disease 50, male infertility 51, depression 52, aging 53, and many types of cancers 54-56, even though it may not always correlate with mitochondrial content and activity 57.
Multiple biological and environmental sources can contribute to variation in mtDNA copy number. However, the design of our study minimizes these. For instance, even though mtDNA copy number is known to vary among tissues 58, this could not have contributed to our estimates because we made our measurements in cell lines of the same type – LCLs. mtDNA copy number is also known to vary depending on the physiological conditions of an individual, which may be driven by the environment (e.g. stress 59). However, the use of cell lines, which are cultured under standard laboratory conditions, should largely remove mtDNA copy number variation originally present among samples due to these factors. Having said that, the observed correlation between mito-nuclear DNA discordance and mtDNA copy number that we found should be replicated and explored in future studies, ideally with mtDNA copy number measured across biological and technical replicates, to minimize biological variation and measurement error. It would also be interesting to test the effect of mito-nuclear DNA discordance on other mitochondrial phenotypes such as mitochondrial morphology and rate of ATP production in this and other cell types. However, the caveat of using cell lines is that it is difficult to say whether or not the effects of mito-nuclear ancestry discordance exist in vivo in the individuals from whom they are derived. Therefore, future in vivo studies will also be of great value.
Corroborating previous studies 36-39, we found a significant sex bias in admixture for all six admixed populations studied. In most admixed populations we found a high level of concordance between the female contribution from each source population estimated from nuclear genetic markers and that estimated from mtDNA. However, in Colombians and Puerto Ricans, the frequency of Native American ancestry in mtDNA is significantly higher than expected based on the ancestry information in the nuclear genome. This deviation cannot be explained by genetic drift experienced by mtDNA since admixture due to its smaller effective population size, and could be due to selection for the Native American haplogroups in Colombians and Puerto Ricans, if the Native American mtDNA was better adapted to the environment. Indeed, climate adaptation may have played a role in mtDNA diversification across human populations 60,61. This observation, which has not been reported previously, requires further investigation.
In line with the hypothesis that selection against mito-nuclear incompatibility should disfavor European ancestry at these loci given that the mtDNA haplogroups in admixed American populations are predominantly African or Native American, we also tested for an enrichment for non-European ancestry at nuclear-encoded mitochondrial genes in admixed populations. We found that the ancestry profiles at nuclear-encoded mitochondrial genes in three out of the six admixed populations studied (African Caribbeans, Colombians, and Peruvians) do not deviate from neutral expectation, i.e. are indistinguishable from the genomic background (Fig. 5). This could mean that there has been no selection on mito-nuclear interactions in these populations. We observed an enrichment of Native American ancestry in Puerto Ricans, and an enrichment of African ancestry in African Americans, at nuclear-encoded mitochondrial genes. Because Puerto Ricans predominantly carry Native American mtDNA haplogroups, whereas African Americans primarily carry African haplogroups, this observation is consistent with our expectation of selection against mito-nuclear incompatibility. However, we also found a significant enrichment of European ancestry in Mexicans – another population with predominantly Native American mtDNA haplogroups. This observation is inconsistent with selection against mito-nuclear incompatibility in admixed populations. This complex picture among different admixed populations suggests that other competing selective pressures on mitochondrial genes might be involved and it may not be straightforward to detect signatures of mito-nuclear incompatibility from deviations in local ancestry alone.
Our ability to detect a signature of ancestry enrichment driven by mito-nuclear incompatibility is influenced by several factors including degree of sex bias, time since admixture, effective population size, selection strength, and the number of loci under selection. One potential limitation of our analysis is that we assume that all nuclear-encoded mitochondrial genes have equal effect sizes for mitochondrial function. Since mitochondrial function is a highly complex trait, this assumption is likely incorrect. A more powerful way of testing for ancestry enrichment across these genes would be to weigh the contribution of each locus by its effect size, as we demonstrate using simulations (Fig. S12). Unfortunately, we do not know what these effect sizes are because genome-wide association studies of mitochondrial phenotypes, such as mtDNA copy number and rate of ATP production, are yet to be conducted in humans. While systematic collection of such data for large cohorts of individuals is pending, it would also be highly informative to explore the effects of mito-nuclear interactions on various health-related phenotypes in large-scale datasets such as the UK Biobank 62.
In conclusion, our results demonstrate that discordance between mtDNA and nuclear ancestry for mitochondrial genes might contribute to phenotypic variation in admixed individuals. Mito-nuclear DNA discordance might contribute to disease phenotypes of non-admixed individuals (reviewed in 23). Therefore, we expect this phenomenon to contribute even more to disease variation in admixed individuals, which needs to be evaluated in future studies. Such evaluation is also critical for making advances in mitochondrial replacement therapy (MRT), a technique in which the mtDNA carrying disease-associated mutations in a patient’s oocyte is replaced with mtDNA from a healthy donor oocyte 63-65. Despite the success of MRT, many human and non-human primate embryos created via mitochondrial replacement do not develop normally 63. Mitochondrial replacement can also lead to detrimental effects on growth, development, respiration, metabolism, aging, fertility, and survival in non-primate animals 63. Human cybrid lines also show variation in mtDNA copy number, ATP turnover rates, reactive oxygen species production, and expression of OXPHOS genes 66. Despite these observations, the degree to which the mitochondrial haplogroup of a donor should match the genetic background of the ‘nuclear’ parents in MRT in humans remains unanswered. The answer to this question is even less clear for admixed individuals, or for individuals whose parents belong to different ethnic groups 67, as their nuclear genomes originate from different populations. Our results highlight the potential of studying admixed individuals to better understand phenotypic effects of mito-nuclear DNA discordance, which will be useful in elucidating MRT-associated risks and evaluating disease susceptibility in contemporary admixed and non-admixed populations 23,68.
Materials and Methods
Global ancestry and mtDNA haplogroup
We downloaded the 1000 Genomes phase 3 vcf files and retained individuals who belonged to one of the six admixed populations: (1) African Americans from the Southwest (ASW); (2) African Caribbeans from Barbados (ACB); (3) Colombians from Medellin, Colombia (CLM); (4) Mexicans from Los Angeles (MXL); (5) Peruvians from Lima, Peru (PEL); and (6) Puerto Ricans from Puerto Rico (PUR). We also used data from Utah residents of Northern and Western European ancestry (CEU) and from Yorubans from Ibadan, Nigeria (YRI), who serve as proxies for the European and African source populations, respectively. To represent the Native American ancestry component, we analyzed previously published genotype data 69 from the following four groups: (1) Aymara; (2) Nahuan; (3) Quechua; and (4) Maya. The data set is summarized in Table 1. We determined global ancestry – the overall contribution of African, European, and Native American ancestry to the nuclear genome – of each analyzed individual using ADMIXTURE 35 (Fig. 2). For this purpose, we merged the 1000 genomes genotype data with the genotype data from Native American groups published by Mao and colleagues 69. We included SNPs that overlapped across both datasets (a total of 691,435 SNPs) and converted the genotypes to binary format for use with PLINK 70,71. We further removed palindromic (A/T, G/C) SNPs to ensure strand consistency across both datasets. Subsequently, the two datasets were merged and SNPs were pruned for LD (PLINK’s --indep-pairwise function with window size of 50, step size of 5, and r2 threshold of 0.1 72,73), which resulted in 88,442 SNPs. We ran unsupervised ADMIXTURE 35 analysis on this genotype dataset for k = 1, 2, 3, 4, and 5 and used the ancestry proportions for k = 3 for all downstream analyses, since it had the lowest cross-validation error (Fig. S5 and Fig. S6). In agreement with previous studies 37,39, the individuals from admixed populations derive their genetic ancestry from three primary source populations: Native American, European, and African. The proportion of ancestry from each source population varies among the admixed populations (Fig. 2 and Fig. S5; Table S2 and S3) because of differences in admixture histories 36,37.
We used Haplogrep version 2.02 74 to determine the mtDNA haplogroup, for individuals from the 1000 Genomes Project only, as the individuals from Mao et al. 69 were not genotyped for mitochondrial variants. All mtDNA haplogroups were called with high accuracy (minimum posterior probability of 0.78). To increase statistical power, we grouped together haplogroups belonging to the same major haplogroup (e.g. L1b1a3 was grouped with L3d1b1 under the L major haplogroup Table S4). We further grouped major haplogroups into regional groups, corresponding to pre-colonization origins (L: African; A, B, C, D: Native American; H, J, K, T, U, V, W: European) (Fig. 2C and Table S4). We excluded two individuals (HG01272 and NA19982), whose mtDNA was predicted to belong to the M haplogroup, which is most frequently found in South Asia. This way, both nuclear and mitochondrial ancestry was categorized into only three regional groups: Native American, European, and African (Fig. 2D and Table S4). Among the admixed individuals, African (L) and Native American (A, B, C, and D) mtDNA haplogroups are more frequent than European haplogroups (H, J, K, T, U, V, and W; Fig. S7), consistent with female bias in the non-European contribution 75.
We calculated mito-nuclear discordance in ancestry as the fraction of the nuclear genome from a different geographical origin than the mtDNA haplogroup. For instance, for individuals with Native American mtDNA haplogroups, mito-nuclear DNA discordance is the proportion of nuclear ancestry that is not Native American (i.e. African plus European).
mtDNA copy number estimation
Given the average sequencing depth of the autosomes and mtDNA, and the fact that there are two autosomal chromosome copies per cell, we can compute the number of copies of mtDNA per cell:
| (1) |
| (2) |
In the equation for average depth, N is the total number of reads aligning to the chromosome, L is the average length of a read, and G is the size of the chromosome in base pairs. Similar approaches have been used previously to accurately estimate mtDNA copy number per cell 58,76. We first calculated mtDNA copy number for each autosomal chromosome separately, and then calculated the mean across all chromosomes.
A subset of samples (a total of 24) in the 1000 Genomes Project Data were sequenced at both low (2–4x) and high coverage (20–40x). We used these to validate whether mtDNA copy numbers calculated from the low- vs. high-coverage alignments agree with each other. As shown in Fig. S8, the copy numbers generally agree, with a few exceptions. Some samples show appreciable mtDNA copy numbers when calculated using the high-coverage alignments but low copy numbers when calculated using the low-coverage alignments (Fig. S8). The source annotation of these samples indicates that some of them (samples for which this information is available) were sequenced from peripheral blood mononuclear cells (PBMCs), instead of Lymphoblastoid Cell Lines (LCLs), the only two types of cell lines used to carry out sequencing for the 1000 Genomes Project (Fig. S8, Table S1). The difference in mtDNA copy number seen between the two cell lines is consistent with previous observations that LCLs are known to carry significantly higher mtDNA copy numbers than PBMCs 77-79. Because annotation for the source DNA is not available for all samples (Table S1), we plotted the density of mtDNA copy number calculated from the low-coverage alignments and observed a clear separation between samples sequenced from PBMCs and LCLs (Fig. S9). Based on this separation, we inferred that samples with more than 250 mtDNA copies per cell are LCLs (Table S1) and only retained these for analysis with mtDNA copy number to exclude samples which were sequenced from PBMCs in an effort to limit variation due to DNA source. After removing such samples, the correlation coefficient between mtDNA copy number from low-coverage and high-coverage alignments is 0.71, as opposed to 0.66 before removing them.
An advantage of using LCLs to study mtDNA copy number variation is that they exhibit high mtDNA content and elevated expression of genes involved in mtDNA replication and transcription, as well as of respiratory genes, consistent with elevated mitochondrial biogenesis 80. Furthermore, because they are maintained following standard protocols in a laboratory, variation due to differences in individuals’ environments, from whom the LCLs are derived, is unlikely to systematically confound our analysis of mtDNA copy number.
Local ancestry
Local ancestry for autosomes was generated using RFMix 81 by the 1000 Genomes Project admixture working group as described by Martin and colelagues 39 https://personal.broadinstitute.org/armartin/tgp_admixture/snp_pos/). For downstream analyses, we masked out regions of the genome where local ancestry was called with less than 0.9 maximum posterior probability. Global ancestry calculated from these filtered local ancestry calls are highly correlated with global ancestry estimated using ADMIXTURE 35(Fig. S10).
Sex-biased admixture
Males and females may contribute different proportions of ancestry from the source populations during admixture. This sex bias can lead to a situation where different chromosomes give different estimates of ancestry proportions (Fig. 1B). For example, for every three X chromosomes, two are contributed by females while one is contributed by males. This is in contrast to autosomes, which are contributed from both sexes equally. Therefore, any sex bias in the admixture composition of the ancestral population will influence the ancestry fraction of the X chromosome more than that of autosomes (Fig. 1B). We took advantage of the difference in admixture fraction between the X chromosome and autosomes to estimate the relative contribution of males and females from each of the three relevant ancestral groups (African, European, and Native American) using the approach described in 82. If is the proportion of ancestors of the admixed group, who were female and from population i, and who were male and from population i, we assume that for each admixed group (e.g. PUR, CLM etc.), , the mean autosomal ancestry fraction from population i, where i ϵ {African, Native American, European}. Furthermore, we assume that . Thus, for values of and , the expected ancestry fraction for the X chromosome in the population, is 83:
| (3) |
We performed a grid search for the values of and that equal and minimize the squared deviation between , predicted using equation (3), and the mean ploidy-adjusted X-chromosomal ancestry fraction inferred from genotype data. Estimated values of and and confidence intervals around these estimates, generated by bootstrapping (sampling individuals with replacement), are shown in Fig. S11.
Simulation of drift in mtDNA since admixture
Our finding that the frequency of Native American mtDNA ancestry in Puerto Ricans and African mtDNA ancestry in African Americans can be explained by at least two factors. First, since mtDNA represents a single genealogical history, it yields a ‘noisier’ estimate of the proportion of females from each parental population than the estimate based on autosomal and X-chromosomal loci, which represent multiple genealogical histories because of recombination and independent assortment. Second, we expect larger fluctuations in mtDNA ancestry as a result of drift because of its smaller effective population size compared to that of autosomal or X-chromosomal loci 84. To test whether genetic drift since the occurrence of admixture can account for the deviation in mtDNA frequency in Puerto Ricans and Colombians, we simulated the expected amount of drift in local ancestry since admixture in Puerto Ricans and Colombians using a simple hybrid-isolation demographic model 85,86. In this model, all three parental groups mix at some time in the past in proportions equal to the mean global ancestry averaged across individuals in the population. In subsequent generations, 2N autosomes and N/2 copies of mtDNA were drawn from a multinomial distribution with the probability of drawing European, African, or Native American ancestry determined by the relative ancestry proportions in the previous generation. After g generations, the final ancestry frequency at each locus, averaged across individuals in the population, was recorded. This process was repeated 10,000 times to simulate the amount of drift experienced by 10,000 independent loci. We used 17 generations for g in PUR and 14 generations for g in CLM, similar to values estimated for these populations in 37, and 1,250 for N following 87. We show in Fig. S3 that the simulated distribution of local ancestry using these parameters matches the observed distribution of local ancestry quite well.
The assumed model of admixture dynamics has several limitations, which could lead to biased estimates of the female and male contributions. Specifically, we assume a hybrid-isolation model with equal reproductive variance and similar generation times between males and females. First, models that incorporate continuous gene flow are likely to yield slightly different estimates, especially if admixture occurred recently, i.e. within the last five generations 88. Because admixture for the populations used in this study started much earlier (>10 generations ago, 36,37,39), this is not a major concern in our case. Second, shorter generation times in females than in males would result in a smaller effective population size for the mtDNA compared to the nuclear genome 89. This would lead to stronger drift in mtDNA, which might explain the observed discrepancy in ancestry proportions between mtDNA and the nuclear genome in Colombians and Puerto Ricans without invoking selection. Third, men typically tend to have higher variance in reproductive success relative to women 90,91, which would increase the effective population size of the mtDNA compared to the nuclear genome. A detailed discourse of how these competing processes affect the inference of sex bias in admixture dynamics is beyond the scope of this paper, but needs to be explored in future studies.
Local ancestry enrichment in nuclear-encoded mitochondrial genes
Strong deviations in local ancestry frequency in a population can be indicative of post-admixture selection (e.g., Tang et al. 200787). Our ability to detect such a signature at individual loci is limited, as we explain in the main text of the manuscript. A more feasible approach is to test for significant deviations in ancestry across a number of loci relative to the genomic background. This is the underlying basis for the approach used in 28, as well as our approach to detecting selection on nuclear-encoded mitochondrial genes. For each admixed population, we begin by calculating the frequency of Native American, European, and African ancestry at every SNP in the genome by averaging across all individuals in the population. These were subtracted from the mean ancestry fraction across all SNPs (the expectation) to calculate the deviation in local ancestry at each SNP. We used the gene annotation provided in MitoCarta 2.0 and generated a list of mitochondrial and non-mitochondrial genes 43. We further split the mitochondrial genes into ‘High-mt’ (N = 167) and ‘Low-mt’ (N = 793) subsets, as curated in 92). An unweighted block bootstrap approach was used to generate the distribution of average deviation in local ancestry for each gene category. We generated windows of 5 Mb spanning each gene (±2.5 Mb on either side of a gene’s midpoint) to take into account LD among SNPs. Subsequently, we used bedtools 93 to intersect SNPs, at which local ancestry deviation was calculated previously, with these windows. For each gene category, we sampled 167 windows with replacement, to match the number of genes in the smallest category (i.e. High-mt), and calculated the mean ancestry deviation first for each window, and then across windows. This process was repeated 1,000 times to generate a distribution of mean deviation in local ancestry for each gene category.
Statistical power to detect selection
We carried out forward simulations to test whether we have adequate statistical power to detect selection with our approach. We started by simulating 167 independent biallelic loci, where, at each locus, the favored allele has a starting frequency of fi and an effect size, βi, randomly drawn from a standard normal distribution. At each locus, the relative fitness of the favored allele is wi = 1 + s*βi, where s is the selection coefficient. In every generation, the allele frequency after a round of genic selection on survival, fi*, was calculated as follows 94:
In the next generation, we sampled 2N (2 × 1250 = 2500) chromosomes at random from a binomial distribution where the probability of drawing a chromosome with a favored allele is equal to fi*. We repeated this until the gth generation, at which point we sampled 2n chromosomes at random, where n is the sample size (i.e. the number of diploid individuals). This last step was carried out to take into account sampling error.
We chose 0.13 for fi, equal to the mean Native American ancestry proportion in Puerto Ricans and used a range of values for s = {0.001, 0.002, 0.003, 0.004, 0.005, 0.01}, the selection coefficient. We set g at 17 generations and N at 1250, to match the degree of drift experienced by Puerto Ricans, supported by the comparison of simulated vs observed distribution of local ancestry (Fig. S3). We carried out 1,000 simulations and, for each simulation, generated the distribution of the mean deviation in ancestry using an unweighted bootstrap approach similar to that used for the real data. Briefly, for every bootstrap, 167 loci were sampled with replacement and the deviation for each locus was calculated by subtracting the expected allele frequency, equal to the frequency at the start of the simulation, from the observed frequency, equal to the frequency at the gth generation. The deviation was averaged across loci and the sampling was repeated 1,000 times to generate a distribution around the unweighted mean. To test the idea that knowledge of effect sizes might provide more statistical power to detect selection as mentioned in the discussion section, we also calculated weighted means by multiplying the deviation at each locus by its effect size prior to summation.
We show in Fig. S12 that even at relatively low selection pressures (s ≈ 0.005), we have more than 80% power to detect selection at the 0.05 level of significance using the unweighted approach with a sample size of 100, which is comparable to the sample sizes used in this study (Table 1). We also show that the weighted approach provides more statistical power to detect selection than using an unweighted approach. Lastly, we show that in the absence of selection (s = 0), the false positive rate is equal to the level of significance (~0.05).
Data and code availability
All analyses were conducted using publicly available data. The 1000 Genomes Project data are available on their ftp site . Intermediate files and code have been made publicly available on github: (https://github.com/makovalab-psu/Mito_nuclear_incompatibility).
Supplementary Material
Acknowledgments
We thank Rasmus Nielsen, Mark Shriver, William Chase, and Sarah Craig for their comments on the manuscript. This project was supported by a seed grant awarded to AAZ and KDM from the Center of Human Evolution and Development (CHED) at The Pennsylvania State University, and by a grant from NIH (R01GM116044). Additional funding was provided by Penn State Eberly College of Sciences, The Huck Institute of Life Sciences at Penn State, and the Penn State Institute for CyberScience, as well as, in part, under grants from the Pennsylvania Department of Health using Tobacco Settlement and CURE Funds. The department specifically disclaims any responsibility for any analyses, responsibility, or conclusions.
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Sackton TB, Haney RA & Rand DM Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution 57, 2315–2325 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Mossman JA et al. Mitonuclear Interactions Mediate Transcriptional Responses to Hypoxia in Drosophila. Mol. Biol. Evol. 34, 447–466 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meiklejohn CD et al. An Incompatibility between a mitochondrial tRNA and its nuclear-encoded tRNA synthetase compromises development and fitness in Drosophila. PLoS Genet. 9, e1003238 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.James AC & Ballard JWO Mitochondrial genotype affects fitness in Drosophila simulans. Genetics 164, 187–194 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Montooth KL, Meiklejohn CD, Abt DN & Rand DM Mitochondrial-nuclear epistasis affects fitness within species but does not contribute to fixed incompatibilities between species of Drosophila. Evolution 64, 3364–3379 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dowling DK, Friberg U, Hailer F & Arnqvist G Intergenomic epistasis for fitness: within-population interactions between cytoplasmic and nuclear genes in Drosophila melanogaster. Genetics 175, 235–244 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hoekstra LA, Siddiq MA & Montooth KL Pleiotropic Effects of a Mitochondrial–Nuclear Incompatibility Depend upon the Accelerating Effect of Temperature in Drosophila. Genetics 195, 1129–1139 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ellison CK, Niehuis O & Gadau J Hybrid breakdown and mitochondrial dysfunction in hybrids of Nasonia parasitoid wasps. J. Evol. Biol 21, 1844–1851 (2008). [DOI] [PubMed] [Google Scholar]
- 9.Niehuis O, Judson AK & Gadau J Cytonuclear genic incompatibilities cause increased mortality in male F2 hybrids of Nasonia giraulti and N. vitripennis. Genetics 178, 413–426 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koevoets T, Niehuis O, van de Zande L & Beukeboom LW Hybrid incompatibilities in the parasitic wasp genus Nasonia: negative effects of hemizygosity and the identification of transmission ratio distortion loci. Heredity 108, 302–311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Immonen E, Rönn J, Watson C, Berger D & Arnqvist G Complex mitonuclear interactions and metabolic costs of mating in male seed beetles. J. Evol. Biol 29, 360–370 (2016). [DOI] [PubMed] [Google Scholar]
- 12.Ellison CK & Burton RS Disruption of mitochondrial function in interpopulation hybrids of Tigriopus californicus. Evolution 60, 1382–1391 (2006). [PubMed] [Google Scholar]
- 13.Ellison CK & Burton RS Interpopulation hybrid breakdown maps to the mitochondrial genome. Evolution 62, 631–638 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Barreto FS & Burton RS Elevated oxidative damage is correlated with reduced fitness in interpopulation hybrids of a marine copepod. Proc. Biol. Sci 280, 20131521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rawson PD & Burton RS Functional coadaptation between cytochrome c and cytochrome c oxidase within allopatric populations of a marine copepod. Proc. Natl. Acad. Sci. U. S. A 99, 12955–12958 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chou J-Y & Leu J-Y Speciation through cytonuclear incompatibility: insights from yeast and implications for higher eukaryotes. Bioessays 32, 401–411 (2010). [DOI] [PubMed] [Google Scholar]
- 17.Lee H-Y et al. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135, 1065–1073 (2008). [DOI] [PubMed] [Google Scholar]
- 18.Chou J-Y, Hung Y-S, Lin K-H, Lee H-Y & Leu J-Y Multiple Molecular Mechanisms Cause Reproductive Isolation between Three Yeast Species. PLoS Biol. 8, e1000432 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ellison CK & Burton RS Cytonuclear conflict in interpopulation hybrids: the role of RNA polymerase in mtDNA transcription and replication. J. Evol. Biol 23, 528–538 (2010). [DOI] [PubMed] [Google Scholar]
- 20.Baris TZ et al. Evolved genetic and phenotypic differences due to mitochondrial-nuclear interactions. PLoS Genet. 13, e1006517 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morales HE et al. Concordant divergence of mitogenomes and a mitonuclear gene cluster in bird lineages inhabiting different climates. Nat Ecol Evol 2, 1258–1267 (2018). [DOI] [PubMed] [Google Scholar]
- 22.Bar-Yaacov D et al. Mitochondrial involvement in vertebrate speciation? The case of mito-nuclear genetic divergence in chameleons. Genome Biol. Evol 7, 3322–3336 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Levin L, Blumberg A, Barshad G & Mishmar D Mito-nuclear co-evolution: the positive and negative sides of functional ancient mutations. Front. Genet 5, 448 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gershoni M et al. Disrupting Mitochondrial–Nuclear Coevolution Affects OXPHOS Complex I Integrity and Impacts Human Health. Genome Biol. Evol 6, 2665–2680 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sloan DB, Fields PD & Havird JC Mitonuclear linkage disequilibrium in human populations. Proc. Biol. Sci 282, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rosenberg NA Genetic Structure of Human Populations. Science 298, 2381–2385 (2002). [DOI] [PubMed] [Google Scholar]
- 27.Cann HM A Human Genome Diversity Cell Line Panel. Science 296, 261b–262 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Sharbrough J, Havird JC, Noe GR, Warren JM & Sloan DB The Mitonuclear Dimension of Neanderthal and Denisovan Ancestry in Modern Human Genomes. Genome Biol. Evol 9, 1567–1581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Serre D et al. No evidence of Neandertal mtDNA contribution to early modern humans. PLoS Biol. 2, E57 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krings M et al. Neandertal DNA sequences and the origin of modern humans. Cell 90, 19–30 (1997). [DOI] [PubMed] [Google Scholar]
- 31.1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.1000 Genomes Project Consortium et al. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bailey LJ & Doherty AJ Mitochondrial DNA replication: a PrimPol perspective. Biochem. Soc. Trans 45, 513–529 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ciesielski GL, Oliveira MT & Kaguni LS Animal Mitochondrial DNA Replication. Enzymes 39, 255–292 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexander DH, Novembre J & Lange K Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Homburger JR et al. Genomic Insights into the Ancestry and Demographic History of South America. PLoS Genet. 11, e1005602 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moreno-Estrada A et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 9, e1003925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Linares A et al. Admixture in Latin America: geographic structure, phenotypic diversity and self-perception of ancestry based on 7,342 individuals. PLoS Genet. 10, e1004572 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Martin AR et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martin AR et al. Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am. J. Hum. Genet 100, 635–649 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Long JC The genetic structure of admixed populations. Genetics 127, 417–428 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hartl DL, Clark AG & Clark AG Principles of population genetics. 116, (Sinauer associates Sunderland, 1997). [Google Scholar]
- 43.Calvo SE, Clauser KR & Mootha VK MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 44, D1251–D1257 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pagliarini DJ et al. A mitochondrial protein compendium elucidates complex I disease biology. Cell 134, 112–123 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schatz G The protein import machinery of mitochondria. Protein Sci. 2, 141–146 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bhatia G, Patterson N, Sankararaman S & Price AL Estimating and interpreting FST: the impact of rare variants. Genome Res. 23, 1514–1521 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Holt IJ & Reyes A Human mitochondrial DNA replication. Cold Spring Harb. Perspect. Biol 4, (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Trounce I, Neill S & Wallace DC Cytoplasmic transfer of the mtDNA nt 8993 T-->G (ATP6) point mutation associated with Leigh syndrome into mtDNA-less cells demonstrates cosegregation with a decrease in state III respiration and ADP/O ratio. Proc. Natl. Acad. Sci. U. S. A 91, 8334–8338 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee HK et al. Decreased mitochondrial DNA content in peripheral blood precedes the development of non-insulin-dependent diabetes mellitus. Diabetes Res. Clin. Pract 42, 161–167 (1998). [DOI] [PubMed] [Google Scholar]
- 50.Pyle A et al. Reduced mitochondrial DNA copy number is a biomarker of Parkinson’s disease. Neurobiol. Aging 38, 216.e7–216.e10 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang M et al. Increased Total mtDNA Copy Number Cures Male Infertility Despite Unaltered mtDNA Mutation Load. Cell Metab. 26, 429–436.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 52.Cai N et al. Molecular signatures of major depression. Curr. Biol 25, 1146–1156 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mengel-From J et al. Mitochondrial DNA copy number in peripheral blood cells declines with age and is associated with general health among elderly. Hum. Genet 133, 1149–1159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hosgood HD 3rd et al. Mitochondrial DNA copy number and lung cancer risk in a prospective cohort study. Carcinogenesis 31, 847–849 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu M Generation, function and diagnostic value of mitochondrial DNA copy number alterations in human cancers. Life Sci. 89, 65–71 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Shen J, Platek M, Mahasneh A, Ambrosone CB & Zhao H Mitochondrial copy number and risk of breast cancer: a pilot study. Mitochondrion 10, 62–68 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Larsen S et al. Biomarkers of mitochondrial content in skeletal muscle of healthy young human subjects. J. Physiol. 590, 3349–3360 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wachsmuth M, Huebner A, Li M, Madea B & Stoneking M Age-related and heteroplasmy-related variation in human mtDNA copy number. PLoS Genet. 12, e1005939 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cai N et al. Genetic Control over mtDNA and Its Relationship to Major Depressive Disorder. Curr. Biol 25, 3170–3177 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mishmar D et al. Natural selection shaped regional mtDNA variation in humans. Proc. Natl. Acad. Sci. U. S. A 100, 171–176 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Balloux F, Handley L-JL, Jombart T, Liu H & Manica A Climate shaped the worldwide distribution of human mitochondrial DNA sequence variation. Proc. Biol. Sci 276, 3447–3455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sudlow C et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 12, e1001779 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reinhardt K, Dowling DK & Morrow EH Medicine. Mitochondrial replacement, evolution, and the clinic. Science 341, 1345–1346 (2013). [DOI] [PubMed] [Google Scholar]
- 64.Wolf DP, Mitalipov N & Mitalipov S Mitochondrial replacement therapy in reproductive medicine. Trends Mol. Med 21, 68–76 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gemmell N & Wolff JN Mitochondrial replacement therapy: Cautiously replace the master manipulator. Bioessays 37, 584–585 (2015). [DOI] [PubMed] [Google Scholar]
- 66.Kenney MC et al. Molecular and bioenergetic differences between cells with African versus European inherited mitochondrial DNA haplogroups: implications for population susceptibility to diseases. Biochim. Biophys. Acta 1842, 208–219 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fu VK Interracial-Interethnic Unions and Fertility in the United States. J. Marriage Fam. Couns 70, 783–795 (2008). [Google Scholar]
- 68.Ballinger SW Beyond retrograde and anterograde signalling: mitochondrial-nuclear interactions as a means for evolutionary adaptation and contemporary disease susceptibility. Biochem. Soc. Trans 41, 111–117 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mao X et al. A genomewide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet 80, 1171–1178 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Purcell S et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang CC et al. Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4, 7 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kloss-Brandstätter A et al. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum. Mutat 32, 25–32 (2011). [DOI] [PubMed] [Google Scholar]
- 75.Rishishwar L & Jordan IK Implications of human evolution and admixture for mitochondrial replacement therapy. BMC Genomics 18, 140 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ding J et al. Assessing Mitochondrial DNA Variation and Copy Number in Lymphocytes of ~2,000 Sardinians Using Tailored Sequencing Analysis Tools. PLoS Genet. 11, e1005306 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Joesch-Cohen LM & Glusman G Differences between the genomes of lymphoblastoid cell lines and blood-derived samples. Adv. Genomics Genet 7, 1–9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chakrabarty S et al. Upregulation of TFAM and mitochondria copy number in human lymphoblastoid cells. Mitochondrion 15, 52–58 (2014). [DOI] [PubMed] [Google Scholar]
- 79.Nickles D et al. In depth comparison of an individual’s DNA and its lymphoblastoid cell line using whole genome sequencing. BMC Genomics 13, 477 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeon J-P et al. Copy number increase of 1p36.33 and mitochondrial genome amplification in Epstein–Barr virus-transformed lymphoblastoid cell lines. Cancer Genet. Cytogenet 173, 122–130 (2007). [DOI] [PubMed] [Google Scholar]
- 81.Maples BK, Gravel S, Kenny EE & Bustamante CD RFMix: a discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet 93, 278–288 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bryc K, Durand EY, Michael Macpherson J, Reich D & Mountain JL The Genetic Ancestry of African Americans, Latinos, and European Americans across the United States. Am. J. Hum. Genet 96, 37–53 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lind JM et al. Elevated male European and female African contributions to the genomes of African American individuals. Hum. Genet 120, 713–722 (2007). [DOI] [PubMed] [Google Scholar]
- 84.Jobling M, Hollox E, Hurles M, Kivisild T & Tyler-Smith C Human Evolutionary Genetics, Second Edition. (Garland Science, 2013). [Google Scholar]
- 85.Long JC The genetic structure of admixed populations. Genetics 127, 417–428 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Pfaff CL et al. Population structure in admixed populations: effect of admixture dynamics on the pattern of linkage disequilibrium. Am. J. Hum. Genet 68, 198–207 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Tang H et al. Recent genetic selection in the ancestral admixture of Puerto Ricans. Am. J. Hum. Genet 81, 626–633 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Goldberg A & Rosenberg NA Beyond 2/3 and 1/3: The Complex Signatures of Sex-Biased Admixture on the X Chromosome. Genetics 201, 263–279 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Jobling M, Hurles M & Tyler-Smith C Human Evolutionary Genetics: Origins, Peoples & Disease. (Garland Science, 2013). [Google Scholar]
- 90.Brown GR, Laland KN & Mulder MB Bateman’s principles and human sex roles. Trends Ecol. Evol 24, 297–304 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Betzig L Means, variances, and ranges in reproductive success: comparative evidence. Evol. Hum. Behav 33, 309–317 (2012). [Google Scholar]
- 92.Sloan DB, Fields PD & Havird JC Mitonuclear linkage disequilibrium in human populations. Proc. Biol. Sci 282, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Quinlan AR BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics 47, 11.12.1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Nielsen R & Slatkin M An introduction to population genetics: theory and applications. (Sinauer Associates Sunderland, 2013). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All analyses were conducted using publicly available data. The 1000 Genomes Project data are available on their ftp site . Intermediate files and code have been made publicly available on github: (https://github.com/makovalab-psu/Mito_nuclear_incompatibility).