Abstract
Charcot-Marie-Tooth (CMT) neuropathies are amongst the most common inherited diseases in neurology. While great strides have been made to identify the genesis of these diseases, a diagnostic gap of 30–60% remains. Classic models of genetic causation may be limited to fully close this gap and, thus, we review the current state and future role of alternative, non-Mendelian forms of genetics in CMT. Promising synergies exist to further define the full genetic architecture of inherited neuropathies, including affordable whole-genome sequencing, increased data aggregation and clinical collaboration, improved bioinformatics and statistical methodology, and vastly improved computational resources. Given the recent advances in genetic therapies for rare diseases, it becomes a matter of urgency to diagnose CMT patients with great fidelity. Otherwise, they will not be able to benefit from such therapeutic options, or worse, suffer harm when pathogenicity of genetic variation is falsely evaluated. In addition, the newly identified modifier and risk genes may offer alternative targets for pharmacotherapy of inherited and, potentially, even acquired forms of neuropathies.
Genetics of Charcot-Marie-Tooth Disease
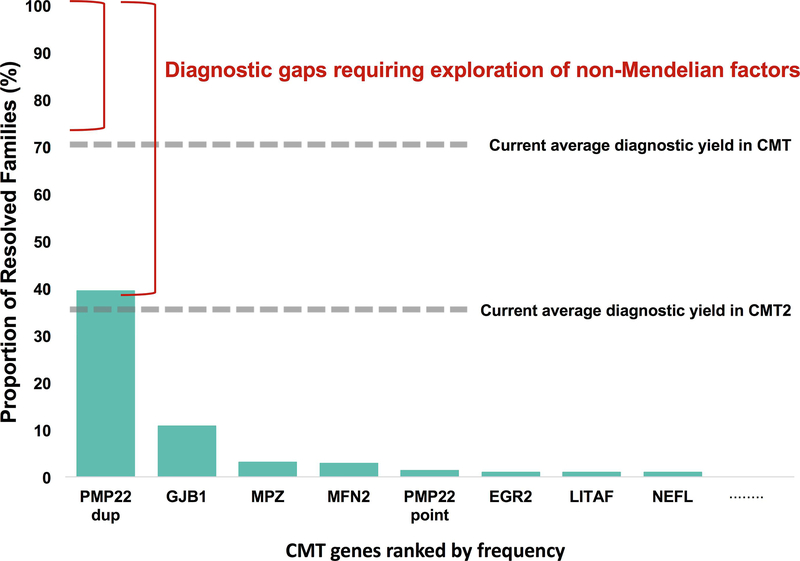
As with many other Mendelian diseases, the introduction of next generation sequencing (NGS) revolutionized the genetic diagnosis of Charcot-Marie-Tooth disease with now over 90 known genes causing CMT.1,2 Though CMT can be caused by an overwhelming amount of genetic defects, it is noteworthy that a handful of genes are responsible for the majority of cases. More than half of all CMT cases are caused by five genetic mutations: PMP22 duplication (39.5%), PMP22 point mutation (1.4%), GJB1 (10.8%), MFN2 (2.8%), and MPZ (3.1%).3 For autosomal dominant (AD) demyelinating CMT (CMT1), the most commonly mutated genes are: GJB1, PMP22, MPZ, EGR2, LITAF, NEFL, or PMP2.3 Unlike CMT1, AD axonal CMT (CMT2) and autosomal recessive (AR) axonal and/or demyelinating forms (CMT4) are caused by many, individually rare, genes that typically affect only a handful of families.3,4 The most common cause of CMT2 (~20%) is mutations within the outer mitochondrial membrane protein, MFN25, while the most common CMT4 genes are GDAP16 and SH3TC2.4,7
In the past 5 years alone, >20 CMT genes have been reported. However, the number of families identified in the initial and follow-on papers is typically low; for example, Tey et al recently identified AHNAK2 to be the causative gene for AR CMT in a single family, and analysis of whole exome sequencing (WES) from 115 unsolved families led to one additional family demonstrating segregation of a different variant in the same gene.8 In 2013, Gonzalez et al reported a single family with a mutation in MARS causative for CMT2 with no additional evidence in 400 additional families. Three additional families have since been identified harboring likely pathogenic variants in this gene.9,10 This has led to the concern that the field is in an asymptotic phase where, even with many new genes, the diagnostic yield may not reach 100%. This potential diagnostic gap in heritability is observed in other rare disorders as well and might be referred to as “dark matter” of clinical genomics.
The “Dark Matter” of Clinical Genomics
It is usually assumed that CMT is caused by Mendelian mechanisms and that eventually nearly all patients will receive a single-gene diagnosis. However, this is not necessarily true. Related motorneuron disorders presently illustrate a diverse situation: inherited ataxias and spastic paraplegias are also highly heterogeneous Mendelian disorders whereas amyotrophic lateral sclerosis (ALS) is largely not explained by Mendelian genes. The proportion of Mendelian genes is even lower in late onset neurodegenerative diseases, such as Parkinson and Alzheimer disease.
The reported diagnostic yield for exome sequencing in the general clinical setting ranges from 25 to 50%.11,12 In CMT, Fridman et al reported a 60.4% (997/1652 patients) genetic diagnosis rate: while 91.2% of CMT1 patients received a genetic diagnosis, only 42% of CMT2 cases received genetic confirmation.13 Bacquet et al report an overall diagnosis rate of 40% (49/123 patients), with 87% of CMT1 cases and 34% of CMT2 cases being solved.14 Finally, genetic testing of 1,206 patients at a laboratory in Aachen, Germany revealed a genetic diagnosis in 56% of demyelinating CMT cases and 17% of axonal CMT cases.15
Several valid pro-Mendelian hypotheses exist as to how to close the diagnostic gap, and the coming years will allow us to test these ideas (Fig. 1). These include non-coding regions of the genome, unorthodox types of mutations (such as repeat expansions) and digenic inheritance models (Fig. 2). However, while the genetics of certain neurodegenerative diseases are deemed ‘complex’, similar models of inheritance are largely unexplored in CMT. Although it is too early to tell the extent of non-Mendelian effects in CMT, increasing observations of non-Mendelian inheritance support further consideration and investigation.
Figure 1.
The gaps in genetic diagnosis in CMT. While novel gene discovery for CMT has been very successful over the past decade, the average diagnostic yield remains under 40% for CMT2 and under 75% for all CMT. Close to 60% of genetically resolved CMT families can be attributed to less than 10 genes, yet over 90 loci are associated with the disease. Many of these loci resolve a small percentage of CMT – sometimes as few as one or two families. Since additional Mendelian disease genes alone have not closed these diagnostic gaps, exploration of non-Mendelian factors is necessary.
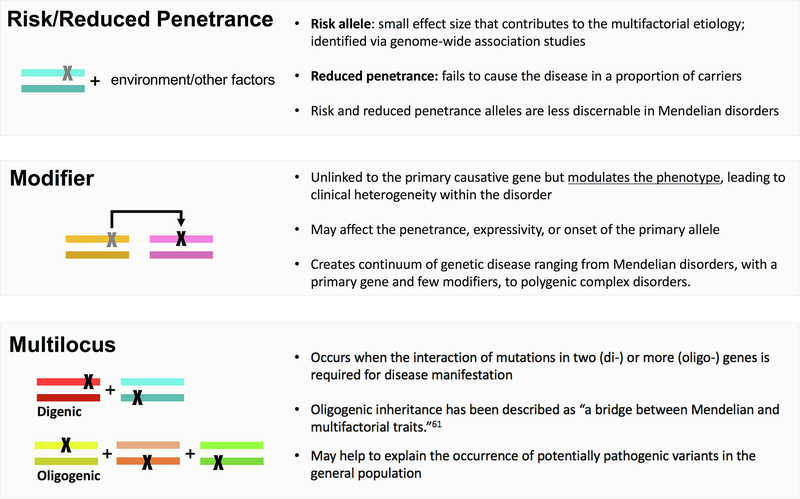
Figure 2.
Each box defines a non-Mendelian phenomenon that is currently investigated in CMT and covered within this review, including risk/reduced penetrance alleles, modifier alleles, and mutlilocus inheritance. Colored rectangles represent diploid genes and X’s represent pathogenic variation.
Beyond Mendelian Inheritance
Based on the assumption of fully penetrant alleles, traditional Mendelian disease analysis focuses on the rare DNA variation that segregates within a family. However, these locus-specific family studies treat Mendelian traits as distinct entities and disregard a more comprehensive genetic model for human disease in which variants of varying effect size as well as environmental influences contribute to disease.16 The challenge is the unexpected large amount of variation in the human genome on a population level, where >99% of all variants show a minor allele frequency of <1%.17 While many of these variants are without phenotypic consequence, some certainly are very harmful, and a considerable number must have effect sizes that are below the threshold of a Mendelian gene, but contribute significantly to phenotypic expression. Identification of strong effect sizes in the background of mostly minor effects is a rising challenge in human genetics. Recent method developments in statistical genetics allow for unbiased genome-wide screens for non-Mendelian alleles, and surprisingly, are able to re-identify bona fide Mendelian genes as well.18 The application of such methods to CMT genomics will eventually generate a more complete genetic architecture of the disease.
Reduced penetrant and risk alleles
Contrary to general expectations for CMT families, asymptomatic carriers are not infrequent, in which case, the genotype is said to be incompletely penetrant (Fig. 2).19,20 Reduced and age-dependent penetrance is a diagnostically challenging situation observed in autosomal dominant CMT, which can lead to misinterpretation of inheritance patterns due to asymptomatic carriers and exclusion of the disease-causing allele.21 Incomplete penetrance can also manifest in autosomal recessive disorders when the primary mutation leads to varying phenotypic effects depending on the secondary mutation.19 For example, recessive mutations in SCO2 typically result in fatal infantile cardioencephalomyopathy; however, Rebelo et al reported a less severe CMT phenotype resulting from the specific location and compound combination of observed biallelic pathogenic variants.22,23
Risk alleles are another form of variation that does not conform to standard Mendelian inheritance (Fig. 2). Risk alleles have been defined as variants with smaller effect sizes that are part of a multifactorial model of disease causation.19 However, since the possibility of risk alleles is only recently recognized in rare Mendelian disease, the line between penetrance and risk is often blurred. In this context, risk alleles more broadly refer to rare variants that may lead to a less severe, later-onset form of disease or contribute to an individual’s susceptibility to disease, likely through an oligogenic or gene-environment model. For example, heterozygous mutations in MME were recently shown to predispose carriers to late-onset axonal neuropathy.24 Late-onset axonal neuropathy is an autosomal dominant disorder with an age-of-onset in the second half of life.24 In MME, the ‘rare variant load’ of missense and loss of function changes in late-onset CMT compared to the general population showed a significant enrichment of such variation, indicating MME may act as a risk gene.24 This gene-wide statistical measure, however, does not easily translate to the assessment of pathogenicity of individual MME alleles. A recent manuscript by Senderek et al aimed at exploring individual allelic pathogenicity in MME (Lancet Neurology, under review). In addition, specific heterozygous variants in MME demonstrated penetrance in central European families, but non-penetrance in Spanish and Japanese families. Further work remains to explain this diversity in penetrance amongst different ethnic groups, which could possibly be due to genetic modifiers or environmental effects.47
Systematic identification of rare variant associations are usually limited by low statistical power unless sample sizes or variant effect sizes are very large.25 To illustrate, >60,000 cases (and an equal number of controls) would be necessary to detect a disease association for an individual rare variant (0.1% frequency) with an odds ratio of 2.0 for a disease with a 5% population prevalence.25 Fortunately, powerful study designs can alleviate the sample size requirement to more reasonable numbers.26 One approach that can be explored in CMT is the gene-based variant burden test, which collapses the number of minor alleles into one genetic score per gene, thus reducing multiple testing and increasing power.27 One successful example of this approach was the identification of a new ALS gene, TBK1, in 2,869 sporadic ALS patients.18 Remarkably, other known ALS genes showed strong associations, indicating that additional variation in known familial ALS genes also contribute to sporadic ALS forms.18 This exome-wide study design was recently applied in a cohort of 343 CMT cases and resulted in the identification of a strong association with EXOC4 (p-value = 6.9 × 10−6, OR = 2.1).28 EXOC4 is involved in vesicle transport and membrane tethering in polarized cells, is expressed in Schwann cells, and is involved in myelination in a CMT4B1 mouse model.29 Similar to the ALS study, bona fide CMT genes were also nominally associated despite an effort to exclude patients with common forms of CMT from exome analysis.28 The gene-based burden tests is not without limitations. Systematic evaluations have shown that they are sensitive to several variables including analysis unit (e.g. exon versus whole gene), the number and functional class of variants tested within an aggregation, and the magnitude of linkage disequilibrium between variants.
Modifier alleles
An increasing number of exceptions to the fundamental “one gene, one phenotype” paradigm are being published across Mendelian phenotypes.30 The oversimplified view that phenotypic expression, even for classically monogenic disorders, is driven exclusively by mutations at a single locus is being replaced by the concept of genetic modification (Fig. 2).31 Though several types of genetic modification are possible, the simple definition is the effect of one allele on the phenotypic outcome of a second allele.31 If the primary allele is sufficient to cause disease, then the secondary allele is a “modifier” that modulates phenotypic expression, such as disease severity or progression. Modifier alleles can interact both directly or indirectly with the primary gene to exacerbate or reduce the phenotype in a non-additive manner. The resulting phenotype is caused by the primary gene, modifier gene, environmental factors, and their interactions.
Given the high clinical variability among CMT cases with the same genetic subtype, genetic modification of the primary allele was anticipated and has been observed in several instances. For example, additional copies of the PMP22 gene act as a genetic modifier in several CMT1A cases, resulting in a severe phenotype.32–34 Additionally, missense variants in the LITAF gene contribute to a more severe and earlier onset form of CMT1A.35,36 About 80% of newly formed PMP22 is degraded due to incorrect folding, and LITAF is thought to play a role in protein degradation, which could explain the exacerbation of the CMT1A phenotype in patients with defective LITAF.35,36 Nam et al reported a polymorphism in microRNA 149 was associated with onset age and severity in CMT1A.37 Finally, variants in JPH1 lead to a more severe clinical expression of CMT2K, as the modifier and GDAP1 share a common cellular pathway.38
Most of these examples were observed in individual families and thus represent a small portion of phenotypic variability. Systematic identification of genetic modifiers in rare disease is limited by the challenges of collecting a large enough sample for genomic studies. Study designs that include more informative individuals can improve the power of genomic studies within rare disease. One study design that increases the statistical power for association testing of rare variants is the extreme phenotype sampling (EPS) approach.26 Based on the assumption that rare causal variants are more likely found in the extremes of a quantitative trait such as age of onset or severity of a symptom, EPS can increase the power to detect rare variants over random sampling.26 For example, Tao et al identified SIPA1L2 as a genetic modifier of muscle strength impairment in CMT1A.39 In vitro knock down of SIPA1L2 in Schwannoma cells lead to a significant reduction in PMP22 expression, offering a potential pathway for therapeutic strategies.39 However, simulations have shown that EPS is not necessarily more powerful than a random sample when other environmental covariates have a strong impact on the phenotype, so it is important to consider the phenotype of interest when using this study design.64
Animal model studies also have large potential in contributing to the discovery of modifier genes in CMT and circumvent the challenge of requiring detailed phenotypic data for human genetic studies. Yeast genetic screens have already been successful in identifying modifiers in Parkinson’s disease (PD) and amyotrophic lateral sclerosis (ALS). For example, PBP1, the yeast ortholog of human ATXN2 (a polyglutamine protein mutated in spinocerebellar ataxia type 2), was identified as a dose-sensitive modulator of TDP-43 toxicity (a major cause of ALS).40 This enhanced toxicity was next confirmed in transgenic Drosophila lines expressing mutant TDP-43, where ATXN2 overexpression in motor neurons resulted in loss of motility.40 Treatment of mice with ATXN-2 anti-sense oligonucleotides dramatically increased survival, indicating that modifiers can be important therapeutic targets for human disease.51 Genetic modifiers of CMT have been suggested in several mouse models. In a CMT2D mouse model, mutated glycyl-transfer RNA synthetase led to aberrant binding of the neuropilin 1 (Nrp1) receptor, an essential receptor for motor neuron axon guidance and cell body migration. Genetic reduction of Nrp1 worsened the CMT phenotype within the model.41 In both CMT2D and CMT4C mouse models, homozygous neuronal cell adhesion molecule (NRCAM) or heterozygous sodium voltage gate channel 8A (SCN8A) mutations exacerbated the neuropathy phenotype. Both proteins are found at the nodes of Ranvier, and although these specific genes may not translate as modifiers in human disease, Morelli et al argue that any variants that affect the physiology at nodes could potentially affect the severity of CMT in humans.42 Nicholson et al report that individuals with identical genotypes in FIG4 causing CMT4J display vast differences in age of onset as well as in disease severity, suggesting a role of modifier alleles.62 Studies in Drosophila recently identified Hippo (hpo), the drosophila ortholog of MST1, as a modifier of CMT4J. Knockdown of dFIG4 in this model resulted in aberrant motor neuron morphology, a phenotype that was improved through the downregulation of hpo.63
Multilocus inheritance
Multilocus inheritance refers to instances when one primary allele is insufficient to cause disease, instead requiring the combined consequence of multiple alleles at multiple loci (Fig. 2).30 Multilocus inheritance is common in complex genetic diseases in which many genes with small effect sizes contribute to disease (polygenic), whereas occurrence of disease-causing variants in two distinct genes (digenic) is well-documented in Mendelian disease.43 Oligogenic inheritance affects more than two genes – but less than a polygenic disease – thus serving as a bridge between Mendelian and complex disease etiologies.
Digenic inheritance has been documented in CMT. In a Japanese cohort, digenic variants were identified in five cases: SETX and ARHGEF10, SH3TC2 and SACS, LRSAM1 and MARS, HARS and ARHGEF10, and MFN2 and PMP22.44 Chung et al reported a patient with a severe clinical presentation of CMT resulting from digenic inheritance of mutations in two known CMT genes: GJB1 and EGR2.45 Brusse et al described a three generation distal hereditary motor neuropathy (dHMN) family with both a BSCL2 mutation and a second disease locus on chromosome 16p.46 Affected individuals carried both alleles while one individual with sub-clinical motor neuron damage carried only the 16p locus. Lastly, possible digenic inheritance of dHMN and CMT2 was identified in one family. One parent transmitted a purely motor phenotype caused by a novel gene mutation while the other parent transmitted a mild CMT2 caused by a MFN2 mutation. The four affected offspring carrying both mutations were more severely affected with an earlier age-at-onset.47
Demonstrating oligogenic inheritance from family studies is challenging without experimental models; however, evidence of oligogenic inheritance has emerged in several neurological disorders. One trending approach to assessing oligogenic inheritance – which has been explored in Parkinson, ALS, Frontotemporal Dementia, Congenital Hypothyroidism, Inherited Neuropathy, and more – is to evaluate the mutational burden across known disease genes through Fisher’s exact test or logistic regression.48–52 For example, in both sporadic and familial ALS cases, patients harboring two or more rare variants had lower survival or earlier age at onset, suggesting that the combined effect of rare variants affects ALS development and progression.53–55 Similarly, over 30% of PD patients carried additional rare variants in Mendelian PD genes and had younger ages at onset.48 In CMT, an increased rare variant burden was observed in two cohorts of inherited neuropathy cases, which was followed up with in vivo zebrafish experiments.49 In zebrafish, more severe phenotypic outcomes were observed as a consequence of increased mutational burden in neuropathy genes, consistent with a positive genetic interaction mechanism of oligogenic inheritance.49 It is important to note that caution should be used with the ‘mutational burden’ approach as systematic bias can lead to the apparent enrichment of ‘oligogenic’ variants in familial cases, and control of such bias is essential for investigating an oligogenic role in neurodegenerative diseases.56
Multilocus variation has been shown to generate unusually severe phenotypes and apparent phenotypic expansion (clinical features beyond those typically reported with a known disease gene).57,58 In 2006, Hodapp et al identified three families with multiple neuromuscular diseases.57 In addition to PMP22 duplications/deletions, each family harbored mutations in other neuromuscular disease-related genes: (1) missense mutations in GJB1 causing a severe demyelinating CMT, (2) trinucleotide repeat expansion of DMPK causing myotonic muscular dystrophy and a more severe neuropathy, and (3) a mutation in ABCD1 causing adrenomyeloneuropathy and severe peripheral neuropathy. The authors termed this augmentative digenic effect as “double trouble” due to the unique phenotypic manifestations and concluded that individuals with mutations in multiple neuromuscular disease-related genes may develop more severe phenotypes.57 Høyer et al also observed this type of dual pathology in a sporadic case with CMT2 and spasticity who carried ‘likely pathogenic’ variants in SETX and REEP1.59 As phenotypic expansion and multiple molecular diagnoses have become more frequent, Karaca et al explored whether apparent phenotypic expansion at one known disease locus was actually the result of blended phenotypes from different loci.58 In a cohort of well-characterized neurodevelopmental phenotypes, the authors identified multilocus variation in 31.6% of families with phenotypic expansion and 2.3% of families without phenotypic expansion, emphasizing the importance of considering multilocus inheritance in apparent phenotypic expansion cases.
Concluding Remarks
Traditional family studies in Mendelian diseases have been very successful in identifying highly penetrant and medically actionable alleles, and next generation sequencing (NGS) has led to an explosion of novel disease genes. Despite great overall advancement, the diagnostic yield for particular genetic subtypes (e.g. simplex axonal CMT) remains surprisingly low. With the increasingly low numbers of additional affected families following a novel gene discovery, we advocate for the exploration of non-Mendelian contributors to the CMT genetic etiology in order to close the diagnostic gap. In this review, we summarized examples of reduced penetrance, risk alleles, modifier alleles, and multilocus inheritance in CMT and related disorders. Observation of such non-Mendelian factors in monogenic disorders provides further support for a “unified genetic model for human disease” to coalesce previously distinct disease entities as part of a continuum of genetic disease.16 Unbiased genomic interrogation has revealed how truly personal each genome is – containing common variants within the population, rare variants from more recent population substructure, new combinations of variants from both parents, and novel de novo variation in each individual.16 Though it may seem apparent that interactions between variants at multiple loci will impact phenotypic expression, these inheritance models still remain largely unexplored in many monogenic disorders. Recent efforts for data aggregation60 and collaboration13 are removing the common limitation of small sample size, which will allow the community to apply statistical approaches to CMT and rare disease overall. Though the extent to which non-Mendelian elements will contribute to the CMT genetic architecture remains unclear – and other possibilities exist to close the diagnostic gap not covered in this review, including the non-coding space and structural variation – the reviewed examples demonstrate that non-Mendelian inheritance will likely continue to emerge as a relevant factor.
Acknowledgements
The writing of this review was support by NIH grants (R01NS075764, U54NS065712), the Charcot-Marie-Tooth Association, the Hereditary Neuropathy Foundation, and the Muscular Dystrophy Association.
Footnotes
Disclosure
Nothing to disclose.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Reilly MM, Murphy SM & Laurá M Charcot-Marie-Tooth disease. J. Peripher. Nerv. Syst 16, 1–14 (2011). [DOI] [PubMed] [Google Scholar]
- 2.Timmerman V, Strickland A & Zuchner S Genetics of Charcot-Marie-Tooth (CMT) Disease within the Frame of the Human Genome Project Success. Genes 5, 13–32 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossor AM, Tomaselli PJ & Reilly MM Recent advances in the genetic neuropathies. Curr. Opin. Neurol 29, 537–548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tazir M, Hamadouche T, Nouioua S, Mathis S & Vallat J-M Hereditary motor and sensory neuropathies or Charcot–Marie–Tooth diseases: An update. Journal of the Neurological Sciences 347, 14–22 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Zuchner S et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat. Genet 36, 449–451 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Cuesta A et al. The gene encoding ganglioside-induced differentiation-associated protein 1 is mutated in axonal Charcot-Marie-Tooth type 4A disease. Nat. Genet 30, 22–25 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Senderek J et al. Mutations in a gene encoding a novel SH3/TPR domain protein cause autosomal recessive Charcot-Marie-Tooth type 4C neuropathy. The American Journal of Human Genetics 73, 1106–1119 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tey S et al. Linkage analysis and whole exome sequencing reveals AHNAK2 as a novel genetic cause for autosomal recessive CMT in a Malaysian family. Neurogenetics 20, 117–127 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Gonzalez M et al. Exome sequencing identifies a significant variant in methionyl-tRNA synthetase (MARS) in a family with late-onset CMT2. J. Neurol. Neurosurg. Psychiatr 84, 1247–1249 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sagi-Dain L et al. Whole-exome sequencing reveals a novel missense mutation in the MARS gene related to a rare Charcot-Marie-Tooth neuropathy type 2U. J. Peripher. Nerv. Syst 23, 138–142 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Chong JX et al. The Genetic Basis of Mendelian Phenotypes: Discoveries, Challenges, and Opportunities. The American Journal of Human Genetics 97, 199–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eldomery MK et al. Lessons learned from additional research analyses of unsolved clinical exome cases. Genome Medicine 9, 26 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fridman V et al. CMT subtypes and disease burden in patients enrolled in the Inherited Neuropathies Consortium natural history study: a cross-sectional analysis. J. Neurol. Neurosurg. Psychiatr 86, 873–878 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bacquet J et al. Molecular diagnosis of inherited peripheral neuropathies by targeted next-generation sequencing: molecular spectrum delineation. BMJ Open 8, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rudnik-Schöneborn S et al. Diagnostic algorithms in Charcot-Marie-Tooth neuropathies: experiences from a German genetic laboratory on the basis of 1206 index patients. Clin Genet 89, 34–43 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Lupski JR, Belmont JW, Boerwinkle E & Gibbs RA Clan genomics and the complex architecture of human disease. Cell 147, 32–43 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lek M et al. Analysis of protein-coding genetic variation in 60,706 humans. Nature 536, 285–291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cirulli ET et al. Exome sequencing in amyotrophic lateral sclerosis identifies risk genes and pathways. Science 347, 1436–1441 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper DN, Krawczak M, Polychronakos C, Tyler-Smith C & Kehrer-Sawatzki H Where genotype is not predictive of phenotype: towards an understanding of the molecular basis of reduced penetrance in human inherited disease. Hum. Genet 132, 1077–1130 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zlotogora J Penetrance and expressivity in the molecular age. Genet Med 5, 347–352 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Nakhro K, Park J-M, Choi B-O & Chung KW Missense mutations of mitofusin 2 in axonal Charcot-Marie-Tooth neuropathy: polymorphic or incomplete penetration? Animal Cells and Systems 17, 228–236 (2013). [Google Scholar]
- 22.Giannoccaro MP et al. Atypical late-onset hereditary spastic paraplegia with thin corpus callosum due to novel compound heterozygous mutations in the SPG11 gene. J. Neurol 261, 1825–1827 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Rebelo AP et al. SCO2 mutations cause early-onset axonal Charcot-Marie-Tooth disease associated with cellular copper deficiency. Brain 141, 662–672 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Auer-Grumbach M et al. Rare Variants in MME, Encoding Metalloprotease Neprilysin, Are Linked to Late-Onset Autosomal-Dominant Axonal Polyneuropathies. The American Journal of Human Genetics 99, 607–623 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kiryluk K Challenges in Rare Variant Association Studies for Complex Kidney Traits: CFHR5 and IgA Nephropathy. J. Am. Soc. Nephrol 27, 2547–2551 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barnett IJ, Lee S & Lin X Detecting rare variant effects using extreme phenotype sampling in sequencing association studies. Genet. Epidemiol. 37, 142–151 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tesson C, Koht J & Stevanin G Delving into the complexity of hereditary spastic paraplegias: how unexpected phenotypes and inheritance modes are revolutionizing their nosology. Hum. Genet. 1–28 (2015). doi: 10.1007/s00439015-1536-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bis DM et al. Rare variant burden analysis deciphers genetic architecture of inherited peripheral neuropathies. in [Google Scholar]
- 29.Bolis A et al. Dlg1, Sec8, and Mtmr2 Regulate Membrane Homeostasis in Schwann Cell Myelination. Journal of Neuroscience 29, 8858–8870 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katsanis N The continuum of causality in human genetic disorders. Genome Biology 17, 233–5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kousi M & Katsanis N Genetic Modifiers and Oligogenic Inheritance. Cold Spring Harbor Perspectives in Medicine 5, a017145–a017145 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sturtz FG et al. Clinical and electrophysiological phenotype of a homozygously duplicated Charcot-Marie-Tooth (type 1A) disease. Eur. Neurol 38, 26–30 (1997). [DOI] [PubMed] [Google Scholar]
- 33.LeGuern E et al. Patients homozygous for the 17p11.2 duplication in CharcotMarie-Tooth type 1A disease. Ann Neurol. 41, 104–108 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Liu P et al. Mechanism, prevalence, and more severe neuropathy phenotype of the Charcot-Marie-Tooth type 1A triplication. Am. J. Hum. Genet 94, 462–469 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meggouh F et al. Early onset neuropathy in a compound form of Charcot-Marie-Tooth disease. Ann Neurol 57, 589–591 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Sinkiewicz-Darol E et al. The LITAF/SIMPLE I92V sequence variant results in an earlier age of onset of CMT1A/HNPP diseases. Neurogenetics 16, 27–32 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nam SH et al. Association of miR-149 polymorphism with onset age and severity in Charcot-Marie-Tooth disease type 1A. Neuromuscul. Disord 28, 502–507 (2018). [DOI] [PubMed] [Google Scholar]
- 38.Pla-Martin D et al. Junctophilin-1 is a modifier gene of GDAP1-related Charcot-Marie-Tooth disease. Hum. Mol. Genet. 24, 213–229 (2015). [DOI] [PubMed] [Google Scholar]
- 39.Tao F et al. Variation in SIPA1L2is correlated with phenotype modification in Charcot- Marie- Tooth disease type 1A. Ann Neurol 85, 316–330 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Elden AC et al. Ataxin-2 intermediate-length polyglutamine expansions are associated with increased risk for ALS. Nature 466, 1069–U77 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.He W et al. CMT2D neuropathy is linked to the neomorphic binding activity of glycyl-tRNA synthetase. Nature 526, 710–714 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Morelli KH et al. Severity of Demyelinating and Axonal Neuropathy Mouse Models Is Modified by Genes Affecting Structure and Function of Peripheral Nodes. CellReports 18, 3178–3191 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schäffer AA Digenic inheritance in medical genetics. J. Med. Genet 50, 641–652 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yoshimura A et al. Genetic profile and onset features of 1005 patients with Charcot-Marie-Tooth disease in Japan. J. Neurol. Neurosurg. Psychiatr 90, 195–202 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chung KW et al. Two missense mutations of EGR2 R359W and GJB1 V136A in a Charcot-Marie-Tooth disease family. Neurogenetics 6, 159–163 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Brusse E et al. A novel 16p locus associated with BSCL2 hereditary motor neuronopathy: a genetic modifier? Neurogenetics 10, 289–297 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Drew AP Genetics of distal hereditary motor neuropathies. (2012). [Google Scholar]
- 48.Lubbe SJ et al. Additional rare variant analysis in Parkinson’s disease cases with and without known pathogenic mutations: evidence for oligogenic inheritance. Hum. Mol. Genet. 25, 5483–5489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gonzaga-Jauregui C et al. Exome Sequence Analysis Suggests that Genetic Burden Contributes to Phenotypic Variability and Complex Neuropathy. CellReports 12, 1169–1183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dols-Icardo O et al. Analysis of known amyotrophic lateral sclerosis and frontotemporal dementia genes reveals a substantial genetic burden in patients manifesting both diseases not carrying the C9orf72 expansion mutation. J. Neurol. Neurosurg. Psychiatr 89, 162–168 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Ferrari R et al. Genetic architecture of sporadic frontotemporal dementia and overlap with Alzheimer’s and Parkinson’s diseases. J. Neurol. Neurosurg. Psychiatr 88, 152–164 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Filippis T et al. A frequent oligogenic involvement in congenital hypothyroidism. Hum. Mol. Genet 26, 2507–2514 (2017). [DOI] [PubMed] [Google Scholar]
- 53.Pang SY-Y et al. Burden of rare variants in ALS genes influences survival in familial and sporadic ALS. Neurobiol. Aging 58, 238.e9–238.e15 (2017). [DOI] [PubMed] [Google Scholar]
- 54.Cady J et al. Amyotrophic lateral sclerosis onset is influenced by the burden of rare variants in known amyotrophic lateral sclerosis genes. Ann Neurol. 77, 100–113 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pang SY-Y et al. The role of gene variants in the pathogenesis of neurodegenerative disorders as revealed by next generation sequencing studies: a review. 1–11 (2017). doi: 10.1186/s40035-017-0098-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Keogh MJ et al. Oligogenic genetic variation of neurodegenerative disease genes in 980 postmortem human brains. J. Neurol. Neurosurg. Psychiatr 89, 813–816 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hodapp JA et al. Double trouble in hereditary neuropathy - Concomitant mutations in the PMP-22 gene and another gene produce novel phenotypes. Arch. Neurol. 63, 112–117 (2006). [DOI] [PubMed] [Google Scholar]
- 58.Karaca E et al. Phenotypic expansion illuminates multilocus pathogenic variation. Genet Med 20, 1528–1537 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Høyer H et al. Genetic diagnosis of Charcot-Marie-Tooth disease in a population by next-generation sequencing. BioMed Research International 2014, 210401–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gonzalez M et al. Innovative Genomic Collaboration Using the GENESIS (GEM.app) Platform. Hum. Mutat. 36, 950–956 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eichers E et al. Triallelic inheritance: a bridge between Mendelian and multifactorial traits. Ann. of Med. 36(4), 262–272 (2004). [DOI] [PubMed] [Google Scholar]
- 62.Nicholson G et al. (2011). Distinctive genetic and clinical features of CMT4J: a severe neuropathy caused by mutations in the PI(3,5)P2 phosphatase FIG4. Brain 134, 1959–1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kushimura Y et al. (2018). Loss-of-function mutation in Hippo suppressed enlargement of lysosomes and neurodegeneration caused by dFIG4 knockdown. Neuroreport 29, 856–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bjørnland T et al. Powerful extreme phenotype sampling designs and score tests for genetic association studies. Statistics in Medicine 37(28), 2018. [DOI] [PubMed] [Google Scholar]