Abstract
Background
The treatment of latent tuberculosis infection (LTBI) in individuals at risk of reactivation is essential for tuberculosis control. However, blood biomarkers associated with LTBI treatment have not been identified.
Methods
Blood samples from tuberculin skin test (TST) reactive individuals were collected before and after one and six months of isoniazid (INH) therapy. Peripheral mononuclear cells (PBMC) were isolated, and an in-house interferon-γ release assay (IGRA) was performed. Expression of chemokine ligand 4 (CCL4), chemokine ligand 10 (CXCL10), chemokine ligand 11 (CXCL11), interferon alpha (IFNA), radical S-adenosyl methionine domain-containing 2 (RSAD2), ubiquitin-specific peptidase 18 (USP18), interferon-induced protein 44 (IFI44), interferon-induced protein 44 like (IFI44L), interferon-induced protein tetratricopeptide repeats 1(IFIT1), and interleukin 2 receptor subunit alpha (IL2RA) mRNA levels were assessed by qPCR before, during, and after INH treatment.
Results
We observed significantly lower relative abundances of USP18, IFI44L, IFNA, and IL2RA transcripts in PBMC from IGRA-positive individuals compared to levels in IGRA-negative individuals before INH therapy. Also, relative abundance of CXCL11 was significantly lower in IGRA-positive than in IGRA-negative individuals before and after one month of INH therapy. However, the relative abundance of CCL4, CXCL10, and CXCL11 mRNA was significantly decreased and that of IL2RA and USP18 significantly increased after INH therapy, regardless of the IGRA result. Our results show that USP18, IFI44L, IFIT1, and IL2RA relative abundances increased significantly, meanwhile the relative abundance of CCL4, CXCL11, and IFNA decreased significantly after six months of INH therapy in TST-positive individuals.
Conclusions
Changes in the profiles of USP18, IL2RA, IFNA, CCL4, and CXCL11 expressions during INH treatment in TST-positive individuals, regardless of IGRA status, are potential tools for monitoring latent tuberculosis treatment.
1. Introduction
Despite substantial efforts, tuberculosis (TB) remains a significant health care problem worldwide, as TB is one of the top 10 causes of death and the leading cause of infection by a single infectious agent [1]. Of the 1.7 billion people infected with Mycobacterium tuberculosis (M. tuberculosis) worldwide, a relatively small proportion (5-10%) develop TB, and in most cases, the disease occurs within 2 years [2]. Among the remaining infected people, the immune system can control the infection, but fails to eliminate the bacteria. Patients with this condition lack clinical symptoms, and the isolation of the bacteria from infected individuals is not feasible. This condition is called latent tuberculosis infection (LTBI) and provides a reservoir for future active TB cases [3]. LTBI is considered a state of a persistent immune response to stimulation with M. tuberculosis antigens with no evidence of clinically active TB [3]. The immunological factors involved in the host immune response during LTBI and its reactivation have not been completely elucidated. However, LTBI can persist throughout a person's lifetime, and factors that reduce immunity, such as human immunodeficiency virus (HIV) infection, antitumor necrosis factor treatment, corticosteroid medication, chemotherapy, and diabetes increase the risk of developing active TB [4, 5]. Therefore, individuals with an increased risk of progression to active TB disease would benefit from testing and treatment of LTBI [6].
Although a gold standard test for LTBI is not available, two methods are used to infer LTBI: the tuberculin skin test (TST) and interferon-γ release assays (IGRA). The TST was the only screening tool for LTBI for over a century [7], though one of the major disadvantages of the TST is its lack of specificity, particularly in populations vaccinated with bacille Calmette-Guérin (BCG) [8]. IGRAs, including the QuantiFERON-TB test (QFT) and the T-SPOT.TB test, use the early secretory antigen-6 (ESAT-6) and culture filtrate protein-10 (CFP-10) proteins that are present in M. tuberculosis but not in most nontuberculous mycobacteria, particularly BCG [9]. The use of the TST and IGRA in a two-step approach has revealed discrepancies between the two tests [10]. Although most discordant results have been detected in the BCG-vaccinated population, in some cases, the discordance occurs independently of prior BCG vaccination [11].
The decision to perform a test for LTBI is accompanied by the intention to treat because the goal of LTBI treatment is to decrease the risk of developing active TB disease [6, 12]. The guidelines for LTBI treatment recommend that isoniazid (INH) should be administered to both adults and children daily at a dose of 5 mg/kg with a maximum dose of 300 mg/day over 6 months in countries with high and low TB incidence rates [3]. Recently, new schemes for LTBI treatment in children and adults from countries with high and low incidence rates of TB have been recommended, including rifampicin plus INH or rifapentine plus INH for three months. Additionally, the new recommendation includes an individualized risk assessment for preventive treatment of high-risk household contacts among patients with multidrug-resistant TB [3]. Several studies have reported changes in M. tuberculosis-specific T-cell responses during preventive therapy; however, the results are inconsistent [13–15]. Additionally, neither IFNγ levels nor the number of IFNγ-producing cells detected after INH treatment can be reliably associated with LTBI treatment [16–18]. Therefore, despite the efficacy of INH treatment, correlates indicating an effective response to LTBI treatment are lacking. Nonetheless, blood-based markers would be useful for both evaluating new regimens and developing of new treatments. Hence, to identify the treatment-associated transcript profile, we selected IFNα-inducible genes previously reported in active TB [19, 20] and genes assessed in LTBI and in models of infection [21–23] and during treatment of TB [24–27] and evaluated the expression profile of chemokine ligand 4 (CCL4), chemokine ligand 10 (CXCL10), chemokine ligand 11 (CXCL11), interferon alpha (IFNA), radical S-adenosyl methionine domain-containing 2 (RSAD2), ubiquitin-specific peptidase 18 (USP18), interferon-induced protein 44 (IFI44), interferon-induced protein 44 like (IFI44L), interferon-induced protein tetratricopeptide repeats 1(IFIT1), and interleukin 2 receptor subunit alpha (IL2RA) mRNAs in peripheral blood mononuclear cells (PBMC) from individuals with LTBI before, during, and after INH treatment. We found that changes in the expression of USP18, IL2RA, IFNA, CCL4, and CXCL11 during and after INH treatment represent a potentially useful tool for monitoring LTBI treatment.
2. Materials and Methods
2.1. Study Population and Inclusion Criteria
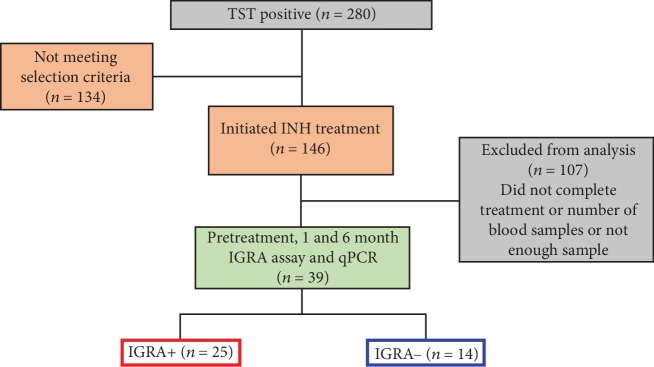
From 2008-2009, we recruited healthy, asymptomatic volunteers (n = 280) who were in close contact with patients with pulmonary TB, without clinical or radiological evidence of TB disease, aged 21 years or older, and reactive to TST (>10 mm) from reference health centers in Orizaba, Veracruz, México. The TB incidence of this region ranked among the ten highest in Mexico in 2016, with 27.4 cases per 100,000 inhabitants [28].
We excluded volunteers who were pregnant, immunosuppressed, HIV-positive, TST-negative, or diagnosed with other clinical conditions (n = 134). The participants received INH (5 mg per kg of body weight, up to 300 mg per day, for 6 months). We also excluded volunteers who did not complete treatment or refused to donate all samples or whose samples were insufficient (n = 107, Figure 1). Participants underwent clinical evaluation. Adherence was periodically monitored by measurement of INH metabolites in urine using Taxo-INH strips (Becton Dickinson, Sparks, MD, USA) and pill counting. Ten milliliters of blood was obtained for PBMC isolation at the following time points: pretreatment (t0), at one month (t1), and six months (t6) after the initiation of treatment. The blood samples were applied to a Lymphoprep (Axis-Shield PoC AS, Oslo, Norway) gradient and centrifuged to obtain PBMC. These cells were used for IGRA and preserved at -70°C until the qPCR assay was performed. Based on the results of the in-house IGRA, the participants were subclassified into IGRA-positive and IGRA-negative groups.
Figure 1.
Flow diagram of the study population.
All assays were performed by personnel blinded to the participant's status. Ethical approval for the protocol was obtained from the Ethics Committees of the Instituto Nacional de Enfermedades Respiratorias Ismael Cosío Villlegas and the Instituto Nacional de Salud Pública, Mexico, and written informed consent was obtained from all participants. The study was registered at https://ClinicalTrials.gov under NCT00293228.
2.2. LTBI
LTBI was defined by the reactivity of TST (>10 mm), and individuals were subclassified according to the in-house IGRA result for ESAT-6, CFP10, or both antigens.
2.3. TST
This test was performed using 0.1 ml (5 tuberculin units, UT) of tuberculin purified protein derivative (PPD) (RT-23; Statens Serum Institut, Copenhagen, Denmark). The result was evaluated at 48 h post-injection. A positive or reactive TST indicated TB infection and was defined as ≥10 mm diameter of induration according to the official Mexican policy for the prevention and control of TB (NOM-006-SSA2-1993) (http://www.salud.gob.mx/unidades/cdi/nom/006ssa23.html).
2.4. IGRA
The in-house IGRA was performed as previously described [29]. Briefly, PBMC were obtained from heparinized peripheral blood and resuspended in RPMI 1640 culture medium containing 2 mM L-glutamine (BioWhittaker, Radnor, PA, USA). The cells were counted using the trypan blue exclusion method, and cell viability was typically close to 93%. PBMC were plated in flat-bottomed, 96-well culture plates at a density of 2 × 105 cells per well and incubated with custom-designed overlapping synthetic peptides covering the protein sequences of the antigens ESAT-6 or CFP-10 (5 μg/ml, Peptide Synthetic UK and ProImmune, Oxford, UK), PPD (10 μg/ml, Statens Serum Institute,), phytohemagglutinin (PHA) (10 μg/ml, Sigma-Aldrich, St. Louis, MO, USA), and culture medium. After 6 days of cultivation at 37°C in a 5% CO2 atmosphere, the cell-free supernatants were collected, and IFNγ secretion was analyzed by ELISA. The detection limit of the IFNγ ELISA was 8 pg/ml. ESAT-6- or CFP10-specific IFNγ responses were considered positive when the values were > 100 pg/ml after subtracting the values obtained from unstimulated cells, as previously described [29]. A value of >100 pg/ml was defined as three-fold higher than the median (34 pg/ml) of unstimulated cells.
2.5. Real-Time Quantitative PCR (qPCR)
qPCR was used to evaluate the effect of INH therapy on the expression of selected genes (Table 1) [19–27]. Total RNA of 1 × 106 PBMC from individuals with LTBI before and after INH therapy was extracted with the RNeasy Mini Kit (Qiagen, CA, USA), and cDNAs were synthesized using a Superscript First-Strand set (Invitrogen, CA, USA). TaqMan Gene Expression Assays (Applied Biosystems, CA, USA), the TaqMan Universal Master Mix (Applied Biosystems, NJ, USA), and TaqMan probes for IFIT1 (Hs03027069_s1), IFI44 (Hs00951349_m1), IFI44L (Hs00915292_m1), IFNA (Hs003406429_gH), CXCL10 (Hs01124252_g1), CXCL11 (Hs04187682_g1), USP18 (Hs00276441_m1), IL2RA (Hs00907778_m1), RSAD2 (Hs00369813_m1), and CCL4 (Hs04421399_gH) and the endogenous control ribosomal 18S RNA(Hs999999001_s1) were used for qPCR. A StepOnePlus Real-Time PCR Systems (Applied Biosystems) was used to amplify the genetic material. Expression values for target genes were normalized to 18S expression, Ct values from all samples were obtained with the same software, and relative gene expression analysis was performed using the delta Ct method (2-ΔΔCt). The mRNA relative abundance was calculated as the Ct relative to 18S expression.
Table 1.
Genes selected for qPCR analysis.
| No | Gene | Name | Signaling pathway | Reported in TB | Reference |
|---|---|---|---|---|---|
| 1 | CCL-4 | C-C motif chemokine ligand 4 | Cytokine signaling in immune system, Toll-like receptor signaling pathway | Yes | [25] |
| 2 | CXCL-10 | C-X-C motif chemokine ligand 10 | Peptide ligand-binding receptors, Toll-like receptor signaling pathway, type II interferon signaling (IFNG) | Yes | [26, 27] |
| 3 | CXCL-11 | C-X-C motif chemokine ligand 11 | Peptide ligand-binding receptors, Toll-like receptor signaling pathway, type II interferon signaling (IFNG) | Yes | [24] |
| 4 | IFI44 | Interferon-induced protein 44 | Immune response IFN alpha/beta signaling pathway | Yes | [19] |
| 5 | IFI44L | Interferon-induced protein like 44 | Immune response IFN alpha/beta signaling pathway | Yes | [19, 20] |
| 6 | IFIT1 | Interferon-induced with tetraticopeptide repeats 1 | Immune response IFN alpha/beta signaling pathway | Yes | [23] |
| 7 | IFNA | Interferon alpha 10 | Immune response IFN alpha/beta signaling pathway | Yes | [19] |
| 8 | IL2RA | Interleukin 2 receptor subunit alpha | TGF-beta pathway, Th17 cell differentiation | Yes | [22] |
| 9 | RSAD2 | Radical S-adenosyl methionine domain-containing 2 | Immune response IFN alpha/beta signaling pathway | Yes | [23] |
| 10 | USP18 | Ubiquitin-specific peptidase 18 | Immune response IFN alpha/beta signaling pathway, antiviral mechanism by IFN-stimulated genes | Yes | [21] |
2.6. Statistical Analysis
GraphPad Prism software version 6.0 for Mac (GraphPad Software, San Diego, CA, USA) was used for statistical analyses. The details of the statistical tests are provided in the figure legends.
3. Results
3.1. Recruitment of Individuals with LTBI
We included thirty-nine participants who completed six months of INH treatment (5 mg/kg daily), as depicted in Figure 1. All individuals had a positive TST result averaging 12.8 mm. Table 2 shows the demographic data of the participants. The majority of the population (60.5%) was female. The average age was 39.2 years. The participants were divided into two groups based on the results of the in-house IGRA response to ESAT-6 or CFP-10 or both overlapping peptides as follows: IGRA-positive (n = 25) and IGRA-negative (n = 14) (Table 2).
Table 2.
Demographic data of the study population.
| IGRA+ | IGRA– | p | |
|---|---|---|---|
| n | 25 | 14 | |
| Gender/women (%) | 56.0 14/25 | 64.3 9/14 | NS |
| Age (years) | 39.9 ± 2.1 | 38.4 ± 3.4 | NS |
| TST induration (mm) | 13.1 ± 0.5 | 12.6 ± 0.6 | NS |
| IFNγ production to ESAT-6 (pg/mL) | 840 ± 351 | 4.7 ± 2.0 | 0.0033 |
| IFNγ production to CFP-10 (pg/mL) | 4779 ± 2798 | 17.7 ± 5.9 | 0.0063 |
| IFNγ production to PPD (pg/mL) | 19694 ± 3758 | 7246 ± 2543 | 0.0579 |
| IFNγ production to PHA (pg/mL) | 42816 ± 12169 | 26019 ± 6659 | 0.3167 |
3.2. Differentially Expressed Genes in IGRA-Positive and IGRA-Negative Individuals before INH Therapy Assessed by qPCR
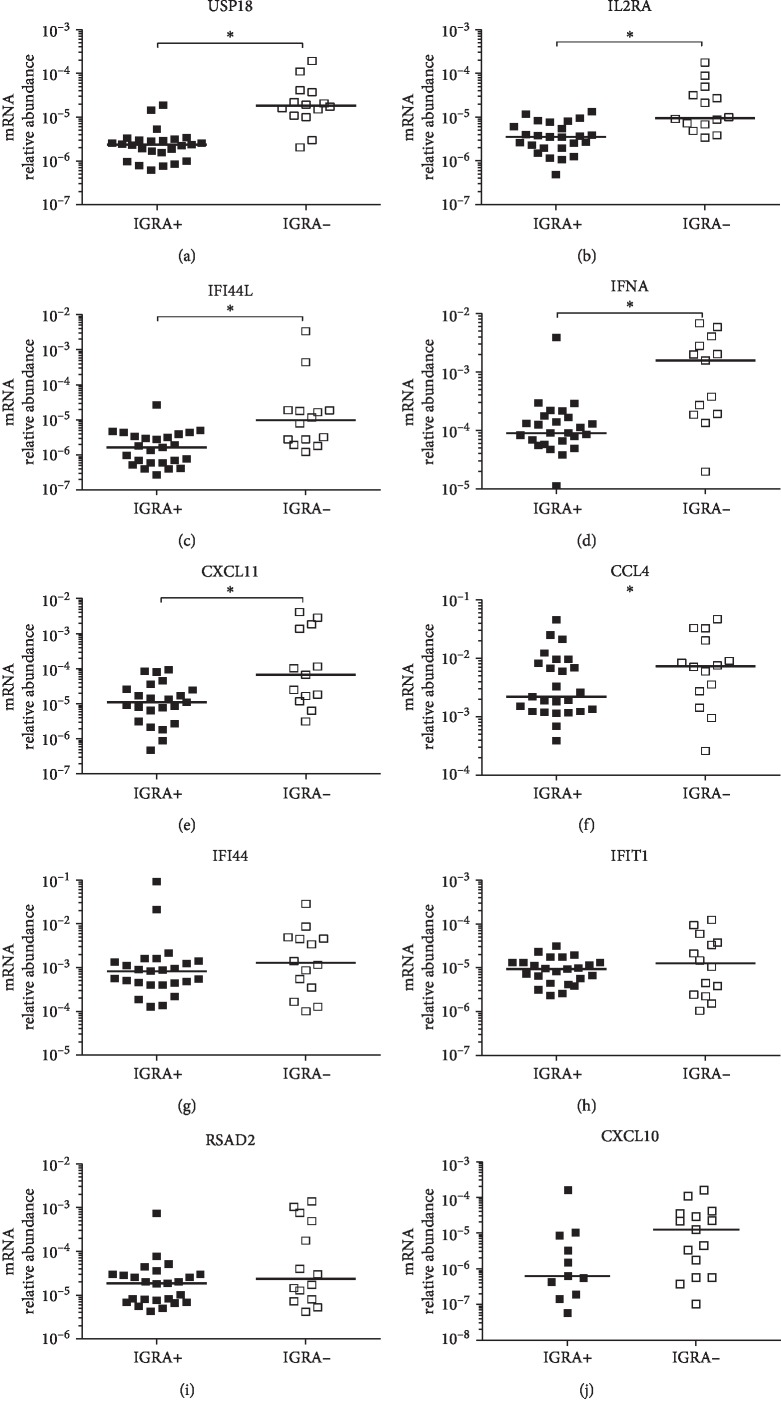
We first investigated whether there were differences in gene expression associated with IGRA status. Therefore, we determined the relative abundances of CCL4, CXCL10, CXCL11, IFNA, RSAD2, USP18, IFI44, IFI44L, IFIT1, and IL2RA transcripts in PBMC from IGRA-positive (n = 25) and IGRA-negative (n = 14) individuals before INH therapy. We observed significantly lower relative abundances of USP18, IL2RA, IFI44L, IFNA, and CXCL11 mRNAs among the IGRA-positive individuals compared to IGRA-negative individuals before INH therapy (t0) (Figures 2(a)–2(e)). Conversely, no significant differences in the relative abundances of the CCL4, IFI44, IFIT1, and RSAD2 mRNAs were observed between the IGRA-positive and IGRA-negative individuals before INH therapy (Figures 2(f)–2(i)). Interestingly, no significant difference in the relative abundance of CXCL10 mRNA was observed either; however, only 44% of IGRA-positive individuals expressed detectable levels of CXCL10 mRNA, whereas 93.75% of IGRA-negative individuals expressed this chemokine before INH therapy (Figure 2(j)).
Figure 2.
Relative levels of the CXCL11, IFNA, RSAD2, USP18, IFI44, IFI44L, IFITI, CCL4, and IL2RA mRNAs in PBMC from IGRA-positive (n = 25, black square) or IGRA-negative (n = 14, open square) individuals before INH treatment. Gene expression was determined by real-time qPCR. Values are presented as levels relative to 18S RNA. Individual results are displayed, and the line indicates the median. ∗ p < 0.05, Mann-Whitney U test.
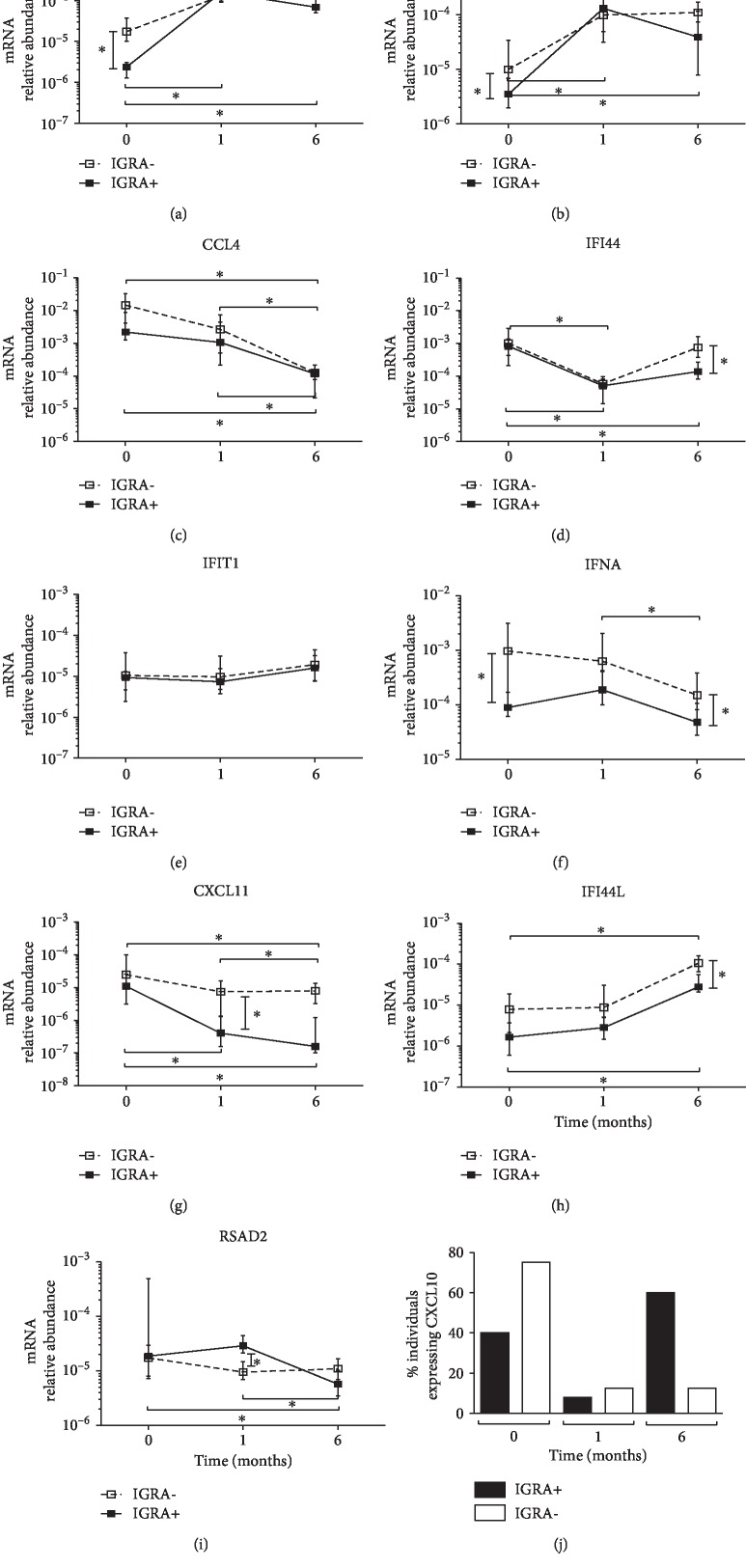
3.3. Differentially Expressed Genes in IGRA-Positive and IGRA-Negative Individuals after One and Six Months of INH Therapy Assessed by qPCR
We used qPCR to analyze the relative abundance of CCL4, CXCL10, CXCL11, IFNA, RSAD2, USP18, IFI44, IFI44L, IFIT1, and IL2RA mRNAs in PBMC from IGRA-positive and IGRA-negative individuals before initiation and after one and six months of INH therapy. USP18 and IL2RA expressions significantly increased in both IGRA-positive and IGRA-negative individuals after one and six months of INH therapy compared to the levels before INH therapy (Figures 3(a) and 3(b)). In contrast, CCL4 expression significantly decreased among all individuals regardless of their IGRA status after six months of INH therapy (Figure 3(c)). IFI44 expression significantly decreased among all individuals after one month of INH therapy (Figure 3(d)). Expression of IFIT1 did not show significant changes after INH therapy (Figure 3(f)). Interestingly, IFNA expression was significantly higher in IGRA-negative individuals than in IGRA-positive individuals before INH therapy, and this difference was also significant after six months of INH therapy (Figure 3(e)). CXCL11 expression was decreased in both IGRA-positive and IGRA-negative individuals after six months of INH therapy, but the decrease was significantly higher among the IGRA-positive individuals after one month of INH therapy (Figure 3(g)). Strikingly, CXCL10 expression dynamics were challenging to follow in the same manner because the percentage of individuals of both groups expressing this chemokine diminished dramatically after one month of treatment, and only IGRA-positive individuals showed increased CXCL10 expression after six months of treatment (Figure 3(j)).
Figure 3.
CXCL11, IFNA, RSAD2, USP18, IFI44, IFI44L, IFITI, CCL4, and IL2RA expressions in PBMC from IGRA-positive individuals (n = 25, black square) or IGRA-negative (n = 14, open square) individuals before and after one and six months of INH treatment. Gene expression was assessed by qPCR. (a–i) The median interquartile range of gene expression relative to that of 18S RNA after one and six months of treatment is reported. The statistical analysis was performed using the nonparametric Kruskal-Wallis ANOVA followed by Dunn's test; ∗p < 0.05. (j) CXCL10 expression detected in PBMC from IGRA-positive individuals (n = 11) and IGRA-negative individuals (n = 12) before and after one and six months of INH treatment is presented as the percentage of individuals expressing the gene.
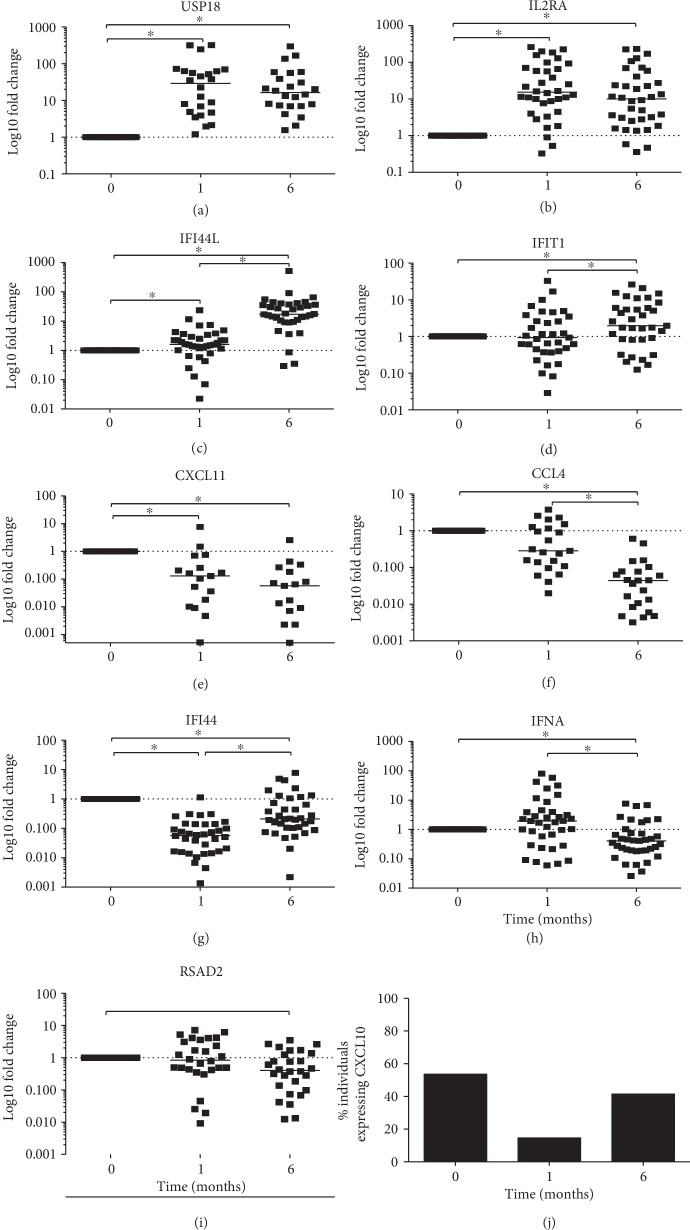
3.4. qPCR Analysis of the 10 Selected Genes in TST-Positive Individuals after INH Therapy
Because several genes were consistently modified following INH treatment regardless of the IGRA status and taking into consideration that TST positivity is the only indicator of LTBI in some individuals, we examined the fold change in gene expression of the above genes after one and six months of INH treatment in all individuals relative to t0 (Figure 4 and Table 3). USP18, IL2RA, and IFI44L expression significantly increased among all participants after one month of INH therapy (Figures 4(a)–4(c)). In contrast, CXCL11 and IFI44 expressions decreased significantly during the same time frame (Figures 4(e) and 4(g)).
Figure 4.
CXCL11, IFNA, RSAD2, USP18, IFI44, IFI44L, IFITI, CCL4, and IL2RA mRNA expressions before and after one and six months of INH treatment in TST-positive individuals. (a–i) The fold changes after one and six months are reported relative to the expression levels prior to INH treatment (t0) using the comparative delta Ct method (n = 39). Individual results are displayed, and the line indicates the median. statistical analysis was performed using non-parametric Friedman's ANOVA followed by Dunn's test; ∗ p < 0.05. (j) CXCL10 expression detected in PBMC from TST-positive individuals (n = 39) before and one and six months after INH treatment is presented as the percentage of individuals expressing the gene.
Table 3.
qPCR analysis of differentially expressed genes in PBMC from TST-positive individuals after 1 and 6 months of INH therapy.
| No. | Gene | Comparison | ∗ p ≤ 0.05 | ||
|---|---|---|---|---|---|
| t1 vs. t0 | t6 vs. t0 | t1 vs. t6 | |||
| 1 | USP18 | Up | Up | — | ∗a, b |
| 2 | IL2RA | Up | Up | — | ∗a, b |
| 3 | IFI44L | Up | Up | Up | ∗a, b, c |
| 4 | IFIT1 | — | Up | Up | ∗b, c |
| 5 | CXCL-11 | Down | Down | — | ∗b |
| 6 | CCL4 | — | Down | Down | ∗b, c |
| 7 | IFI44 | Down | Down | Up | ∗a, b, c |
| 8 | IFNA | — | Down | Down | ∗b, c |
| 9 | RSAD2 | — | Down | — | ∗c |
| 10 | CXCL-10 | — | — | — | NS |
Mann-Whitney Test; NS: not significant; a = t1 vs. t0; b = t6 vs. t0; c = t1 vs. t6.
USP18, IL2RA, IFI44L, and IFIT1 expressions increased significantly after six months of INH therapy; however, only the increase in IFI44L, IFI44, and IFIT1 was significant in comparison with levels at one month of INH treatment (Figures 4(a)–4(d)).
Conversely, CXCL11, CCL4, IFI44, IFNA, and RSAD2 expressions decreased significantly after six months of treatment, and CCL4 and IFNA expression also showed a significant decrease in comparison to one month of INH therapy (Figures 4(e)–4(i) and Table 3). Overall, USP18 and IL2RA expressions significantly increased after one and six months of INH therapy (Figures 4(a) and 4(b)). Also, CXCL10 expression decreased to undetectable levels after one month of INH therapy in most of the participants in comparison to before treatment (Figure 4(j)).
4. Discussion
Few studies have assessed the effect of INH therapy on the immune response in blood cells from individuals with LTBI [13–16]. In the present study, we defined LTBI based on a positive result of the TST. Among the participants, 64% were IGRA-positive as determined by long-incubation in-house IGRA that allowed us to detect IFNγ from both effector memory cells and central memory T-cells, both of which are involved in TB infection [30]. Although TST specificity is limited among BCG-vaccinated populations, and neither test predicts subsequent development of active TB among household contacts of pulmonary TB patients. However, TST remains the preferred method for LTBI diagnosis in resource-limited, high TB burden settings due to its low cost and straightforward implementation.
Since our population was vaccinated at birth, and the mean age was 39.2 years, BCG cross-reactivity is unlikely to explain the discordance observed in the TST-positive individuals who are IGRA negative. In addition, it has been reported that vaccination in infancy has minimal effect on TST results after 10 or more years of vaccination [31]. Different studies have shown the importance of interpreting both TST and IGRA results in the context of prevalence and exposure [32–34]. We studied individuals who had been exposed to pulmonary tuberculosis patients and were, therefore, more likely infected. In addition, it was reported that household contacts that were TST positive but IGRA negative had less exposure to the index case compared to the concordant group [35]. These data suggest that following M. tuberculosis exposure, IGRA conversion may take longer than TST [35]. Therefore, TST-positive individuals with negative IGRA results as the ones that occurred in our study should not be interpreted as the absence of infection since previous studies in Mexican population and in high tuberculosis burden settings have shown that TST results correlates well with M. tuberculosis infection [36, 37].
When we analyzed the expression of the selected genes in individuals according to IGRA results before INH treatment, we found that expression of IFI44L, IFNA, IL2RA, and USP18 was significantly lower in IGRA-positive individuals than in IGRA-negative individuals. These results suggest that concerning the assessed genes before INH therapy, TST-reactive individuals harbor immunological differences according to their IGRA result.
Among other genes identified as relevant in our study, expression of CXCL10 (also known as IP-10), CXCL11, and CCL4 was significantly decreased after six months of INH therapy compared to baseline levels and after one month of treatment, regardless of the IGRA status. CXCL10 has been used to distinguish between TB and LTBI [26]. In addition, a decrease in plasma CXCL10 levels after two weeks of TB treatment has been reported [27]. Although the role of CXCL10 in LTBI treatment has not been studied, decreased CXCL10 expression after a few weeks of INH treatment may indicate reduced antigen load. CCL4 is overexpressed in lung tissues of mice with TB and in patients with late-stage TB [25], suggesting that the significant decrease in CCL4 expression observed in individuals with LTBI after INH treatment might be associated with a decreased risk of developing active TB and could, therefore, be useful as a biomarker of treatment efficacy.
USP18 and IL2RA expressions significantly increased after one and six months of INH therapy in both IGRA-negative and IGRA-positive individuals. These results suggest that INH therapy is not associated with a differential gene expression profile between IGRA-negative and IGRA-positive individuals, but rather with a general immunomodulatory effect of the drug. Therefore, upregulation of both genes might be a useful tool to monitor the response of INH therapy and might be associated with the efficacy of INH therapy. However, the evaluation of the efficacy of INH treatment was beyond the scope of this study.
Overall, the qPCR analysis revealed an association between INH treatment and significant changes in the expression of nine of the ten selected genes among TST-positive individuals. We hypothesize that these changes may be associated with the INH bactericidal effect.
Among TST-positive individuals, USP18, IL2RA, and CXCL11 expression levels changed significantly after one month of INH therapy, remaining without significant changes at six months of therapy. These initial change may be associated with an early INH bactericidal effect, and prolonged INH administration seems not to produce subsequent modification of expression levels of these genes. It is also possible that the time frame of our study was unable to assess the kinetics of the expression of these genes.
Additionally, we also observed that the expression of ISGs such as IFIT1, IFI44, and IFI44L increased significantly after six months in comparison to one month of INH treatment among TST-positive individuals. In this regard, it has been described that a variety of stimuli, such as infection with RNA or DNA viruses and viral and bacterial molecular patterns (PAMPs), directly induce transcription of the IFIT1/ISG56 family [38]. Therefore, mycobacterial antigens may be continuously released during INH treatment and induce the expression of these genes.
In contrast, IFNA expression was significantly decreased after six months in comparison to one month of INH therapy in TST-positive individuals. This result suggests that six months of INH treatment induced an increase in USP18 and IFI44L expressions that negatively regulated IFNA [39, 40] explaining its low levels after six months of INH therapy. So, our observation that there is an increase in the expression of genes that regulate IFNA after six months of INH treatment indicates that USP18 and IFI44L expressions might be relevant as biomarkers of the effectiveness of INH therapy. These results also support the suitability of prolonged INH administration.
In particular, IFNA expression decreased significantly in PBMC of TST-positive individuals after six months of INH therapy compared to the expression levels recorded before treatment and after one month of treatment. Because viral and bacterial persistence are associated with deleterious immunoregulatory effects of sustained type I IFN expression [41], this decrease in IFNA expression might be due to a reduction in the latent bacterial load caused by INH therapy. Therefore, we speculate that INH therapy kills the bacteria in LTBI and decreases the bacterial burden necessary to sustain IFNA expression. In addition, IFNα production is associated with suppression of the innate immune response [42, 43]. Thus, an increase in type I IFN signaling may lead to increased susceptibility of the host to bacterial infections [44, 45]. Type I IFN is also associated with TB susceptibility, as has been demonstrated by the observation that IFNα treatment in patients with hepatitis D infection exacerbates TB [46]. One of the most prevalent biosignatures in the blood of patients with TB is the elevated expression of transcripts involved in type I and type II IFN signaling, mainly driven by neutrophils [19]. Moreover, IFNα signaling has been proposed as a valuable predictive biomarker of the progression of LTBI toward active TB disease [47]. Accordingly, we suggest that the decrease in IFNA expression observed after INH therapy represents a potential biomarker of INH efficacy treatment in individuals with LTBI.
We also noted that the USP18 expression was increased in PBMC from TST-positive individuals after INH therapy. Such upregulation of USP18 expression and the decrease in IFNA expression after six months of INH therapy are consistent with the function of USP18, which is a potent negative regulator of IFNA signaling [40]. USP18 regulates the IFN type I signaling pathway by inhibiting IFN type I-induced JAK/STAT activation and removes interferon-stimulated genes 15 (ISG15) adducts from substrate proteins through its independent function as a protease [48]. USP18 might play a role in controlling bacterial infections, as suggested in a study using mice with a mutation in USP18 that showed increased bacterial loads, increased inflammatory responses, and increased activity of the type I IFN signaling pathway [21]. The mechanism regulating USP18 during INH therapy is unknown, but patients with a deficiency in ISG15 are susceptible to mycobacterial diseases, and the absence of intracellular ISG15 prevents the accumulation of USP18, resulting in amplification of IFNα/β responses [49]. Consequently, the upregulation of USP18 during INH therapy observed in the present study suggests that it has an essential role in controlling M. tuberculosis infection and may constitute a potential biomarker for LTBI treatment.
Additionally, expression of other interferon-stimulated genes (ISGs), such as IFIT1, increased significantly in PBMC from TST-positive individuals after six months of INH therapy. IFIT1 and RSAD2 expressions were recently shown to be induced in macrophages infected with M. tuberculosis [23]. Therefore, such an increase in the expression of these ISGs may be involved in the antimycobacterial activity of INH therapy, though, the precise mechanisms involved remain unknown and are worthy of exploration. Our results showed that the expression of IFI44L was significantly increased in PBMC from TST-positive individuals after six months of INH therapy. In contrast, other authors have reported that the expression of IFIT44L in patients with active TB is significantly reduced after six months of TB treatment [20].
IFI44 is an IFNα-inducible protein that is associated with several viral infections, but its role in bacterial infections is unknown [50]. IL2RA expression, which is associated with T-cell activation and regulatory cell profiles, was significantly increased in PBMC from TST-positive individuals after one and six months of INH therapy. In contrast, it has been reported that treatment of active TB is associated with significantly decreased levels of soluble IL2R after six months of anti-TB therapy for pleural TB, though these reduced levels are still higher than those in controls [51]. In addition, it has been reported that s-IL2R levels are higher in TST-positive individuals than in TST-negative controls [22].
Thus, changes in gene expression profiles associated with INH therapy in TST-positive individuals, regardless of the IGRA status, represent potentially useful tools to monitor the response to INH treatment and to evaluate new alternative LTBI regimens.
5. Limitations
The main limitation of our study is the unavailability of a hard-clinical endpoint of bacterial clearance in individuals with LTBI that prevented us from determining the association between differential expression of USP18, IL2RA, IFNA, CXCL11, or CCL4 mRNAs and the efficacy of INH therapy. Another limitation is that we did not control for intraindividual changes in gene expression over time.
In conclusion, in the present study, we observed increased USP18 and IL2RA expressions after one and six months of INH therapy and decreased IFNA, CCL4, and CXCL11 expressions after six months of LTBI treatment. These results might facilitate the use of mRNA expression as a tool for monitoring INH treatment and may have the potential for monitoring new alternative LTBI regimens.
Acknowledgments
We thank all subjects who participated in this study. We thank S. Pedraza-Sánchez for carefully revising the manuscript. Eleane de Oyarzabal is a doctoral student from the Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autonoma de Mexico (UNAM) and has received CONACYT fellowship 270421. This work was supported by the Mexican Secretariat of Health, the Mexican Council of Science and Technology (CONACYT) Fondo Sectorial de Salud #00000000140416, and the Bill and Melinda Gates Foundation (grant GCGH #11, reference 37222). RJW is funded by Wellcome Trust (104803 and 203135), the Francis Crick Institute (10218), and the NIH (U19AI 111276).
Data Availability
The data used to support the findings of this study are included in supplementary information files ().
Disclosure
Eduardo Sada present address Hospital ABC, Mexico City.
Conflicts of Interest
The authors declare that there is no conflict of interest regarding the publication of this paper.
Authors' Contributions
EdO performed the experiments, participated in the data mining and interpretation and wrote the manuscript. CR analyzed and wrote the manuscript. LFR, EJ, and EdO analyzed the qPCR data. LO and LGG made valuable contributions to the study design. CC, EJ, and MTH analyzed and interpreted the data and drafted the manuscript. LGG, LFR, ES, APC, JSO, and RJW participated in the study design and data interpretation. MT participated in the study design and wrote the manuscript. All authors reviewed and approved the final manuscript.
Supplementary Materials
Data set of IGRA-positive and IGRA-negative individuals, Time of sampling and qPCR data set for all the assessed genes.
References
- 1.World Health Organization. Global tuberculosis report 2018. Geneva: 2018. http://apps.who.int/bookorders. [Google Scholar]
- 2.Raviglione M., Sulis G. Tuberculosis 2015: burden, challenges and strategy for control and elimination. Infectious Disease Reports. 2016;8(2):33–37. doi: 10.4081/idr.2016.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization. Global TB Programme, Latent Tuberculosis Infection-executive Summary. Geneva, Switzerland: World Health Organization; 2018. https://www.who.int/tb/publications/2018/latent-tuberculosis-infection/en/ [Google Scholar]
- 4.Ferrara G., Murray M., Winthrop K., et al. Risk factors associated with pulmonary tuberculosis. Current Opinion in Pulmonary Medicine. 2012;18(3):233–240. doi: 10.1097/MCP.0b013e328351f9d6. [DOI] [PubMed] [Google Scholar]
- 5.Christopoulos A. I., Diamantopoulos A. A., Dimopoulos P. A., Goumenos D. S., Barbalias G. A. Risk factors for tuberculosis in dialysis patients: a prospective multi-center clinical trial. BMC Nephrology. 2009;10(1):1–7. doi: 10.1186/1471-2369-10-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burgos J. L., Kahn J. G., Strathdee S. A., et al. Targeted screening and treatment for latent tuberculosis infection using QuantiFERON®-TB gold is cost-effective in Mexico. The International Journal of Tuberculosis and Lung Disease. 2009;13(8):962–968. [PMC free article] [PubMed] [Google Scholar]
- 7.Amicosante M., Ciccozzi M., Markova R. Rational use of immunodiagnostic tools for tuberculosis infection: guidelines and cost effectiveness studies. The New Microbiologica. 2010;33(2):93–107. [PubMed] [Google Scholar]
- 8.Seddon J. A., Paton J., Nademi Z., et al. The impact of BCG vaccination on tuberculin skin test responses in children is age dependent: evidence to be considered when screening children for tuberculosis infection. Thorax. 2016;71(10):932–939. doi: 10.1136/thoraxjnl-2015-207687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arend S. M., Geluk A., Van Meijgaarden K. E., et al. Antigenic equivalence of human T-cell responses to mycobacterium tuberculosis-specific RD1-encoded protein antigens ESAT-6 and culture filtrate protein 10 and to mixtures of synthetic peptides. Infection and Immunity. 2000;68(6):3314–3321. doi: 10.1128/iai.68.6.3314-3321.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machado A., Jr., Emodi K., Takenami I., et al. Analysis of discordance between the tuberculin skin test and the interferon-gamma release assay. The International Journal of Tuberculosis and Lung Disease. 2009;13(2008):446–453. [PubMed] [Google Scholar]
- 11.Al Hajoj S., Varghese B., Datijan A., et al. Interferon gamma release assay versus tuberculin skin testing among healthcare workers of highly diverse origin in a moderate tuberculosis burden country. PLoS One. 2016;11(5, article e0154803) doi: 10.1371/journal.pone.0154803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez Sanchís A., Calpe Calpe J. L., Llavador Ros G., Ena Muñoz J., Calpe Armero A. Primary prevention and treatment of latent tuberculosis infection with isoniazid: efficacy of a control program, 1997-2002. Archivos de Bronconeumología (English Edition) 2005;41(1):27–33. doi: 10.1016/S1579-2129(06)60391-1. [DOI] [PubMed] [Google Scholar]
- 13.Chee C. B. E., Khinmar K. W., Gan S. H., Barkham T. M. S., Pushparani M., Wang Y. T. Latent tuberculosis infection treatment and T-cell responses to Mycobacterium tuberculosis-specific antigens. American Journal of Respiratory and Critical Care Medicine. 2007;175(3):282–287. doi: 10.1164/rccm.200608-1109OC. [DOI] [PubMed] [Google Scholar]
- 14.Wilkinson K. A., Kon O. M., Newton S. M., et al. Effect of treatment of latent tuberculosis infection on the T cell response to mycobacterium tuberculosis antigens. The Journal of Infectious Diseases. 2006;193(3):354–359. doi: 10.1086/499311. [DOI] [PubMed] [Google Scholar]
- 15.Higuchi K., Harada N., Mori T. Interferon‐γ responses after isoniazid chemotherapy for latent tuberculosis. Respirology. 2008;13(3):468–472. doi: 10.1111/j.1440-1843.2008.01244.x. [DOI] [PubMed] [Google Scholar]
- 16.Lee S. W., Lee S. H., Yim J.-J. Serial interferon-gamma release assays after chemoprophylaxis in a tuberculosis outbreak cohort. Infection. 2012;40(4):431–435. doi: 10.1007/s15010-012-0265-2. [DOI] [PubMed] [Google Scholar]
- 17.Chiappini E., Bonsignori F., Mangone G., et al. Serial T-Spot.Tb and quantiferon-tb-gold in-tube assays to monitor response to antitubercular treatment in Italian children with active or latent tuberculosis infection. The Pediatric Infectious Disease Journal. 2012;31(9):974–977. doi: 10.1097/INF.0b013e31825d0d67. [DOI] [PubMed] [Google Scholar]
- 18.Johnson J. L., Geldenhuys H., Thiel B. A., et al. Effect of isoniazid therapy for latent tb infection on quantiferon-tb gold in- tube responses in adults with positive tuberculin skin test results in a high TB incidence area. Chest. 2014;145(3):612–617. doi: 10.1378/chest.13-1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berry M. P. R., Berry M., Graham C. M., et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature. 2010;466(7309):973–977. doi: 10.1038/nature09247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sambarey A., Devaprasad A., Mohan A., et al. Unbiased Identification of blood-based biomarkers for pulmonary tuberculosis by modeling and mining molecular interaction networks. EBioMedicine. 2017;15:112–126. doi: 10.1016/j.ebiom.2016.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dauphinee S. M., Richer E., Eva M. M., et al. Contribution of increased ISG15, ISGylation and deregulated type I IFN signaling in Usp18 mutant mice during the course of bacterial infections. Genes & Immunity. 2014;15(5):282–292. doi: 10.1038/gene.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shitrit D., Izbicki G., Bar-Gil Shitrit A., Raz M., Sulkes J., Kramer M. R. Role of soluble interleukin-2 receptor levels in patients with latent tuberculosis. Lung. 2006;184(1):21–24. doi: 10.1007/s00408-005-2558-2. [DOI] [PubMed] [Google Scholar]
- 23.Andreu N., Phelan J., De Sessions P. F., Cliff J. M., Clark T. G., Hibberd M. L. Primary macrophages and J774 cells respond differently to infection with Mycobacterium tuberculosis . Scientific Reports. 2017;7:1–12. doi: 10.1038/srep42225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung W. Y., Yoon D., Lee K. S., et al. The usefulness of serum CXCR3 ligands for evaluating the early treatment response in tuberculosis. Medicine. 2016;95(17):1–6. doi: 10.1097/md.0000000000003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangel-Santiago J. F., Baay-Guzman G. J., Duran-Padilla M. A., et al. A novel role of Yin-Yang-1 in pulmonary tuberculosis through the regulation of the chemokine CCL4. Tuberculosis. 2016;96:87–95. doi: 10.1016/j.tube.2015.10.013. [DOI] [PubMed] [Google Scholar]
- 26.Wergeland I., Pullar N., Assmus J., et al. IP-10 differentiates between active and latent tuberculosis irrespective of HIV status and declines during therapy. The Journal of Infection. 2015;70(4):381–391. doi: 10.1016/j.jinf.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 27.Tonby K., Ruhwald M., Kvale D., Dyrhol-Riise A. M. IP-10 measured by dry plasma spots as biomarker for therapy responses in Mycobacterium tuberculosis infection. Scientific Reports. 2015;5:1–6. doi: 10.1038/srep09223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CENAPRECE. CENAPRECE Dir Micobacterioris; 2010. Mortalidad por Tuberculosis Todas Formas Estados Unidos Mexicanos 1990-2010. May 2019, http://www.cenaprece.salud.gob.mx/programas/interior/micobacteriosis/descargas/pdf/9MorbiTbTodas16.pdf. [Google Scholar]
- 29.Torres M., García-García L., Cruz-Hervert P., et al. Effect of isoniazid on antigen-specific interferon-γ secretion in latent tuberculosis. European Respiratory Journal. 2015;45(2):473–482. doi: 10.1183/09031936.00123314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butera O., Chiacchio T., Carrara S., et al. New tools for detecting latent tuberculosis infection: evaluation of RD1-specific long-term response. BMC Infectious Diseases. 2009;9, article 182 doi: 10.1186/1471-2334-9-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farhat M. False-positive tuberculin skin tests: what is the absolute effect of BCG and nontuberculous mycobacteria? The International Journal of Tuberculosis and Lung Disease. 2006;10(11):1192–1204. [PubMed] [Google Scholar]
- 32.Detjen A. K., Loebenberg L., Grewal H. M. S., et al. Short-term reproducibility of a commercial interferon gamma release assay. Clinical and Vaccine Immunology. 2009;16(8):1170–1175. doi: 10.1128/CVI.00168-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringshausen F. C., Nienhaus A., Costa J. T., et al. Within-subject variability of Mycobacterium tuberculosis-specific gamma interferon responses in German health care workers. Clinical and Vaccine Immunology. 2011;18(7):1176–1182. doi: 10.1128/CVI.05058-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zwerling A., van den Hof S., Scholten J., Cobelens F., Menzies D., Pai M. Interferon-gamma release assays for tuberculosis screening of healthcare workers: a systematic review. Thorax. 2012;67(1):62–70. doi: 10.1136/thx.2010.143180. [DOI] [PubMed] [Google Scholar]
- 35.Ribeiro-Rodrigues R., Kim S., Coelho Da Silva F. D., et al. PLoS One. 2014;9(5, article e96564) doi: 10.1371/journal.pone.0096564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.García-Sancho F. M. C., García-García L., Jiménez-Corona M. E., et al. Is tuberculin skin testing useful to diagnose latent tuberculosis in BCG-vaccinated children? International Journal of Epidemiology. 2006;35(6):1447–1454. doi: 10.1093/ije/dyl213. [DOI] [PubMed] [Google Scholar]
- 37.Dheda K., van Zyl Smit R., Badri M., Pai M. T-cell interferon-γ release assays for the rapid immunodiagnosis of tuberculosis: clinical utility in high-burden vs. low-burden settings. Current Opinion in Pulmonary Medicine. 2009;15(3):188–200. doi: 10.1097/mcp.0b013e32832a0adc. [DOI] [PubMed] [Google Scholar]
- 38.Fensterl V., Sen G. C. The ISG56/IFIT1 gene family. Journal of Interferon & Cytokine Research. 2011;31(1):71–78. doi: 10.1089/jir.2010.0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.ML D. D., Martinez-Sobrido L., Topham D. J. Novel functions of IFI44L as a feedback regulator of host antiviral responses. Journal of Virology. 2019;93(21) doi: 10.1128/jvi.01159-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.François-Newton V., de Freitas Almeida G. M., Payelle-Brogard B., et al. USP18-based negative feedback control is induced by type I and type III interferons and specifically inactivates interferon α response. PLoS One. 2011;6(7, article e22200) doi: 10.1371/journal.pone.0022200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murira A., Lamarre A. Type-I interferon responses: from friend to foe in the battle against chronic viral infection. Frontiers in Immunology. 2016;7:p. 609. doi: 10.3389/fimmu.2016.00609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trinchieri G. Type I interferon: friend or foe? The Journal of Experimental Medicine. 2010;207(10):2053–2063. doi: 10.1084/jem.20101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Decker T., Müller M. Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nature Reviews Immunology. 2005;5(9):675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- 44.Richer E., Prendergast C., Zhang D.-E., Qureshi S. T., Vidal S. M., Malo D. N-ethyl-N-nitrosourea-induced mutation in ubiquitin-specific peptidase 18 causes hyperactivation of IFN-αβ signaling and suppresses STAT4-induced IFN-γ production, resulting in increased susceptibility toSalmonellaTyphimurium. Journal of Immunology. 2010;185(6):3593–3601. doi: 10.4049/jimmunol.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan R., Sancho-Shimizu V., Prendergast C., Roy M.-F., Loredo-Osti J.-C., Malo D. Refinement of the genetics of the host response to Salmonella infection in MOLF/Ei: regulation of type 1 IFN and TRP3 pathways by Ity2. Genes & Immunity. 2012;13(2):175–183. doi: 10.1038/gene.2011.69. [DOI] [PubMed] [Google Scholar]
- 46.Telesca C., Angelico M., Piccolo P., et al. Interferon-alpha treatment of hepatitis D induces tuberculosis exacerbation in an immigrant. Journal of Infection. 2007;54(4):e223–e226. doi: 10.1016/j.jinf.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 47.Maertzdorf J., Kaufmann S. H. E., Weiner J. Molecular signatures for vaccine development. Vaccine. 2015;33(40):5256–5261. doi: 10.1016/j.vaccine.2015.03.075. [DOI] [PubMed] [Google Scholar]
- 48.Malakhov M. P., Malakhova O. A., Il Kim K., Ritchie K. J., Zhang D. E. UBP43 (USP18) specifically removes ISG15 from conjugated proteins. Journal of Biological Chemistry. 2002;277(12):9976–9981. doi: 10.1074/jbc.M109078200. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X., Bogunovic D., Payelle-Brogard B., et al. Human intracellular ISG15 prevents interferon-α/β over-amplification and auto- inflammation. Nature. 2015;517(7532):89–93. doi: 10.1038/nature13801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.ML D. D., Nogales A., Martinez-Sobrido L., Topham D. J. Interferon-induced protein 44 interacts with cellular FK506-binding protein 5, negatively regulates host antiviral responses, and supports virus replication. MBio. 2019;10(4):1–20. doi: 10.1128/mBio.01839-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Porcel J. M., Gazquez I., Vives M., Perez B., Rubio M., Rivas M. C. Diagnosis of tuberculous pleuritis by the measurement of soluble interleukin 2 receptor in pleural fluid. The International Journal of Tuberculosis and Lung Disease. 2000;4(10):975–979. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data set of IGRA-positive and IGRA-negative individuals, Time of sampling and qPCR data set for all the assessed genes.
Data Availability Statement
The data used to support the findings of this study are included in supplementary information files ().