Abstract
In 2009, the Clinical Pharmacogenetics Implementation Consortium (CPIC; www.cpicpgx.org), a shared project between Pharmacogenomics Knowledge Base (PharmGKB, http://www.pharmgkb.org) and the National Institutes of Health (NIH), was created to provide freely available, evidence-based, peer-reviewed, and updated pharmacogenetic clinical practice guidelines. To date, CPIC has published 23 guidelines (of which 11 have been updated), covering 19 genes and 46 drugs across several therapeutic areas. CPIC also now provides additional resources to facilitate the implementation of pharmacogenetics into routine clinical practice and the electronic health record. Furthermore, since its inception, CPIC’s interactions with other resources, databases, websites and genomic communities have grown. This purpose of this paper is to highlight the progress of CPIC over the past 10 years.
Keywords: pharmacogenetics, pharmacogenomics, CPIC, PharmGKB, guideline
One of the major barriers to implementing pharmacogenetic results into routine clinical care had been the lack of clinical guidance on how to use genetic test results to adjust the use or dose of medications (1). To overcome this barrier, as a shared project between the Pharmacogenomics Knowledge Base (PharmGKB, http://www.pharmgkb.org) and the National Institutes of Health (NIH), the Clinical Pharmacogenetics Implementation Consortium (CPIC) was created with the mission of facilitating the translation of research findings into clinical actions for those gene/drug pairs with sufficient evidence. CPIC’s main goal is to provide freely available, evidence-based, peer-reviewed, and updated pharmacogenetic clinical practice guidelines (1). CPIC guidelines have always focused on how to use available pharmacogenetics test results. CPIC guidelines do not discuss whether to order genetic tests and this approach is increasingly relevant as availability of genetic testing is increasing, cost of testing is decreasing, and direct-to-consumer testing is on the horizon. Members are welcome from academia, industry, patient advocacy groups, government and non-governmental organizations with credentials and an interest in pharmacogenetics. CPIC has grown from 60 members in 2012 to over 350 members in 2019 from 245 institutions and 33 countries with observers from the Food and Drug Administration (FDA) and NIH. CPIC has published 23 guidelines (of which 11 have been updated), covering 19 genes and 46 drugs across several therapeutic areas (Table 1) (see https://cpicpgx.org/guidelines/ for a list of current guidelines). The CPIC website is frequently accessed by users; in the past year there have been approximately 398,000 total pageviews over 123,000 sessions with 3.23 pages/session (with an average session duration of 4 min 9 sec).
Table 1.
Genes and associated drugs included in CPIC guidelines as of July 2019
| Gene | Drugs | References |
|---|---|---|
| CACNA1S | volatile anesthetic agents, succinylcholine | (13) |
| CFTR | ivacaftor | (14) |
| CYP2B6 | efavirenz | (15) |
| CYP2C19 | clopidogrel, voriconazole, SSRIs, TCAs, PPIsa | (16–19) |
| CYP2C9 | phenytoin, warfarin, NSAIDsa | (20, 21) |
| CYP2D6 | atomoxetine, codeine, ondansetron, tramadol, tropisetron, tamoxifen, SSRIs, TCAs | (17, 22–25) |
| CYP3A5 | tacrolimus | (26) |
| CYP4F2 | warfarin | (21) |
| DPYD | capecitabine, fluorouracil | (27) |
| G6PD | rasburicase | (28) |
| HLA-A | carbamazepine | (20, 29) |
| HLA-B | carbamazepine, oxcarbazepine, abacavir, allopurinol, phenytoin | (29–31) |
| IFNL3 | peginterferon alfa | (32) |
| mtRNR1 | aminoglycosides | underway |
| NUDT15 | azathioprine, mercaptopurine, thioguanine | (18) |
| RYR1 | volatile anesthetic agents, succinylcholine | (13) |
| SLCO1B1 | simvastatin | (33) |
| TPMT | azathioprine, mercaptopurine, thioguanine | (18) |
| UGT1A1 | atazanavir | (34) |
| VKORC1 | warfarin | (21) |
SSRI, selective serotonin reuptake inhibitors; TCA, tricyclic antidepressants; PPIs, proton pump inhibitors; NSAIDs, non-steroidal anti-inflammatory drugs
Guideline underway
Over the past ten years, CPIC has evolved to meet the needs of guideline users and now provides tables for guideline-specific information that enable implementation of pharmacogenetic test results into electronic health records with clinical decision support. CPIC coordinates projects with the goal of standardizing elements to facilitate information exchange and enable interoperability of pharmacogenetic test results among disparate systems (2). As such, CPIC has become widely recognized as the gold standard resource for the clinical implementation of pharmacogenetics, and CPIC guidelines are used internationally (https://cpicpgx.org/implementation/).
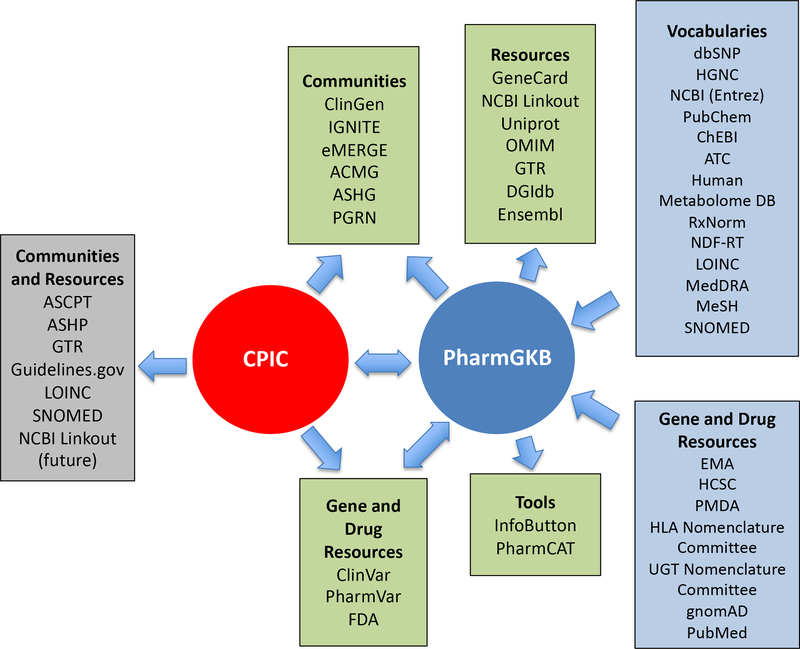
Since its inception, CPIC has partnered closely with PharmGKB, the internationally renowned pharmacogenetics database and website. CPIC’s interactions with other resources, databases, websites and genomic communities have also grown over the past 10 years (Figure 1). CPIC and PharmGKB share contents with, and benefit from, genomics communities and NIH-funded resources such as PharmVar (https://www.pharmvar.org/), ClinVar (https://www.ncbi.nlm.nih.gov/clinvar/) and ClinGen (https://clinicalgenome.org/) (3–5). CPIC also contributes pharmacogenetics summaries to the Genetic Testing Registry (https://www.ncbi.nlm.nih.gov/gtr/) and pharmacogenetics terminology to the Logical Observation Identifiers Names and Codes (LOINC) and the Systematized Nomenclature of Medicine-Clinical Terms (SNOMED CT). CPIC guidelines and terminology are used by the eMERGE (https://emerge.mc.vanderbilt.edu/) and IGNITE (https://ignite-genomics.org) communities, and CPIC is working to further standardize functional classification systems for pharmacogenetics alleles that will facilitate use by other genomics resources. Furthermore, CPIC is working to deposit pharmacogenetics information into ClinVar and ClinGen and is coordinating with PharmVar (the centralized variation data repository) to collect, assign and define gene haplotype nomenclature. CPIC also has growing interactions with other pharmacogenetics guideline developers and resources (e.g., Dutch Pharmacogentics Working Group and the European Pharmocogenetics Implementation Consortium) and other key stakeholders, including the FDA, PharmCAT.(https://www.pharmgkb.org/page/pharmcat), and other partners.
Figure 1. CPIC interactions with PharmGKB and other resources, databases, websites and genomics communities. (Image used with permission from PharmGKB).
ACMG, American College of Medical Genetics and Genomics; ASCPT, American Society for Clinical Pharmacology & Therapeutics; ASHG, American Society of Human Genetics; ASHP, American Society of Health-System Pharmacists; ClinGen, Clinical Genome Resource; eMERGE, Electronic Medical Records and Genomics; FDA, Food and Drug Administration; GTR, Genetic Testing Registry; HGNC, HUGO Gene Nomenclature Committee; IGNITE, Implementing Genomics in Practice; LOINC, Logical Observation Identifiers Names and Codes; NCBI, National Center for Biotechnology Information; NCBO, National Center for Biomedical Ontology; PGRN, Pharmacogenomics Research Network; PharmVar, Pharmacogene Variation Consortium; SNOMED CT, Systematized Nomenclature of Medicine-Clinical Terms
Another challenge is the need to prioritize which pharmacogenetic tests and drugs should be the subject of clinical implementation (1). By assigning CPIC levels of actionability (levels A and B are actionable; levels C or D are not actionable) to gene/drug pairs and providing criteria for prioritizing guidelines (https://cpicpgx.org/prioritization-of-cpic-guidelines/), CPIC provides guideposts to indicate which drugs and genes are most likely to be clinically useful based on current evidence. The criteria that are used to evaluate evidence and assign genes and drugs to different levels have been described (6) and are posted https://cpicpgx.org/genes-drugs/; this is a highly accessed page on the CPIC website (> 52,000 views in the past year). CPIC regularly updates assignment levels as evidence becomes available and is open to input from the user community. To address the need to clearly state that there is insufficient evidence, confidence, or agreement to provide a recommendation to guide clinical practice at the time the guideline is written, CPIC added a “no recommendation” option for authors to use. There are currently still a number of level A or B gene/drug pairs that require guidelines, and this constitutes a substantial “to do” list for CPIC to tackle. Furthermore, having evidence-based interpretations that a genetic test result should not be used to guide prescribing (CPIC levels C and D) for specific drugs is highly valuable to the CPIC user community, particularly when these genetic tests are offered by commercial laboratories; CPIC has plans to develop such guidelines as well. It should be noted that final assignment of genes/drugs to CPIC levels is often not made until after a CPIC guideline authorship group has systematically evaluated the evidence and determined whether their prescribing recommendations will be strong/moderate, optional, or no recommendation, so the CPIC assignments for gene/drug pairs without guidelines should be considered provisional.
With the growing concerns of the impact of financial interests in guideline development, CPIC has continued to adhere to the Institute of Medicine (now The National Academies of Medicine) Clinical Practice Guidelines We Can Trust Standards for Developing Trustworthy Clinical Practice Guidelines (7). As such, all authors must declare any funding interests and activities potentially resulting in conflict of interest (COI) by written disclosure providing statements as to why the relationship could and/or will not influence the guideline development process or specific recommendations. The CPIC Steering Committee weighs the disclosed COI with the need for an author with specific expertise before approving the authorship plan. Though it is not feasible to exclude all authors with any COI, the CPIC Steering Committee has successfully managed COIs for the writing committees and, consistent with the COI policy, authors with substantial COIs may not serve as lead authors and have constituted a minority of authors on any given CPIC guideline.
CPIC is frequently asked by guideline users to provide a list of clinically actionable variants. A clinically actionable variant is a variant that if present in the right gene combination (e.g., usually as part of a diplotype with another similarly actionable variant), prescribing decisions would be altered from the standard course of action. Therefore, to assume that no actionable variants are present, the genome must be interrogated with good enough coverage for actionable variants in order to assume with confidence that the individual is wild-type at the respective potentially actionable loci; there is controversy surrounding how common an actionable variant must be in order to be required for inclusion in a genetic test. CPIC authors are tasked with deciding if there is adequate evidence to assign a “clinical function” to each allele which will be used to generate lists of clinically actionable variants for each gene subject to a CPIC guideline. Downloading a list of actionable variants for all CPIC genes will be facilitated by the current project to build a relational database with an application programming interface (API) for CPIC content. Of note, the Association for Molecular Pathology has published recommendations for the key attributes of CYP2C9 and CYP2C19 alleles (more genes are planned) recommended for clinical testing and a minimum set of variants that should be included in clinical PGx genotyping assays (8, 9).
In 2013, a formal working group within CPIC was formed to focus on informatics aspects of CPIC guidelines. The CPIC Informatics Working Group has recognized that successful adoption of pharmacogenetics into routine clinical care will be greatly facilitated by open access to machine readable tables that accompany CPIC guidelines, currently downloadable via the supplemental tables posted on the CPIC website for each guideline (2). These tables use a standardized format to provide fundamental knowledge for implementation in an EHR-agnostic format, including standardized terminology for describing allele function, phenotype, and also systems for translating genotype to phenotype. Phenotype is linked to clinical decision support language for each drug. Based on the collective experience developing these resources and implementing pharmacogenetics in the EHR with CDS, CPIC Informatics also provided five principles to define the key features of future knowledge bases for pharmacogenetics (2). To further increase the utility of the CPIC implementation resources, all of these tables will be used to populate the new CPIC database, providing greater ability to interrogate and analyze CPIC knowledge in both machine and human readable formats.
To address the need for standardization within pharmacogenetics, CPIC has sponsored and organized specific projects to develop consensus among experts. For example, the Term Standardization Project, in which CPIC brought together 58 experts representing multiple organizations, used the Delphi method to formally develop consensus terms for pharmacogenetic phenotypes and allele function (10). Besides being incorporated into CPIC guidelines and implementation resources, these terms have already been widely adopted by the informatics community, laboratories, and genomics communities (10–12) and are now available as SNOMED CT and LOINC identifiers. A similar working group, the CYP2D6 Genotype-Phenotype Standardization Project, was formed to find consensus among experts to tackle inconsistency across guidelines and laboratories in translating CYP2D6 genotype to phenotype (https://cpicpgx.org/resources/cyp2d6-genotype-to-phenotype-standardization-project/), and the results of this project will be incorporated into CPIC guideline content.
As the development of guidelines and resources does not guarantee its uptake in practice without targeted dissemination (13), a formal working group within CPIC, PharmGKB, PharmVar, and the PGRN was formed in 2018 to focus on the dissemination of these resources and implementation strategies of pharmacogenetics. The goal of the PGx Dissemination Working Group is to support the dissemination of these resources to clinicians, clinical laboratories, payors, and disease specific groups/societies by identifying opportunities to raise awareness of these resources (https://cpicpgx.org/dissemination/).
CPIC guidelines have come to be a widely used and trusted source of unbiased information on how to use genetic test information to guide prescribing (14). Increased scrutiny on genetic testing in general has meant that the standards for maintaining high quality guidelines continue to be of utmost importance. Constantly evolving information on new variants and functional assignments necessitates continual updating of guidelines. The fact that CPIC will likely never be “finished” in creating and updating unbiased guidelines for implementing pharmacogenetics in the clinic necessitates that creative funding mechanisms continue to be considered for maintaining CPIC in the future.
ACKNOWLEDEMENTS
We acknowledge the critical input of members of the CPIC especially Andrea Gaedigk, Ph.D. (PharmVar), Andrew Monte, MD (Dissemination Workgroup Chair), Daniel Mueller, MD, Ph.D, and Andria del Tredici, Ph.D. (Dissemination Workgroup co-chairs) and the CPIC guideline authors.
Funding:
This work was funded by the National Institutes of Health (NIH) for CPIC (R24GM115264; U24HG010135–01) and PharmGKB (R24GM61374) and ALSAC.
Footnotes
Conflicts of interest:
The authors declared no competing interests for this work.
References
- (1).Relling MV & Klein TE CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin Pharmacol Ther 89, 464–7 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Hoffman JM et al. Developing knowledge resources to support precision medicine: principles from the Clinical Pharmacogenetics Implementation Consortium (CPIC). J Am Med Inform Assoc 23, 796–801 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Gaedigk A et al. The Evolution of PharmVar. Clin Pharmacol Ther 105, 29–32 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Landrum MJ & Kattman BL ClinVar at five years: Delivering on the promise. Hum Mutat 39, 1623–30 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Rehm HL et al. ClinGen--the Clinical Genome Resource. N Engl J Med 372, 2235–42 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Caudle KE, Gammal RS, Whirl-Carrillo M, Hoffman JM, Relling MV & Klein TE Evidence and resources to implement pharmacogenetic knowledge for precision medicine. Am J Health Syst Pharm 73, 1977–85 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Caudle KE et al. Incorporation of pharmacogenomics into routine clinical practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline development process. Curr Drug Metab 15, 209–17 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Pratt VM et al. Recommendations for Clinical CYP2C9 Genotyping Allele Selection: A Joint Recommendation of the Association for Molecular Pathology and College of American Pathologists. J Mol Diagn, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Pratt VM et al. Recommendations for Clinical CYP2C19 Genotyping Allele Selection: A Report of the Association for Molecular Pathology. J Mol Diagn 20, 269–76 (2018). [DOI] [PubMed] [Google Scholar]
- (10).Caudle KE et al. Standardizing terms for clinical pharmacogenetic test results: consensus terms from the Clinical Pharmacogenetics Implementation Consortium (CPIC). Genet Med 19, 215–23 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Aronson S, Nolen J & Wood G DIGITizE: Displaying and Integrating Genetic Information Through the EHR. <http://www.nationalacademies.org/hmd/Activities/Research/GenomicBasedResearch/Innovation-Collaboratives/DIGITizE.aspx>. Accessed June 26, 2019.
- (12).Caudle KE, Keeling NJ, Klein TE, Whirl-Carrillo M, Pratt VM & Hoffman JM Standardization can accelerate the adoption of pharmacogenomics: current status and the path forward. Pharmacogenomics 19, 847–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Grimshaw J et al. Toward evidence-based quality improvement. Evidence (and its limitations) of the effectiveness of guideline dissemination and implementation strategies 1966–1998. J Gen Intern Med 21 Suppl 2, S14–20 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Volpi S et al. Research Directions in the Clinical Implementation of Pharmacogenomics: An Overview of US Programs and Projects. Clin Pharmacol Ther 103, 778–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Gonsalves SG et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for the Use of Potent Volatile Anesthetic Agents and Succinylcholine in the Context of RYR1 or CACNA1S Genotypes. Clin Pharmacol Ther 105, 1338–44 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Clancy JP et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for ivacaftor therapy in the context of CFTR genotype. Clin Pharmacol Ther 95, 592–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Desta Z et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2B6 and Efavirenz-containing Antiretroviral Therapy. Clin Pharmacol Ther, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Moriyama B et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP2C19 and Voriconazole Therapy. Clin Pharmacol Ther 102, 45–51 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Hicks JK et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and CYP2C19 Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin Pharmacol Ther 98, 127–34 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Relling MV et al. Clinical Pharmacogenetics Implementation Consortium Guideline for Thiopurine Dosing Based on TPMT and NUDT15 Genotypes: 2018 Update. Clin Pharmacol Ther 105, 1095–105 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Scott SA et al. Clinical Pharmacogenetics Implementation Consortium guidelines for CYP2C19 genotype and clopidogrel therapy: 2013 update. Clin Pharmacol Ther 94, 317–23 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Caudle KE et al. Clinical pharmacogenetics implementation consortium guidelines for CYP2C9 and HLA-B genotypes and phenytoin dosing. Clin Pharmacol Ther 96, 542–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Johnson JA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin Pharmacol Ther 102, 397–404 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Crews KR et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450 2D6 genotype and codeine therapy: 2014 update. Clin Pharmacol Ther 95, 376–82 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Bell GC et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin Pharmacol Ther 102, 213–8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Goetz MP et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for CYP2D6 and Tamoxifen Therapy. Clin Pharmacol Ther 103, 770–7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Hicks JK et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin Pharmacol Ther 102, 37–44 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (28).Birdwell KA et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guidelines for CYP3A5 Genotype and Tacrolimus Dosing. Clin Pharmacol Ther 98, 19–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Amstutz U et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin Pharmacol Ther 103, 210–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Relling MV et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for rasburicase therapy in the context of G6PD deficiency genotype. Clin Pharmacol Ther 96, 169–74 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Phillips EJ et al. Clinical Pharmacogenetics Implementation Consortium Guideline for HLA Genotype and Use of Carbamazepine and Oxcarbazepine: 2017 Update. Clin Pharmacol Ther 103, 574–81 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Saito Y et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for human leukocyte antigen B (HLA-B) genotype and allopurinol dosing: 2015 update. Clin Pharmacol Ther 99, 36–7 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Martin MA et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for HLA-B Genotype and Abacavir Dosing: 2014 update. Clin Pharmacol Ther 95, 499–500 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Muir AJ et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for IFNL3 (IL28B) genotype and PEG interferon-alpha-based regimens. Clin Pharmacol Ther 95, 141–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Ramsey LB et al. The clinical pharmacogenetics implementation consortium guideline for SLCO1B1 and simvastatin-induced myopathy: 2014 update. Clin Pharmacol Ther 96, 423–8 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gammal RS et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for UGT1A1 and Atazanavir Prescribing. Clin Pharmacol Ther 99, 363–9 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]