Abstract
A refined liver cancer staging system and effective prognostic prediction can help clinicians make optimized treatment decisions, which is essential in our fight against cancer and for improving the unsatisfying survival rate of liver cancer globally. The prognosis of liver cancer is not only related to tumor status, it is also affected by the patients' liver functions and the chosen treatment. Currently, several staging systems are being tested. Herein, we analyzed RNA-seq data from the TCGA database and identified a newly annotated lncRNA, ACVR2B-AS1, whose expression is upregulated in liver cancer. Higher ACVR2B-AS1 expression is an independent adverse prognostic factor for overall survival (OS) and relapse-free survival (RFS) in liver cancer patients. Our work suggests that the lncRNA ACVR2B-AS1 could be a candidate biomarker for liver cancer prognosis. Furthermore, ACVR2B-AS1 might serve as a potential therapeutic target, which is a possibility that is worthy of further study.
1. Introduction
Liver cancer is one of the leading causes of cancer-related deaths worldwide [1]. Although numerous resources have been invested into basic research and clinical trials, the survival rate of liver cancer patients remains dismal. For example, in the US, the latest five-year relative survival rate for liver cancer patients is only 18% [2]. This low rate is largely due to the aggressive nature of liver cancer and the fact that most patients diagnosed are already at advanced stages [3]. While the current staging system utilizes both patient assessment and proposed treatment responses, it remains important to refine the risk classification system, which is crucial for optimizing decision making and reducing cancer mortality [1].
Long noncoding RNAs (lncRNAs) represent a class of genes that are not translated into proteins and whose functions have only begun to emerge in the past few years [4, 5]. They are evolutionarily less conserved than coding genes, and it is estimated that the number of lncRNA genes is far larger than that of protein coding genes [5], although their exact abundance is not clear. Increasing evidence has demonstrated that lncRNAs have important biological functions and that their dysregulation is associated with diseases, including cancers [6]. However, the majority of annotated lncRNAs have not been functionally characterized, and their biological significance remains elusive [6].
ACVR2B-AS1 (ACVR2B-antisense RNA1) is a recently annotated lncRNA [7]. It is located on 3p22.2 and is transcribed from the opposite strand of ACVR2B. The function of ACVR2B-AS1 has not been studied. Here, using data extracted from the TCGA database, we identified ACVR2B-AS1 as an independent prognostic factor for liver cancer patient clinical outcomes. This may help us to refine liver cancer prognostic prediction and to further studies of ACVR2B-AS1.
2. Materials and Methods
2.1. Data Mining
RNA-seq data and patients' clinical characteristics were downloaded from the TCGA database (https://portal.gdc.cancer.gov/). Transcript abundances were quantified using the RNA-Seq data via the Expectation-Maximization (RSEM) software [8].
2.2. Statistical Analysis
All statistical tests were performed in R [9]. Nonparametric Wilcoxon and Kruskal–Wallis tests were used for differential expression analysis among different subgroups. The diagnostic value of ACVR2B-AS1 was estimated using a receiver operating characteristic (ROC) curve drawn with the pROC package [10], and the area under curve (AUC) was calculated as previously described [10]. A cut-off value was determined by utilizing the ROC curve, and the overall population was then divided into two groups (ACVR2B-AS1 High and ACVR2B-AS1 Low) for the subsequent analysis. The associations between ACVR2B-AS1 and patient clinical features were analyzed via Fisher's exact or Chi-squared tests. Overall survival and relapse-free survival were determined via Kaplan–Meier curves using the survival package in R [11]. The statistical significance of the differences noticed above was calculated using the log-rank test. The prognostic capabilities of ACVR2B-AS1 were assessed via univariate and multivariate Cox regression analysis using the Cox proportional hazard model. The ggplot2 package in R was used for data visualization [12].
3. Results
3.1. Patient Characteristics
RNA-seq data from a total of 371 patients diagnosed with liver cancer were extracted from the TCGA database for analysis. The clinical characteristics of the patients, including age, sex, tumor histological type, grade, stage, and vital status, are listed in Table 1.
Table 1.
Clinical characteristic of the included patients.
| Characteristics | Number of pts (%) |
|---|---|
| Age | |
| <55 | 117 (31.62) |
| ≥55 | 253 (68.38) |
| NA | 1 (0) |
|
| |
| Gender | |
| Female | 121 (32.61) |
| Male | 250 (67.39) |
|
| |
| Histological type | |
| Fibrolamellar carcinoma | 3 (0.81) |
| Hepatocellular carcinoma | 361 (97.3) |
| Hepatocholangiocarcinoma | 7 (1.89) |
|
| |
| Histologic grade | |
| G1 | 55 (14.82) |
| G2 | 177 (47.71) |
| G3 | 122 (32.88) |
| G4 | 12 (3.23) |
| NA | 5 (1.35) |
|
| |
| Stage | |
| I | 171 (46.09) |
| II | 86 (23.18) |
| III | 85 (22.91) |
| IV | 5 (1.35) |
| NA | 24 (6.47) |
|
| |
| T classification | |
| T1 | 181 (48.79) |
| T2 | 94 (25.34) |
| T3 | 80 (21.56) |
| T4 | 13 (3.5) |
| TX | 1 (0.27) |
| NA | 2 (0.54) |
|
| |
| N classification | |
| N0 | 252 (67.92) |
| N1 | 4 (1.08) |
| NX | 114 (30.73) |
| NA | 1 (0.27) |
|
| |
| M classification | |
| M0 | 266 (71.7) |
| M1 | 4 (1.08) |
| MX | 101 (27.22) |
|
| |
| Radiation therapy | |
| No | 338 (91.11) |
| Yes | 8 (2.16) |
| NA | 25 (6.74) |
|
| |
| Residual tumor | |
| R0 | 324 (87.33) |
| R1 | 17 (4.58) |
| R2 | 1 (0.27) |
| RX | 22 (5.93) |
| NA | 7 (1.89) |
|
| |
| Vital status | |
| Deceased | 130 (35.04) |
| Living | 241 (64.96) |
|
| |
| Relapse | |
| No | 179 (48.25) |
| Yes | 139 (37.47) |
| NA | 53 (14.29) |
|
| |
| ACVR2B-AS1 | |
| High | 211 (56.87) |
| Low | 160 (43.13) |
3.2. ACVR2B-AS1 Expression Is Upregulated in Liver Cancer
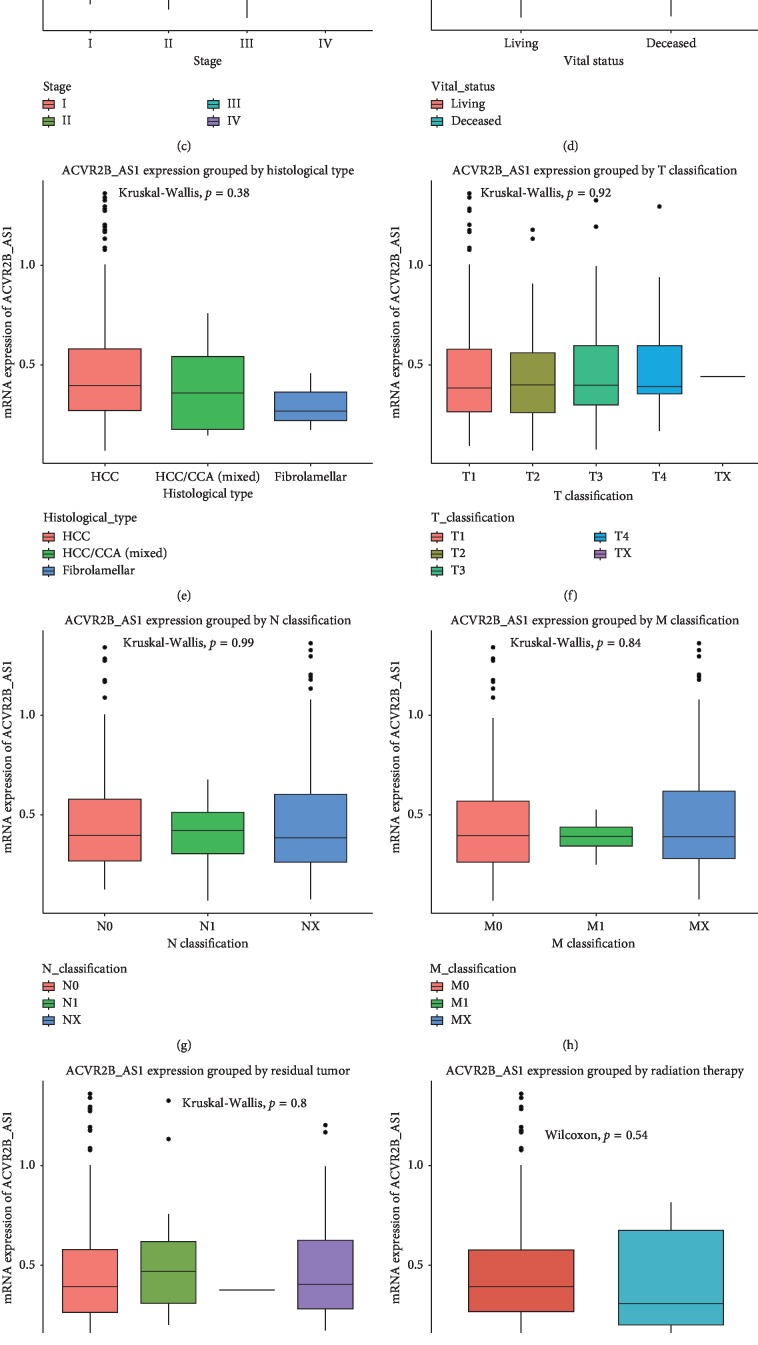
ACVR2B-AS1 levels were significantly elevated in liver cancer specimens compared with their normal counterparts (p=0.034) (Figure 1). A subgroup analysis revealed that ACVR2B-AS1 was differentially expressed in tumors of different histological grades (p=0.02), and grade IV tumors showed the highest ACVR2B-AS1 levels. Furthermore, by categorizing the patients based on vital status, we found that deceased patients showed higher ACVR2B-AS1 expression levels than did patients who were still alive at the time of sampling (p=0.0043). No significant differential expression was detected in the other subgroups analyzed.
Figure 1.
ACVR2B-AS1 is overexpressed in liver cancer and is differentially expressed in the corresponding subtypes. The significance was calculated based on nonparametric Wilcoxon and Kruskal–Wallis tests. Note: the subgroups include tumors versus normal liver tissue, histological grade, stage, vital status, histological type, T classification, N classification, M classification, residual tumor, radiation therapy, age, and sex.
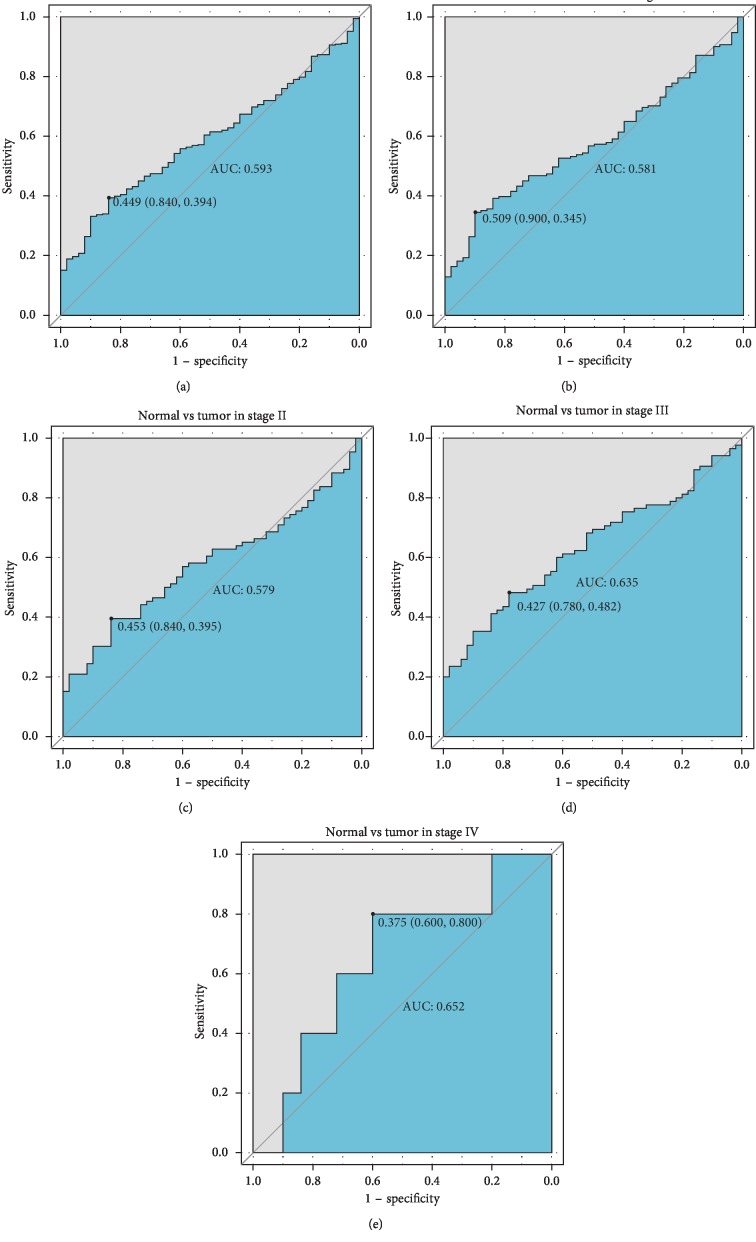
To further examine these findings, we generated an ROC curve to test the diagnostic abilities of ACVR2B-AS1 level for liver cancer diagnosis and histological grade classification. The results of the AUC analysis did not show any significance (Figure 2), indicating that ACVR2B-AS1 alone may not be an accurate diagnostic parameter.
Figure 2.
The ROC curves of ACVR2B-AS1 in liver cancer cohorts and different stages. Abbreviations: AUC, area under the curve.
3.3. ACVR2B-AS1 Levels Correlate with Patient Vital Status
Consistent with the previous results, correlation analysis confirmed that the vital status of the patients is correlated with ACVR2B-AS1 levels (p=0.003) (Table 2). The correlation between ACVR2B-AS1 levels and histological grade hardly reached statistical significance (p=0.054).
Table 2.
Correlation between the clinicopathologic variables and ACVR2B-AS1 expression.
| Clinical characteristics | ACVR2B-AS1 expression | |||||||
|---|---|---|---|---|---|---|---|---|
| Variable | No. of patients | High | % | Low | % | χ 2 | p value | |
| Age | <55 | 117 | 67 | (31.75) | 50 | (31.45) | 0 | 1.000 |
| ≥55 | 253 | 144 | (68.25) | 109 | (68.55) | |||
|
| ||||||||
| Gender | Female | 121 | 71 | (33.65) | 50 | (31.25) | 0.1417 | 0.6557 |
| Male | 250 | 140 | (66.35) | 110 | (68.75) | |||
|
| ||||||||
| Histological type | Fibrolamellar | 3 | 1 | (0.47) | 2 | (1.25) | 1.2706 | 0.515 |
| Hepatocellular | 361 | 207 | (98.1) | 154 | (96.25) | |||
| Hepatocholangiocarcinoma | 7 | 3 | (1.42) | 4 | (2.5) | |||
|
| ||||||||
| Histologic grade | G1 | 55 | 28 | (13.46) | 27 | (17.09) | 7.2563 | 0.054 |
| G2 | 177 | 97 | (46.63) | 80 | (50.63) | |||
| G3 | 122 | 72 | (34.62) | 50 | (31.65) | |||
| G4 | 12 | 11 | (5.29) | 1 | (0.63) | |||
|
| ||||||||
| Stage | I | 171 | 92 | (46.23) | 79 | (53.38) | 2.8209 | 0.458 |
| II | 86 | 50 | (25.13) | 36 | (24.32) | |||
| III | 85 | 53 | (26.63) | 32 | (21.62) | |||
| IV | 5 | 4 | (2.01) | 1 | (0.68) | |||
|
| ||||||||
| T Classification | T1 | 181 | 97 | (46.19) | 84 | (52.83) | 2.7575 | 0.647 |
| T2 | 94 | 56 | (26.67) | 38 | (23.9) | |||
| T3 | 80 | 47 | (22.38) | 33 | (20.75) | |||
| T4 | 13 | 9 | (4.29) | 4 | (2.52) | |||
| TX | 1 | 1 | (0.48) | 0 | (0) | |||
|
| ||||||||
| N classification | N0 | 252 | 147 | (70) | 105 | (65.62) | 1.588 | 0.528 |
| N1 | 4 | 3 | (1.43) | 1 | (0.62) | |||
| NX | 114 | 60 | (28.57) | 54 | (33.75) | |||
|
| ||||||||
| M Classification | M0 | 266 | 154 | (72.99) | 112 | (70) | 1.1272 | 0.597 |
| M1 | 4 | 3 | (1.42) | 1 | (0.62) | |||
| MX | 101 | 54 | (25.59) | 47 | (29.38) | |||
|
| ||||||||
| Radiation therapy | No | 338 | 191 | (98.45) | 147 | (96.71) | 0.5046 | 0.307 |
| Yes | 8 | 3 | (1.55) | 5 | (3.29) | |||
|
| ||||||||
| Residual tumor | R0 | 324 | 183 | (88.83) | 141 | (89.24) | 0.8406 | 1.000 |
| R1 | 17 | 10 | (4.85) | 7 | (4.43) | |||
| R2 | 1 | 1 | (0.49) | 0 | (0) | |||
| RX | 22 | 12 | (5.83) | 10 | (6.33) | |||
|
| ||||||||
| Vital status | Deceased | 130 | 88 | (41.71) | 42 | (26.25) | 8.8834 | 0.003 |
| Living | 241 | 123 | (58.29) | 118 | (73.75) | |||
3.4. ACVR2B-AS1 Is an Independent Adverse Prognostic Factor for OS in Liver Cancer
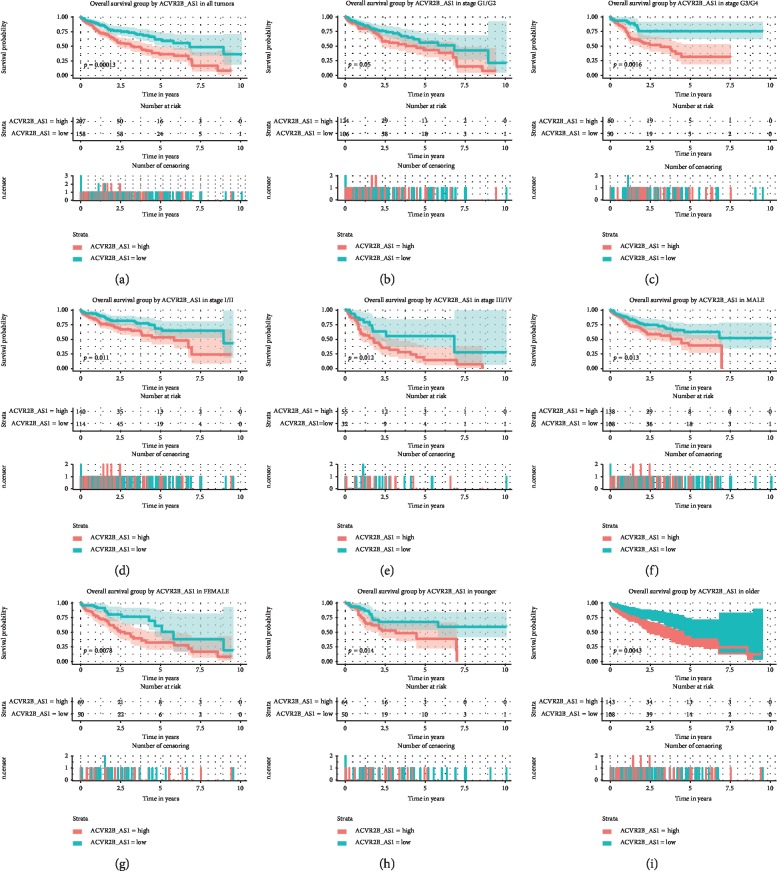
We next sought to verify the prognostic usefulness of ACVR2B-AS1 in liver cancer. A Kaplan–Meier curve was plotted against different ACVR2B-AS1 levels within the different subgroups (previously defined via the ROC curve against vital status) (Figure 3). The log-rank test revealed that patients with lower ACVR2B-AS1 expression levels had a significantly longer overall survival (OS) than those with higher ACVR2B-AS1 expression levels (p=0.0013). A subgroup analysis showed that ACVR2B-AS1-low patients displayed better OS among all of the subgroups analyzed.
Figure 3.
Overall survival outcomes based on ACVR2B-AS1 expression levels in different subgroups. Notes: The subgroups include tumor grades G1/G2, G3/G4, stage I/II, stage III/IV, males, females, and younger and older patients.
To better verify these results, we also performed a Cox regression analysis (Table 3). A univariate Cox regression analysis showed that patients with high ACVR2B-AS1 expression levels had significantly poorer outcomes (HR = 2.03, 95% CI (1.4–2.94)) compared with those with low ACVR2B-AS1 expression levels. Other common clinical parameters were also analyzed, and tumor clinical stage, together with T classification, and residual tumor were shown to be unfavorable factors for patient survival. To rule out possible interference among the variables, we performed a multivariate Cox regression analysis, which confirmed that the adverse effects of high ACVR2B-AS1 expression, T classification, and residual tumor remained significant. Together, these data demonstrated that high ACVR2B-AS1 expression (HR = 1.90, 95% CI (1.31–2.76)) is an independent prognostic factor for OS in liver cancer patients.
Table 3.
Univariate and multivariate analysis of overall survival in patients with liver cancer.
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95%CI (lower∼upper) | p value | Hazard ratio | 95% CI (lower-upper) | p value | |
| Age | 1.02 | 0.7–1.48 | 0.926 | |||
| Gender | 0.82 | 0.57–1.16 | 0.263 | |||
| Histological type | 0.98 | 0.27–3.63 | 0.982 | |||
| Histologic grade | 1.05 | 0.85–1.31 | 0.651 | |||
| Stage | 1.38 | 1.15–1.65 | 0.001 | 0.90 | 0.72–1.11 | 0.325 |
| T Classification | 1.65 | 1.38–1.98 | ≤0.001 | 1.72 | 1.36–2.16 | ≤0.001 |
| N classification | 0.71 | 0.5–1.03 | 0.071 | |||
| M Classification | 0.70 | 0.48–1.02 | 0.061 | |||
| Radiation therapy | 0.52 | 0.26–1.03 | 0.061 | |||
| Residual tumor | 1.42 | 1.12–1.79 | 0.004 | 1.46 | 1.14–1.87 | 0.003 |
| ACVR2B-AS1 | 2.03 | 1.4–2.94 | ≤0.001 | 1.90 | 1.31–2.76 | 0.001 |
3.5. High ACVR2B-AS1 Expression Predicts Shorter RFS in Liver Cancer
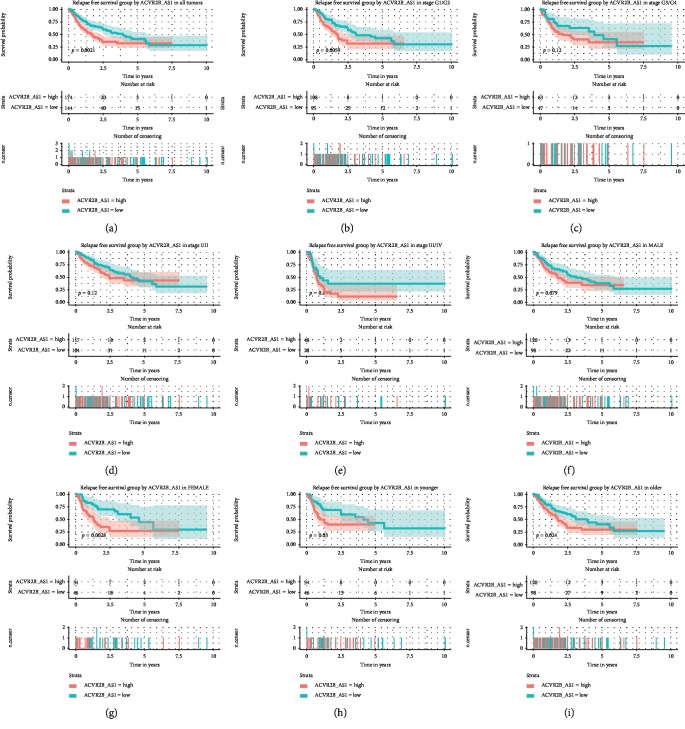
We next evaluated the prognostic value of ACVR2B-AS1 expression level for relapse-free survival (RFS) (Figure 4). The log-rank test showed that patients with high ACVR2B-AS1 expression levels had significantly shorter RFS (p=0.0021). When we analyzed this parameter within the different categories, we found that although high ACVR2B-AS1 expression levels predicted poorer RFS in patients at histological stage I (p=0.0059), this effect failed to reach statistical significance in stage III patients (p=0.12). There were no significant differences in RFS between high and low ACVR2B-AS1 patients in either the stage I or stage III groups, respectively. It was interesting to see that when taking sex into consideration, female patients with higher ACVR2B-AS1 expression levels had much shorter RFS than those females with lower expression levels (p=0.0028), although no difference was found in males. In both the young and old subgroups, patients with higher ACVR2B-AS1 expression levels had shorter times before relapse.
Figure 4.
Relapse-free survival outcomes based on different ACVR2B-AS1 expression levels in different subgroups. Notes: The subgroups include tumor grades G1/G2, G3/G4, stage I/II, stage III/IV, males, females, and younger and older patients.
Consistent with the aforementioned findings, a univariate Cox regression analysis showed that high ACVR2B-AS1 expression is an unfavorable prognostic factor for RFS (HR = 1.71, 95% CI [1.21–2.41]) (Table 4). An additional multivariate Cox analysis confirmed that ACVR2B-AS1 is an independent adverse prognostic factor for RFS, and the adjusted hazard ratio was 1.66 (p=0.005).
Table 4.
Univariate and multivariate analysis of relapse-free survival in patients with liver cancer.
| Parameters | Univariate analysis | Multivariate analysis | ||||
|---|---|---|---|---|---|---|
| Hazard ratio | 95%CI (lower∼upper) | p value | Hazard ratio | 95% CI (lower-upper) | p value | |
| Age | 0.89 | 0.63–1.27 | 0.521 | |||
| Gender | 0.98 | 0.69–1.4 | 0.919 | |||
| Histological type | 2.03 | 0.66–6.29 | 0.218 | |||
| Histologic grade | 0.98 | 0.8–1.21 | 0.873 | |||
| Stage | 1.66 | 1.38–1.99 | ≤0.001 | 1.16 | 0.9–1.5 | 0.259 |
| T Classification | 1.78 | 1.49–2.12 | ≤0.001 | 1.57 | 1.2–2.04 | 0.001 |
| N classification | 0.98 | 0.68–1.42 | 0.926 | |||
| M Classification | 1.19 | 0.8–1.78 | 0.394 | |||
| Radiation therapy | 0.75 | 0.26–2.17 | 0.592 | |||
| Residual tumor | 1.27 | 1.01–1.61 | 0.042 | 1.39 | 1.09–1.76 | 0.007 |
| ACVR2B-AS1 | 1.71 | 1.21–2.41 | 0.002 | 1.66 | 1.17–2.37 | 0.005 |
4. Discussion
Liver cancer is one of the most life threatening cancers, and most people are diagnosed at later stages when surgical resection is not an option [3]. A refined staging system and more effective prognostic assessment is not only necessary for the selection of the best treatment for individual patients, but also important for designing clinical trials and coordinating the exchange of information between researchers with comparable criteria [1]. The old TNM staging system has gone out of fashion because establishing prognosis in liver cancer patients is highly sensitive to the patients' liver functions, the chosen treatment, and other factors. Although there are several staging systems currently being tested, most of which include tumor status, liver function, patient status, and treatment responses, a more comprehensive system is urgently needed. Our previous work revealed several RNA biomarkers that are associated with cancer prognosis and can help us refine cancer staging systems [13–18]. Here, using data extracted from the TCGA database, we identified a novel lncRNA, ACVR2B-AS1, whose upregulation is common in liver cancer, and found that higher ACVR2B-AS1 expression predicted poorer OS and RFS in liver cancer patients.
ACVR2B-AS1 is a newly annotated lncRNA. So far, only two studies using TCGA data have mentioned potential roles of ACVR2B-AS1 in cancers. One of these studies that focused on endometrial cancer (UCEC) found that ACVR2B-AS1 was upregulated in cancer samples. A combined lncRNA-focus expression signature (LFES) that included 10 additional lncRNAs functioned as a superior unfavorable prognostic factor (AUC = 0.887) [19]. The other study, however, identified ACVR2B-AS1 as an independent favorable prognostic factor in breast cancer [20]. Our study revealed that ACVR2B-AS1 is an adverse factor in liver cancer. It is possible that ACVR2B-AS1 may have different roles in distinct biological contexts.
Although controversial results have been reported from genome-wide studies indicating possible regulatory roles of ACVR2B-AS1, its functions have not been experimentally dissected [19, 20]. Recent accumulating evidence has indicated that neighboring/overlapping genes tend to show correlated expression in eukaryotic genomes, indicating possible local regulatory mechanisms [21, 22]. Thus, as ACVR2B-AS1 is an intragenic antisense long noncoding RNA that overlaps with the promoter region of the ACVR2B protein coding gene, it is possible that ACVR2B-AS1 might regulate its neighbor gene ACVR2B in a cis manner. For example, ACVR2B-AS1 transcription might disturb the activation of ACVR2B transcription via “transcription interference” [23]. Alternatively, ACVR2B-AS1 activation might promote the transition of the local chromatin into a relatively open state to enhance ACVR2B transcription [24]. ACVR2B encodes a protein called activin receptor type IIB, which is a major receptor of the transforming growth factor beta (TGF-β) signaling pathway with many functions. For example, binding of ACVR2B to its ligand activin can activate the transcription of genes that inhibit muscle growth and is probably responsible for cancer-related cachexia [25, 26]. ACVR2B has also been reported to be a member of the MALAT1/miR-194-5p/ACVR2B signaling axis, which promotes clear cell kidney carcinoma (KIRC) progression [27]. In our study, ACVR2B-AS1 was associated with poorer prognosis, although it remains unknown if and how ACVR2B-AS1 might regulate tumorigenesis and progression via the mechanisms we mentioned above. Further experiments are required to answer these remaining questions.
It was worth noting that liver functions, another prognostic factor, are not included into the multivariate COX regression analysis, since such data are not available in the TCGA database. Nonetheless, our work here has demonstrated the prognostic value of ACVR2B-AS1 in multivariate COX regression analysis and managed to raise the potential possibility of incorporating ACVR2B-AS1 into liver cancer staging. To better testify its independent value as a prognosticator, further studies are encouraged, for example, in another patients cohort where more detailed information is available.
5. Conclusions
Overall, our data reported here identified the lncRNA ACVR2B-AS1 as an independent adverse prognostic factor in liver cancer. Attempts to decipher and identify the clinical significance of the enormous information hidden in RNA-seq data and to characterize the roles of ACVR2B-AS1 are encouraged, which might help refine the staging system in the future, especially at a time when RNA-seq are becoming more prevalent and more data will become available. In addition, more attention should be paid to basic research on ACVR2B-AS1, which might be a potential therapeutic target for liver cancer.
Acknowledgments
This work was supported in part by the National Natural Science Foundation (81670143, 81071920, 30370439, and 81372835) in China.
Contributor Information
Yan Jiao, Email: lagelangri1@126.com.
Wei Li, Email: jdyylw@163.com.
Data Availability
The data used to support the findings of this study are included within the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Galle P. R., Forner A., Llovet J. M. EASL clinical practice guidelines: management of hepatocellular carcinoma. Journal of Hepatology. 2018;69(1):182–236. doi: 10.1016/j.jhep.2018.03.019. [DOI] [PubMed] [Google Scholar]
- 2.Siegel R. L., Miller K. D., Jemal A. Cancer statistics, 2019. CA: A Cancer Journal for Clinicians. 2019;69(1):7–34. doi: 10.3322/caac.21551. [DOI] [PubMed] [Google Scholar]
- 3.Liu C.-Y., Chen K.-F., Chen P.-J. Treatment of liver cancer. Cold Spring Harbor Perspectives in Medicine. 2015;5(9) doi: 10.1101/cshperspect.a021535.a021535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kopp F., Mendell J. T. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Djebali S., Davis C. A., Merkel A., et al. Landscape of transcription in human cells. Nature. 2012;489(7414):101–108. doi: 10.1038/nature11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huarte M. The emerging role of lncRNAs in cancer. Nature Medicine. 2015;21(11):1253–1261. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 7. National Center for Biotechnology Information, https://www.ncbi.nlm.nih.gov/
- 8.Li B., Dewey C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):p. 323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.R Foundation for Statistical Computing. Team RDCJC. R.: A Language and Environment for Statistical Computing. Vol. 14. Vienna, Austria: R Foundation for Statistical Computing; 2009. [Google Scholar]
- 10.Robin X., Turck N., Hainard A., et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12:p. 77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Therneau T. M., Grambsch P. M. Modeling Survival Data: Extending the Cox Model. Vol. 97. New York, NY, USA: Springer; 2000. [Google Scholar]
- 12.Wickham H. Ggplot2: elegant graphics for data analysis. Journal of the Royal Statistical Society. 2011;174(1):245–246. [Google Scholar]
- 13.Jiao Y., Fu Z., Li Y., Meng L., Liu Y. High EIF2B5 mRNA expression and its prognostic significance in liver cancer: a study based on the TCGA and GEO database. Cancer Management and Research. 2018;10:6003–6014. doi: 10.2147/cmar.s185459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiao Y., Fu Z., Li Y., Zhang W., Liu Y. Aberrant FAM64A mRNA expression is an independent predictor of poor survival in pancreatic cancer. PLoS One. 2019;14(1) doi: 10.1371/journal.pone.0211291.e0211291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao Y., Li Y., Lu Z., Liu Y. High trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Digestive Diseases and Sciences. 2019;64(1):137–143. doi: 10.1007/s10620-018-5315-x. [DOI] [PubMed] [Google Scholar]
- 16.Jiao Y., Li Y., Jiang P., Han W., Liu Y. PGM5: a novel diagnostic and prognostic biomarker for liver cancer. PeerJ. 2019;7 doi: 10.7717/peerj.7070.e7070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao Y., Li Y., Liu S., Chen Q., Liu Y. ITGA3 serves as a diagnostic and prognostic biomarker for pancreatic cancer. OncoTargets and Therapy. 2019;12:4141–4152. doi: 10.2147/ott.s201675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Jiao Y., Fu Z., Luo Z., Su J., Li Y. High miR-454-3p expression predicts poor prognosis in hepatocellular carcinoma. Cancer Management and Research. 2019;11:2795–2802. doi: 10.2147/cmar.s196655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou M., Zhang Z., Zhao H., Bao S., Sun J. A novel lncRNA-focus expression signature for survival prediction in endometrial carcinoma. BMC Cancer. 2018;18(1):p. 39. doi: 10.1186/s12885-017-3983-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pang B., Wang Q., Ning S., et al. Landscape of tumor suppressor long noncoding RNAs in breast cancer. Journal of Experimental & Clinical Cancer Research. 2019;38(1):p. 79. doi: 10.1186/s13046-019-1096-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghanbarian A. T., Hurst L. D. Neighboring genes show correlated evolution in gene expression. Molecular Biology and Evolution. 2015;32(7):1748–1766. doi: 10.1093/molbev/msv053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ning Q., Li Y., Wang Z., Zhou S., Sun H., Yu G. The evolution and expression pattern of human overlapping lncRNA and protein-coding gene pairs. Scientific Reports. 2017;7(1):p. 42775. doi: 10.1038/srep42775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Latos P. A., Pauler F. M., Koerner M. V., et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science. 2012;338(6113):1469–1472. doi: 10.1126/science.1228110. [DOI] [PubMed] [Google Scholar]
- 24.Wang K. C., Yang Y. W., Liu B., et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi: 10.1038/nature09819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nissinen T. A., Hentila J., Penna F., et al. Treating cachexia using soluble ACVR2B improves survival, alters mTOR localization, and attenuates liver and spleen responses. Journal of Cachexia, Sarcopenia and Muscle. 2018;9(3):514–529. doi: 10.1002/jcsm.12310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao X., Zhao P., Hu J., et al. MicroRNA-194 protects against chronic hepatitis B-related liver damage by promoting hepatocyte growth via ACVR2B. Journal of Cellular and Molecular Medicine. 2018;22(9):4534–4544. doi: 10.1111/jcmm.13714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ye Y., Zhang F., Chen Q., Huang Z., Li M. LncRNA MALAT1 modified progression of clear cell kidney carcinoma (KIRC) by regulation of miR-194-5p/ACVR2B signaling. Molecular Carcinogenesis. 2019;58(2):279–292. doi: 10.1002/mc.22926. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article.