Introduction
Testing, reporting and translation of pharmacogenetics (PGx) into clinical recommendations requires vast knowledge resources. The Pharmacogene Variation (PharmVar) Consortium catalogs pharmacogene variation and provides standardized nomenclature that is utilized by the Pharmacogenomics Knowledgebase (PharmGKB) and the Clinical Pharmacogenetic Implementation Consortium (CPIC). PharmVar allele definitions are also widely used for test design and reporting. This perspective paints a landscape of PGx resources that are needed to facilitate implementation of PGx into clinical practice.
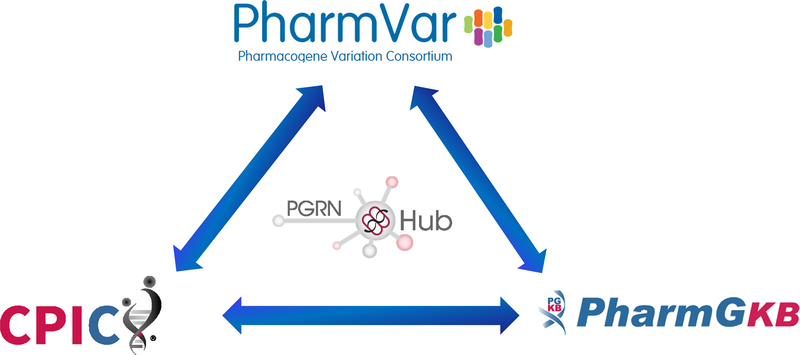
The PharmVar, PharmGKB and CPIC triangle
PharmVar serves as a centralized pharmacogene variation data repository. Its major focus is to catalogue high-quality variation data of genes involved in drug metabolism, disposition and response in order to provide the global PGx communities with a unifying allele designation system (or nomenclature). Cytochrome P450 nomenclature was initially hosted by the Human Cytochrome P450 (CYP) Allele Nomenclature Database and transitioned to PharmVar in 2017 (1), and the database was launched in early 2018. The first non-CYP gene, NUDT15, was introduced into the PharmVar database in 2018 (2). PharmVar offers a growing number of features and tools facilitating easy and intuitive access to relevant gene information (3).
In order to provide standardized and consistent information, PharmVar works closely with the PharmGKB (4) and CPIC (5) (Figure 1). All three are part of the Pharmacogenomics Research Network (PGRN; https://www.pgrn.org), which provides a hub for the PGx community (the PGRN is “catalyzing and leading research in precision medicine for the discovery and translation of genomic variation influencing therapeutic and adverse drug effects”). For example, star allele definitions utilized by the PharmGKB are based on those defined by PharmVar, PharmVar gene expert panels include PharmGKB representatives and a PharmVar representative is participating in CPIC guidelines ensuring consistent use of allele definitions and terminology. Furthermore, specific information is cross-posted to facilitate easy access to information and avoid redundancy in efforts. This is exemplified by PharmVar displaying CPIC clinical allele function assignments. Other examples of collaborative efforts include the PharmVar/PharmGKB initiative developing core allele definitions (i.e. collapsing the growing number of suballeles into a single definition) (6) that will be used by CPIC in future guidelines and all accompanying PharmGKB gene reference tables and the CPIC/PharmVar/PharmGKB initiative to standardize CYP2D6 genotype to phenotype translation (7).
Figure 1. The PharmVar, PharmGKB and CPIC Triangle.
PharmVar, PharmGKB and CPIC are closely collaborating maximizing efforts to provide clinicians and researchers standardized information across all three endeavors.
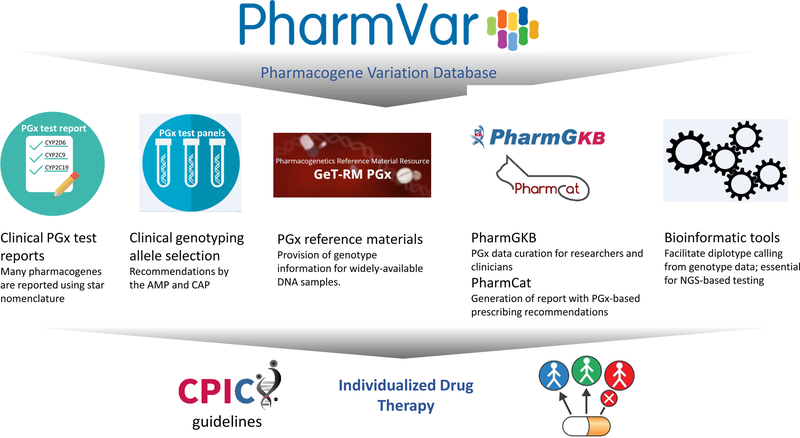
In the following sections we describe other efforts that directly involve PharmVar and/or utilize information that is provided by PharmVar (Figure 2).
Figure 2. Overview of Resources, Applications and Tools that Rely on PharmVar.
Standardized and unified pharmacogene nomenclature facilitates test development, validation and reporting as well as the development of tools to facilitate the interpretation of test results.
PharmVar GeneReviews
PharmVar is launching a series of gene-centric review articles. The inaugural article covers CYP2D6, which is a prominent pharmacogene as evidenced by the numerous drugs that are metabolized through this pathway and six CPIC guidelines covering CYP2D6 gene-drug pairs. CYP2D6 is also a highly polymorphic and structurally complex gene (6). Each gene-centered review covers history, clinical relevance, genetic variability, allele frequency, function and ethnic differences, gene nomenclature, PharmVar curation efforts and particular gene-specific challenges. Recently implemented PharmVar features and tools (e.g. core allele definitions and the graphic allele comparison tool (CAVE)) are exemplified. In addition, new variants that have been defined by PharmVar, methods suitable for haplotype characterization and templates recommended for standardized reporting, genotyping methods and translation into phenotype are also provided.
PharmVar Submissions
PharmVar encourages investigators to submit new allelic variants for designation before publication. Information of how to submit and criteria that need to be fulfilled are provided at https://www.pharmvar.org under the ‘submission’ menu tab. PharmVar also accepts submissions for known haplotypes to raise their evidence level (6) (also see the Allele Criteria and Evidence Level Document for more detailed information). Submission of functional information is optional unless the submitter requests that the allele receives a new star allele designation based on a non-coding and/or synonymous sequence variation. At this point in time PharmVar mostly relies on investigator-initiated submissions to grow its catalog of variants and does not systematically screen the literature or databases for novel allelic variants. On occasion, PharmVar may contact authors to submit published data.
PharmVar and the Pharmacogenetics Reference Materials Resource (Get-RM PGx)
Although PGx testing is increasingly offered by clinical laboratories, reference materials (commonly referred to as quality controls) may be limited or not available for certain allelic variants. Therefore, the analytical accuracy in detecting alleles without reference materials could be questioned. Such sample materials are also critical for PGx research, tool and methods development. The Centers for Disease Control and Prevention–based Genetic Testing Reference Material Coordination Program (GeT-RM), in collaboration with the Coriell Cell Repositories, reagent and assay manufacturers, and members of the PGx testing community continue to develop reference materials (https://www.pgrn.org/get-rm_pgx.html). The most recent project expanded the number of samples as well as the number of alleles for which comprehensive CYP2D6 genotype data are now available (8). Not only were samples with rare allelic variants and/or diplotypes discovered, resequencing of selected alleles led to the confirmation or revision of allele definitions raising their evidence level from ‘Limited’ to ‘Definitive’. PharmVar will continue to closely work with the GeT-RM initiative and plans to cross-link allele definitions with Coriell IDs in the future. This feature will be particularly valuable for the clinical testing community.
PharmVar and Clinical Genotyping Allele Selection
The Association for Molecular Pathology (AMP) Clinical Practice Committee’s Pharmacogenomics (PGx) Working Group are developing minimum panels of variant alleles (“Tier 1”) and an extended panel of variant alleles (“Tier 2”) that will aid clinical laboratories when designing assays for PGx testing through expert consensus. The AMP PGx Working Group considers allele definitions (using PharmVar), functional impact of the variants (using CPIC), allele frequencies in multiethnic populations (e.g. https://gnomad.broadinstitute.org/ and CPIC/PharmGKB frequency tables), the availability of reference materials (RMs, see GeT-RM Program above), as well as other technical considerations for PGx testing when developing these recommendations. These recommendations are not to be interpreted as restrictive but to provide a reference guide to promote standardization of PGx gene/allele testing across clinical laboratories.
Clinical PGx Test Reports
Ideally, PGx test results and interpretations should be consistent regardless of which clinical laboratory performs the test and whether the results were obtained by genotyping or by massively parallel sequencing (commonly referred to as next generation sequencing or NGS). However, laboratories differ with respect to the PGx variants/alleles that are tested and the way in which results are interpreted and communicated to the requesting provider (9). The College of American Pathologists (CAP) requires accredited laboratories to use standard nomenclature to designate genes and variants. In PGx testing, the star allele nomenclature is widely used. CAP states (MOL.49630, Molecular Pathology Checklist, version 08.22.2018) ”…where a common name is also in wide use in the medical literature, it may also be given in the report to improve clarity and prevent misunderstanding.” For clinical laboratories, having PharmVar as an online reference to define the alleles is an essential resource for providing the clarity.
Bioinformatic Tools and Algorithms
PGx test platforms such as the PharmacoScan (Thermo Fisher Scientific), xTAG Nucleic Acid Assay System (Luminex) or iPLEX PGx panels (Agena Biosciences) to name a few, offer software tools to automatically translate results from the tested variants into haplotypes, diplotypes and phenotypes with logic derived from star allele definitions. Testing laboratories and interpretive service providers may also use custom algorithms for calling star alleles for reporting. However, since star nomenclature has been widely adopted for test reporting, it is empirical that standardized nomenclature, such as that provided by PharmVar, is utilized to avoid any confusion of what a test means.
With the increased usage of NGS-based tests such as whole genome, whole exome and targeted panel sequencing, there is a growing need for translation of sequence-based data into pharmacogenetic allele calls. A number of software tools (6) have been developed for calling star alleles from NGS variant data including Astrolabe, Stargazer, Aldy and the CYP2D6 VCF Translator. While the underlying algorithms for mapping sequence variants to star alleles vary between these tools, all share a common requirement for high quality allele definitions that are consistent between multiple reference sequences and genome builds. PharmVar makes allele definitions available in multiple text-based data formats including standard file formats such as VCF and FASTA to facilitate easy downloading and incorporation of data into software applications. Additionally, PharmVar utilizes a strict versioning scheme and defines unique, consistent identifiers with an associated update history to allele definitions which allows applications to unambiguously record which version of definitions was used at a given time.
Pharmacogenomics Clinical Annotation tool (PharmCAT)
The Pharmacogenomics Clinical Annotation Tool, or PharmCAT, is software that generates a report containing clinically-relevant genotype-based information, including CPIC drug prescribing recommendations, from genotype or sequencing data provided as input (10). The tool extracts genetic variants from a VCF file and predicts haplotypes and diplotypes for the majority of genes with CPIC guidelines. PharmCAT then uses CPIC diplotype to phenotype mapping to produce metabolizer phenotypes and provide corresponding CPIC drug prescribing recommendations. PharmCAT is a prototype software pipeline that needs to be developed into production level code to include the generation of batch reporting, not just one report at a time.
Dissemination of PGx Resources
CPIC has created a PGx Dissemination Working Group focused on the dissemination of information about pharmacogenetic resources including CPIC, PharmVar, PharmGKB and the PGRN, as well as implementation strategies for PGx in general. The group’s goal is to identify opportunities to engage with clinicians, payors, clinical labs and professional associations and societies to make them aware of these existing resources. This group offers prepared materials describing the resources and works to increase resource presence on society websites and in social media, with each resource having their own Twitter account to tweet news and updates. Information about resources is also publicized via the PharmGKB Blog. Additionally, the Dissemination Working Group publishes commentaries on existing CPIC guidelines in domain specific journals to raise awareness of their existence.
Conclusion
The PGx resources covered in this perspective article each have their own mission while complementing each other. Continuing efforts ensure harmonized data interpretation and display to provide the research community and clinical implementers up-to-date PGx information that is essential to move the field forward.
Funding
This work was funded by the National Institutes of Health for the Pharmacogene Variation Consortium (R24GM123930; PI, A.G.) and PharmGKB (R24GM61374; PI, T.E.K.). V.M.P. is supported by the Implementing Genomics in Practice (IGNITE) project grants (U01HG007762 and HG010245).
HHS | NIH | National Institute of General Medical Sciences (NIGMS): Andrea Gaedigk, Neil (A) Miller R24GM123930; HHS | NIH | National Institute of General Medical Sciences (NIGMS): Michelle Whirl-Carrillo, Teri (E.) Klein R24GM61374; HHS | NIH | National Human Genome Research Institute (NHGRI): Victoria (M) Pratt U01HG007762; HHS | NIH | National Human Genome Research Institute (NHGRI): Victoria (M) Pratt HG010245 none
Footnotes
Conflict of Interest
The Indiana University School of Medicine Pharmacogenomics Laboratory is a fee-for-service clinical laboratories that offer clinical pharmacogenetic testing (V.M.P.). A.G., M.W.C, N.A.M. and T.E.K. do not declare any conflicts of interest.
References
- (1).Gaedigk A et al. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin Pharm Ther 103, 399–401 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Yang JJ et al. Pharmacogene Variation Consortium Gene Introduction: NUDT15. Clin Pharmacol Ther 105, 1091–4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gaedigk A et al. The Evolution of PharmVar. Clin Pharmacol Ther 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Whirl-Carrillo M et al. Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92, 414–7 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89, 464–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Nofziger C et al. PharmVar GeneReView: CYP2D6. Clin Pharm Ther submitted (in revision). [Google Scholar]
- (7).Caudle KE et al. Standardizing CYP2D6 Genotype to phenotype translation: Consensus recommendations from CPIC and DPWG. CTS accepted, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gaedigk A & et al. Characterization of Reference Materials for Genetic Testing of CYP2D6 Alleles: A GeT-RM Collaborative Project. J Mol Diagn, (2019). doi 10.1016/j.jmoldx.2019.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Kalman LV et al. Pharmacogenetic allele nomenclature: International workgroup recommendations for test result reporting. Clin Pharmacol Ther 99, 172–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sangkuhl K et al. Pharmacogenomics Clinical Annotation Tool (PharmCAT). Clin Pharmacol Ther, (2019). doi doi: 10.1002/cpt.1568 [DOI] [PMC free article] [PubMed] [Google Scholar]