Abstract
Macroautophagy (referred to hereafter as autophagy) is an intracellular degradation pathway in which the formation of a double-membrane vesicle called the autophagosome is a key event in the transport of multiple cytoplasmic cargo (e.g. proteins, protein aggregates, lipid droplets or organelles) to the vacuole (lysosome in mammals) for degradation and recycling. During this process, autophagosomes are formed de novo by membrane fusion events leading to phagophore formation initiated at the phagophore assembly site (PAS). In yeast, Atg11 and Atg17 function as protein scaffolds, essential for selective and non-selective types of autophagy, respectively. While Atg17 functions in non-selective autophagy are well-defined in the literature, less attention is concentrated on recent findings regarding the roles of Atg11 in selective autophagy. Here, we summarize current knowledge about the Atg11 scaffold protein and review recent findings in the context of its role in selective autophagy initiation and autophagosome formation.
Keywords: Atg11, Atg17, autophagosome, membrane tethering, scaffold protein, selective autophagy, selective autophagy receptor
Introduction
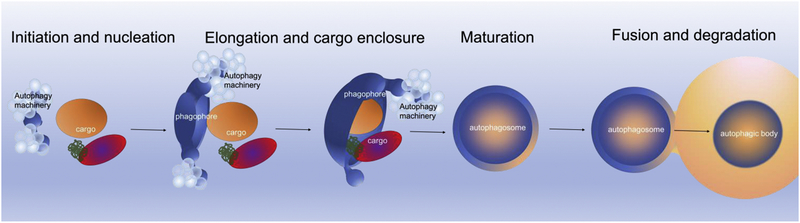
Autophagy is a conserved, intracellular, eukaryotic degradation and recycling process that helps maintain cellular homeostasis - one of the most prerequisites for cell survival. In response to different stimuli, as well as in non-induced conditions, cells use this process to degrade and recycle a wide range of cellular components from individual macromolecules such as proteins or lipids, to entire organelles. In yeast, as a consequence of stress, non-selective autophagy is often initiated for “self-consumption” of non-essential, intracellular macromolecules from the cytoplasm, whose building blocks are recycled for cellular use. Similar to its non-selective counterpart, selective autophagy can also be activated to turn over superfluous and damaged organelles or other unwanted cytoplasmic components, such as protein aggregates or lipid droplets, but in a highly selective way. In both non-selective and selective processes, cargos are sequestered within a double-membrane vesicle, named the autophagosome. The formation of autophagosome is initiated de novo at the phagophore assembly site (PAS), followed by expansion of a double membrane around the cargo in the form of a cup-shaped phagophore (also termed isolation membrane) through membrane addition. Whereas in non-selective autophagy the phagophore engulfs bulk cytoplasm (with or without resident organelles), the targets of selective autophagy are specifically recognized by selective autophagy receptors (SARs), which engage simultaneously with the general autophagic machinery. After vesicle maturation, the cellular content captured in the autophagosome is delivered to the vacuole (or lysosome in mammals) via fusion of the outer membrane of the autophagosome with the vacuolar membrane. Consequently, the cargo, surrounded by the inner membrane of autophagosome (called autophagic body), is degraded in the vacuole lumen by vacuolar hydrolases and the ensuing building blocks are then reused by the cells (Fig. 1).
Figure 1. Schematic diagram of the steps of autophagy.
An autophagy induction signal leads to the formation of a crescent-shaped, sequestering membrane called the phagophore or isolation membrane (initiation and nucleation step). During this process, the autophagy core machinery proteins are activated and recruited to the PAS. The concerted action of these proteins leads to the expansion of the phagophore, during which cytoplasmic constituents, including organelles, aggregated proteins etc. are enwrapped (elongation and cargo enclosure step) by the phagophore. During this step, a phagophore can engulf bulk cytoplasm nonspecifically, including entire organelles, or specifically targeted cargos. At the end of elongation, the phagophore closes, resulting in the formation of a double-membrane vesicle called the autophagosome (maturation step). Once the autophagosome is mature, its outer membrane fuses with the vacuole/lysosome membrane and the autophagic body/autolysosome is released for vacuolar/lysosomal hydrolases degradation (fusion and degradation steps).
The formation of autophagosomes is orchestrated by well-conserved, autophagy-related (Atg) proteins, encoded by ATG genes. In yeast, around 41 Atg proteins have been identified so far, which are responsible for the different steps of autophagy. Of these, approximately 19 Atgs are referred to as “the core autophagic machinery” that is shared between non-selective and selective autophagys.[1] These proteins are divided in the literature into five, multifunctional modules regulating different stages of autophagy: the Atg1 kinase complex (Atg1, Atg13, Atg17, Atg29 and Atg31), Atg9-containing vesicles, the Atg2–Atg18 complex, the Atg8 and the Atg5-Atg12 conjugation systems (Atg3, Atg4, Atg5, Atg7, Atg8, Atg10, Atg12 and Atg16) and the class III phosphoinositide 3-kinase (PI3K) complex I (Atg6, Atg14, Atg38 and two vacuolar protein-sorting proteins, Vps15 and Vps34).[2, 3]
To initiate autophagosome formation in yeast, scaffold proteins and membrane-tether components, like Atg11 and Atg17, are required. These proteins serve as hubs or platforms for the recruitment of the core autophagic machinery. While Atg17 is sufficient to drive non-selective autophagy, the involvement of Atg11 is required for selective autophagy, which, besides playing a similar function to Atg17, plays a critical role in organizing and activating the selective autophagy-specific PAS (selective PAS) by interacting with the SAR/cargo complexes.
Due to the increasing numbers of reports revealing new functions of Atg11 as a principal component of the selective PAS and its crucial role in the molecular mechanisms of autophagosome formation, we review here current knowledge regarding Atg11 and its roles in selective autophagy. Based on data obtained for the Atg11 protein from yeast studies, we summarize structural and functional properties of this protein and we emphasize its currently known roles at each step of selective autophagy. We also describe the evolutionary conservation of Atg11 in plants and mammals, drawing attention to its structural and functional similarities within the eukaryotic kingdoms.
Atg11 - a scaffold protein for selective autophagy
The spatial and temporal organization of molecules within a cell is critical for coordinating various biological processes and pathways carried out by the cell. One mechanism by which such coordination is achieved is by using scaffold proteins, which recruit and juxtapose multiple protein partners involved in specific functions and simultaneously assemble them into a functional unit where their concentrations are vastly enriched at specific locations to enhance and/or regulate signaling specificity, efficiency and fidelity.[4, 5] However, to ensure dynamic regulation, coordination and signaling within the cell, scaffold proteins can also exert complex allosteric control over their partners and are themselves the target of regulation.[4–7]
The modular nature of their structures is an indispensable part of the ability of scaffold proteins to associate with a multitude of proteins via protein-protein interaction domains, motifs or regions. Such modular structures and a large number of established interaction partners are characteristics of the Atg11 protein. These properties, together with its tendency to homo-dimerize, defines Atg11 as a scaffold protein and selective autophagy adaptor that binds to a variety of SARs to mediate different types of selective autophagy. For example, Atg11 binds to Atg19, Atg32, as well as to Atg36 or Atg30, which recognize various cargos delivered by the Cvt (Cytoplasm to vacuole), mitophagy (selective mitochondria degradation by autophagy) and pexophagy (selective peroxisome degradation by autophagy) pathways in S. cerevisiae or P. pastoris, respectively.[8–13] Atg11 also interacts with the Atg1 kinase and the Atg17-Atg31-Atg29 complex, which connects the steps of cargo selection to the initiation of autophagosome biogenesis.[11] Additionally, by its interaction with Atg9, Ypt1, Atg20 and with Atg8, a key protein decorating autophagosomes, as well as with the Atg5-Atg12 conjugation system required for Atg8 lipidation, Atg11 plays a role in membrane-trafficking, stabilization of membrane curvature, in membrane tethering and in the expansion of autophagosomal membrane.[11, 14–17] Furthermore, it has been reported that Atg11 resides on mitochondria or peroxisomes prior to mitophagy or pexophagy, respectively, where it interacts with the membrane-fission GTPase, Dynamin-related protein 1 (Dnm1 or Drp1), Mitochondrial fission 1 protein (Fis1) and Vacuolar protein sorting-associated protein 1 (Vps1) - components of organelle fission machineries.[18, 19] Finally, during the termination of selective autophagy, a role of Atg11 as a scaffold for the autophagosome-vacuole fusion step has also been suggested.[20] These numerous interactions make Atg11 a critical protein for selective autophagy and an important hub that coordinates several steps of selective autophagy from cargo selection to autophagosome maturation and the termination of selective autophagy.
Atg11 - from structure to function
Structure and coiled-coil domain organization
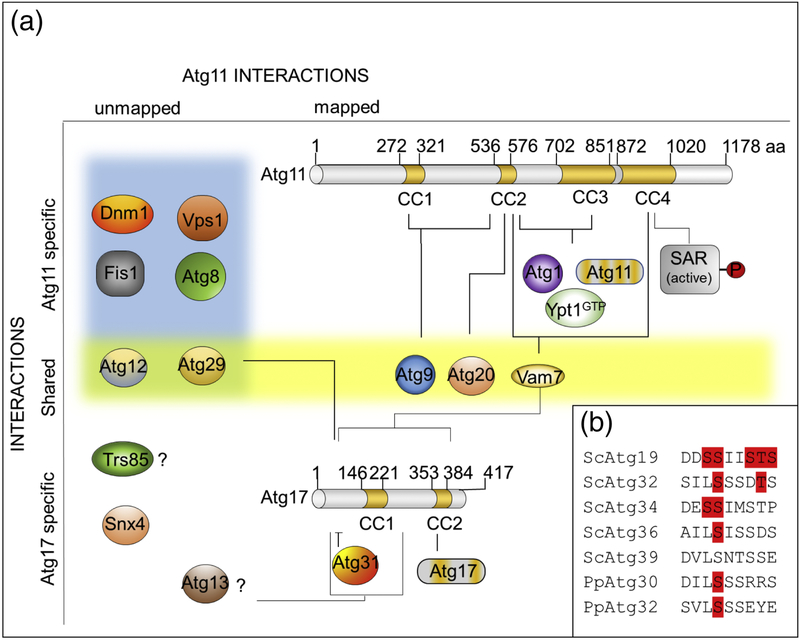
Atg11 with 1178 aa (135 kDa) is the second largest protein when compared with other core Atg proteins. Despite its size, the only structural characteristic suggested by homology domain searches is the abundance in Atg11 of coiled-coil (CC) modules that are distributed in its sequence (Fig. 2A). CCs constitute one of the principal subunit oligomerization motifs in proteins.[21] Naturally-occurring CCs have between two and five amphipathic α-helices that twist around one another to form a supercoil.[22] CCs can be arranged in either parallel or antiparallel orientations. Recent studies demonstrate that in contrast to Atg17, which has antiparallel CCs and forms S-shaped dimers,[23, 24] Atg11 has a parallel CC architecture.[25] Both N-terminal and C-terminal halves of Atg11 (containing CC1 and CC2 or CC3 and CC4, respectively) form stable extended homo-dimers.[25] Suzuki and Noda suggest that these architectural differences between Atg11 and Atg17 (Fig. 2A) reflect the variation in the selective and non-selective PAS organization.[25] In the selective PAS, binding of multiple Atg11 dimers to the surface of the large cargo via SARs might be sufficient to adopt the characteristic cup-shape of the phagophore, which subsequently surrounds and encloses the cargo within the autophagosome (Fig. 3). In the non-selective PAS, where the cargo has no conserved shape/structure, assembling the cup-shaped phagophore (Fig. 3, inset) requires repeated three-dimensional interactions with 34 nm long, S-shaped Atg17 complexes to overcome a substantial kinetic barrier in this membrane shape transition.[26]
Figure 2. Scheme of Atg11 structure and its interacting proteins.
A. Atg11, an 1178 aa protein, contains 4 coiled coils regions (CC1–4, marked in yellow). The proteins that interact with specific regions of Atg11 are indicated. Other known and suggested (labeled with a question mark) Atg11 protein partners, whose interactions with Atg11 have not been mapped or exactly determined yet, are shown on the left of the figure. Note, that Atg11 interactions with Trs85 and Atg13 were reported but since there is no evidence for their direct binding to Atg11 in comparison with Atg17, they are shown as being specific for Atg17. Positions of CCs within Atg11 and Atg17 are drawn to scale. The Atg17 protein with its coiled coils regions and interacting partners is show to highlight similarities and differences between these two scaffold proteins. The AIMs in Atg11 predicted by iLIR software (27 in total) are sites that could interact with Atg8. Note that the SAR proteins need to be properly phosphorylated (P) prior to their binding to Atg11. Also Atg9 binding to Atg17 is not mapped and could be mostly indirect via Atg13. B. The alignment of Atg11 binding regions (A11BR) from different SARs. Confirmed phosphorylation sites are marked in red.
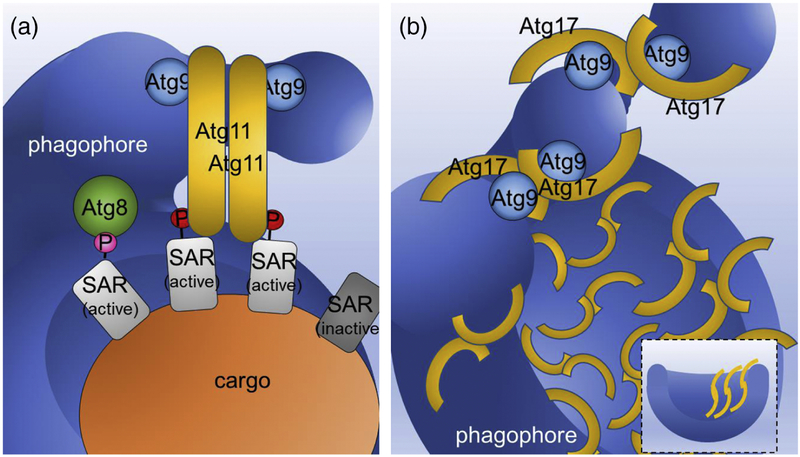
Figure 3. Comparison of proposed mechanisms of scaffolding the cup-shaped double membrane of the autophagosome by Atg11 and Atg17.
A. Proposed model for the role of Atg11 in membrane tethering at the selective PAS. Only SARs (light gray) activated by phosphorylation at their Atg11-binding regions (A11BRs) (A11BR phosphorylation is indicated by a red circle labelled and the pink circle labelled P shows AIM phosphorylation) interact with Atg11. Binding to activated SAR stimulates Atg11 dimerization in a parallel manner. Atg11 also interacts with Atg9-vesicles and Atg9-decorated phagophore membranes. In this state, Atg9-vesicles are brought into close physical proximity to the phagophore, which along with other components of the autophagy machinery (not shown for simplicity) initiates their tethering and drives their fusion. Atg8 binds to SARs phosphorylated at their AIMs. B. Proposed model for the role of Atg17 in membrane tethering at the non-selective PAS. Note that non-selective cargo is not shown for simplicity. Due to the antiparallel dimerization of Atg17, this protein, together with the Atg31-Atg29 subcomplex (not shown for simplicity) forms the Atg17–Atg31–Atg29 double crescent. In induced conditions, Atg17 dimer formation is activated, leading to the subsequent binding of Atg9-vesicles to the double crescents and their localization next to one another. Next, as described for Atg11 tethering, with the assistance of the autophagy machinery (not shown for simplicity), Atg9-vesicle tethering and subsequent fusion is initiated. Note that computational integration of coarse-grained protein complexes into a model of membrane shape dynamics[26] revealed that the Atg17 dimer also matches the profile of the bowl-shaped phagophore membrane and therefore does not need the cargo for scaffolding and shaping the autophagosome (inset at bottom right). For more information regarding Atg17 scaffolding, the cup-shaped double membrane in autophagy and how multiple Atg17 dimers cooperate to remove the barriers for the transition from tubular to disk shapes, and then to phagophore shapes, see Ref. 26.
The CC4 region of Atg11 serves as the SAR recognition module
Besides Atg11, CCs are found among other core Atg proteins that also form homo- (Atg6, Atg16 and Atg17) or hetero-dimers (Atg6-Atg14).[24, 27–29] These structures commonly facilitate oligomerization or serve as scaffolds for large macromolecular complexes needed, for instance, for membrane dynamics during autophagy.[30] CCs regions of Atg11 were shown to be required for self-assembly into an oligomeric complex, as well as for interaction with other proteins.[11] Yeast two-hybrid (Y2H) studies followed by biophysical characterization of Atg11 pointed to CC2 and CC3 as regions required for its dimerization at the PAS, but CC4 is also essential for this process, as shown by fluorescence microscopy. While the truncated protein, Atg11ΔCC4, was unable to form perivacuolar puncta known as the selective PAS, expression of full-length Atg11 re-established puncta formation.[11] Interestingly, the CC4 domain of Atg11 is responsible for the binding of SARs (Fig. 2A).[11, 31] This observation implicates SARs (and indirectly the cargo) in the process of Atg11 homo-oligomerization. When the interactions of Atg11 with the SARs, Atg19 and Atg32, for the Cvt and mitophagy pathways, respectively, were examined, it was found that the truncated mutant, Atg11851–1178, containing mainly CC4, was sufficient for interaction with the Cvt and mitophagy SARs.[11, 31] Additionally, both full-length Atg11 and the truncated mutant, Atg11851–1178, colocalized with Atg19 and its Cvt-pathway cargo, ApeI.[11] Binding of Atg19 and Atg32 to the same region of Atg11 suggests that the C-terminal fragment of Atg11 comprising CC4 contains a common binding site for SARs.[11] Allocation of the SAR interaction module to the CC4 of Atg11 is additionally supported by the finding that the pexophagy receptors, Atg36 and Atg30 from S. cerevisiae and P. pastoris, respectively, also bind to the CC4 region of Atg11.[13] Such high conservation of the Atg11-binding region (A11BR) within known yeast SARs suggests a common mechanism for the binding of SARs to the CC4 region in Atg11. Interestingly, the CC4 region of Atg11, together with its CC2, interacts with the vacuolar SNARE, Vam7, in the late steps of autophagy, when the outer membrane of the autophagosome fuses with the vacuole. During this SNARE-mediated process, Atg11, in coordination with the Atg17-Atg31-Atg29 complex and Vam7, mediates autophagosome-vacuole fusion.[20] Since Atg11 is itself not degraded during autophagy (unlike SARs), it is tempting to speculate that recruitment of Vam7 to the autophagosome might support release Atg11 from Atg11-SAR complexes and draw it away from SARs, which might help orchestrate other events at the SAR (e.g. Atg8 recruitment). CC4 is also conserved in the C-terminal domain of the mammalian protein FIP200 (Focal Adhesion Kinase Family–Interacting Protein of 200 kDa, also known as RB1CC1), whose function is similar to that of Atg11, suggesting the evolutionary conservation of the role of the CC4 domain in autophagy.
CC2 and CC3 regions in Atg11 and their interaction partners
Between cargo recruitment to the selective PAS and the termination of selective autophagy by autophagosome-vacuole fusion, Atg11 interacts via its CCs with many other proteins to organize and orchestrate the autophagy machinery during autophagosome formation. The same CC regions of Atg11 important for self-assembly also control its interaction with the Atg1 kinase complex that initiates autophagosome formation (Fig. 2A). This interaction was disrupted in strains missing either the CC2 or CC3 of Atg11.[11] In the selective PAS, Atg11 seems to be an integral component of the Atg1 kinase complex,[32] with which it interacts through its CC2 and CC3 regions.[11, 33] However, recent discoveries show that Atg13, another component of the Atg1 kinase complex, regulates the interaction between the Atg1 complex and Atg11.[33, 34] Atg11 interacts also with Atg20, which is disrupted when CC2 of Atg11 is deleted (Fig. 2A),[11] and with Atg29, which needs to be phosphorylated for it to interact with Atg11.[35]
During autophagy, the Atg1 kinase complex closely associates with Ypt1 (a GTP-binding protein) and its autophagy-specific, guanine-nucleotide exchange factor (GEF), the TRAPPIII Trs85 subunit, which activates Ypt1.[36] It was established that GTP-bound, active Ypt1 recruits the Atg1 kinase to the PAS.[37] It is quite intriguing that Atg11 also directly interacts with Ypt1 and the binding sequence maps to the same CC2 and CC3 regions of Atg11 that interact with Atg1.[15] Moreover, while Atg11-Trs85 interaction is unclear by combining single- and multi-color bimolecular fluorescence complementation (BiFC) assays and co-localization analyses, Ypt1 interacts with Trs85 on Atg9-marked compartments, forming several fluorescent puncta, one of which co-localizes with Atg1, Atg8 and the Ypt1-Atg11 BiFC complex.[11] It is possible that Ypt1 bringing Atg11 with attached cargo into close proximity of Atg1 is what locally activates the Atg1 kinase and initiates autophagosome formation.
Atg9-binding module - CC1 and CC2 in Atg11
The initiation of the phagophore depends on the recruitment and fusion of Atg9-vesicles. Atg9 was demonstrated to interact with Atg11 by co-immunoprecipitation and Y2H assays of truncated partner proteins narrowed down the interaction site to the CC2 of Atg11 and Atg9132–255.[14] However, another study found that CC1 of Atg11 is required as well (Fig. 2A).[38]
In summary, the interaction of Atg11 with Atg1, Ypt1 and itself are all mediated by the CC2 and CC3 regions, while the interactions with Atg17, Atg20 and Atg9 are all mediated via CC2, and with CC1 also being required in the case of Atg9 (Fig. 2A). The binding of Atg11 to SARs via its CC4 seems to be spatially separated from these interactions, while the interaction with Vam7 required both CC4 and CC2 (Fig. 2A). Interestingly, after deletion of the genes encoding many of the Atg11-interacting proteins mentioned above, Atg11-GFP fusions form a bright cytoplasmic punctum, suggesting that these other interactions of Atg11 with its partners limit its ability to homo-oligomerize. This observation might suggest that binding these proteins to particular CCs of Atg11 may not be simultaneous, but rather mutually exclusive and probably highly coordinated by mechanisms that are still unclear.
Atg11 acts as a cargo adaptor - interaction of Atg11 with SARs
The ability of Atg11 to link cargo-bound SARs to the core autophagy machinery, such as the Atg1 kinase complex, makes it a critical scaffold protein for selective autophagy. This binding is essential for subsequent autophagosome formation around the cargo and for further delivery of targeted cellular components to the vacuole for degradation. With the exception of Cue5 (SAR for protein aggregates), all yeast SARs known so far bind to Atg11 protein.[39] The recognition of the SARs by Atg11 is regulated by the phosphorylation of SARs within their A11BRs (Fig. 2B).[13] Only phospho-activated SARs bind to Atg11 to initiate selective PAS formation (Fig. 4A–C).[13, 31, 40]
Figure 4. Model of Atg11 roles in selective autophagy.
A. SAR activation by kinases during selective autophagy - In selective autophagy conditions, SARs bound to cargos are activated by phosphorylation by a kinase, such as Hrr25 or CK2, in their Atg11-binding regions (A11BRs) (phosphorylation is indicated by a red circle labelled P). Prior to SAR activation, Atg11 is mostly monomeric and cytosolic. Note that under nutrient-rich conditions (vegetative growth), Atg13 hyper-phosphorylated by the mTOR kinase, has only a weak affinity for Atg1, which corelates with low activity of Atg1 kinase. B. Atg11 recruitment to the activated SAR and cargo recruitment to the PAS - A phospho-activated SAR binds Atg11 to initiate the selective PAS formation. Binding SARs to the C-terminal CC4 of Atg11 promotes its dimerization, allowing recruitment of the cargo to the PAS, which is located adjacent to the vacuole. Independently, Atg1 is recruited to the vacuole by Atg13. Simultaneously, via the interaction between Trs85 and Atg9, the Trs85-containing TRAPPIII complex resides on Atg9-vesicles and facilitates the association of Ypt1 (GTP-bound and active) on Atg9-vesicles. C. Atg11-mediated scaffolding events at the PAS - Atg11 activated by the SAR also recruits to the PAS the following: 1) Atg9-vesicles with active Ypt1 via its interactions with Atg9, Ypt1 and possibly Trs85. 2) The Atg17 scaffold complex, composed of Atg17, Atg31 and Atg29, via its interaction with phosphorylated Atg29. 3) At the vacuole, due to Atg1-Atg11 association, Atg1 can bind to and cluster on the cargo to become active. These events allow the spatial and temporal activation of selective autophagy. D. Both Atg11 and the Atg17 complexes scaffold the core autophagy machinery - It remains unclear whether Atg11 and Atg17 form mutually-exclusive complexes with Atg1. E. Steps orchestrated at the PAS by active Atg1 kinase, including initiation of phagophore membrane formation - Active Atg1 kinase recruits other core autophagy proteins, including Atg8 conjugated to phosphatidylethanolamine (PE) on the phagophore membrane. By using pre-existing scaffolding of Atg11 and Atg17, Atg1 contributes to tight vesicle tethering, initiating phagophore formation. F. COPII vesicle recruitment to the PAS for phagophore expansion - In starvation-induced autophagy, efficient phagophore expansion requires recruitment to the PAS of not only Atg9-vesicles, but also COPII vesicles. Hrr25 can also be recruited to the PAS in an Atg9-independent manner on COPII vesicles. It is unknown if Atg11 recruits Hrr25 also on COPII vesicles, as Atg17 does. The precise role of Hrr25 at the PAS is enigmatic. G. Initiation of membrane curvature and expansion of the phagophore membrane – The interaction of Atg11 with the Atg20-Snx4 complex recruits the PtdIns3P-enriched membranes and stabilizes phagophore curvature. The Atg17 complex and Atg11-Atg20-Snx4 at the PAS contribute to the phagophore expansion. H. Phagophore membrane engulfment of cargo - Phagophore expansion continues around the cargo via engaging other activated SARs, which can bind Atg8-PE (the phosphorylation within the AIM is indicated by a pink circle labeled P). At the same time, Atg11 coordinates with the Atg17-Atg31-Atg29 complex (not shown for simplicity) to recruit the Vam7 SNARE and prevent premature membrane fusion. I. Autophagosome completion, dissociation of protein components and autophagosome-vacuole fusion - In the final step, detachment of Atg11 and other autophagy machinery components initiates autophagosome-vacuole fusion using autophagosomal and vacuolar SNARE proteins described in the main text.
Activation of SARs by phosphorylation prior to their recognition by Atg11
Based on a comparison of the A11BRs in yeast SARs identified so far, Farre et al. proposed that the A11BR has a consensus sequence of two hydrophobic residues, followed by a serine residue crucial for the Atg11-bindng motif activation (Fig. 2B).[13] These residues are surrounded by a several serines (S) and/or threonines (T) and/or acidic residues (D or E), such as in Atg19 (DDSSIISTS) or in Atg34 (DESSIMSTP). For the membrane-associated receptors such as Atg30, Atg32, Atg36 and Atg39, the consensus sequence for the A11BR is even further narrowed to three crucial residues I/VLS. In all known SARs (except Atg40 for which the Atg11 binding region was not determined), the A11BRs are cryptic and require activation by phosphorylation of serine prior to Atg11 binding (Fig. 2B). Because only the phospho-activated SAR binds to Atg11, such phospho-regulation of Atg11 recruitment is strongly required and allows precise control of which and when cargo is targeted for selective autophagy. Two kinases phosphorylate the crucial serine within the A11BR of Atg11. Casein kinase 2 (CK2) phospho-activates the mitophagy receptor, Atg32,[41] and the Hrr25 kinase (a CK1) phospho-activates Atg19 and Atg34 involved in the Cvt pathway,[42–44] as well as the pexophagy receptors, Atg30 and Atg36 from P. pastoris and S. cerevisiae, respectively.[43, 45]
As established by Farre et al.[13], the A11BRs in yeast SARs are usually positioned close (separated by 0–62 amino acids) to their Atg8-family interacting motifs (AIMs), facilitating interactions with Atg8s. Atg8 proteins tether the cargo-bound SARs to the growing phagophore during selective autophagy and ensure tight adhesion of the cargo to autophagosomes. Such close proximity of the A11BR and AIM within Atg11 would seem to preclude the simultaneous binding of both Atg8 and Atg11 to a given SAR.[13] However, since any given SAR needs to interact with both Atg11 and Atg8 proteins, but which can be separated in time and place, the mutually-exclusive recruitment of Atg11 and Atg8 to the SAR could be a mechanism to orchestrate events at SARs, such as the recruitment of multiple SARs, or even releasing Atg11 from the SAR-bound cargo at the selective PAS, prior to cargo tethering to the phagophore.
Atg11-dependent cargo recruitment to Atg1 at the PAS
For quite a long time it was unclear which autophagy component was needed to recruit cargo to the PAS. Among the possibilities mentioned were: cargo by itself, SARs and Atg11 protein. Use of the induced-bypass approach (iPASS) in yeast[33] revealed that SARs can be bypassed completely by directly tethering Atg11 to the cargo. The authors clearly demonstrated that Atg11 recruits the cargo to the PAS, where the Atg1 kinase complex resides. This key event subsequently leads to the initiation of selective autophagy (Fig. 4C).
Moreover, they also established that Atg13 regulates Atg1-Atg11 association at the PAS by recruiting Atg1 to the vacuole, but does not mediate this interaction. Furthermore, Atg19 and its cargo ApeI form a complex without Atg11, but this complex is not localized at the PAS.[33, 38, 46] Similarly, the Atg1-Atg19 fusion protein in atg11Δ atg13Δ atg19Δ cells clusters with the Ape1 cargo, pulling Atg1 away from the vacuole in the absence of Atg13.[33] Thus, Atg11 is a critical factor for cargo recruitment to the selective PAS that appears in the perivacuolar region,[47] where the Atg1 kinase is brought by Atg13 via its interaction with Vam8. However, Atg1 and Atg13 by themselves are not factors that regulate Atg11 presence at the PAS. In cells deleted for ATG1, the Cvt complex of Atg19 and prApe1 is seen at the PAS in the perivacuolar region[46, 48] and Atg11 concentrates at the PAS even in the absence of Atg1.[11] Also, Atg11 co-localizes with Atg1 tethered to the vacuole by the iPASS technique in atg13Δ cells.[33]
Previous studies pointed out that PAS localization of Atg11 under nutrient-rich conditions depends on the cargo/SAR recognition, as well as on the actin cytoskeleton.[11] Atg11 was observed not to localize in a punctate structure in the absence of Ape1 cargo or Atg19. Similarly, the integrity of the actin cytoskeleton was essential for correct targeting of Atg11 to the PAS.[14] Interaction between Atg11 and Smy1, Kinesin-like myosin passenger-protein, which interacts with Myosin-2 (Myo2) and controls actin cable structure and dynamics, was reported.[49, 50]
The dimerization of Atg11 also plays a role in Atg11 targeting to the PAS.[11] Because removal of the CC4 of Atg11 prevents its accumulation as a punctum in cells, and only the presence of full-length Atg11, but not Atg11ΔCC2, allowed Atg11ΔCC4 to assemble into a punctate structure,[11] it was hypothesized that SAR binding to Atg11 initiates some conformational changes further promoting the dimerization of Atg11 and its recruitment to the PAS. In more recent studies, Atg11 was suggested to be an effector of Ypt1 for the Cvt pathway as the Ypt1–Atg11 interaction is required for PAS assembly and Atg11, Atg8 and Atg1 localization is altered in ypt1–1 mutant cells.[15] Since the same region within Atg11 was shown to be crucial for Atg1 and Ypt1 interactions as for dimerization (Fig. 2A), SAR binding to Atg11 may also stimulate Atg11 complex formation with Atg1 and/or Ypt1. Thus, interaction of Atg11 with SARs might be a general mechanism for Atg11 activation as a scaffold and its recruitment to the PAS.
Atg11 functions as a scaffold to activate Atg1 in selective autophagy
Over the years, the binding of Atg13 to Atg1 to activate Atg1 kinase activity has been extensively studied in the context of selective autophagy.[51–56] However, until now, it was quite enigmatic how selective autophagy is triggered since Atg1 kinase activation is generally repressed in nutrient-rich conditions by TORC1 kinase signaling, which inhibits non-selective autophagy.[57–59] It has also been a question if there is any role for Atg11 and the cargo in Atg1 activation.
Recently, two independent groups clearly proved that Atg11 is more than just a passive scaffold during early step of selective autophagy, but additionally it dynamically controls the Atg1 kinase activity locally by probably transducing signal from receptor-bound cargo to Atg1, even under conditions favoring autophagy inhibition, rather than initiation. Using cell-free, genetic and biochemical approaches, it was established that Atg1 locally promotes selective autophagy under nutrient-rich conditions (where autophagy would be inhibited) and this Atg1 catalytic activity requires both Atg11 and SARs (such as Atg19 or Atg36 for Cvt or pexophagy, respectively).[34] Studies by Torggler et al.[33] shed some light on how at the molecular level, SAR/cargo complexes use Atg11 to activate Atg1. Using an elegant set of experiments, both groups converged upon a model in which during selective autophagy, Atg11 with attached cargo complexes is recruited to the vacuole, separately from the recruitment of Atg1 via Atg13[33], which requires functional Atg11[56] and Ypt1-Atg11 interactions[15] (Fig. 4B). Then, at the vacuole, Atg11, by interacting with the Atg1-Atg13 complex, presents bound cargo to the Atg1 kinase (Fig. 4C). Since dimerization or oligomerization, followed by trans-autophosphorylation, is a common mechanism of kinase activation,[60] it can be hypothesized that accumulation of Atg1 on the cargo can itself promote Atg1 dimerization or clustering, which subsequently leads to Atg1 activation. Thus Atg11, by presenting the cargo to Atg1, regulates the Atg1 activation in space and time (Fig. 4C). Such a mechanism of activation can explain why Atg1 only clusters on cargo complexes driven by Atg11 to the vacuole and why loss of Atg11 (or the cargo and Atg19) attenuates the Atg1 kinase activity to a similar level as loss of Atg13 itself.[33] In other words, the receptor-bound cargo (for instance Atg19-Ape1) uses the Atg11 scaffold as a switch to activate Atg1 kinase and drive autophagosome formation, promoting selective autophagy of the cargo. In this way Atg11 locally orchestrates Atg1 kinase activity on the vacuole under nutrient-rich conditions, despite the existence of other cellular mechanisms (like TORC1) that would suppress Atg1 activity. Because dependence of the Atg1 kinase activity on Atg11-Atg19 and Atg13 interactions can be bypassed when Atg1 is fused to Atg19 via a GFP or protein A linker in atg11Δ atg13Δ atg19Δ, but not in ape1Δ cells,[33] in this model Atg13 (or Atg11) does not directly activate Atg1, in contrast to the commonly accepted belief.
The role of Atg11 in PAS organization and isolation membrane expansion
Upon engulfment of cargo by the phagophore, SARs are not only in contact with Atg11, but also directly interact with the Atg8 protein decorating autophagosomal membranes from their early stages of formation. In yeast, Atg8 and Atg11 puncta co-localize to some extent.[11] Such partial co-localization suggests that: 1) Atg11 can be one of the first autophagic proteins to reach the selective PAS and that it acts as a scaffold for the core autophagy machinery, prior to phagophore membrane formation, 2) Atg11 remains at the PAS as an adaptor between cargo and the autophagosomal membrane,[61] and additionally, 3) Atg11 can be involved in phagophore expansion (Fig. 4B–G).[16]
Atg11 recruits Ypt1 and Atg9-vesicles to the PAS
Recruitment of Atg9, a membrane carrier for vesicle formation, to the PAS is essential for initiation and expansion of phagophore. During selective autophagy, this process is mediated by Atg11 which governs Atg9 cycling through the PAS (Fig. 4C, 4D).[14] However, like all intracellular trafficking pathways in yeasts, selective autophagy is also regulated by the Ypt GTPases - proteins that organize membrane micro-domains required for membrane trafficking.[62] Ypts, when stimulated by GEFs, bind intracellular trafficking machinery components, like motors and tethers. Recently, Lipatova et al.[15] established that Atg11 interacts with Ypt1 in a GTP-dependent manner as its downstream effector and this interaction is required for Atg11 localization to the PAS (Fig. 4C). Ypt1 is a well-characterized protein that plays roles in multiple membrane-trafficking processes. It recruits distinct tethers to both COPI and COPII vesicles and plays an undefined role in autophagy. Specificity is provided by the various multi-subunit tethering TRAPP complexes that also act as GEFs to activate Ypt1. COPII vesicle trafficking requires the small TRAPPI complex, whereas COPI requires the larger TRAPPII complex. In contrast, the moderate-sized TRAPPIII complex, with its specific Trs85 component, and the more recently-discovered TRAPPIV complex, containing Trs33, are proposed as GEFs for Ypt1 during autophagy.[15, 63, 64]
The Ypt1-Atg11 interaction was shown to be required for PAS assembly in the Cvt pathway under normal growth conditions and to regulate shuttling of tagged, overexpressed, and likely misfolded proteins from the ER to the PAS.[15, 64] Lipatova et al.[15] showed that Ypt1 and Trs85 interact on Atg9-containing membrane compartments and these complexes co-localize with Atg11. Similar observations were made for Ypt1-Trs85 in the context of Atg17 and starvation-induced autophagy.[37] But while direct interaction between Trs85 and Atg17 was proven,[37] it is unclear if Trs85 by itself binds to Atg11. Nevertheless, based on concurrent data for Atg9, Atg11 and the Trs85-containing TRAPPIII complex, it can be suggested that Atg11 recruits Atg9-vesicles and Ypt1 to the PAS. However, the single point mutation in Atg9 (Atg9H192L), which disrupts the Cvt pathway by blocking the Atg11 interaction, does not affect bulk autophagy,[14] indicating that Atg9-vesicles are recognized differently in selective and bulk autophagy.
Ypt1 regulates Atg9-vesicle tethering by modulating the delivery of Atg1 to the PAS.[37] Therefore, GTP-bound Ypt1 might facilitate the interaction between Atg11 and Atg9 for the initial steps in the PAS assembly. Kakuta et al.[65] showed that via the interaction between Trs85 and Atg9, the Trs85-containing TRAPPIII complex can reside on Atg9-vesicles and facilitate the association of Ypt1 with Atg9-vesicles (Fig. 4B). Atg9 movement to the PAS depends on this interaction. In the trs85Δ vps51Δ double mutant (TRAPPIII mutant) cells, Atg9 is trapped in the vesicle and its movement to the PAS is severely affected.[66] Through possible connections of Atg11 with Atg9 and with Ypt1 (and maybe Trs85) this Atg9-vesicle can be recognized by Atg11 molecules that have been activated via interaction with SARs phosphorylated at their A11BR site (Fig. 4C). Subsequently, the Trs85-Ypt1-Atg11 module in turn targets Atg9 and its associated vesicular membrane to the PAS and there stimulates Atg1 puncta formation (Fig. 4C, 4D).[15, 37, 67] The precise role of TRAPPIV in this process is unclear. However, because the trs33Δ atg11Δ double mutant affecting TRAPPIV, like the trs85Δ mutant affecting TRAPPIII, does not cause a more severe autophagy phenotype than that of atg11Δ cells, Lipatova et al.[63] suggested that Trs33 functions in autophagy also in the context of the Ypt1-Atg11 module. Interestingly, both the TRAPPIII and TRAPPIV-sorting pathways are sufficient to provide Atg9 reservoirs for the PAS under nutrient-rich conditions when mainly only the Cvt pathway operates, but the assistance of the late endosome-to-Golgi sorting pathway is required when nutrients are limited (Fig. 4E, 4F).[63]
Role of Atg11 as a vesicle tether
After recruitment of Atg9-vesicles to the PAS, their tethering is initiated (Fig. 4D–F). While the details of the nucleation process in selective autophagy are still not clear, a recent study on starvation-induced, non-selective autophagy shed some light on this process.[55] In the initial step, an average of three Atg9-vesicles is required to merge at the PAS to form an early phagophore.[68] In non-selective autophagy, the Atg9-vesicles were recruited to the PAS in an Atg17-dependent manner, making Atg17 the earliest protein arriving at the non-selective PAS, as defined by the Atg17-Atg31-Atg29 complex[24, 69] Upon induction of non-selective autophagy, factors arriving early (Atg1 and Atg13) are efficiently recruited via the Atg17 scaffold.[55] Two complexes, Atg17-Atg31-Atg29, which by itself is defective in Atg9-vesicle tethering, and Atg1-Atg13 assemble together into a higher order complex that activates the vesicle-tethering activity of Atg17. Because the functions of Atg17 and Atg11 are conserved, similar mechanisms of action might be involved for selective autophagy. Based on this similarity, it is plausible that the Atg1-Atg13 complex interacting with Atg11 activates the Atg9-vesicle tethering activity of Atg11, in a similar manner as it does with Atg17.[55]
Recent studies on phagophore formation revealed that the Atg17 dimers are necessary for membrane tethering.[24, 26] While the monomeric mutant of Atg17 was able to bind Atg9, it failed to tether vesicles.[55] Since dimerization of Atg11 is required for it localization at the PAS[11] it is plausible that SAR binding also initiates structural changes in Atg11 to activate the tethering activity of Atg11 (Fig. 3).
Atg11 acts as a scaffold also for autophagosome formation
We have discussed earlier the multiple lines of evidence proving that Atg11 not only delivers cargo and membranes to the selective PAS, but also organizes it. As stated earlier, Atg11 interacts with Atg1, but the co-precipitation of these proteins was severely depleted in atg13Δ strains.[33, 34] These data, together with the colocalization of Atg11 with Atg13 puncta, indicates their simultaneous presence in the Atg1 kinase complex.[70] Also, other Atg11 interactions, like those with the Atg17-Atg31-Atg29 complex, Atg20 or Atg8 proteins, support a role for Atg11 as a scaffold for the expanding phagophore membrane.
The Atg17-Atg31-Atg29 complex
Atg17 was characterized as a component of the Atg1 kinase complex that is specific for non-selective autophagy.[71, 72] This protein scaffold forms a complex composed of Atg17-Atg31-Atg29 dimers and displays an elongated S-shaped structure, reminiscent of two, antiparallel crescents, allowing it to bind to curved membranes.[23, 24, 26, 35] Atg17, like Atg11, acts in the initial steps of autophagosome biogenesis and is required for the further recruitment of the other components of the core autophagy machinery. Despite the fact that Atg11 has been suggested to act exclusively during selective autophagy and Atg17 does this primarily during the non-selective autophagy, recent findings point to a contribution of Atg17 to both pathways (Fig. 4D–F).[35] For example, selective pexophagy requires both proteins, and the Cvt pathway requires Atg11 only in nutrient-rich, but not starvation, conditions.[73] Additionally, Y2H studies showed direct interaction between Atg11 and Atg17,[11] and recent studies on Atg29 revealed that phosphorylation of this protein is required for its binding to Atg11 and subsequent targeting of the Atg17-Atg31-Atg29-Atg11 complex to the PAS (Fig. 4C, 4D).[35] The Atg17-Atg31-Atg29 complex comprises the first set of proteins localized to the PAS.[74, 75] However, in the absence of Atg11, this complex is diffuse in the cytosol, rather than accumulating at the PAS. Based on published studies,[35] in the absence of Atg17 or Atg29, the presence of Atg11 permits cells to retain residual autophagy activity, even though the deletion of the ATG11 gene alone does not have an obvious effect on autophagy activity.[32, 56] How Atg11 and Atg17 molecularly contribute to the shaping of the phagophore during selective autophagy remains unknown. It also remains unclear whether Atg11 and Atg17 form mutually exclusive complexes with the Atg1 kinase complex or together form a bigger, multi-component scaffold complex. Furthermore, we do not know if the Atg17-Atg31-Atg29-Atg11 complex dissociates after reaching the PAS and if Atg9-vesicles can be recruited by this complex.
Atg20
Recently, Atg11 was also shown to be a part of the membrane-sensing and membrane-remodeling Atg11-Atg20-Snx4 complex.[11, 16] Atg11 binds only to Atg20, not to Snx4 (also called Atg24) (Fig. 2, 4G).[11, 16] Both Snx4 and Atg20 are peripheral membrane proteins localizing to the endosome and the PAS[76–78]. It has been suggested that Atg20-Snx4 role in autophagy is to mediate trafficking of Atg9 from the endosome to the Golgi apparatus, from where it is packaged into a vesicle later required for phagophore assembly at the PAS (Atg9-vesicle, see above).[68, 79, 80] It was also proposed that the binding of Atg20-Snx4 hetero-dimers to PtdIns3P-enriched membranes,[77] as well as their involvement in lipid export from the endosome and/or vacuole via retrograde vesicular trafficking,[78, 81–84] might contribute to the recruitment of a membrane source to the selective PAS. Why Atg11 binds to the Atg20-Snx4 complex and whether it regulates the function of the complex is unknown. However, it was proposed that the elongated Atg11-Atg20-Snx4 complex could stabilize a certain membrane curvature during the formation of the phagophore. Both Atg20 and Snx4 interact with Atg17 as well, but it is unclear if Snx4-Atg20 fulfills the same functions in both selective and non-selective autophagy.[76, 85–87]
The Atg8 and Atg12 ubiquitin-like conjugation systems
There is only preliminary evidence pointing to an association of Atg11 with the Atg8 ubiquitin-like conjugation system in yeasts, such as: co-localization of Atg11 with Atg8[11] and the presence of several canonical AIMs within the Atg11 sequence. However, these results, together with recent description of Atg8-Atg11 interaction detected via BiFC in A. thaliana[88] suggest a plausible interaction between these two proteins.
However, it is an open question how these proteins interact. Based on current mechanisms regarding how Atg8 interacts with other proteins, several possibilities can be considered. First, Atg11, may use classical AIM(s), which are abundant within its sequence (iLIR software predicted 27 AIMs within Atg11). Thus, Atg11 like Atg19,[89, 90] might interact with Atg8 via not one but perhaps several AIMs. Second, the interaction might occur through an unconventional AIM, as described for Atg7[91] and for a vacuole membrane protein, Hfl1 (homolog of human TMEM184 proteins).[92] Third, the AIM in Atg11 might not be required for this interaction because it is mediated by a different non-canonical mechanism, for example, similar to that described for Atg21[93] and Shp1/Ubx1 (Suppressor of high-copy PP1 protein/UBX domain-containing protein 1), which bind to the N-terminal helical domain (NHD) of Atg8.[94] Finally, another protein, for instance from the Atg5-Atg12 conjugation system, might bridge Atg8-Atg11 complex formation. In fact, Ho et al.[95] identified Atg11 in a complex with Atg12 by affinity-capture MS and mammalian ATG16L1 directly interacts with FIP200,[17] which is proposed to be an Atg11 ortholog in higher eukaryotes.[96] However, the regions of Atg11/FIP200 required for these interactions were not mapped. Unfortunately, at this point, the role of Atg11 in the context of the Atg8, and the Atg12 ubiquitin-like conjugation system, is enigmatic and requires further elaboration.
The role of Atg11 in autophagosome-vacuole fusion and the termination of selective autophagy
After formation of a complete autophagosome, the outer membrane of the double-membrane autophagosome fuses with the vacuolar membrane releasing the autophagic body into the vacuolar lumen for subsequent degradation. Autophagy-specific membrane fusion with the vacuole requires the action of the GTP-binding protein, Ypt7, together with the Mon1–Ccz1 GEF complex present at the vacuole and the autophagosome,[97–99] the HOPS (homotypic vacuole fusion and protein sorting)-tethering complex preparing SNARE proteins for fusion, and SNARE proteins (R-SNARE Ykt6 on the autophagosome, together with the Q-SNAREs Vam3, Vam7, and Vti1 on the vacuole).[97, 100]
Recently it was demonstrated that Atg11, together with Atg17, is also involved in the regulation of autophagosome-vacuole fusion by interacting with vacuolar SNARE protein, Vam7.[20] It was established that Vam7 is recruited to the PAS at an early stage of autophagy by the Atg17-Atg31-Atg29 complex, or independently of this complex by the Atg11 scaffold. Interaction between Vam7 and Atg11, but not Atg17, was observed under nutrient-rich conditions when the Cvt pathway operates. Although, Vam7 interaction with Atg11 was not as thoroughly researched as its interaction with Atg17, the fact that Atg11 is an effective equivalent in organizing the selective PAS in nutrient-rich conditions, as Atg17 is in organizing the starvation-induced PAS, together with the fact that both proteins recruit Vam7 via similar modules (although Atg11-Vam7 interaction is weaker than that of Atg17-Vam7 at least in the Y2H system) suggests that Atg11 and Atg17 act similarly as regulators of autophagosome fusion with the vacuole. Indeed, when the Atg17F317D mutant, partially deficient in Vam7-binding but competent in maintaining its other interactions, was expressed in the atg17Δ strain and in the atg11Δ atg17Δ double-deletion strain, autophagy activity was completely restored in the atg17Δ strain and only to approximately 70% in the atg11Δ atg17Δ strain, in comparison to a wild-type strain. These data indicate that Atg11 can compensate for the partial defect in the Atg17F317D protein in engaging Vam7.[20, 101]
In contrast to classical SNARE proteins, Vam7 lacks a transmembrane domain but binds to PtdIns3P via its PX domain (Fig. 4H).[102] It is known that autophagosomal membranes are enriched in PtdIns3P.[103] If Vam7 is attracted to autophagosomes by PtdIns3P, it could explain its association with Atg17 and Atg11 during the early, but not later stages of autophagy, as is the case with the mammalian STX17 that is recruited to the sealed autophagosome prior to autophagosome-lysosome fusion.[104, 105] However, cytosolic Vam7 lacking its PX domain, Vam7ΔPX, is still recruited to the PAS by the Atg17-Atg31-Atg29 complex.[20] Recruiting SNARE components at such an early step could theoretically initiate premature fusion of the incomplete autophagosome/phagophore with the vacuolar membrane. However, Vam7 also interacts with two CCs of Atg17 via the same SNARE core domain required for SNARE complex formation with Vam3.[49, 50] Thus, Vam7 binding to Atg17 (and Atg11) seems to prevent the premature initiation of membrane fusion between the autophagosome/phagophore and the vacuole (Fig. 4H). After autophagosome completion, Vam7 needs to be released from Atg17 and Atg11 to terminate autophagy, since unlike Ypt7, Vam7 is required to act on the vacuolar side during autophagosome-vacuole fusion (Fig. 4I).[97] It is plausible that the accumulation of Vam7 during the growth of autophagosomes and its subsequent release (together with the autophagy machinery proteins) through the clearance of PtdIns3P from the outer membrane of autophagosomes by the Ymr1 protease,[106, 107] stimulates autophagy-specific fusion by concentrating Vam7 at the autophagosome-vacuole interface. Interestingly, the PX domain of Vam7 is not required for Atg17-Vam7 interaction, but is still important for autophagy activity[20] suggesting that the PX domain of Vam7 might play a role in its dissociation from Atg17 (and perhaps from Atg11).
Atg11 - not a sole scaffold protein
Handover from Atg11 to Atg17 – the molecular switch from selective to non-selective autophagy
S. cerevisiae cells employ separate membrane tethers for selective and non-selective autophagy. The transition from one to another mode of autophagy seems to involve a switch from Atg11 to Atg17.[55] Atg11 is active during vegetative growth, but its function is handed over to Atg17 in starvation conditions. In vegetative conditions, Atg17 is maintained in an inactive state by its sequestration within the Atg17-Atg31-Atg29 subcomplex,[55] which also works as an allosteric inhibitor for Atg9-vesicle recruitment to Atg17.[51, 55]
Conversely, to effectively switch from selective to non-selective autophagy, yeast cells seem to activate Atg17 by changing the phosphorylation status of Atg13, which is hyper-phosphorylated in vegetative conditions and is dephosphorylated during starvation, when non-selective autophagy is induced. Dephosphorylation of specific serines (Ser428 and Ser429) in Atg13 enhances its interaction with Atg17.[108] It is tempting to speculate that during vegetative growth, the Atg1 kinase complex with Atg13 in its hyper-phosphorylated state preferentially binds to Atg11 because phosphorylation of Atg13 at Ser428 and Ser429 prevents its interaction with Atg17. But dephosphorylation of Atg13 during starvation conditions would increase the affinity of Atg17 for the Atg1-Atg13 complex because dephosphorylated Atg13 interacts more strongly with both Atg1 and Atg17.[53, 57, 72]
Atg11 and Atg17 - competitors or cooperators during selective autophagy?
If Atg11 and Atg17 perform similar scaffolding functions during growth in nutrient-rich and starvation conditions, respectively, and do so by interactions with similar partner proteins, it is relevant to inquire whether Atg11 and Atg17 compete for interaction partners such as Atg9-vesicles and the Atg1-Atg13 complex. Atg17 physically interacts with Atg13 which as described earlier, regulates Atg1 kinase activity.[53] Atg11 binds directly to Atg1, but while Atg13 regulates Atg1-Atg11 association at the vacuole it does not mediate this interaction per se.[33] Since Atg11 and Atg17 bind to different components of the Atg1-Atg13 complex and Atg17 is sensitive to the phosphostatus of Atg13, they may not compete for this complex after all. In view of the supportive function of Atg17 in selective autophagy,[109] the recognition of phosphorylated Atg29 by Atg11 and the necessity of Atg11 for the recruitment of the Atg17-Atg31-Atg29 complex to the PAS could trigger dynamic conformational changes within the Atg17-Atg31-Atg29 complex to promptly regulate autophagy induction in response to specific stimuli.[35] However, another possibility is that while Atg11 initiates a selective PAS (orchestrating in time and place the local accumulation at the vacuole of Atg1, Atg11, Atg13 and cargo, which subsequently promotes the binding of Atg11 to the Atg1-Atg13 complex (Fig. 4F)), it is later replaced by Atg17, leading to elevated levels of selective autophagy.
Based on current studies it seems that Atg9-vesicles are recruited to the PAS differently in nutrient-rich and starvation conditions. It was shown that a missense mutation within Atg9 that affects Atg9-Atg11 interaction results in a defect in transporting Atg9 to the PAS, causing a block in the Cvt pathway, without affecting bulk autophagy induced during starvation. Because Atg11 is needed for the Cvt pathway but not for bulk autophagy[32] it is reasonable to conclude that the anterograde transport of Atg9 during bulk autophagy may be mediated by a different mechanism that is at least relatively independent of Atg11. Indeed, binding of Atg9 to Atg17 (which is mostly bridged by Atg13[110]) is an independent event from the Atg9‐Atg11 interaction, as this interaction was not affected in atg11Δ cells.[69] It needs to be stressed here that Atg9 interacts with Atg17 in a manner highly dependent on Atg1 and Atg13, which efficiently binds Atg9 via its HORMA domain.[110] Atg9 was not co-precipitated with Atg17 in atg13Δ or atg1Δ cells.[69, 110] It was suggested that the Atg17–Atg9 interaction is not sufficient for recruitment of Atg9 to the PAS since the atg13 mutant, which exhibited impaired PAS localization of Atg9 did not affect Atg17 localization at the PAS. Thus, under nutrient-rich conditions, Atg11 recruits Atg9 vesicles for the Cvt pathway. However, under starvation conditions, Atg9 vesicles are recruited to the PAS primarily via the interaction with the Atg13.
Atg11 is required for organelle fission during selective autophagy
Recently another role of Atg11 as a scaffold for selective autophagy was established. Atg11 recruits the membrane fission machinery during mitophagy and pexophagy. Deletion of the VPS1 or DNM1 genes resulted in substantially lower efficiency of pexophagy (for both VPS1 and DNM1 genes) or mitophagy (only upon deletion of the DNM1 gene).[18, 19, 111, 112] Vps1 and Dnm1 are 2 dynamin-related GTPases that together with cytosolic Mitochondrial division protein 1 (Mdv1), and CCR4-associated factor 4 (Caf4) and with the membrane protein, Fis1, are the essential proteins for mitochondrial and peroxisome fission in yeast.[112, 113] Thus, Atg11 colocalization with Dnm1 and Fis1 on mitochondria or with Dnm1, Fis1 and Vps1 on peroxisomes[18, 19] suggests that during mitophagy and pexophagy the fission proteins are recruited to these organelles targeted for turnover (Fig. 5). Fascinatingly, during mitophagy, only a small portion of the total Dnm1 population forms a complex with Atg11, suggesting the existence of additional populations of fission complexes: perhaps one involved in general mitochondrial division and another assembled in response to mitophagy induction and Atg11 recruitment to the mitochondria destined for degradation. Consistent with this idea, Atg11 by itself is not a part of fission machinery and is dispensable for general fission. It was demonstrated that lack of binding of Atg11 to Dnm1 point mutants (Dnm1E728R and Dnm1D729R) affects mitophagy, but not general mitochondrial fission.[18]
Figure 5. Hypothetical model of simultaneous phagophore formation around organelles and their fission.
Proposed mechanism of mitochondrial or peroxisomal fission during selective autophagy. A. Activation of SARs bound to mitochondria or peroxisomes targeted for degradation initiates recruitment of the Atg11 and also PAS formation at the vacuole (see Fig. 4). Meanwhile, Dnm1 and other organelle fission machinery are recruited to these organelles from the cytosol by activated Atg11 residing on these organelles. B. In this hypothetical model, Atg11-Dnm1 contacts mark the site of mitochondrial or peroxisomal fission. Due to the cup-shaped structure of the phagophore and Atg11 localization on its edges, the fission machinery recruited by Atg11 assembles into spiral/ring- shaped oligomers. C. Expansion of the phagophore around the cargo and its enclosure is coordinated with fission. Assembly of Dnm1 onto the mitochondria or peroxisome simultaneously causes membrane constriction at the fission site. D. Organellar membranes are not able to exist as separate units inside the Dnm1 rings, which leads to the final step of fission and enclosure of a portion of mitochondria or peroxisomes inside autophagosomes, which then fuse with the vacuole.
It is possible that during selective autophagy large organelles need to be fragmented into smaller ones to be effectively engulfed by phagophores. In such an instance, Atg11, by recruiting the organelle fission machinery, may facilitate autophagy of large organelle cargos by reducing their size. Mao et al.[18] proposed that when mitophagy is induced, the activated mitophagy SAR, Atg32, recruits Atg11, Dnm1 and other mitochondrial fission proteins (Fig. 5A). The presence of this fission complex then promotes division of mitochondria destined for clearance and subsequent transport of these small fragments of mitochondria to the PAS (Fig. 5B–D). This mechanism is reminiscent of the removal of large intraperoxisomal protein aggregates via an asymmetric peroxisome fission in the methylotrophic yeast H. polymorpha driven by Dnm1 and Vps1[114] or the formation of Rubisco-containing bodies (RCBs) in plants.[115–117] RCBs are a type of autophagosome that deliver a portion of the stromal proteins of plant chloroplasts into the vacuole. However, the fission machinery seems to regulate selective autophagy also independently of organelle size. Atg11 and the Dnm1-containing fission complex are shared by peroxisomes and mitochondria. In yeast, peroxisomes use the same molecular fission complex, Fis1-Mdv1-Dnm1, for fission during pexophagy[19] but the small peroxisomes induced by oleate treatment (an average diameter of ~150 nm) and dispersed in the cytoplasm should easily be engulfed by autophagosomes (~900 nm in diameter). In this scenario, it is an open question why Atg11 attracts the fission machinery to smaller organelles that could be entirely enclosed in autophagosomes.
Functional conservation of Atg11
Mammalian Atg11 counterparts
The protein sequences of yeast Atg11 and Atg17 are not evolutionarily conserved in mammals. Instead, the FIP200 (also known as RB1CC1) scaffold protein is proposed to be the mammalian counterpart of yeast Atg11 and Atg17.[80, 118] An analysis of the domain organization of FIP200 revealed that this protein contains an Atg17-like region, multiple CC regions and an Atg11-homology domain corresponding to CC4 of Atg11.[119] Based on its domain architecture and size, FIP200 is more similar to Atg11 than to Atg17.
FIP200, as a scaffold, has many mammalian binding partners such as ULK1, C9orf72, SEC12, ASK1, FAK, p53, Pyk2, TRAF2 and TSC1.[120–122] Besides autophagy, it is involved in various processes including cell migration, proliferation, cell size control and apoptosis. However, several lines of evidence point to FIP200 as an Atg11 counterpart in mammals.
First, FIP200 is essential for autophagosome formation and the proper function of ULK1, the mammalian homolog of yeast Atg1 kinase. Autophagosome formation and the phosphorylation status of ULK1 were strongly affected in FIP200-deficient cells.[118] Additionally, in mammals, FIP200, ATG9A and ULK1, localize to the phagophore[123] and this localization of ULK1, as well as its auto-phosphorylation activity, depends on the FIP200 interaction. These are analogous to the requirement of Atg11 (and Atg17) for Atg1 kinase activity and its localization in yeast.
Secondly, like Atg11, which is recruited to the PAS independently of Atg1 and Atg13, FIP200 can also associate with the membranes independently of the other ULK complex components.[124] Moreover, FIP200 associates with SEC12 and facilitates the remodeling of ER exit sites (ERES) associated with SEC12 (SEC12-ERES).[121] COPII vesicles, which transport regulated and constitutive secretory cargos,[125] bud from ERES and are linked with autophagosome biogenesis[66, 67, 126] by serving as precursors of the phagophore membrane.[126] Thus FIP200, by acting at the ERES, could regulate the early steps of autophagosome biogenesis.[121] Considering that Atg11 and Atg17 form homo-dimers that serve as a scaffold for tethering vesicles,[24, 55] FIP200 may perform a similar function and cluster SEC12 molecules at the ERES, leading to the increase of the membrane size.
Finally, FIP200 binds directly to the ER membrane-resident protein, CCPG1, to promote ER-phagy in a manner similar to the Atg11 recognition of yeast SARs. The FIP200-interacting regions (FIRs) of CCPG1, resembling the yeast Atg11BR motifs (a core containing hydrophobic residues surrounded by negative charges and S/T amino acids, often preceded by AIM/LIR), were required for binding to the C-terminal, Atg11-homology domain of mammalian FIP200.[127] In the context of mechanistic conservation, it would be interesting to know if FIP200 preserves the prototypical Atg11-SAR interaction paradigm, in which Atg8 and Atg11 binding to the SAR coordinates and facilitates selective autophagy.
Unfortunately, it is unknown if FIP200 alone has all the properties of Atg11 or whether other proteins have taken over some of the functions of the Atg11 protein. For instance, C9orf72 functions as a Rab1a effector and mediates the interaction between the active ULK1 initiation complex and the GTP-Rab1a (Ypt1 homolog) in mammals and in this way enables the Rab1a-dependent trafficking of the ULK1 initiation complex to the phagophore.[120] It is also an open question if BCL2L13, a mammalian homolog of the yeast Atg32, mediates both mitochondrial fragmentation and mitophagy in mammalian cells by itself or via binding to FIP200 or some other unidentified counterpart of Atg11.[128–130]
Another suggested counterpart of Atg11 in mammals is the Huntingtin protein (HTT).[131] This large protein shares sequence similarities with three different components of the yeast autophagy system: 1) Atg23, which acts at the early stages of Atg9 trafficking, 2) Vac8, which generates contacts between the autophagosome and the vacuole and is required for vacuolar homotypic fusion and 3) Atg11. The N-terminal part of HTT is similar to Atg23, the central region of HTT is similar to Vac8, and the C-terminal part resembles Atg11. Additionally, the more important fact is that HTT interacts with the ULK1 kinase complex, autophagic receptor proteins and mammalian Atg8-family members (LC3/GABARAP proteins).[131, 132] The possibility that HTT plays a role as an Atg11-like scaffold protein for the mammalian autophagy machinery is supported by the accumulation of p62- and ubiquitin-containing aggregates, which usually reflect a loss of aggrephagy (the selective degradation of protein aggregates by the autophagy machinery[133]) in HTT knockout CNS mice.[131]
Plant Atg11
Plants also possess an Atg11 counterpart, which in Arabidopsis is represented by a 1148 aa protein ATG11. However, this protein contains traces of similarity to both Atg11 and Atg17 and contains similar domain architecture to mammalian FIP200, including a short Atg17-like domain followed by CC regions and a C-terminal, Atg11-homology domain. AtATG11 colocalizes with the Arabidopsis ATG1 kinase complex indirectly through ATG13 and also interacts directly with ATG13 and ATG101.[88] Like yeast Atg11, AtATG11 acts at early stages of autophagosome formation, upstream of ATG2. It was established that AtATG11, together with ATG9 and phosphatidylinositol 3-kinase, controls ATG2-mediated formation of autophagosomes.[134] However, AtATG11 is also required at later steps of autophagy when it promotes vesicle delivery to the vacuole, without being essential for the ATG8/ATG12 conjugation.[88] It was also shown that AtATG11 binds to ATG8 on autophagic vesicles and is eventually deposited inside the vacuole, along with ATG1 and ATG13. This starvation-induced turnover of the ATG1-ATG13 complex is probably mediated through the interaction of AtATG11 with ATG8.[88]
Evolutionary conservation of the Atg11 scaffold protein
It is intriguing that the functions of Atg11 appear to differ among eukaryotes. For instance, although the requirement of Atg11 for Atg1 phosphorylation is shared by all eukaryotes, it is required for ATG1-ATG13 turnover during starvation only in Arabidopsis.[88, 118] Interestingly, the lipidation of LC3 requires FIP200, as it is greatly reduced in mammalian FIP200-deficient cells,[118] but in yeast or Arabidopsis atg11 mutants, the synthesis of Atg12-Atg5 and Atg8-PE conjugates does not seem to be affected.[88, 135] Also, while loss of yeast Atg11 does not affect non-selective bulk autophagy or the deposition of autophagic bodies in the vacuole,[77] autophagosome formation is mostly abolished in the absence of mammalian FIP200,[118] but not in the Arabidopsis atg11–1 mutant, where the lack of ATG11 significantly affects only the deposition of autophagic bodies.[88] In summary, phylogenetic analyses of Atg11 and Atg17-related proteins, along with their molecular characteristics, suggest a complex evolution of Atg11 among eukaryotes with substantial divergence of their roles within the autophagy system. Metazoans and plants appear to lack canonical Atg11 proteins, but instead encode a hybrid Atg17/Atg11 protein which might also properties of other autophagy components, such as Vac8 or Atg23. It is plausible that hybrids of Atg17/Atg11 may represent the ancestral scaffolds shared by general and selective autophagy. In such a case, the Atg17 protein seems to be a relatively new component of the fungal autophagy machinery, which likely arose by duplication of the Atg11 locus and loss of the Atg11 homology domain that led to Atg11 and Atg17 specialization over time.[136]
Highlights.
Atg11 a critical protein for selective autophagy.
Atg11 serves as a platform for the recruitment of the core autophagic machinery and a hub for numerous protein-protein interactions during selective autophagy.
Atg11 coordinates several steps of selective autophagy from cargo selection and PAS organization to autophagosome maturation and the termination of selective autophagy.
Atg11 scaffold has functional counterparts in higher eukaryotes
Acknowledgements
This work was supported by NIH grants 2RO1 DK41737 and 1R03 AI42388 to SS, who holds a Tata Chancellor’s Endowed Professorship in Molecular Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References:
- [1].Ohsumi Y Historical landmarks of autophagy research. Cell Res. 2014;24:9–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Wen X, Klionsky DJ. An overview of macroautophagy in yeast. J Mol Biol. 2016;428:1681–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Suzuki H, Osawa T, Fujioka Y, Noda NN. Structural biology of the core autophagy machinery. Curr Opin Struct Biol. 2017;43:10–7. [DOI] [PubMed] [Google Scholar]
- [4].Good MC, Zalatan JG, Lim WA. Scaffold proteins: hubs for controlling the flow of cellular information. Science. 2011;332:680–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Zeke A, Lukacs M, Lim WA, Remenyi A. Scaffolds: interaction platforms for cellular signalling circuits. Trends Cell Biol. 2009;19:364–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lim WA. Designing customized cell signalling circuits. Nat Rev Mol Cell Biol. 2010;11:393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Bashor CJ, Helman NC, Yan S, Lim WA. Using engineered scaffold interactions to reshape MAP kinase pathway signaling dynamics. Science. 2008;319:1539–43. [DOI] [PubMed] [Google Scholar]
- [8].Motley AM, Nuttall JM, Hettema EH. Pex3-anchored Atg36 tags peroxisomes for degradation in Saccharomyces cerevisiae. EMBO J. 2012;31:2852–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Suzuki K, Kondo C, Morimoto M, Ohsumi Y. Selective transport of alpha-mannosidase by autophagic pathways: identification of a novel receptor, Atg34p. J Biol Chem. 2010;285:30019–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kanki T, Wang K, Cao Y, Baba M, Klionsky DJ. Atg32 is a mitochondrial protein that confers selectivity during mitophagy. Dev Cell. 2009;17:98–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Yorimitsu T, Klionsky DJ. Atg11 links cargo to the vesicle-forming machinery in the cytoplasm to vacuole targeting pathway. Mol Biol Cell. 2005;16:1593–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Farre JC, Manjithaya R, Mathewson RD, Subramani S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev Cell. 2008;14:365–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Farre JC, Burkenroad A, Burnett SF, Subramani S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013;14:441–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].He C, Song H, Yorimitsu T, Monastyrska I, Yen WL, Legakis JE, et al. Recruitment of Atg9 to the preautophagosomal structure by Atg11 is essential for selective autophagy in budding yeast. J Cell Biol. 2006;175:925–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Lipatova Z, Belogortseva N, Zhang XQ, Kim J, Taussig D, Segev N. Regulation of selective autophagy onset by a Ypt/Rab GTPase module. Proc Natl Acad Sci U S A. 2012;109:6981–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Popelka H, Damasio A, Hinshaw JE, Klionsky DJ, Ragusa MJ. Structure and function of yeast Atg20, a sorting nexin that facilitates autophagy induction. Proc Natl Acad Sci U S A. 2017;114:E10112–E21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Gammoh N, Florey O, Overholtzer M, Jiang X. Interaction between FIP200 and ATG16L1 distinguishes ULK1 complex-dependent and -independent autophagy. Nat Struct Mol Biol. 2013;20:144–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Mao K, Wang K, Liu X, Klionsky DJ. The scaffold protein Atg11 recruits fission machinery to drive selective mitochondria degradation by autophagy. Dev Cell. 2013;26:9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Mao K, Liu X, Feng Y, Klionsky DJ. The progression of peroxisomal degradation through autophagy requires peroxisomal division. Autophagy. 2014;10:652–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Liu X, Mao K, Yu AYH, Omairi-Nasser A, Austin J 2nd, BS Glick, et al. The Atg17-Atg31-Atg29 complex coordinates with Atg11 to recruit the Vam7 SNARE and mediate autophagosome-vacuole fusion. Curr Biol. 2016;26:150–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Truebestein L, Leonard TA. Coiled-coils: The long and short of it. Bioessays. 2016;38:903–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Burkhard P, Stetefeld J, Strelkov SV. Coiled coils: a highly versatile protein folding motif. Trends Cell Biol. 2001;11:82–8. [DOI] [PubMed] [Google Scholar]
- [23].Chew LH, Setiaputra D, Klionsky DJ, Yip CK. Structural characterization of the Saccharomyces cerevisiae autophagy regulatory complex Atg17-Atg31-Atg29. Autophagy. 2013;9:1467–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Suzuki H, Noda NN. Biophysical characterization of Atg11, a scaffold protein essential for selective autophagy in yeast. FEBS Open Bio. 2018;8:110–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Bahrami AH, Lin MG, Ren X, Hurley JH, Hummer G. Scaffolding the cup-shaped double membrane in autophagy. PLoS Comput Biol. 2017;13:e1005817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Li X, He L, Che KH, Funderburk SF, Pan L, Pan N, et al. Imperfect interface of Beclin1 coiled-coil domain regulates homodimer and heterodimer formation with Atg14L and UVRAG. Nat Commun. 2012;3:662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. J Biol Chem. 2010;285:1508–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Obara K, Sekito T, Ohsumi Y. Assortment of phosphatidylinositol 3-kinase complexes--Atg14p directs association of complex I to the pre-autophagosomal structure in Saccharomyces cerevisiae. Mol Biol Cell. 2006;17:1527–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–9. [DOI] [PubMed] [Google Scholar]
- [31].Aoki Y, Kanki T, Hirota Y, Kurihara Y, Saigusa T, Uchiumi T, et al. Phosphorylation of Serine 114 on Atg32 mediates mitophagy. Mol Biol Cell. 2011;22:3206–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Kim J, Kamada Y, Stromhaug PE, Guan J, Hefner-Gravink A, Baba M, et al. Cvt9/Gsa9 functions in sequestering selective cytosolic cargo destined for the vacuole. J Cell Biol. 2001;153:381–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Torggler R, Papinski D, Brach T, Bas L, Schuschnig M, Pfaffenwimmer T, et al. Two independent pathways within selective autophagy converge to activate Atg1 kinase at the vacuole. Mol Cell. 2016;64:221–35. [DOI] [PubMed] [Google Scholar]
- [34].Kamber RA, Shoemaker CJ, Denic V. Receptor-bound targets of selective autophagy use a scaffold protein to activate the Atg1 kinase. Mol Cell. 2015;59:372–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Mao K, Chew LH, Inoue-Aono Y, Cheong H, Nair U, Popelka H, et al. Atg29 phosphorylation regulates coordination of the Atg17-Atg31-Atg29 complex with the Atg11 scaffold during autophagy initiation. Proc Natl Acad Sci U S A. 2013;110:E2875–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Lynch-Day MA, Bhandari D, Menon S, Huang J, Cai H, Bartholomew CR, et al. Trs85 directs a Ypt1 GEF, TRAPPIII, to the phagophore to promote autophagy. Proc Natl Acad Sci U S A. 2010;107:7811–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wang J, Menon S, Yamasaki A, Chou HT, Walz T, Jiang Y, et al. Ypt1 recruits the Atg1 kinase to the preautophagosomal structure. Proc Natl Acad Sci U S A. 2013;110:9800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Chang CY, Huang WP. Atg19 mediates a dual interaction cargo sorting mechanism in selective autophagy. Mol Biol Cell. 2007;18:919–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Farre JC, Subramani S. Mechanistic insights into selective autophagy pathways: lessons from yeast. Nat Rev Mol Cell Biol. 2016;17:537–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nakatogawa H Hrr25: an emerging major player in selective autophagy regulation in Saccharomyces cerevisiae. Autophagy. 2015;11:432–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kanki T, Kurihara Y, Jin X, Goda T, Ono Y, Aihara M, et al. Casein kinase 2 is essential for mitophagy. EMBO Rep. 2013;14:788–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Pfaffenwimmer T, Reiter W, Brach T, Nogellova V, Papinski D, Schuschnig M, et al. Hrr25 kinase promotes selective autophagy by phosphorylating the cargo receptor Atg19. EMBO Rep. 2014;15:862–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tanaka C, Tan LJ, Mochida K, Kirisako H, Koizumi M, Asai E, et al. Hrr25 triggers selective autophagy-related pathways by phosphorylating receptor proteins. J Cell Biol. 2014;207:91–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Mochida K, Ohsumi Y, Nakatogawa H. Hrr25 phosphorylates the autophagic receptor Atg34 to promote vacuolar transport of alpha-mannosidase under nitrogen starvation conditions. FEBS Lett. 2014;588:3862–9. [DOI] [PubMed] [Google Scholar]
- [45].Zientara-Rytter K, Ozeki K, Nazarko TY, Subramani S. Pex3 and Atg37 compete to regulate the interaction between the pexophagy receptor, Atg30, and the Hrr25 kinase. Autophagy. 2018:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Shintani T, Huang WP, Stromhaug PE, Klionsky DJ. Mechanism of cargo selection in the cytoplasm to vacuole targeting pathway. Dev Cell. 2002;3:825–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Suzuki K, Kirisako T, Kamada Y, Mizushima N, Noda T, Ohsumi Y. The pre-autophagosomal structure organized by concerted functions of APG genes is essential for autophagosome formation. EMBO J. 2001;20:5971–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Suzuki K, Kamada Y, Ohsumi Y. Studies of cargo delivery to the vacuole mediated by autophagosomes in Saccharomyces cerevisiae. Dev Cell. 2002;3:815–24. [DOI] [PubMed] [Google Scholar]
- [49].Wang Y, Zhang X, Zhang H, Lu Y, Huang H, Dong X, et al. Coiled-coil networking shapes cell molecular machinery. Mol Biol Cell. 2012;23:3911–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Zhang H, Chen J, Wang Y, Peng L, Dong X, Lu Y, et al. A computationally guided protein-interaction screen uncovers coiled-coil interactions involved in vesicular trafficking. J Mol Biol. 2009;392:228–41. [DOI] [PubMed] [Google Scholar]
- [51].Stjepanovic G, Davies CW, Stanley RE, Ragusa MJ, Kim DJ, Hurley JH. Assembly and dynamics of the autophagy-initiating Atg1 complex. Proc Natl Acad Sci U S A. 2014;111:12793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Yeh YY, Shah KH, Herman PK. An Atg13 protein-mediated self-association of the Atg1 protein kinase is important for the induction of autophagy. J Biol Chem. 2011;286:28931–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Kabeya Y, Kamada Y, Baba M, Takikawa H, Sasaki M, Ohsumi Y. Atg17 functions in cooperation with Atg1 and Atg13 in yeast autophagy. Mol Biol Cell. 2005;16:2544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Noda NN, Fujioka Y. Atg1 family kinases in autophagy initiation. Cell Mol Life Sci. 2015;72:3083–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rao Y, Perna MG, Hofmann B, Beier V, Wollert T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat Commun. 2016;7:10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Cheong H, Nair U, Geng J, Klionsky DJ. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol Biol Cell. 2008;19:668–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Kamada Y, Yoshino K, Kondo C, Kawamata T, Oshiro N, Yonezawa K, et al. Tor directly controls the Atg1 kinase complex to regulate autophagy. Mol Cell Biol. 2010;30:1049–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Chang YY, Neufeld TP. Autophagy takes flight in Drosophila. FEBS Lett. 2010;584:1342–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011;124:161–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Pelech S Dimerization in protein kinase signaling. J Biol. 2006;5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011;12:9–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ao X, Zou L, Wu Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014;21:348–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Lipatova Z, Majumdar U, Segev N. Trs33-containing TRAPP IV: a novel autophagy-specific Ypt1 GEF. Genetics. 2016;204:1117–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lipatova Z, Shah AH, Kim JJ, Mulholland JW, Segev N. Regulation of ER-phagy by a Ypt/Rab GTPase module. Mol Biol Cell. 2013;24:3133–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Kakuta S, Yamamoto H, Negishi L, Kondo-Kakuta C, Hayashi N, Ohsumi Y. Atg9 vesicles recruit vesicle-tethering proteins Trs85 and Ypt1 to the autophagosome formation site. J Biol Chem. 2012;287:44261–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Shirahama-Noda K, Kira S, Yoshimori T, Noda T. TRAPPIII is responsible for vesicular transport from early endosomes to Golgi, facilitating Atg9 cycling in autophagy. J Cell Sci. 2013;126:4963–73. [DOI] [PubMed] [Google Scholar]
- [67].Wang J, Davis S, Menon S, Zhang J, Ding J, Cervantes S, et al. Ypt1/Rab1 regulates Hrr25/CK1delta kinase activity in ER-Golgi traffic and macroautophagy. J Cell Biol. 2015;210:273–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, et al. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. J Cell Biol. 2012;198:219–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Sekito T, Kawamata T, Ichikawa R, Suzuki K, Ohsumi Y. Atg17 recruits Atg9 to organize the preautophagosomal structure. Genes Cells. 2009;14:525–38. [DOI] [PubMed] [Google Scholar]
- [70].Stephan JS, Yeh YY, Ramachandran V, Deminoff SJ, Herman PK. The Tor and PKA signaling pathways independently target the Atg1/Atg13 protein kinase complex to control autophagy. Proc Natl Acad Sci U S A. 2009;106:17049–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Kofinger J, Ragusa MJ, Lee IH, Hummer G, Hurley JH. Solution structure of the Atg1 complex: implications for the architecture of the phagophore assembly site. Structure. 2015;23:809–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Nazarko TY, Farre JC, Subramani S. Peroxisome size provides insights into the function of autophagy-related proteins. Mol Biol Cell. 2009;20:3828–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Suzuki K, Kubota Y, Sekito T, Ohsumi Y. Hierarchy of Atg proteins in pre-autophagosomal structure organization. Genes Cells. 2007;12:209–18. [DOI] [PubMed] [Google Scholar]
- [75].Kawamata T, Kamada Y, Kabeya Y, Sekito T, Ohsumi Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol Biol Cell. 2008;19:2039–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Nice DC, Sato TK, Stromhaug PE, Emr SD, Klionsky DJ. Cooperative binding of the cytoplasm to vacuole targeting pathway proteins, Cvt13 and Cvt20, to phosphatidylinositol 3-phosphate at the preautophagosomal structure is required for selective autophagy. J Biol Chem. 2002;277:30198–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Reggiori F, Klionsky DJ. Autophagic processes in yeast: mechanism, machinery and regulation. Genetics. 2013;194:341–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Ma M, Kumar S, Purushothaman L, Babst M, Ungermann C, Chi RJ, et al. Lipid trafficking by yeast Snx4 family SNX-BAR proteins promotes autophagy and vacuole membrane fusion. Mol Biol Cell. 2018;29:2190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. J Cell Biol. 2010;190:1005–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Ohashi Y, Munro S. Membrane delivery to the yeast autophagosome from the Golgi-endosomal system. Mol Biol Cell. 2010;21:3998–4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Hettema EH, Lewis MJ, Black MW, Pelham HR. Retromer and the sorting nexins Snx4/41/42 mediate distinct retrieval pathways from yeast endosomes. EMBO J. 2003;22:548–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Bean BD, Davey M, Conibear E. Cargo selectivity of yeast sorting nexins. Traffic. 2017;18:110–22. [DOI] [PubMed] [Google Scholar]
- [83].Ma M, Burd CG, Chi RJ. Distinct complexes of yeast Snx4 family SNX-BARs mediate retrograde trafficking of Snc1 and Atg27. Traffic. 2017;18:134–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Suzuki SW, Emr SD. Membrane protein recycling from the vacuole/lysosome membrane. J Cell Biol. 2018;217:1623–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Vollert CS, Uetz P. The phox homology (PX) domain protein interaction network in yeast. Mol Cell Proteomics. 2004;3:1053–64. [DOI] [PubMed] [Google Scholar]
- [86].Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, et al. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623–7. [DOI] [PubMed] [Google Scholar]
- [88].Li F, Chung T, Vierstra RD. AUTOPHAGY-RELATED11 plays a critical role in general autophagy- and senescence-induced mitophagy in Arabidopsis. Plant Cell. 2014;26:788–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Abert C, Kontaxis G, Martens S. Accessory interaction motifs in the Atg19 cargo receptor enable strong binding to the clustered ubiquitin-related Atg8 protein. J Biol Chem. 2016;291:18799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Sawa-Makarska J, Abert C, Romanov J, Zens B, Ibiricu I, Martens S. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat Cell Biol. 2014;16:425–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Noda NN, Satoo K, Fujioka Y, Kumeta H, Ogura K, Nakatogawa H, et al. Structural basis of Atg8 activation by a homodimeric E1, Atg7. Mol Cell. 2011;44:462–75. [DOI] [PubMed] [Google Scholar]
- [92].Liu XM, Yamasaki A, Du XM, Coffman VC, Ohsumi Y, Nakatogawa H, et al. Lipidation-independent vacuolar functions of Atg8 rely on its noncanonical interaction with a vacuole membrane protein. Elife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Juris L, Montino M, Rube P, Schlotterhose P, Thumm M, Krick R. PI3P binding by Atg21 organises Atg8 lipidation. EMBO J. 2015;34:955–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [94].Krick R, Bremer S, Welter E, Schlotterhose P, Muehe Y, Eskelinen EL, et al. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J Cell Biol. 2010;190:965–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, et al. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–3. [DOI] [PubMed] [Google Scholar]
- [96].Mizushima N The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010;22:132–9. [DOI] [PubMed] [Google Scholar]
- [97].Bas L, Papinski D, Licheva M, Torggler R, Rohringer S, Schuschnig M, et al. Reconstitution reveals Ykt6 as the autophagosomal SNARE in autophagosome-vacuole fusion. J Cell Biol. 2018;217:3656–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Hegedus K, Takats S, Boda A, Jipa A, Nagy P, Varga K, et al. The Ccz1-Mon1-Rab7 module and Rab5 control distinct steps of autophagy. Mol Biol Cell. 2016;27:3132–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Gao J, Reggiori F, Ungermann C. A novel in vitro assay reveals SNARE topology and the role of Ykt6 in autophagosome fusion with vacuoles. J Cell Biol. 2018;217:3670–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Reggiori F, Ungermann C. Autophagosome maturation and fusion. J Mol Biol. 2017;429:486–96. [DOI] [PubMed] [Google Scholar]
- [101].Liu X, Klionsky DJ. The Atg17-Atg31-Atg29 complex and Atg11 regulate autophagosome-vacuole fusion. Autophagy. 2016;12:894–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–8. [DOI] [PubMed] [Google Scholar]
- [103].Obara K, Ohsumi Y. Dynamics and function of PtdIns(3)P in autophagy. Autophagy. 2008;4:952–4. [DOI] [PubMed] [Google Scholar]
- [104].Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–69. [DOI] [PubMed] [Google Scholar]
- [105].Kumar S, Jain A, Farzam F, Jia J, Gu Y, Choi SW, et al. Mechanism of Stx17 recruitment to autophagosomes via IRGM and mammalian Atg8 proteins. J Cell Biol. 2018;217:997–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Cebollero E, van der Vaart A, Zhao M, Rieter E, Klionsky DJ, Helms JB, et al. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr Biol. 2012;22:1545–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Zhou F, Zou S, Chen Y, Lipatova Z, Sun D, Zhu X, et al. A Rab5 GTPase module is important for autophagosome closure. PLoS Genet. 2017;13:e1007020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fujioka Y, Suzuki SW, Yamamoto H, Kondo-Kakuta C, Kimura Y, Hirano H, et al. Structural basis of starvation-induced assembly of the autophagy initiation complex. Nat Struct Mol Biol. 2014;21:513–21. [DOI] [PubMed] [Google Scholar]
- [109].Cheong H, Yorimitsu T, Reggiori F, Legakis JE, Wang CW, Klionsky DJ. Atg17 regulates the magnitude of the autophagic response. Mol Biol Cell. 2005;16:3438–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Suzuki SW, Yamamoto H, Oikawa Y, Kondo-Kakuta C, Kimura Y, Hirano H, et al. Atg13 HORMA domain recruits Atg9 vesicles during autophagosome formation. Proc Natl Acad Sci U S A. 2015;112:3350–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Lefevre SD, Kumar S, van der Klei IJ. Inhibition of peroxisome fission, but not mitochondrial fission, increases yeast chronological lifespan. Cell Cycle. 2015;14:1698–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Motley AM, Ward GP, Hettema EH. Dnm1p-dependent peroxisome fission requires Caf4p, Mdv1p and Fis1p. J Cell Sci. 2008;121:1633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Westermann B Molecular machinery of mitochondrial fusion and fission. J Biol Chem. 2008;283:13501–5. [DOI] [PubMed] [Google Scholar]
- [114].Manivannan S, de Boer R, Veenhuis M, van der Klei IJ. Lumenal peroxisomal protein aggregates are removed by concerted fission and autophagy events. Autophagy. 2013;9:1044–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Izumi M, Wada S, Makino A, Ishida H. The autophagic degradation of chloroplasts via rubisco-containing bodies is specifically linked to leaf carbon status but not nitrogen status in Arabidopsis. Plant Physiol. 2010;154:1196–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Ishida H, Yoshimoto K, Izumi M, Reisen D, Yano Y, Makino A, et al. Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 2008;148:142–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Wada S, Ishida H, Izumi M, Yoshimoto K, Ohsumi Y, Mae T, et al. Autophagy plays a role in chloroplast degradation during senescence in individually darkened leaves. Plant Physiol. 2009;149:885–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Hara T, Takamura A, Kishi C, Iemura S, Natsume T, Guan JL, et al. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J Cell Biol. 2008;181:497–510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [119].Noda NN, Mizushima N. Atg101: Not just an accessory subunit in the autophagy-initiation complex. Cell Struct Funct. 2016;41:13–20. [DOI] [PubMed] [Google Scholar]
- [120].Webster CP, Smith EF, Grierson AJ, De Vos KJ. C9orf72 plays a central role in Rab GTPase-dependent regulation of autophagy. Small GTPases. 2018;9:399–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Ge L, Zhang M, Kenny SJ, Liu D, Maeda M, Saito K, et al. Remodeling of ER-exit sites initiates a membrane supply pathway for autophagosome biogenesis. EMBO Rep. 2017;18:1586–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Gan B, Guan JL. FIP200, a key signaling node to coordinately regulate various cellular processes. Cell Signal. 2008;20:787–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Hara T, Mizushima N. Role of ULK-FIP200 complex in mammalian autophagy: FIP200, a counterpart of yeast Atg17? Autophagy. 2009;5:85–7. [DOI] [PubMed] [Google Scholar]
- [124].Nishimura T, Tamura N, Kono N, Shimanaka Y, Arai H, Yamamoto H, et al. Autophagosome formation is initiated at phosphatidylinositol synthase-enriched ER subdomains. EMBO J. 2017;36:1719–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [125].Budnik A, Stephens DJ. ER exit sites--localization and control of COPII vesicle formation. FEBS Lett. 2009;583:3796–803. [DOI] [PubMed] [Google Scholar]
- [126].Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Smith MD, Harley ME, Kemp AJ, Wills J, Lee M, Arends M, et al. CCPG1 is a non-canonical autophagy cargo receptor essential for ER-phagy and pancreatic ER proteostasis. Dev Cell. 2018;44:217–32 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Murakawa T, Yamaguchi O, Hashimoto A, Hikoso S, Takeda T, Oka T, et al. Bcl-2-like protein 13 is a mammalian Atg32 homologue that mediates mitophagy and mitochondrial fragmentation. Nat Commun. 2015;6:7527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Otsu K, Murakawa T, Yamaguchi O. BCL2L13 is a mammalian homolog of the yeast mitophagy receptor Atg32. Autophagy. 2015;11:1932–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Levchenko M, Lorenzi I, Dudek J. The degradation pathway of the mitophagy receptor Atg32 is rerouted by a posttranslational modification. PLoS One. 2016;11:e0168518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Ochaba J, Lukacsovich T, Csikos G, Zheng S, Margulis J, Salazar L, et al. Potential function for the Huntingtin protein as a scaffold for selective autophagy. Proc Natl Acad Sci U S A. 2014;111:16889–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Rui YN, Xu Z, Patel B, Cuervo AM, Zhang S. HTT/Huntingtin in selective autophagy and Huntington disease: A foe or a friend within? Autophagy. 2015;11:858–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Lamark T, Johansen T. Aggrephagy: selective disposal of protein aggregates by macroautophagy. Int J Cell Biol. 2012;2012:736905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Kang S, Shin KD, Kim JH, Chung T. Autophagy-related (ATG) 11, ATG9 and the phosphatidylinositol 3-kinase control ATG2-mediated formation of autophagosomes in Arabidopsis. Plant Cell Rep. 2018;37:653–64. [DOI] [PubMed] [Google Scholar]
- [135].Shintani T, Klionsky DJ. Autophagy in health and disease: a double-edged sword. Science. 2004;306:990–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [136].Li F, Vierstra RD. Arabidopsis ATG11, a scaffold that links the ATG1-ATG13 kinase complex to general autophagy and selective mitophagy. Autophagy. 2014;10:1466–7. [DOI] [PMC free article] [PubMed] [Google Scholar]