Significance
When and how long we sleep is determined by the time of day and how long we have been awake, which are tracked molecularly by a circadian and a sleep–wake-driven process, respectively. We measured the long-term consequences of a short-term sleep deprivation on gene expression and regulation in the mouse brain, and used mathematical models to determine the relative contributions of the circadian and sleep–wake-driven processes. We find that many genes, including most of the genes that constitute the molecular circadian clock, are perturbed by sleep deprivation long after the mice ceased showing behavioral signs of sleep loss. Our results have implications for human health, given the high prevalence of insufficient and poor-quality sleep in our contemporary society.
Keywords: circadian, sleep, gene expression, epigenetics, long-term effects
Abstract
The timing and duration of sleep results from the interaction between a homeostatic sleep–wake-driven process and a periodic circadian process, and involves changes in gene regulation and expression. Unraveling the contributions of both processes and their interaction to transcriptional and epigenomic regulatory dynamics requires sampling over time under conditions of unperturbed and perturbed sleep. We profiled mRNA expression and chromatin accessibility in the cerebral cortex of mice over a 3-d period, including a 6-h sleep deprivation (SD) on day 2. We used mathematical modeling to integrate time series of mRNA expression data with sleep–wake history, which established that a large proportion of rhythmic genes are governed by the homeostatic process with varying degrees of interaction with the circadian process, sometimes working in opposition. Remarkably, SD caused long-term effects on gene-expression dynamics, outlasting phenotypic recovery, most strikingly illustrated by a damped oscillation of most core clock genes, including Arntl/Bmal1, suggesting that enforced wakefulness directly impacts the molecular clock machinery. Chromatin accessibility proved highly plastic and dynamically affected by SD. Dynamics in distal regions, rather than promoters, correlated with mRNA expression, implying that changes in expression result from constitutively accessible promoters under the influence of enhancers or repressors. Serum response factor (SRF) was predicted as a transcriptional regulator driving immediate response, suggesting that SRF activity mirrors the build-up and release of sleep pressure. Our results demonstrate that a single, short SD has long-term aftereffects at the genomic regulatory level and highlights the importance of the sleep–wake distribution to diurnal rhythmicity and circadian processes.
According to the 2-process model (1, 2), sleep regulation results from an interaction between a sleep homeostatic process and a circadian process often referred to as “process S” and “process C,” respectively. The sleep homeostat tracks the need or pressure for sleep as it increases during wake and decreases during sleep, while the circadian process dictates the optimal time of day for sleep to occur. Their fine-tuned interaction assures optimal timing, duration, and quality of both wakefulness and sleep, and even minor changes in either of these processes or their alignment cause performance decrements and clinically significant sleep disruption (3, 4).
The circadian clock is described as self-sustained ∼24-h oscillations involved in a variety of physiological processes and behaviors, such as sleep (3, 5). It is encoded molecularly through negative feedback loops involving the core clock genes, which are capable of generating oscillations in constant environmental conditions; that is, in the absence of periodically occurring time cues such as the light/dark cycle (6). However, this apparent autonomy does not inevitably imply that the expression of all genes displaying a rhythm with a period of 24 h is directly driven by the circadian clock. For example, the light/dark cycle, besides entraining the circadian clock, directly influences many physiological and behavioral processes (7). Also, the rhythmic organization of sleep–wake behavior and associated feeding and locomotion directly drives gene expression (8). Disentangling the respective contributions of the circadian and sleep–wake-driven processes is experimentally challenging and has been addressed by methods suppressing 1 component (e.g., surgical or genetic ablation of circadian oscillators) or uncoupling their relationship through forced desynchrony or sleep deprivation (SD) (3, 9).
SD experiments aiming at identifying genes associated with the sleep homeostatic process follow the rationale that causing mice to stay awake during a time when they normally sleep will induce an acute response in sleep–wake-driven genes. Indeed, studies comparing gene-expression levels immediately after SD with controls collected at the same time of day have identified many differentially expressed genes (10–13), and a few studies have probed the punctual effect of SD at different times of the 24-h cycle in mice (13, 14), or expression dynamics in blood during SD in humans (15, 16). However, assessing the respective contributions of the 2 processes requires measuring gene expression over multiple time points, not only under SD (i.e., enforced waking), but also under spontaneous sleep–wake dynamics pre- and post-SD. Furthermore, to systematically link temporal gene expression to the sleep–wake distribution and circadian clock, the analysis should consider the entire time series, rather than only pair-wise differential comparisons. Finally, the regulatory mechanisms underlying such dynamics are largely unexplored (17), particularly in this kind of dynamic context.
To systematically investigate the gene-expression dynamics caused by 1 acute SD episode, as well as the underlying regulatory events, we measured chromatin accessibility alongside mRNA expression in the cerebral cortex of adult C57BL6/J mice over 24 h before, during, and over 48 h following 1 6-h session of total SD, as well as 7 d after the intervention (see study design in Fig. 1A). We modeled the entire time series based on the assumptions of the 2-process model as to the dynamics of each of the 2 contributing processes, to objectively assess whether the mRNA accumulation dynamics of each cortically expressed gene follow process S, process C, or a combination thereof. This setting allowed us to characterize the temporal dynamics of the consequences of SD on gene expression and regulation, and dissect the interaction between processes S and C. Moreover, we identified genomic regulatory elements implicated in the transcriptional response to sleep loss by exploring the hitherto understudied epigenetic landscape of sleep (18).
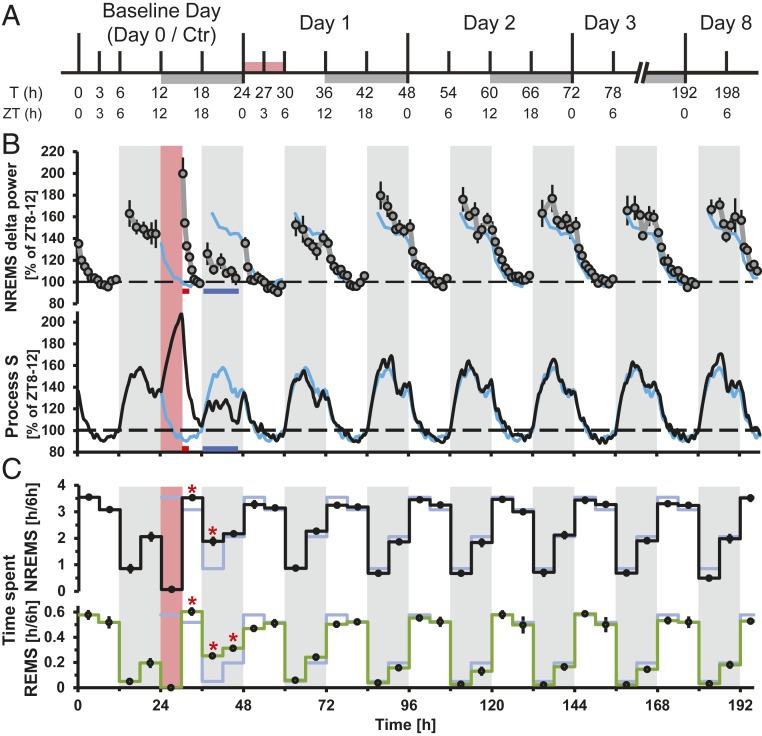
Fig. 1.
Study design and long-term effects of SD on sleep behavior and EEG δ-power. (A) Tissue collection schedule with time in hours from beginning of the experiment (T) and corresponding ZT. White and gray bars are 12-h:12-h light/dark cycle. Red bar is SD. (B) Long-term effects of SD on NREM sleep δ-power (1 to 4 Hz, Upper), and simulation of process S (Lower). White and gray shading is 12-h:12-h light/dark cycle. Red shading is SD. Red and blue bars under the graphs denote significant increase, respectively, and decrease compared to baseline (t test, P < 0.05, n = 6). Baseline data are repeated over the entire time course as a light blue line. Mean δ-power values (±SEM) and process S are expressed as the percentage of intraindividual deviations from the time interval in baseline with the lowest overall power (ZT8 to ZT12, average across 2 d). (C) NREM (Upper) and REM (Lower) sleep quantity. Asterisks denote significant increases compared to baseline (t test, P < 0.05, n = 6). Baseline data and shading as above.
Results
Behavioral Response and Recovery after Sleep Deprivation.
We recorded sleep and the electroencephalogram (EEG) across the 198-h protocol (Methods and Fig. 1A) (n = 6) and observed the typical distribution of sleep over 24 h, with mice spending most of the light periods asleep, while being predominantly awake during the dark periods (Fig. 1C). δ-Power in nonrapid-eye movement (NREM) sleep (Fig. 1 B, Upper), an EEG-derived variable considered to reflect sleep “pressure,” was high after spontaneous waking in the dark phase and decreased during the light phase, and we observed the well-known effects of SD, namely an increase in δ-power within the first hour immediately following SD and a rebound of time spent in NREM sleep observed during the first 12 h of recovery (T30 to T42). We found that values for NREM sleep no longer significantly deviated from baseline levels already after T42 (Fig. 1C, black line). During the dark phase (T36 to T48), δ-power even dropped below the levels reached at this time during baseline. Computer simulation of process S (Methods) based on the sleep–wake distribution demonstrated that this decrease of δ-power below baseline level was a consequence of the increased time spent in NREM sleep during the first 12 h after SD (Fig. 1 B, Lower). REM sleep was affected in the same manner as NREM sleep (Fig. 1 C, Lower).
We asked whether the fast reversal of the phenotype in the EEG data would be paralleled by changes at the gene expression and regulatory levels in the cerebral cortex, or whether novel molecular dynamic patterns could be observed. We therefore measured and analyzed the temporal dynamics of transcriptomes and chromatin accessibility over a total of 78 h, including baseline, SD, and recovery.
Sleep–Wake History Is the Main Driver of Transcriptome Dynamics.
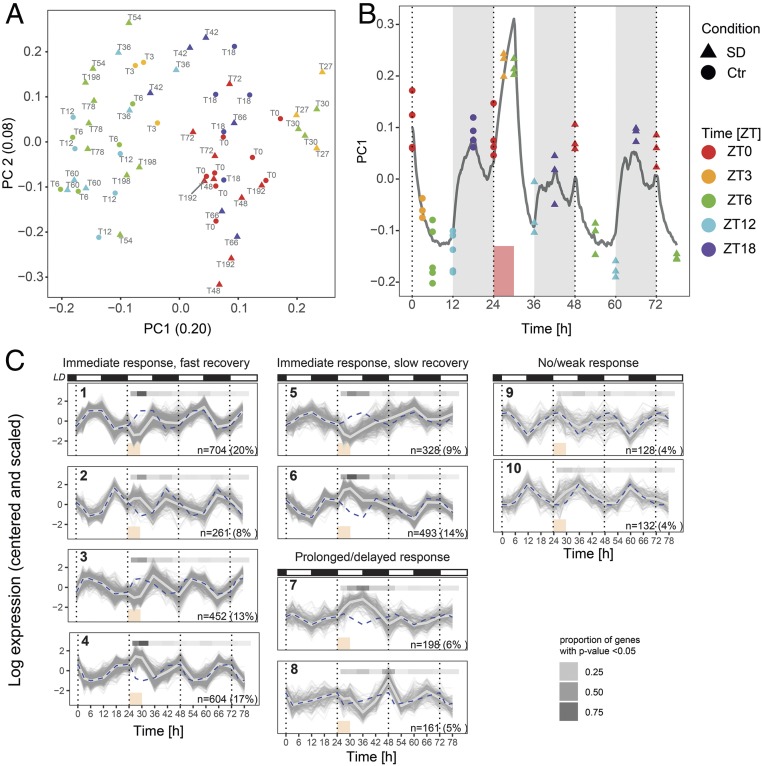
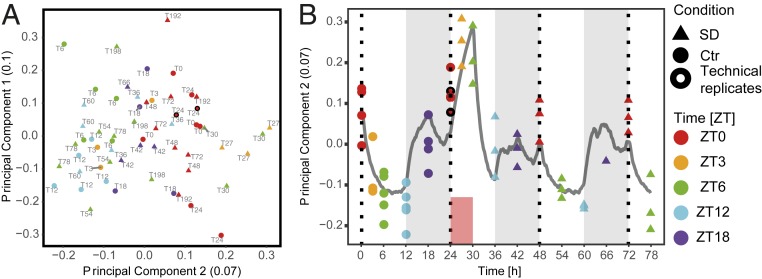
We first examined the overall variability of the detected fraction of the transcriptome (13,842 genes) using principal component analysis (PCA) (Fig. 2A) over all 56 tissue samples (n = 3 to 4 per time point per condition) (Methods). We observed that samples formed 3 groups along the first principal component (PC1) axis. The left-most group gathered time points during the light phase of the light–dark cycle, while the middle group represented time points during the dark phase. Without a forced-wakefulness treatment, this separation could evoke that PC1 separates samples according to time of day (or Zeitgeber time, ZT, where ZT0 denotes the onset of the light phase of the 24-h period). However, this notion is challenged by the shift toward the right of the samples taken at ZT3 and ZT6 during SD (T27 and T30), suggesting that the PC1 axis follows (from left to right) increased time spent awake prior to sampling rather than ZT, as animals spent more time asleep in the light (left) than in the dark period (middle group), while the right-most group did not sleep at all prior to sampling.
Fig. 2.
Sleep–wake history is the main driver of transcriptome dynamics. (A) PCA of the expression of the 13,842 detected genes. The number in parenthesis in the axis label denotes the fraction of the variance explained by the component. Colors denote ZT (ZT0 to ZT12: light period; ZT12 to ZT24/0: dark period). Baseline samples are represented by discs and samples collected during or after SD by triangles. Text labels denote time of sample collection according to the experimental design (Fig. 1A). (B) PC1 plotted over time. Gray line: Sleep–wake-driven simulation of process S with the same time constants used to predict EEG δ-power dynamics (Fig. 1B). (C) k-Means clusters of the 3,461 genes with statistically significant temporal gene expression (FDR-adjusted P < 0.001, likelihood ratio test). Vertical dotted lines are ZT0 of each day. White and black bars are 12-h:12-h light/dark cycle. Blue dashed line is average of the cluster under baseline, repeated for comparison over the 3 d of the experiment. Light gray thick line is cluster average. Red box is SD. Gray shaded bar above each graph: proportion of genes with P < 0.05 according to a likelihood ratio test between SD and baseline conditions at the same ZT.
To further illustrate that PC1 is sleep–wake driven, we overlaid the changes in PC1 with expected levels of homeostatic sleep need based on the sleep–wake distribution of the current experiment (see simulated process S in Fig. 1B). Indeed, PC1 increased during periods of waking, decreased during periods of sleep and, importantly, reached its maximum during SD, paralleling process S (Fig. 2B) and EEG δ-power (19) (Fig. 1B), thus highlighting the pervasive impact of sleep–wake distribution on gene expression, which we further explore below.
Clustering of mRNA Temporal Profiles Highlights Diverse Response and Recovery Kinetics.
To uncover and classify broad temporal patterns in our data, we first performed an exploratory analysis using k-means clustering. With this unsupervised clustering, we grouped the expression time courses of 3,461 conservatively selected genes displaying statistically robust temporal variation from T0 to T78 (false-discovery rate [FDR]-adjusted P < 0.001, likelihood ratio test). We observed distinct profiles of response to SD and subsequent recovery by comparing the cluster average of T24 to T78 with the baseline day (T0 to T18) (Fig. 2C, light gray line and blue dashed line, respectively). The general patterns cover immediate response (clusters 1 to 6), prolonged response (cluster 7), delayed response (cluster 8), as well as slight to absent response (clusters 9 and 10, mean P values across genes >0.24). These patterns are combined with either a fast (clusters 1 to 4) or slow (clusters 5 to 8) reversal to baseline values. Generally, the fast response and fast reversal dynamics, together with a direction of change opposite to what is expected by time of day, suggests that these genes are sleep–wake driven; that is, genes that usually go up when the mouse is predominantly asleep are down-regulated during SD (clusters 1, 3, 5) and vice versa (clusters 2, 4, 6). Furthermore, the slower dynamics of response and reversal (cluster 7, 8) show that the effects of SD can also occur after the immediate response and last beyond the exposure itself. This suggests that the time required for molecular recovery exceeds the time for phenotypic recovery.
Modeling Temporal Transcriptome Dynamics.
To better characterize and distinguish these expression dynamics, we modeled gene expression over the entire time course, including baseline, SD, and the aftermath of SD, taking into account that the gene-expression dynamics can be sleep–wake driven, circadian, or a combination of both, following the assumptions of the 2-process model as to the dynamics of both processes. Specifically, process S is a sleep–wake-driven process in which a homeostatic need for sleep increases according to an exponential saturating function while awake and again exponentially decreases while asleep. The dynamics of process C, on the other hand, are assumed to follow a sinusoidal oscillation not influenced by sleep–wake state (20). Furthermore, because rhythmicity can be suppressed or altered during mistimed or restricted sleep (13, 15, 16, 21), we included the possibility that it could remain perturbed after the end of the SD.
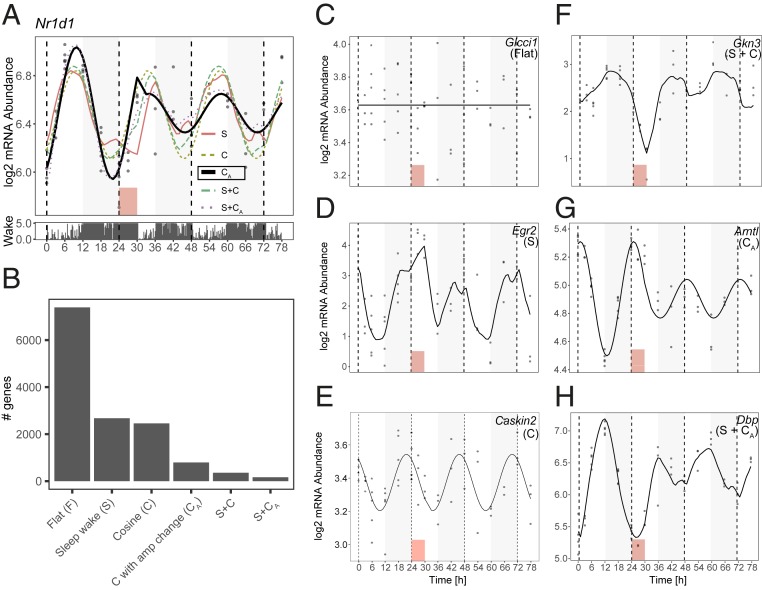
We thus devised 6 models to explain the gene-expression dynamics of the full transcriptome (all 13,842 detected genes) (SI Appendix, Fig. S1): 1) Constant or “flat” model (F); 2) sleep–wake history modeled from sleep–wake data [S, in analogy to process S in the 2-process model (1)]; 3) cosine dynamics with a 24-h rhythm (C); 4) cosine with amplitude change after SD (CA); 5) sleep–wake + cosine (S+C); and 6) sleep–wake + cosine with amplitude change (S+CA). To select the best among competing models, we used the Bayesian Information Criterion (BIC) (22) to balance model fit and model complexity, which we transformed into model probability or weight w (Methods). For each gene, the sum of the weights for all 6 models equals 1, and the model with the highest weight is assigned to the gene. Each gene is assigned 1 of the 6 models. In the example of the core clock gene Nr1d1/RevErbα (Fig. 3A), the selected model was cosine with amplitude change after SD (model CA, represented as a bold line in Fig. 3A), due to a very high weight w = 0.977. Indeed, the baseline pattern (from T0 to T24) is consistent with a circadian oscillation, the amplitude of which is significantly reduced after SD and, surprisingly, not reestablished by T78. All fits are presented in Dataset S1.
Fig. 3.
Modeling temporal transcriptome dynamics. (A) Example of model fitting on Nr1d1/RevErbα. Dots are RNA level data points. Bold line is best-fitting model (here CA with w = 0.977). Barplot under main panel is time spent awake in intervals of 5 min, derived from EEG data. Red box is SD. (B) Number of genes per model. (C–H) Examples of a gene fit to each model.
Model S Recapitulates Known Sleep–Wake-Driven Genes and Closely Parallels EEG δ-Power Dynamics.
We summarized the genes assigned to each model genome-wide (Fig. 3B), and found that, after the flat model, which fit more than half the detected genes (7,391 genes) (see example in Fig. 3C), the sleep–wake-driven model (model S) had the largest number of genes assigned to it (2,677 genes) (see example in Fig. 3D), consistent with the interpretation of PC1 reflecting sleep–wake history and the predominance of fast-response dynamics in our cluster analysis (clusters 1 to 6). Analyzing the parameters of genes associated with model S, we found that the fitted time constants describing the rates at which expression changes during waking and reverts while sleeping (τw, median = 7.05 h) and sleep (τs, median = 1.68 h) were strikingly close to the dynamics of EEG δ-power found for this inbred strain (23) (Fig. 1B) and explains why the time course of PC1 closely follows process S dynamics derived from EEG δ-power (Fig. 2B).
Model S included genes previously described as affected at the end of SD, such as Egr2, Arc, Fos, and Cirbp (12, 13, 24) (Fig. 3D and SI Appendix, Fig. S2) (all w > 0.833), but also, surprisingly, the core clock genes Clock and Npas2 (w = 0.787, respectively w = 0.527) (SI Appendix, Fig. S2). Generally, model S genes were enriched for gene ontology (GO) terms related to the organization of synapses and mitochondria, as well as ion transport (Dataset S2).
Model S genes that were up-regulated during SD, in baseline had their maximum expression during the dark period when waking prevails, while those down-regulated during SD peaked during the baseline light period (Fig. 4A). Thus, many sleep–wake-driven genes appear rhythmic under baseline conditions, and conversely, rhythmic genes could therefore in part be sleep–wake driven. To address this question, we applied a harmonic regression to the baseline (day 0) time points, which yielded a set of 862 oscillating genes (FDR-adjusted P < 0.05). Strikingly, we found that the majority of these genes (578, 67%) were assigned to the S model, and were underrepresented in model C (SI Appendix, Fig. S3A) (P = 2.4e-76, χ2 test). Oscillating genes assigned to model S were enriched for terms related to metabolism, signaling, and molecular transport, while those assigned to other models were enriched for GO terms related to neurogenesis (Dataset S2).
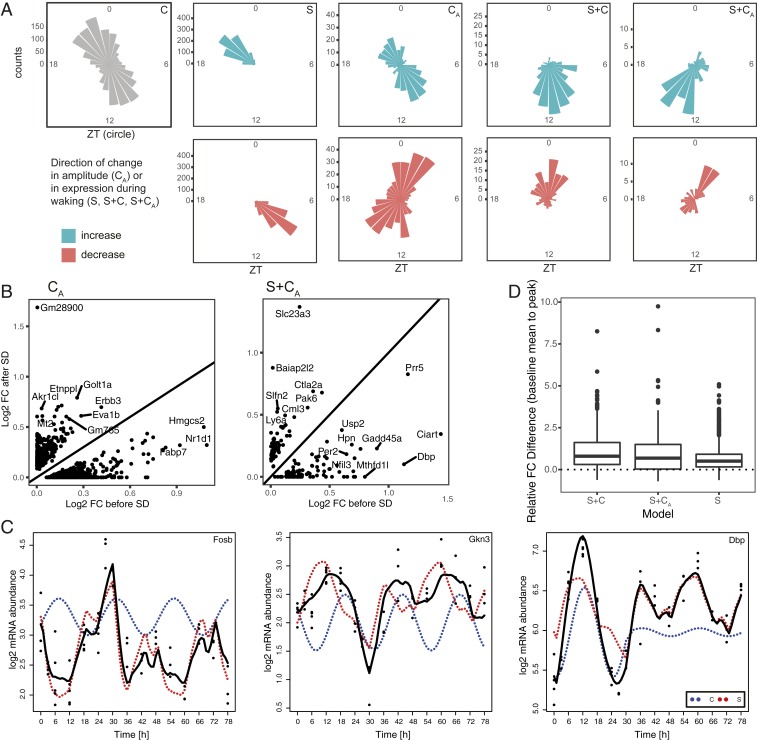
Fig. 4.
Contribution of the S and C components to phase and amplitude of oscillating genes under baseline and after SD. (A) Phase maps of non-F model genes. Each cone reflects the number of genes peaking during 1 h around the ZT clock. Non-C model genes are divided according to the direction of change of the amplitude after SD (CA), or the direction of the change in expression occurring under wakefulness (S, S+C, S+CA). (B) Scatter plots of the log2 amplitude before and after SD of the C component for models CA and S+CA. (C) Contribution of the S and C components (red and blue dotted curves, respectively) to the overall fitted temporal gene expression profile (solid black curve) of 3 example genes. (D) Difference between the highest point before and after SD, both compared to the baseline average, for models S, S+C, and S+CA.
mRNA Expression of All but 1 Core Clock Gene Is Sensitive to SD.
Following closely, the cosine model (model C) was the third most-abundant category with 2,457 genes. This model gathers genes the oscillation of which is largely unaffected by SD as illustrated by the top fit Caskin2 (w = 0.880) (Fig. 3E). The genes with the largest amplitude showing 24-h oscillations in gene expression resistant to SD were Sgk1, a glucocorticoid regulated kinase, and Cldn5, a principal tight junction protein in the blood–brain barrier (SI Appendix, Fig. S2), reaching their peak just before the light-to-dark and dark-to-light transitions, respectively. Generally, examining the oscillation patterns of this group of genes, we find that the phases (i.e., time of day at which expression peaks) of model C genes tend to accumulate in the second half of the light or dark phases (ZT10 and ZT22) (Fig. 4A).
Interestingly, our algorithm assigned Cry1 as the only core clock gene in this model, with model CA second in explaining its mRNA dynamics (w = 0.661 and w = 0.336, respectively) (SI Appendix, Fig. S2). The other clock genes, instead of fitting model C as might be expected, were assigned to model S (see above), the altered amplitude model CA, or the more complex models (models S+C and S+CA, see below). Clock genes assigned to model CA (which contained in total 794 genes) had damped amplitudes after SD, and encompassed the example Nr1d1 (Fig. 3A), as well as Arntl/Bmal1 (Fig. 3G), Per3, Cry2, and Nr1d2/RevErbβ (SI Appendix, Fig. S2). Except for Cry2, where the top weights were w(CA) = 0.645 and w(C) = 0.347, the model C weight w(C) of the other clock genes was negligible [highest w(C) = 0.0003 for Nr1d1], meaning these clock genes were assigned to CA either unequivocally or with a close call to the more complex models S+C and S+CA (Dataset S1). Model CA also contained genes with increased amplitudes (331 of 794; 42%) after SD, such as Akr1cl, Erbb3, Eva1b, and Zfp473 (Fig. 4B and SI Appendix, Fig. S2). Interestingly, the phases of expression differed between the genes with increased vs. decreased amplitudes, the former group having a similar distribution of phases to those of model C genes (ZT10 and ZT22), while the latter group peaked around ZT2 and ZT15 (Fig. 4A). Model C genes were mainly related to morphogenesis, while model CA genes were enriched for axonogenesis (decreased amplitude after SD) and lipid metabolism genes (increased amplitude after SD) (Dataset S2).
Sleep and the Circadian Process Can Work in Opposition to Limit Oscillation Amplitudes in Baseline.
Model S+C (357 genes) incorporated the sleep–wake history and time of day to output a strong response to SD while maintaining modest fold-changes during baseline (e.g., Gkn3, Per1, and Fosb) (Fig. 3F and SI Appendix, Fig. S2). Indeed, this model, together with its altered-amplitude counterpart (S+CA, see below) allowed us to explain complex temporal patterns, notably due to contributions from both the S and C models to the log mRNA abundance. For genes up-regulated during SD, both the S and C components increased concurrently during SD, but discordantly in baseline (e.g., Fosb) (Fig. 4C). Similarly, for genes down-regulated during SD, the S and C components decreased concurrently during SD, but discordantly in baseline (e.g., Gkn3) (Fig. 4C). The discordant action caused dynamics in mRNA levels to be limited during baseline, while the concordant action during SD allows large fold-changes relative to baseline. In comparison, in model S where the buffering by the C component is absent, the difference in fold-change from the highest point after SD and the highest point in baseline, both compared to the baseline average, was smaller than for models S+C and S+CA (Fig. 4D), meaning that expression changes in model S genes are similar whether wakefulness is spontaneous or enforced, and regardless of time of day. We note that S+C genes, which were enriched in genes involved in corticosteroid response (Dataset S2), tend to peak around the light-to-dark and dark-to-light transitions, a shift in comparison with model C and model S genes (Fig. 4A).
SD Represses the Circadian Contribution in Genes with Complex Dynamics, Leaving Them Predominantly under the Control of Process S.
The most complex of our models, model S+CA (166 genes, example Dbp) (Fig. 3H), incorporated sleep–wake history, time of day, and altered amplitudes to model the response to SD and subsequent change in amplitudes after SD. In this model, we see that the additive dynamic process observed in model S+C can be accompanied by altered amplitudes after SD (Fig. 4B) (104 damped, respectively, 62 increased amplitudes), meaning that the contribution of the C component to gene-expression dynamics, relative to that of the S component, is modulated after SD. For example, for Dbp, breaking down the contribution of the S and CA part of the dynamics show that in baseline, the expression is influenced by both sleep–wake history and time of day, while after SD, the contribution of time of day is diminished by 90%, and thus the recovery dynamics are driven mainly by S (Fig. 4C, blue dotted curve). Other examples of genes with damped amplitudes include Per2 (SI Appendix, Fig. S2), a gene known to be subject to complex interactions between processes S and C (25), as well as the clock output genes Nfil3/E4bp4 and Bhlhe41/Dec2 (SI Appendix, Fig. S2). S+CA genes peak during the first half of the light, respectively, and dark phases, at yet a different time than all other models. Still, genes displaying decreased expression under waking had phases overlapping with model CA (Fig. 4A). Globally, model S+CA was enriched for genes involved in rhythmic processes, brain development, and response to hormones and drugs (Dataset S2).
Recovery Time Course Uncovers Hitherto Unnoticed Genes Affected by SD.
Strikingly, a majority of the genes assigned to the amplitude-affected models CA and S+CA (759 of 960 genes, 79%) were not identified when we examined differential expression at the end of SD alone (i.e., T30 vs. T6), as in previous studies (e.g., refs. 13, 24, and 26). For example, fatty acid binding protein 7, Fabp7 (model CA) (SI Appendix, Fig. S2), is not differentially expressed at T27 nor T30; however, its oscillation amplitude displays the strongest reduction from T36 onwards among nondifferentially expressed genes, possibly due to its being a target of Nr1d1 (model CA, differentially expressed at T27 and T30) and thus downstream of the primary response (27). This was especially true for the CA model, where only 53 of 794 (7%) genes were differentially expressed at ZT6, vs. 115 of 166 (69%) for the S+CA model (as a comparison, 1,675 of 2,677 [63%] genes in the S model were significantly differentially expressed at T30, and 172 of 2,457 [7%] genes in the C model).
Conversely, genes that were differentially expressed at T27 or T30 (3,558 genes) were enriched in models S, S+C and S+CA, and underrepresented in models C and CA (SI Appendix, Fig. S3B). Interestingly, 604 of these genes were assigned to model F, representing genes acutely affected by SD, which is not equivalent to sleep–wake-driven, as their expression is systematically perturbed only at 1 or both of these SD time points, but otherwise not modulated by the sleep–wake distribution. These genes are enriched for morphogenesis and metabolism (Dataset S2).
To assess how gene-expression dynamics return to baseline, we examined differential expression at each time point during and after SD compared to baseline (i.e., T27 vs.T3, T30 vs. T6, T36 vs. T12, etc., until T78 vs. T6) (see experimental design in Fig. 1A) and found 210 genes genome-wide that were differentially expressed after phenotypic recovery (i.e., after T42), namely 137 genes at T48 and 75 genes at T60. This was consistent with the observation that the proportion of genes with P < 0.05 in the cluster analysis did not reach 0 for all post-SD time points (Fig. 2C).
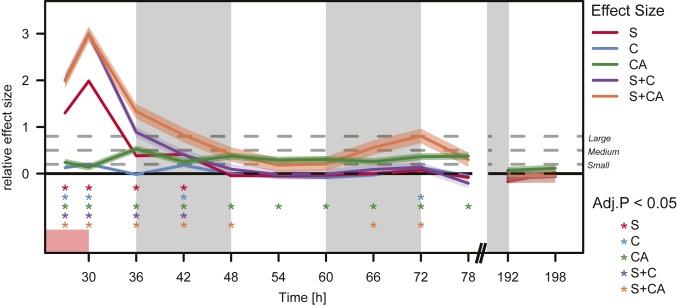
To summarize the long-term changes in gene expression, we quantified—for each of the 5 dynamic models—the effect sizes of the differential expression, time point by time point, and compared them to the effect sizes found for genes in the static model F, which we used as background (Fig. 5). Models S and S+C returned to baseline by T48, while model C showed marginal effects during the dark phase. Strikingly, models CA and S+CA showed long-term effects, the former as a continuously small-to-moderate effect size until and including T78, and the latter as a renewed effect late in the time course following a 12-h period without effect. We note that the effects have ceased for all models by T192 (i.e., 7 d after SD).
Fig. 5.
Effect size of differential expression during and after SD over time in all dynamic models contrasted to model F. Relative effect size by Cohen’s D over time with 95% confidence interval (colored shading), significance (corrected P values by 1-sided nonparametric Wilcoxon test for “greater” effect, asterisks of corresponding colors, bottom). Magnitude of difference (gray stippled lines) is marked as small, medium, and large, corresponding to Cohen’s D > 0.2, >0.5 and >0.8, respectively. White/gray shading is light/dark cycle. Red rectangle is SD.
Genome-Wide Assay for Transposase-Acessible Chromatin Using Sequencing Analysis Shows a Rapid Response and Sleep–Wake-Driven Dynamics in Chromatin Accessibility.
We next asked which regulatory elements are underlying the extensive transcriptome dynamics in our dataset. We used assay for transposase-accessible chromatin using sequencing (ATAC-seq) (28) to identify a union of 130,727 regions of accessible chromatin (hereafter referred to as “peaks”) (Methods) over all time points (i.e., a peak is present in at least 1 time point). The majority of peaks do not change over time and are constantly accessible. Indeed, while 25% of expressed genes showed a time dynamic according to a likelihood ratio test at a 0.001 FDR threshold, only 4% of ATAC-seq peaks display a time dynamic at the same stringent threshold. While PC1 (10%) probably represents experimental noise, 7% of the variance among time points, represented by PC2, could be attributed to the sleep–wake history and follows sleep–wake dynamics, paralleling the RNA-seq data (see PC2 in Fig. 6; see also Fig. 2 A and B).
Fig. 6.
Chromatin accessibility shows sleep–wake-driven dynamics and a rapid response to SD. (A) PCA of all detected ATAC-seq peaks. Plot features as in Fig. 2A. A black symbol outline highlights technical replicates. (B) PC2 plotted over time. Color and shape code as in A. Gray line is sleep–wake-driven simulation of process S with the same time constants used to predict EEG δ-power dynamics (Fig. 1B).
The strongest differential signal relative to baseline occurred during SD with 1,793 peaks differentially accessible (differentially accessible sites, DAS) at ZT3 (T27 vs. T3, after 3-h SD), 2,098 at ZT6 (T30 vs. T6, end of 6-h SD), with 607 peaks in common. Differential signal during SD (ZT3 and ZT6) consisted predominantly of increased accessibility (91% of DAS for ZT3 and 88% of DAS for ZT6), while the 645 late-responding DAS (i.e., differentially accessible at ZT12 only, T36 vs. T12, after 6 h of recovery) (SI Appendix, Fig. S4A) were more likely to be decreased (56%).
The DAS followed the homeostatic process, as we found that the effect of spontaneous waking during the first 6 h of the baseline dark phase (ZT12 to ZT18, when mice are predominantly awake) (Fig. 1C) was similar to the effect of forced wakefulness (SD). Specifically, 83% of the DAS displayed a similar response to spontaneous and forced wakefulness, and are thus likely to be sleep–wake-driven instead of being affected by other factors associated with the SD protocol, such as stress.
Consistent with previous studies (e.g., refs. 29–31), accessibility peaks from all time points and conditions were mainly located in intronic or intergenic regions (SI Appendix, Fig. S4B). When considering only DAS sites (at ZT3, ZT6, or ZT12), the proportion of intergenic regions was increased at the expense of the other regions (Fisher’s exact test, FDR adjusted P < 0.01), suggesting that SD influences the accessibility of distal rather than proximal elements (SI Appendix, Fig. S4 C–E).
Gene Expression Correlates with Chromatin Accessibility at Distal Elements Rather than Promoters.
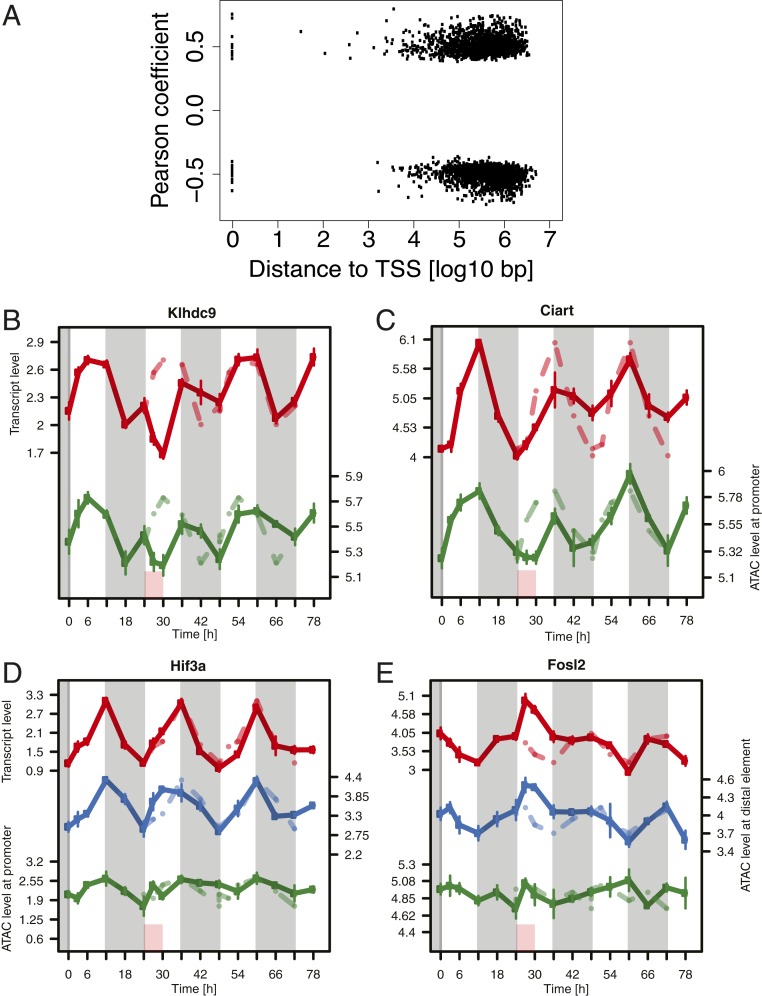
Broadly classifying the dynamic profiles of chromatin accessibility over time by k-means clustering, we found 3 main types of temporal profiles, all of which were reminiscent of sleep–wake-driven dynamics (SI Appendix, Fig. S4F). Clusters 1 to 3 displayed an early response to SD with a fast recovery; clusters 4 to 7 also presented an immediate response, but a slow recovery; while clusters 8 to 10 displayed a late response. Because of this similarity with the RNA-seq clustering (RNA clusters 1 to 8), and because of the similarity in the general dynamics observed by PCA between RNA-seq and ATAC-seq, we sought to connect these changes in accessibility with changes in gene expression and, as both signals originate from the same mouse, correlated ATAC-seq peak signal over time to gene-expression levels over time by calculating the Pearson correlation across samples (Methods). We confined the possible peak-to-gene associations to a single peak per gene within the same topologically associated domain defined from chromatin looping interaction data (HiC) generated from mouse cerebral cortex (32). The rationale of this approach is that any element known to physically interact with the promoter of a gene and shown to correlate in activity with the expression of that gene is a likely regulator of it. In total, the expression level of 3,294 genes (24% of detected genes) was significantly associated with the ATAC-seq signal of 1 peak, either positively or negatively, at distances ranging from the transcription start site (TSS) to 5 Mb away (mean distance for all significant associations at 0.05 FDR: 0.65 Mb) (Fig. 7A).
Fig. 7.
Gene expression predominantly correlates with the dynamics of distal accessible genomic regions rather than promoters. (A) Distance from ATAC-seq peak to the associated TSS. (B–E) Temporal patterns of gene–peak associations (with promoter for genes associated with distal elements). Red: RNA-seq signal. Blue and green: ATAC signal of the distal element, respectively promoter, of the associated gene. Red box is SD. Gray shading is dark phase of the light–dark cycle. Baseline data are repeated over the entire time course as a stippled line.
SD significantly changed chromatin accessibility (DAS at T27 and/or T30) for 24% of all chromatin-gene expression pairs we detected (802 of 3,294). Of these pairs, 529 showed a change in gene expression at T27 or at T30. A large proportion of this set involved sleep–wake-driven genes (354 and 54 pairs involving model S and S+C genes, respectively, 82%). Furthermore, we found that 64 of the 802 pairs were associated to models with altered circadian amplitude after SD (37 and 27 pairs involved genes in model CA and S+CA genes, respectively). This suggests that chromatin changes could be partially responsible for both immediate and delayed dynamic alterations in gene expression induced by SD.
Only 38 of 3,294 significant gene–peak associations were proximal (i.e., the peak spanned the TSS), overlapping with only 2 genes of the top 34 pairs, namely Klhdc9, an interaction partner of CDK2-associated cyclin A1, Ccna1 (33) (model S) (Fig. 7B), and Ciart/Chrono (model S+CA) (Fig. 7C), an interaction partner and suppressor of the ARNTL and PER2 proteins (34).
Thus, the majority of genes correlated more strongly with a distal element than with the accessibility of their promoters. Generally, the average correlation coefficient between the expression and the ATAC signal spanning the TSS of all genes involved in an association was 0.16, while it was 0.5 for the correlation between expression and the ATAC signal of the top associated distal peak for the same genes (Fig. 7A). The top correlations to a distal element involved the environmental sensor Hif3a (model C) (Fig. 7D), and the immediate-early gene (IEG) Fosl2 (model S) (Fig. 7E). In the case of Hif3a, the distal element (Fig. 7D, blue line) displayed an immediate response to SD with a significant increase at T27 and T30, possibly a relative decrease at T36 before resuming the baseline pattern from T42, while the RNA oscillation (Fig. 7D, red line) was largely unperturbed. For Fosl2, we observed a fast response of the distal element together with the mRNA, plateauing already at T27 and followed by a fast recovery by T42, whereas accessibility at the promoter followed a different pattern. We observed similar relationships between gene expression and accessibility of the corresponding promoter and associated distal peak for the remaining genes of the top 34 correlations (SI Appendix, Fig. S5).
The widespread lack of correspondence between promoter accessibility dynamics and transcription hint at a model where transcription happens from an accessible promoter under the regulation of a distal element mediated by transcription factors (TF). A TF motif activity analysis (35, 36) taking advantage of our paired RNA-seq and ATAC-seq data predicted the serum response factor (SRF) motif to be by far the most statistically significant candidate in the entire temporal gene-expression dataset (i.e., the genes assigned to models S, C, CA, S+C and S+CA, z-score = 3.35) (SI Appendix, Fig. S6A). Randomizing the association between TF motif site counts and the expression profile showed that obtaining a z-score of 3.35 was highly statistically significant (P < 10−7). The inferred temporal activity of SRF (Methods) was consistent with sleep–wake-driven dynamics, paralleling the expression of the Srf transcript (SI Appendix, Fig. S6 B and C). The genes with the strongest contribution to the enrichment signal—namely Egr2, Junb, Fos, Arc, and Nr4a1—are IEGs and were all classified under model S, as was Srf itself (model S, w = 0.979). Scanning the open chromatin regions corresponding to <5 kb up- and downstream of the promoters of these genes, we found SRF binding sites that overlapped with chromatin immunoprecipitation sequencing (ChIP-seq) peaks against SRF in mouse fibroblasts (37) (see examples in SI Appendix, Fig. S6D).
Discussion
We have characterized the dynamics in the cerebral cortex of transcriptome and regulatory elements in relation to the sleep–wake distribution before, during, and after an SD, a 1-time and short intervention during the first half of the habitual rest phase of mice. To classify the genes according to their respective temporal expression patterns, we developed a model-selection approach according to sleep–wake-driven dynamics, time-of-day dynamics, interactions between the 2, with the possibility of an alteration of the oscillation amplitude. This set of models allowed us to determine the relative contributions of and interaction between the circadian and sleep–wake processes governing the expression of genes with 24-h rhythms. This approach, which recapitulated previously identified sleep–wake-driven genes (SI Appendix, Fig. S3 C and D), proved more powerful in identifying genes affected by SD than past single time-point differential expression studies.
SD Causes Long-Term Alterations of the Molecular Circadian Clock.
Our most striking finding is the pervasive control sleep–wake dynamics exerted on the expression of rhythmic genes, as well as the widespread and long-lasting impact of SD. Indeed, genes displaying rhythmic expression under baseline conditions were in majority assigned to the S model and even underrepresented in the C model. Moreover, genes perturbed late in the time course were enriched for GO terms related to circadian rhythms (Dataset S2). The molecular perturbations outlast the phenotypic changes, as seen particularly by the long-lasting effects of SD on model CA and S+CA, meaning that the mice have not yet recovered from SD despite behavioral and cortical electrophysiological measures of sleep need having returned to baseline. Although long-term electrophysiological consequences have been observed in the rat hypothalamus (38), the long-term aftereffects of a SD challenge are generally understudied.
Specifically, we found that most clock genes were affected by SD, in the form of a damped amplitude after SD, and conversely, we found an enrichment of rhythmic process genes in model S+CA. The damped amplitude we observed in the rhythmic expression of most of the known clock genes (i.e., Arntl, Per2, Per3, Nr1d1, Nr1d2, Cry2, Ciart, and the clock-output genes Dbp, Nfil3, Bhlhe41, all of which were model CA and S+CA genes) could be an important long-term consequence of SD. Indeed, previous work had already shown that the cortical expression of a number of clock genes is affected during SD (for reviews, see refs. 26, 39, and 40) and that SD acutely suppresses the specific DNA-binding of the circadian transcription factors BMAL1 and NPAS2 to their target genes Per2 and Dbp (41), demonstrating that prolonged wakefulness intervenes directly at the core of the circadian molecular machinery. Because Ciart is sensitive to and affects stress signaling pathways (34, 42), it can be conjectured that the stress associated with SD (24) could be a factor contributing to the sustained circadian dysregulation of the clock gene circuitry. Furthermore, this long-term damping of clock gene rhythms was all the more surprising given the fact that the observations were made under entrained light–dark conditions and in the presence of a largely unaffected diurnal sleep–wake distribution, 2 factors known to contribute to high amplitude clock gene expression. Because disrupted clock gene rhythms have been causally implicated in the etiology of disease like metabolic syndrome (reviewed in ref. 4), clock genes could be a final common molecular pathway underlying the etiology of metabolic syndrome associated both with insufficient good quality sleep and with circadian misalignment (39).
We observed that, for a subset of genes controlled by both the circadian and the sleep–wake-driven process (classified as model S+C and S+CA genes), the 2 processes worked in opposition, thereby limiting gene expression under baseline conditions. This “buffering” interaction provides an intriguing parallel with human and primate studies of cognitive performance under forced desynchrony or SD protocols, where it was found that the phase of the circadian wake-promoting signal is timed in such a way that it opposes the sleep–wake-dependent accumulation of sleep propensity and peaks in the hour prior to habitual sleep onset. This timing is essential for maintaining high and stable levels of attention and cognitive performance during the day, as well a consolidated period of sleep during the night (43–48).
Do Sleep–Wake-Driven Changes in Chromatin Accessibility Predict Transcriptome Dynamics?
Quantifying chromatin accessibility allowed us to discover a set of genomic regions as candidate first actors in a possible repertoire set in motion early during SD and giving rise to differential gene expression. The time course of the accessibility of these regions closely resembles an IEG response with a very fast modulation of the chromatin, detected already after the first 3 h of SD, a striking illustration of the plasticity of this compartment. This IEG-like response of both mRNA and accessibility, and the similarities of the rise and decay rates of the model S genes, raises the possibility that EEG δ-power is linked to, or even preceded by, molecular changes in IEGs in the brain.
Overall, dynamics in chromatin accessibility were most pronounced during SD, and no longer significantly differed from baseline by 12 h after SD (T42). Although we cannot exclude that this seemingly faster recovery is due to a lower sensitivity of the ATAC-seq signal relative to RNA-seq, these results do show that changes in chromatin accessibility start appearing early in the response to SD, confirming that chromatin accessibility is dynamic and can change on short time scales, even faster than observed in circadian oscillations (49).
The increased proportion of distal elements among DAS compared to all regions, together with the correlation of dynamic gene expression with distal elements rather than the respective promoters, is consistent with a scenario where expression is modulated by different enhancers or repressors interacting with an accessible promoter under the influence of regulator proteins. For example, SRF is bound to a constitutively open promoter, ready for an interaction with a distal element that changes its own activity and mediates the changes in gene expression. The implication of SRF as a candidate priming factor in the response to SD is compelling, as it plays a key role in activity-dependent modulation of synaptic strength (50), and its ortholog blistered is required to increase sleep after social enrichment in Drosophila (51, 52).
Conclusion
Our results imply that beyond an apparent recovery from SD lie deeper, more complex, and longer-lasting molecular perturbations, especially among clock genes. We also show that genes can seem unchanged by SD when sampled at a single time point, yet be affected by a profound perturbation later on or, in some cases, before that time point, as was illustrated here for Srf expression and chromatin accessibility. These perturbations eventually recover, as hinted by the absence of differential expression after 7 d; however, until baseline is reached, this temporary regulatory background could possibly cause the response to another exposure (repeated SD or other) to differ from that under the pre-SD baseline background. While it is debated whether repeated SD on subsequent days alter the homeostatic response at the phenotypic level in rats (EEG) (53, 54), recent studies in humans found that even 2 nights of recovery sleep were insufficient to completely reverse the metabolic perturbations caused by multiple nights of restricted sleep (55, 56). Follow-up experiments at the molecular level will show how such a transient “new baseline” due to only a partial recovery would influence the response to a second event occurring before full recovery.
Methods
Animals.
Male C57BL/6J mice were purchased from Charles River France and allowed to acclimate to our sleep study facility for 2 to 4 wk prior to habituation to the experimental setting. Animals were kept in accordance to the Swiss Animal Protection Act, and all experimental procedures were approved by the veterinary authorities of the state of Vaud (i.e., the Service de la Consommation et des Affaires Vétérinaires [SCAV]).
Surgery, EEG Recording, and Analysis.
The EEG cohort consisted of 12 male C57BL/6J mice 10 to 12 wk at the time of SD from another study (12). Surgical implantation of electrodes, EEG recording, data collection, and processing were performed according to our standard procedure (57). EEG was recorded from 2 d prior to SD (which were averaged to constitute a 24-h baseline) until 2 d after SD, and 5 additional days in 6/12 mice. For details on EEG spectral analysis, simulation of process S and statistics, see SI Appendix, Materials and Methods.
SD and Tissue Collection.
Mice for tissue collection were divided into SD and non-SD (controls, Ctr), with 3 to 4 individuals per time point per condition. After habituation to the experimental setting, the SD mice (now 11 to 12 wk) were SD for 6 h starting at light onset (ZT0 to ZT6) as described in ref. 57, and allowed to recover according to the tissue collection schedule (Fig. 1A and SI Appendix, Materials and Methods). Control mice were killed at the same time of day as SD mice. The cortex was rapidly dissected and flash-frozen in liquid nitrogen.
Tissue Processing and Sequencing Library Preparation.
Tissue from each individual was used for both RNA-seq and ATAC-seq. RNA-seq libraries were prepared from total RNA by a standard Illumina protocol. ATAC-seq libraries were generated from 100,000 nuclei according to ref. 58, with minor modifications.
Sequencing Data Analysis.
RNA-seq data were analyzed using kallisto v0.43.0 (59) followed by sleuth (60) with batch effect correction by ComBat [R package sva (61)]. ATAC-seq reads were processed using bowtie2 (62), samtools (63), and bedtools, followed by peak-calling and quantification by Macs2 (64) and HTSeq.
Clustering of mRNA and ATAC-Seq Profiles.
We performed k-means clustering on genes displaying a statistically significant variation over time, as defined by a likelihood ratio test in sleuth for RNA-seq and edgeR for ATAC-seq, and empirically chose k = 10 as a balance between variance explained and generalizability of each cluster.
mRNA Time-Course Analysis.
We used a model selection approach to classify the temporal log mRNA abundance of all 13,842 expressed genes into the scenarios described in Results (see also SI Appendix, Fig. S1), penalizing for model complexity using the BIC. Rationale, equations, and mathematical details are found in SI Appendix, Materials and Methods.
Peak-To-Gene Expression Association.
To associate gene-expression dynamics with chromatin accessibility dynamics, we used a Pearson correlation coefficient across the samples and confined the possible association tests to previously defined topological interaction domains in cortex tissue from ref. 32.
Prediction of TF Binding Site Activity in Promoters.
We inferred TF activity based on the presence of TF motifs within ATAC-seq–positive regions and the abundance of the nearby transcript, using MARA, as previously described (35, 36), with position weight matrices of 179 mouse TFs (http://swissregulon.unibas.ch).
Data Accessibility.
Raw read files (.fastq), RNA transcripts per million, ATAC-seq peak calls, and quantification are publicly available in the Gene Expression Omnibus/Sequence Read Archive repository under accession no. GSE140345 (65).
Code Availability.
Code to run the model selection analysis is publicly available, found at https://github.com/jakeyeung/SleepDepModelSelection (66).
Supplementary Material
Acknowledgments
We thank Shanaz Diessler, Marieke Hoekstra, Konstantinos Kompotis, Dessislava Petrova, and the Lausanne Genomics Technologies Facility for technical assistance. Computations and analyses were performed at Vital-IT (https://www.vital-it.ch/). C.N.H. and M.J. were funded by a Grant 31003A_173182 of the Swiss National Science Foundation (to P.F.). J.Y. benefited from the Natural Sciences and Engineering Research Council of Canada Postgraduate Studies Doctoral scholarship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE140345). Code to run the model selection analysis is available on GitHub https://github.com/jakeyeung/SleepDepModelSelection.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910590116/-/DCSupplemental.
References
- 1.Borbély A. A., A two process model of sleep regulation. Hum. Neurobiol. 1, 195–204 (1982). [PubMed] [Google Scholar]
- 2.Daan S., Beersma D. G., Borbély A. A., Timing of human sleep: Recovery process gated by a circadian pacemaker. Am. J. Physiol. 246, R161–R183 (1984). [DOI] [PubMed] [Google Scholar]
- 3.Dijk D. J., Franken P., “Interaction of sleep homeostasis and circadian rhythmicity: Dependent or independent systems?” in Principles and Practice of Sleep Medicine, Kryger M. H., Roth T., DementR W. C., Eds. (Saunders/Elsevier, 2005) pp. 418–434. [Google Scholar]
- 4.Maury E., Hong H. K., Bass J., Circadian disruption in the pathogenesis of metabolic syndrome. Diabetes Metab. 40, 338–346 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Bass J., Takahashi J. S., Circadian integration of metabolism and energetics. Science 330, 1349–1354 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Takahashi J. S., Transcriptional architecture of the mammalian circadian clock. Nat. Rev. Genet. 18, 164–179 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fisk A. S., et al. , Light and cognition: Roles for circadian rhythms, sleep, and arousal. Front. Neurol. 9, 56 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.da Costa Souza A., Ribeiro S., Sleep deprivation and gene expression. Curr. Top. Behav. Neurosci. 25, 65–90 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Franken P., Dijk D. J., Circadian clock genes and sleep homeostasis. Eur. J. Neurosci. 29, 1820–1829 (2009). [DOI] [PubMed] [Google Scholar]
- 10.Cirelli C., Faraguna U., Tononi G., Changes in brain gene expression after long-term sleep deprivation. J. Neurochem. 98, 1632–1645 (2006). [DOI] [PubMed] [Google Scholar]
- 11.Cirelli C., Tononi G., Gene expression in the brain across the sleep-waking cycle. Brain Res. 885, 303–321 (2000). [DOI] [PubMed] [Google Scholar]
- 12.Diessler S., et al. , A systems genetics resource and analysis of sleep regulation in the mouse. PLoS Biol. 16, e2005750 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Maret S., et al. , Homer1a is a core brain molecular correlate of sleep loss. Proc. Natl. Acad. Sci. U.S.A. 104, 20090–20095 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anafi R. C., et al. , Sleep is not just for the brain: Transcriptional responses to sleep in peripheral tissues. BMC Genomics 14, 362 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Möller-Levet C. S., et al. , Effects of insufficient sleep on circadian rhythmicity and expression amplitude of the human blood transcriptome. Proc. Natl. Acad. Sci. U.S.A. 110, E1132–E1141 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arnardottir E. S., et al. , Blood-gene expression reveals reduced circadian rhythmicity in individuals resistant to sleep deprivation. Sleep 37, 1589–1600 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Massart R., et al. , The genome-wide landscape of DNA methylation and hydroxymethylation in response to sleep deprivation impacts on synaptic plasticity genes. Transl. Psychiatry 4, e347 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Callaghan E. K., Green E. W., Franken P., Mongrain V., Omics approaches in sleep-wake regulation. Handb. Exp. Pharmacol. 253, 59–81 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Franken P., Chollet D., Tafti M., The homeostatic regulation of sleep need is under genetic control. J. Neurosci. 21, 2610–2621 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Borbély A. A., Daan S., Wirz-Justice A., Deboer T., The two-process model of sleep regulation: A reappraisal. J. Sleep Res. 25, 131–143 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Archer S. N., et al. , Mistimed sleep disrupts circadian regulation of the human transcriptome. Proc. Natl. Acad. Sci. U.S.A. 111, E682–E691 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kass R. E., Raftery A. E., Bayes factors. J. Am. Stat. Assoc. 90, 773–795 (1995). [Google Scholar]
- 23.Franken P., et al. , NPAS2 as a transcriptional regulator of non-rapid eye movement sleep: Genotype and sex interactions. Proc. Natl. Acad. Sci. U.S.A. 103, 7118–7123 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mongrain V., et al. , Separating the contribution of glucocorticoids and wakefulness to the molecular and electrophysiological correlates of sleep homeostasis. Sleep 33, 1147–1157 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Curie T., et al. , Homeostatic and circadian contribution to EEG and molecular state variables of sleep regulation. Sleep 36, 311–323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang H., Liu Y., Briesemann M., Yan J., Computational analysis of gene regulation in animal sleep deprivation. Physiol. Genomics 42, 427–436 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Schnell A., et al. , The nuclear receptor REV-ERBα regulates Fabp7 and modulates adult hippocampal neurogenesis. PLoS One 9, e99883 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilken M. S., et al. , DNase I hypersensitivity analysis of the mouse brain and retina identifies region-specific regulatory elements. Epigenetics Chromatin 8, 8 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thurman R. E., et al. , The accessible chromatin landscape of the human genome. Nature 489, 75–82 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fullard J. F., et al. , Open chromatin profiling of human postmortem brain infers functional roles for non-coding schizophrenia loci. Hum. Mol. Genet. 26, 1942–1951 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dixon J. R., et al. , Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Diederichs S., et al. , Identification of interaction partners and substrates of the cyclin A1-CDK2 complex. J. Biol. Chem. 279, 33727–33741 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Anafi R. C., et al. , Machine learning helps identify CHRONO as a circadian clock component. PLoS Biol. 12, e1001840 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Balwierz P. J., et al. , ISMARA: Automated modeling of genomic signals as a democracy of regulatory motifs. Genome Res. 24, 869–884 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yeung J., et al. , Transcription factor activity rhythms and tissue-specific chromatin interactions explain circadian gene expression across organs. Genome Res. 28, 182–191 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Esnault C., et al. , Rho-actin signaling to the MRTF coactivators dominates the immediate transcriptional response to serum in fibroblasts. Genes Dev. 28, 943–958 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fifel K., Meijer J. H., Deboer T., Long-term effects of sleep deprivation on neuronal activity in four hypothalamic areas. Neurobiol. Dis. 109, 54–63 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Franken P., A role for clock genes in sleep homeostasis. Curr. Opin. Neurobiol. 23, 864–872 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Mang G. M., Franken P., Genetic dissection of sleep homeostasis. Curr. Top. Behav. Neurosci. 25, 25–63 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Mongrain V., La Spada F., Curie T., Franken P., Sleep loss reduces the DNA-binding of BMAL1, CLOCK, and NPAS2 to specific clock genes in the mouse cerebral cortex. PLoS One 6, e26622 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goriki A., et al. , A novel protein, CHRONO, functions as a core component of the mammalian circadian clock. PLoS Biol. 12, e1001839 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Edgar D. M., Dement W. C., Fuller C. A., Effect of SCN lesions on sleep in squirrel monkeys: Evidence for opponent processes in sleep-wake regulation. J. Neurosci. 13, 1065–1079 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dijk D. J., Duffy J. F., Czeisler C. A., Circadian and sleep/wake dependent aspects of subjective alertness and cognitive performance. J. Sleep Res. 1, 112–117 (1992). [DOI] [PubMed] [Google Scholar]
- 45.Dijk D. J., Czeisler C. A., Contribution of the circadian pacemaker and the sleep homeostat to sleep propensity, sleep structure, electroencephalographic slow waves, and sleep spindle activity in humans. J. Neurosci. 15, 3526–3538 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dijk D. J., Czeisler C. A., Paradoxical timing of the circadian rhythm of sleep propensity serves to consolidate sleep and wakefulness in humans. Neurosci. Lett. 166, 63–68 (1994). [DOI] [PubMed] [Google Scholar]
- 47.Muto V., et al. , Local modulation of human brain responses by circadian rhythmicity and sleep debt. Science 353, 687–690 (2016). [DOI] [PubMed] [Google Scholar]
- 48.Maire M., et al. , Human brain patterns underlying vigilant attention: Impact of sleep debt, circadian phase and attentional engagement. Sci. Rep. 8, 970 (2018). Erratum in: Sci. Rep.9, 12379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sobel J. A., et al. ; CycliX consortium , Transcriptional regulatory logic of the diurnal cycle in the mouse liver. PLoS Biol. 15, e2001069 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ramanan N., et al. , SRF mediates activity-induced gene expression and synaptic plasticity but not neuronal viability. Nat. Neurosci. 8, 759–767 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Donlea J. M., Ramanan N., Shaw P. J., Use-dependent plasticity in clock neurons regulates sleep need in Drosophila. Science 324, 105–108 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Donlea J. M., Ramanan N., Silverman N., Shaw P. J., Genetic rescue of functional senescence in synaptic and behavioral plasticity. Sleep 37, 1427–1437 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim Y., Laposky A. D., Bergmann B. M., Turek F. W., Repeated sleep restriction in rats leads to homeostatic and allostatic responses during recovery sleep. Proc. Natl. Acad. Sci. U.S.A. 104, 10697–10702 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Leemburg S., et al. , Sleep homeostasis in the rat is preserved during chronic sleep restriction. Proc. Natl. Acad. Sci. U.S.A. 107, 15939–15944 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Depner C. M., et al. , Ad libitum weekend recovery sleep fails to prevent metabolic dysregulation during a repeating pattern of insufficient sleep and weekend recovery sleep. Curr. Biol. 29, 957–967.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ness K. M., et al. , Two nights of recovery sleep restores the dynamic lipemic response, but not the reduction of insulin sensitivity, induced by five nights of sleep restriction. Am. J. Physiol. Regul. Integr. Comp. Physiol. 316, R697–R703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mang G. M., Franken P., Sleep and EEG phenotyping in mice. Curr. Protoc. Mouse Biol. 2, 55–74 (2012). [DOI] [PubMed] [Google Scholar]
- 58.Buenrostro J. D., Wu B., Chang H. Y., Greenleaf W. J., ATAC-seq: A method for assaying chromatin accessibility genome-wide Curr. Protoc. Mouse Biol. 109, 21.29.1–21.29.9 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bray N. L., Pimentel H., Melsted P., Pachter L., Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 34, 525–527 (2016). [DOI] [PubMed] [Google Scholar]
- 60.Pimentel H., Bray N. L., Puente S., Melsted P., Pachter L., Differential analysis of RNA-seq incorporating quantification uncertainty. Nat. Methods 14, 687–690 (2017). [DOI] [PubMed] [Google Scholar]
- 61.Leek J. T., et al. , sva: Surrogate Variable Analysis. R package version 3.34.0. Bioconductor, 10.18129/B9.bioc.sva (2019). [DOI] [Google Scholar]
- 62.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li H., et al. ; 1000 Genome Project Data Processing Subgroup , The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhang Y., et al. , Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hor C. N., et al. , RNA-seq and ATAC-seq data. NCBI Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE140345. Deposited 13 November 2019.
- 66.Yueng J., Model selection code. GitHub. https://github.com/jakeyeung/SleepDepModelSelection. Deposited 7 November 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw read files (.fastq), RNA transcripts per million, ATAC-seq peak calls, and quantification are publicly available in the Gene Expression Omnibus/Sequence Read Archive repository under accession no. GSE140345 (65).