Abstract
Much has been written about the promise of “precision medicine”, especially in oncology, where somatic mutations can influence the response of cancer cells to “targeted therapy”. There have been successful examples of targeted therapy improving the outcome of some childhood cancers, such as the addition of an ABL class tyrosine kinase inhibitor to conventional chemotherapy substantially improving the cure rate for patients with BCR-ABL1 positive acute lymphoblastic leukemia. Although there are other mutations serving as putative targets in various childhood leukemias and solid tumors, effective targeted therapy has yet to be established for them in prospective clinical trials. There are also uncertainties about which “targeted therapy” to use when patients have multiple targetable genomic lesions in their cancer cells, given the paucity of data upon which to develop evidence-based guidelines for selecting and integrating targeted agents for individual patients. There are also multiple examples of inherited germline variants for which evidence-based guidelines have been developed by CPIC to guide the selection and dosing of medications in children with cancer. Clinical pharmacology is poised to play a critical role in both the discovery and development of new targeted anticancer agents and their evidence-based translation into better treatment for children with cancer. To embrace these challenges and opportunities of “precision medicine”, clinical and basic pharmacologists must expand the depth of our science and the bandwidth of our translational capacity, if we are to optimize precision medicine and advance the treatment of cancer in children and adults.
Introduction
Cure rates for most childhood cancers have improved impressively over the last several decades, with the collective cure rate increasing from about 20% in the 1960s to over 80% today. (1, 2) Advances have been even more impressive for the most common childhood cancer, acute lymphoblastic leukemia (ALL), for which cure rates have improved from <10% in 1960 to over 90% today (3). Yet cancer remains the leading cause of death by disease in children in the developed countries, and the toxicity associated with contemporary therapy adversely affects quality of life during and after treatment (4). Thus, it is imperative that we harness the power of today’s science and technology to develop more effective and less toxic treatments for children with cancer.
There has been much written about the potential of “precision medicine” in oncology, using data from whole genome, whole exome, whole transcriptome and/or whole methylome interrogation to select the optimal treatment for individual patients, based on both germline variants and the nature of somatic mutations and not merely the histology and staging of a patient’s tumor (5–7). Indeed, it was on this basis that the US NCI launched the MATCH (Molecular Analysis for Therapy Choice) clinical trial, within which adults whose tumors were found to have mutations in either PIK3CA or FGFR or over-expression of HER2 (excluding breast and gastric cancers) were treated with agents targeting these mutated genes/pathways (i.e., taselisib, AZD4547 or ado-trastuzumab emtansine, respectively). However, the initial results were disappointing with only partial responses in just 10% of patients given the FGFR inhibitor or the HER2 inhibitor and no objective response with the PIK3CA inhibitor (8). The disappointment was offset somewhat by the fact that many of these patients had not responded well to extensive chemotherapy prior to being enrolled on the MATCH trial. Likewise, the initial enthusiasm for using tumor mutation burden (TMB) determined by whole-exome sequencing as a biomarker for identifying non-small cell lung cancer (NSCLC) patients more likely to respond to PD-1 inhibitors (e.g., pembrolizumab) has been dampened by disappointing results in follow-up prospective clinical trials and by the inability to assess TMB in a high percentage (~30–40%) of patients with NSCLC (9). These findings are a clear signal that we are in the early days of “precision oncology”, and this is especially true in pediatric oncology where the number of eligible patients is small and only a few studies have been completed.
Recent attempts to improve precision medicine strategies have included the use of drug combinations based on tumor DNA sequencing (I-PREDICT), sequencing of circulating tumor cells (TARGET) and sequencing tumor DNA coupled with RNA sequencing of adjacent normal tissue (WINTHER) (11). While there were some encouraging responses observed in previously treated patients, only a small minority of patients had objective responses (4–11%). These findings are consistent with the SHIVA trial that found no difference in progression-free survival in previously treated patients with metastatic cancers, after treatment with molecularly targeted therapies compared to physician’s choice of treatment (10).
Nonetheless, the number of MATCH-style trials for childhood cancers is growing, including PROFYLE in Canada, ESMART across Europe, and NCI Pediatric MATCH in the US. The US pediatric version of MATCH currently includes ten targeted therapeutics, inhibiting ALK, BRAF, EZH2, MEK, TRK, PARP, ERK, PI3K/mTOR, CDK4/6, or FGFR signaling pathways (12). In addition to scientific discoveries fueling these potentially exciting trials, legislative initiatives such as RACE for Children Act and the STAR Act are boosting efforts and interest in testing novel agents in pediatric populations, although these studies are not without challenges. There is also great interest in using genomics to guide the “repurposing” of FDA approved medications as a less expensive and more expedient strategy for expanding treatment for many diseases, including childhood cancers (13).
Glass Half Full
Enthusiasm for targeted therapies for childhood cancers is bolstered by promising results in BCR-ABL1 ALL, for which the addition of an ABL tyrosine kinase inhibitor to conventional combination chemotherapy markedly improved cure rates from ~30% in historical controls to ~ 60% (14). This treatment advance was subsequently extended beyond the 2–4% of childhood ALL cases with the BCR-ABL1 fusion, when it was discovered that an additional ~10–15% of pediatric ALL cases have BCR-ABL1-like ALL with a gene expression pattern resembling leukemia with the BCR-ABL1 fusion (15, 16). The underlying genetic mechanisms of BCR-ABL1-like ALL have now been largely elucidated; about 50% of these cases have CRLF2 rearrangements with or without JAK2 mutations and among the remaining cases, 15–20% have ABL1-class fusions, which exhibited in vitro sensitivity to ABL tyrosine kinase inhibitors (17). Another 10 to 15% have JAK2 or EPOR rearrangements or other JAK-STAT mutations and exhibited in vitro sensitivity to JAK inhibitors (17). Ongoing clinical trials are assessing whether the addition of these agents to conventional chemotherapy will translate into improved cure rates for BCR-ABL1-like ALL with targetable lesions, as was observed for BCR-ABL1 ALL. These discoveries nicely exemplify how genomic studies can identify new subtypes of ALL and establish the scientific basis for selectively incorporating new “targeted” agents into the treatment of patients whose cancer harbors specific somatic mutations.
Similarly, genomic studies have identified multiple subtypes of medulloblastoma (MB), a common type of malignant brain tumor in children (18). The WHO has incorporated consensus criteria to define four major subtypes of MB based on genomic characterization: WNT abnormalities, sonic hedgehog (SHH) abnormalities, and two other distinct groups (Group 3 with high MYC amplification and Group 4 that harbors a variety of genetic abnormalities), and treatment today is guided by integration of molecular genetics, histomorphology, and imaging to risk-stratify patients. Clinical trials are currently testing whether escalation of therapy (irradiation, chemotherapy) in high-risk patients or de-escalation of therapy for lower-risk patients can improve treatment outcomes. There are also ongoing studies in patients with SHH-MB to assess the efficacy of targeted SHH inhibitors (that can compromise skeletal development) in skeletally mature patients. Also, inherited germline polymorphisms in GST-M1/T1 have been associated with increased neuropsychological morbidity after craniospinal irradiation in children with MB (19). In a different type of brain tumor, glioblastoma, somatic hyper-methylation of the O6‐methylguanine‐DNA methyltransferase (MGMT) gene promoter has been associated with a better response to alkylating agents, including temozolomide, in both children and adults (20–23).
These are still early days of using genomics to tailor the nature and intensity of treatment for pediatric ALL and brain tumors, and ongoing studies are assessing novel cellular therapies, including CAR T-cell therapy targeting either CD19 in B-lineage ALL or HER2 in a subset of MB expressing this epitope.
Likewise, genomic studies are providing new insights and potential novel therapeutic strategies for several pediatric solid tumors. For example, MYCN, TRK and ALK have pathologic and prognostic relevance in pediatric neuroblastoma, and there are early-stage clinical trials to assess the potential of ALK inhibitors (e.g., crizotinib) for treating ALK-mutated neuroblastomas, with newer generation ALK inhibitors (lorlatinib) showing greater promise in model systems. There have been similar advances with other pediatric solid tumors, including Ewing sarcoma and osteosarcoma, where genomic studies are pointing to new therapeutic targets, some of which are being assessed in early stage clinical trials (24). There have also been promising results using tropomyosin receptor kinase (TRK)-inhibitors (e.g., larotrectinib) in treating a diverse spectrum of pediatric solid tumors with chromosomal rearrangements creating TRK-fusions (25).
It remains to be seen whether treatment advances will emerge from ongoing clinical trials that are deploying various genomic methods to identify additional therapeutic targets in pediatric solid tumors (e.g., INFORM) (26). Early results in some pediatric cancers with a dismal prognosis (e.g., diffuse intrinsic pontine glioma) indicate that this approach can reveal previously unrecognized targets for which medications are currently available (27).
And of course there are very well-established examples of using inherited germline variants to guide the selection of appropriate dosages of chemotherapy (e.g., TPMT and NUDT15 to guide thiopurine dosing), or in guiding the use of ancillary medications in pediatric cancer patients, including CYP2C19 for voriconazole dosing and CYP2D6 for codeine analgesia. (5) There have also been important advances in building active clinical decision support into the electronic health record, to alert clinicians to the importance of pharmacogenomics for high-risk medications, using evidence-based criteria developed by the Clinical Pharmacogenetics Implementation Consortium (CPIC) (5). It is also not uncommon for inherited functional variants to be present in genes encoding the targets of anticancer agents (28).
Somatic genetic and epigenetic analyses are also providing new insights into the genomic mechanisms of cancer cell resistance to conventional and targeted anticancer agents and revealing potential strategies to mitigate resistance (29–31).
Indeed, the glass is approaching half-full at this stage, with genomics providing deeper insights into disease pathogenesis, and offering improved methods for discovering new targets, assigning prognosis, guiding the intensity of treatment and/or selecting more-targeted chemotherapy for some diseases.
Glass half empty
The above recent progress notwithstanding, the reality is that “precision oncology” is in its infancy for both adult and pediatric cancers. Although genomics provides important diagnostic and therapeutic insights to improve treatment outcome for some malignancies, there is currently a paucity of rigorous evidence of this success for most pediatric cancers. There is promise, but scant evidence.
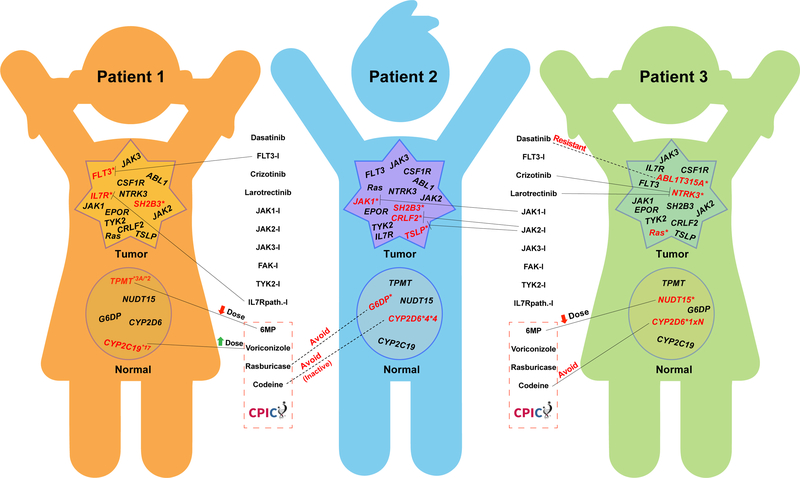
Therefore, in the coming decade precision oncology must continue to move forward within the context of prospective clinical trials. Yet even within clinical trials, there is often reluctance to define a priori the precise genetic abnormalities that will be used to guide treatment decisions, including the selection of medications. Some argue this is understandable, because the body of evidence on which somatic mutations justify changes in treatment is not yet clearly defined. And while there are often “hot spots” for mutations that activate or inactivate critical cancer genes, new driver or cooperative mutations are constantly being discovered as more patient tumors are sequenced and it is extremely challenging to characterize their functional consequences in real-time to determine their impact on drug response. Unfortunately, this often leads to clinical trials with vaguely defined criteria, which risks heterogeneity in treatment decisions depending on who is interpreting the data for any given patient. Furthermore, there are often mutations in multiple genes within the same tumor (Figure 1), making it unclear which mutations should drive treatment decisions and the sequence in which multiple targeted agents should be given has not been rigorously defined. Furthermore, it does not help that most CLIA-compliant genome sequencing services merely report out all mutations in a defined set of “cancer genes” (e.g., COMIC genes), leaving it to the treating clinicians to determine the therapy to be prescribed. This approach is comparable to what is done for tests like blood glucose; almost never do the clinical diagnostic laboratories recommend treatment, they just report out the result and let the treating physician decide what to do. In the case of hyperglycemia, the path toward a precise diagnosis is relatively straightforward, and there is typically little urgency in initiating optimal treatment. But this is not the case when using multiple cancer genome mutations to select optimal cancer therapy for an individual patient. One could argue that CLIA-compliance with genome sequencing is no more important than rigorous evidence-based interpretation of genome sequences, yet the latter is left to flounder outside the CLIA structure, often without strict quality controls. A carefully-designed process for establishing clinical guidelines for using somatic genome variants to direct cancer therapy (similar to CPIC for germline pharmacogenomics) is needed to ensure rational deployment of precision medicine in oncology. Evidence-based artificial intelligence coupled with electronic clinical decision support may hold the answers in the coming decade.
Figure 1. Complexity in selecting optimal medications based on combinations of germline and somatic genome variation in cancer.
Somatic (tumor) and germline (normal) genome variation is reflected for three hypothetical patients with BCR-ABL1-like acute lymphoblastic leukemia (ALL), based on actual genome variants documented from sequencing patients with this disease (17). For each hypothetical patient, genes indicated have already been shown to have functional alterations in ALL (mutations or structural alterations in leukemia cells, inherited variants altering function in germline DNA), and those with mutations in each patient are indicated in red font with an asterisk. Somatic variants are often activating, whereas germline genes are typically loss-of-function (TPMT, NUDT15, CYP2D6) or more rarely gain-of-function (CYP2C19 and CYP2D6 duplication alleles). Potential selection of medications targeting somatic mutations is based on in vitro or in vivo activity of each medication against target proteins. Multiple variants often occur in the same leukemia cell, as documented in prior sequencing studies (17), and often only a subset are treated, as depicted for each patient. All inherited germline variants are essentially always present in the tumor (not depicted). Selection of optimal therapy using inherited germline variants follows evidence-based CPIC guidelines (for medications below the dotted red line). A substantial number of additional somatic and inherited genome variants are known to exist in this disease, adding further complexity to evidenced-based selection of optimal treatment.
Pediatric precision oncology is also being slowed by the lack of interest within the pharmaceutical industry to develop novel agents that target genes commonly mutated in childhood cancers. The reasons are sadly understandable, as these companies prioritize making a profit, and the number of cases of any childhood cancers pales in comparison to the number of cases of lung cancer, breast cancer or most other adult malignancies. Plus, children are smaller, and thus require fewer milligrams of therapy, all of which makes the financial incentives de minimis in the for-profit world. This is nicely exemplified by the development of ALK inhibitors (e.g., crizotinib). ALK mutations (structural variants) were originally discovered in 1994 in a pediatric lymphoma, hence anaplastic lymphoma kinase (32). But pharma had no interest in developing an ALK inhibitor until ALK was found to also be activated via a chromosomal translocation in non-small cell lung cancer, 13 years later (2007). There are now five different ALK inhibitors approved by the FDA, with others under development, although none has yet been approved for childhood cancers. It is unclear whether the coming decade will yield incentives or regulatory requirements for pharmaceutical companies to develop targeted agents for childhood cancers, but little has happened in the decade since the Institute of Medicine report on the failure of pharma to develop new medications for childhood cancers (33). It is hard to be optimistic that this will soon change.
Prospectus
Indeed, in many ways the glass now seems only half full, but the glass is constantly growing due to advances in science and technology that are rapidly expanding our universe of knowledge and challenging our ability to translate this into more effective and less toxic therapy for childhood cancers. Clinical pharmacology is poised to play a critical role in both the discovery and development of new targeted anticancer agents and their translation into better treatment. Over the coming decade, clinical and basic pharmacologists should rise to this challenge (and opportunity) to expand the depth of our science and the bandwidth of our translational capacity, if we are to optimize the use of “precision medicine” to advance the treatment of childhood cancers.
Acknowledgments
Funding: This work was funded in part by funds from NIH Grants R01 CA36401 (W.E.E), P50 GM115279 (W.E.E.), U01 GM92666 (W.E.E.), and R01 GM118578 (J.J.Y). St. Jude Comprehensive Cancer Center grant CA21765 from the National Cancer Institute (C.H.P), V Foundation Translational Research Award (T2015-006) (J.J.Y), Leukemia Lymphoma Society SCOR (7010-14) (J.J.Y), the Children’s Oncology Group HEM ITSC grant (J.J.Y), and by the American Lebanese Syrian Associated Charities. All authors receive funding from NIH, but the content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
CHP has received honoraria from UpToDate and owns stock in Adaptive Biotechnologies Corp. St. Jude Children’s Research Hospital received a major donation from AbbVie pharmaceutical to support the Family Commons, a treatment and research-free space for patients and families.
Footnotes
Conflicts of Interest: WEE and JJY do not have any conflicts of interest to report.
References
- (1).Allemani C. et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet 391, 1023–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Rodriguez-Galindo C. et al. Toward the Cure of All Children With Cancer Through Collaborative Efforts: Pediatric Oncology As a Global Challenge. J Clin Oncol 33, 3065–73 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Heikamp EB & Pui CH Next-Generation Evaluation and Treatment of Pediatric Acute Lymphoblastic Leukemia. J Pediatr 203, 14–24 e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Couzin-Frankel J. Beyond survival. Science 363, 1166–9 (2019). [DOI] [PubMed] [Google Scholar]
- (5).Relling MV & Evans WE Pharmacogenomics in the clinic. Nature 526, 343–50 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).DuBois SG, Corson LB, Stegmaier K & Janeway KA Ushering in the next generation of precision trials for pediatric cancer. Science 363, 1175–81 (2019). [DOI] [PubMed] [Google Scholar]
- (7).Ma X. et al. Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555, 371–6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Health, N.I.o. NCI-MATCH precision medicine clinical trial releases new findings, strengthens path forward for targeted cancer therapies. (National Cancer Institute National Institutes of Health, 2018). [Google Scholar]
- (9).Addeo A, Banna GL & Weiss GJ Tumor Mutation Burden-From Hopes to Doubts. JAMA Oncol, (2019). [DOI] [PubMed] [Google Scholar]
- (10).Le Tourneau C. et al. Molecularly targeted therapy based on tumour molecular profiling versus conventional therapy for advanced cancer (SHIVA): a multicentre, open-label, proof-of-concept, randomised, controlled phase 2 trial. Lancet Oncol 16, 1324–34 (2015). [DOI] [PubMed] [Google Scholar]
- (11).Le Tourneau C, Borcoman E & Kamal M. Molecular profiling in precision medicine oncology. Nat Med 25, 711–2 (2019). [DOI] [PubMed] [Google Scholar]
- (12).Institute, N.C. <https://www.cancer.gov/about-cancer/treatment/clinical-trials/nci-supported/pediatric-match#ui-id-2>. 2019.
- (13).Pushpakom S. et al. Drug repurposing: progress, challenges and recommendations. Nat Rev Drug Discov 18, 41–58 (2019). [DOI] [PubMed] [Google Scholar]
- (14).Schultz KR et al. Improved early event-free survival with imatinib in Philadelphia chromosome-positive acute lymphoblastic leukemia: a children’s oncology group study. J Clin Oncol 27, 5175–81 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Den Boer ML et al. A subtype of childhood acute lymphoblastic leukaemia with poor treatment outcome: a genome-wide classification study. Lancet Oncol 10, 125–34 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Mullighan CG et al. Deletion of IKZF1 and prognosis in acute lymphoblastic leukemia. N Engl J Med 360, 470–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Roberts KG et al. Targetable kinase-activating lesions in Ph-like acute lymphoblastic leukemia. N Engl J Med 371, 1005–15 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Northcott PA et al. Medulloblastoma. Nat Rev Dis Primers 5, 11 (2019). [DOI] [PubMed] [Google Scholar]
- (19).Barahmani N. et al. Glutathione S-transferase M1 and T1 polymorphisms may predict adverse effects after therapy in children with medulloblastoma. Neuro Oncol 11, 292–300 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Hegi ME et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med 352, 997–1003 (2005). [DOI] [PubMed] [Google Scholar]
- (21).Paz MF et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res 10, 4933–8 (2004). [DOI] [PubMed] [Google Scholar]
- (22).Hegi ME et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res 10, 1871–4 (2004). [DOI] [PubMed] [Google Scholar]
- (23).Esteller M. et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med 343, 1350–4 (2000). [DOI] [PubMed] [Google Scholar]
- (24).Grohar PJ, Janeway KA, Mase LD & Schiffman JD Advances in the Treatment of Pediatric Bone Sarcomas. Am Soc Clin Oncol Educ Book 37, 725–35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Drilon A. et al. Efficacy of Larotrectinib in TRK Fusion-Positive Cancers in Adults and Children. N Engl J Med 378, 731–9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Worst BC et al. Next-generation personalised medicine for high-risk paediatric cancer patients-The INFORM pilot study. Eur J Cancer 65, 91–101 (2016). [DOI] [PubMed] [Google Scholar]
- (27).Pfaff E. et al. Brainstem biopsy in pediatric diffuse intrinsic pontine glioma in the era of precision medicine: the INFORM study experience. Eur J Cancer 114, 27–35 (2019). [DOI] [PubMed] [Google Scholar]
- (28).Scharfe CPI, Tremmel R, Schwab M, Kohlbacher O & Marks DS Genetic variation in human drug-related genes. Genome Med 9, 117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Paugh SW et al. NALP3 inflammasome upregulation and CASP1 cleavage of the glucocorticoid receptor cause glucocorticoid resistance in leukemia cells. Nat Genet 47, 607–14 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Watson PA, Arora VK & Sawyers CL Emerging mechanisms of resistance to androgen receptor inhibitors in prostate cancer. Nat Rev Cancer 15, 701–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Shah NP et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell 2, 117–25 (2002). [DOI] [PubMed] [Google Scholar]
- (32).Morris SW et al. Fusion of a kinase gene, ALK, to a nucleolar protein gene, NPM, in non-Hodgkin’s lymphoma. Science 263, 1281–4 (1994). [DOI] [PubMed] [Google Scholar]
- (33).Adamson PC, National Cancer Policy Board (U.S.). Committee on Shortening the Time Line for New Cancer Treatments., Institute of Medicine (U.S.), National Research Council (U.S.) & National Academy of Sciences (U.S.) Making better drugs for children with cancer (National Academies Press: Washington, D.C., 2005). [Google Scholar]