Significance
To develop a cure for HIV-1, it is necessary to understand how infected cells persist despite treatment. Integrated HIV DNA (proviruses) can be distinguished by their sites of integration into the host genome and by their proviral sequence. We applied multiple-displacement amplification (MDA) to single proviruses isolated from blood and lymph nodes to determine their integration sites and full-length sequences. We found that identical subgenomic HIV-1 sequences can result from either clonal expansion or from genetic bottlenecks that occurred with transmission or with the emergence of drug resistance mutations. Furthermore, we show this MDA approach can be used to infer intact ancestral HIV-1 sequences from the archives of defective ones, providing an approach for investigations of HIV-1 evolution in vivo.
Keywords: HIV persistence, proviral structure, integration site analysis
Abstract
Understanding HIV-1 persistence despite antiretroviral therapy (ART) is of paramount importance. Both single-genome sequencing (SGS) and integration site analysis (ISA) provide useful information regarding the structure of persistent HIV DNA populations; however, until recently, there was no way to link integration sites to their cognate proviral sequences. Here, we used multiple-displacement amplification (MDA) of cellular DNA diluted to a proviral endpoint to obtain full-length proviral sequences and their corresponding sites of integration. We applied this method to lymph node and peripheral blood mononuclear cells from 5 ART-treated donors to determine whether groups of identical subgenomic sequences in the 2 compartments are the result of clonal expansion of infected cells or a viral genetic bottleneck. We found that identical proviral sequences can result from both cellular expansion and viral genetic bottlenecks occurring prior to ART initiation and following ART failure. We identified an expanded T cell clone carrying an intact provirus that matched a variant previously detected by viral outgrowth assays and expanded clones with wild-type and drug-resistant defective proviruses. We also found 2 clones from 1 donor that carried identical proviruses except for nonoverlapping deletions, from which we could infer the sequence of the intact parental virus. Thus, MDA-SGS can be used for “viral reconstruction” to better understand intrapatient HIV-1 evolution and to determine the clonality and structure of proviruses within expanded clones, including those with drug-resistant mutations. Importantly, we demonstrate that identical sequences observed by standard SGS are not always sufficient to establish proviral clonality.
Clonal populations of infected CD4+ T cells carrying intact HIV-1 proviruses that can survive and continue to divide for over a decade of suppressive antiretroviral therapy (ART) are a major obstacle to curing HIV-1 infection (1–9). Thus far, the integration sites and full proviral sequences of only a few infectious proviruses within clones of expanded cells have been determined (5, 10). Although there are reports of replication-competent proviruses in “probable” clones based on identical full-length proviral sequences (2, 6, 11), clonality was not proven in these studies by demonstrating identical integration sites in multiple cells. The first replication-competent provirus in an expanded clone with a known integration site, called AMBI-1, was found in a donor who had been on ART for greater than 10 y (1).
The presence of identical subgenomic or near–full-length (NFL) HIV-1 sequences in the plasma of individuals treated in chronic infection (12, 13), in rebound virus after ART interruption (7, 8), and in infectious virus recovery assays (2, 4, 6) is consistent with virus production from clonally expanded cells. It does not, however, prove that these viruses are produced by HIV-1–infected cell clones because identical viral sequences can arise from genetic bottlenecks during infection, including the persistence of transmitted/founder viruses (14), the selection of drug-resistant mutants following incompletely suppressive ART (15, 16), or the selection of immune escape mutations (17–20). Distinguishing among these possibilities is essential to understanding the origin and maintenance of the HIV-1 reservoir.
The widespread distribution of HIV-1 integration sites in the host genome (21) and the finding that the large majority of proviruses have the same viral sequences at the junction of viral and cellular DNA, allows integration site analysis (ISA) to unambiguously identify proviruses that originated from a single infection and integration event (1, 10, 22). Once the integration site of a provirus of interest is known, this information can be used to genetically characterize the provirus, rescue and characterize the virus it encodes, quantify the size of the HIV-1–infected cell clone, and monitor its cellular dynamics, anatomic distribution, and response to therapy (10).
The provirus at the AMBI-1 clonal integration site was characterized using PCR primers designed to amplify overlapping host-proviral sequences (10). This approach, although useful, is inefficient and requires the design and optimization of integration site-specific PCR assays for each individual provirus. The need for an efficient, standardized method to characterize intact proviruses in HIV-1–infected cell clones has been apparent since the discovery of HIV-1–infected, expanded clones in clinical samples (1, 9). Here, we describe an approach that combines multiple-displacement amplification (MDA) with ISA and NFL sequencing, similar to a recently described method (5), to determine the clonality of cells that carry proviruses with identical P6–PR–RT subgenomic sequences and to characterize the intactness of proviruses within confirmed infected cell clones.
After optimizing and validating the methods on proviruses in the well-characterized, HIV-infected ACH-2 cell line, we analyzed proviruses in peripheral blood mononuclear cells (PBMCs) from 5 ART-treated donors and lymph node mononuclear cells (LNMCs) from 2 of the 5 donors. We found 1 intact provirus in a clone of expanded cells and identified several defective proviruses, including those containing drug resistance mutations, within expanded cell clones. We also found proviruses with identical subgenomic sequences but different sites of integration, meaning they were derived from independent integration events. This occurrence likely results from genetic bottlenecks that may have been imposed shortly after the transmission/infection event or following selection for specific mutations. Last, we found different expanded T cell clones carrying identical proviruses except for nonoverlapping deletions, allowing us to reconstruct the intact viral ancestor from the intrapatient HIV-1 proviral landscape. The findings in this study show that multiple-displacement amplification–single-genome sequencing (MDA-SGS) can be used to characterize the HIV-1 reservoir and to perform “viral reconstruction” from the remnants of defective proviruses, informing the evolutionary history of the virus from the time of transmission.
Results
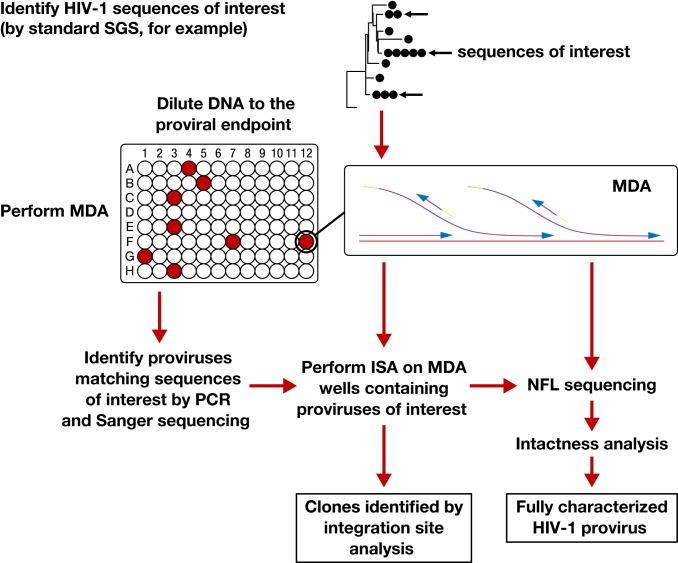
MDA-SGS Workflow.
To determine the origin of clonal proviral DNA sequences observed in HIV-infected individuals during suppressive ART, we developed a combined SGS and ISA approach (Fig. 1). First, SGS encompassing the region encoding P6, protease, and 900 bp of reverse transcriptase (P6–PR–RT) was applied to PBMCs and LNMCs to detect proviral populations with identical subgenomic sequences. Subsequently, we applied a custom statistical test to determine, based on the size and diversity of the observed proviral population, if groups (rakes) of identical subgenomic sequences may be more frequent than expected by chance (details in Methods). Next, genomic DNA was diluted in a microtiter plate such that each well contained, on average, less than 1 provirus with P6–PR–RT, typically, along with DNA from 1,000 to 100,000 uninfected cells. MDA was performed in each well to amplify all input DNA including any proviruses present and their flanking sequences. Individual MDA wells were screened by P6–PR–RT PCR and sequencing to identify wells containing proviruses of interest. Third, integration sites were determined for the proviruses of interest using a previously described ISA method (1, 10). Finally, the same MDA wells were used to fully characterize the proviruses by nested NFL PCR amplification and Sanger or next-generation sequencing (SI Appendix, Fig. S1). A custom “intactness” pipeline was then used to determine whether the sequenced proviruses were intact (detailed definition for intactness in Methods) and potentially replication competent (pipeline available at https://home.ncifcrf.gov/hivdrp/resources.html).
Fig. 1.
Workflow for MDA-SGS. Extracted DNA is aliquoted so that individual proviruses, along with background genomic DNA, can be independently amplified ∼1,000-fold by MDA. This amplification allows the same provirus to be identified by SGS (1–3), analyzed by ISA (integration sites analysis) (1, 10, 22) and NFL (near–full-length) sequencing, followed by further downstream analyses, if needed. Infected cell clones are identified by finding identical integration sites across multiple MDA wells.
Validation of MDA-SGS on Proviruses in ACH-2 Cells.
To validate the MDA-SGS assay, ACH-2 cells, a line of semiclonal, stably HIV-infected CD4+ T cells (23), were spiked (1:1,000) into uninfected CEM cells (also a CD4+ T cell line) to mimic the average ratio of infected to uninfected PBMCs in ART-suppressed HIV+ donors (24). DNA extracted from the mixture of cells was diluted to the point at which less than 30% of wells were positive for HIV-1 DNA by P6–PR–RT nested PCR. The endpoint-diluted DNA was divided into 96 aliquots, each of which was amplified ∼100- to 10,000-fold using MDA (25, 26). The MDA produced DNA segments of >10 kb in 18-h reactions (SI Appendix, Fig. S2A).
Integration sites were obtained for 4 MDA-amplified proviruses in the ACH-2 cells. All were on chromosome 7 at the same position within the NT5C3A gene, identical to the major reported integration site in ACH-2 cells (27, 28). From one MDA well, a 4-fragment PCR amplification strategy (SI Appendix, Fig. S1) generated amplicons that were used for Sanger sequencing. The 9,493-bp sequence of the NFL HIV genome obtained was assembled from the amplicons (accession no. MN691959). The resulting sequence contained a single-base pair difference and a 15-bp insertion/duplication in the V1/V2 loop compared to the ACH-2 proviral sequence reported by Symons et al. (27). The sequence was 92 bp different from the HIV laboratory strain LAV that was reportedly used to generate the ACH-2 cell line (23), likely the combined result of in vitro changes during cell line generation, viral passage, and, perhaps, including some sequencing error.
Validation of MDA-SGS on a Clinical Sample.
Previously, a replication-competent provirus, AMBI-1, was described to be present in a T cell clone from an infected individual (patient 1 of ref. 1). In that report and a subsequent one (10), the sequence at the AMBI-1 integration site was identified and its full-length provirus was sequenced, shown to produce infectious virus, and to match the plasma virus that persisted during ART in this individual (1, 10). We used effector memory CD4+ T cells from patient 1, a subset in which AMBI-1 was highly represented, to validate MDA-SGS on a clinical sample and to ensure that we could isolate the previously characterized AMBI-1 clone and obtain the same integration site and NFL sequence.
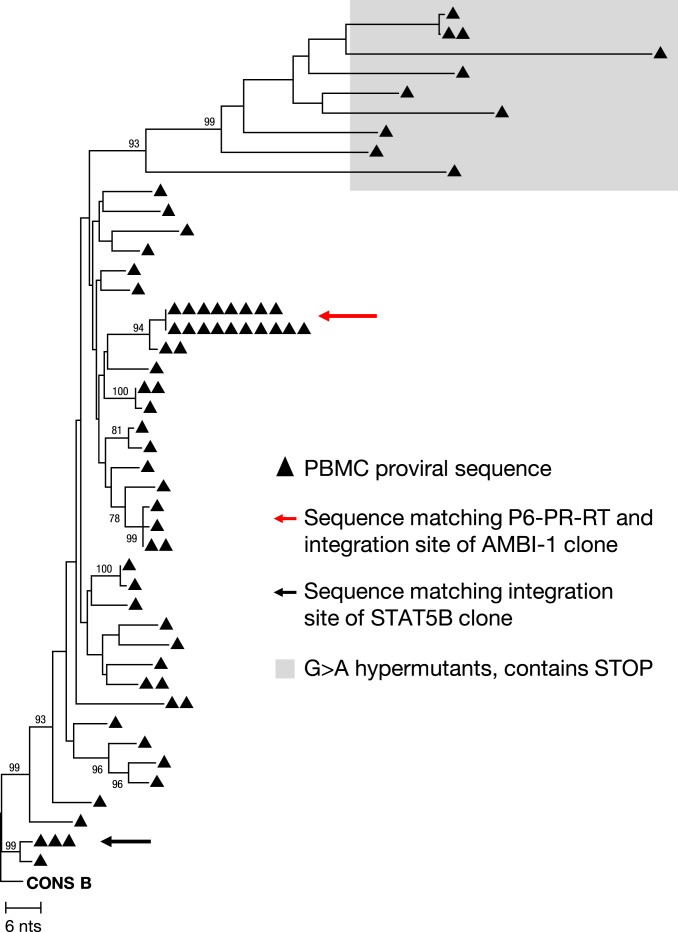
We tested the robustness of the amplification reaction from patient 1 DNA using droplet digital PCR (ddPCR) to quantify HIV long terminal repeat (LTR) sequences following MDA (29). ddPCR results showed that MDA-mediated LTR amplification ranged from 280 to 47,000-fold from one well to the next, providing sufficient material for downstream NFL sequencing and ISA. MDA-SGS on 5 96-well plates resulted in 68 wells with P6–PR–RT sequences, of which 18 matched the sequence of AMBI-1 (Fig. 2, red arrow). Twelve of the 18 yielded the known AMBI-1 integration site, 2 failed ISA, and 4 had integrations other than the AMBI-1 site including 1 well with 3 different integration sites reflecting a mixture of proviruses in that well (Table 1). The 4 wells with sites that did not match the AMBI-1 integration site failed to generate 9-kb NFL amplicons, suggesting defective proviruses. However, one produced a ∼4,500-bp sequence from the gag leader to integrase that matched the AMBI-1 provirus to within 1 bp (accession no. MN692144), suggesting a common ancestor. These results demonstrated that, using MDA-SGS, we could isolate proviruses of interest and identify their sites of integration. Nested NFL PCR amplification (SI Appendix, Fig. S2B) and Sanger and PacBio sequencing of the AMBI-1 provirus from 3 confirmed AMBI-1 MDA wells resulted in 8,977-bp coverage matching the previously reported AMBI-1 sequence within 2 bp by either method of sequencing (10) (accession nos. MN692145–MN692147).
Fig. 2.
Neighbor-joining phylogenetic tree of P6–PR–RT MDA-SGS on effector memory T cells from patient 1. The red arrow indicates sequences matching P6–PR–RT of the provirus in the AMBI-1 clone, a provirus resulting in virus continuously recovered in plasma during ART and shown to be replication competent (1, 10). The black arrow indicates group of identical sequences with integration site matching the STAT5B expanded clone reported in Maldarelli et al. (1). Bootstrap values are shown for nodes greater than 70% confidence.
Table 1.
Integration sites in groups (rakes) of identical P6–PR–RT sequences
| PID | Rake ID* | Drug resistance | Integration site (hg19) (no. of MDA wells) | Chromosome | Gene (or nearest gene) | Provirus orientation relative to gene | Cell clone in rake |
| 1 | AMBI-1 | None | Ambiguous† (12) | Unmapped | Unmapped | Unmapped | Yes |
| 204385823 (1) | 2 | RAPH1 | With | ||||
| 46748345 (1) | 13 | LCP1 | With | ||||
| 153344124 (1) | X | MECP2 | Against | ||||
| 87241304† (1)‡ | 2 | PLGLB1 | Against | ||||
| 155077386 (1)‡ | 6 | SCAF8 | Against | ||||
| 75740900 (1)‡ | 10 | VCL | Intergenic | ||||
| STAT5B | None | 40421711† (2) | 17 | STAT5B | Against | Yes | |
| 1683 | 1 | None | 175452717† (13) | 2 | WIPF1 | Against | Yes (3) |
| 28169752 (2) | 16 | XPO6 | Against | ||||
| 35327305† (1) | 17 | AATF | With | ||||
| 90744698 (1) | 6 | BACH2 | With | ||||
| 101889112 (1) | 9 | TGFBR1 | With | ||||
| 64664850 (1) | 11 | ATG2A | Against | ||||
| 76753364 (1) | 17 | CYTH1 | With | ||||
| 2669 | CYLD§ | K103N, M184V, P225H, K238T | 50797769 (2) | 16 | CYLD | Against | Yes |
| 2 | K103N | 38340281† (5) | 15 | TMCO5A | Intergenic | Yes | |
| 73035568 (1) | 15 | BBS4 | Intergenic | ||||
| 7285669 (1)‡ | 7 | C1GALT1 | Against | ||||
| 98741685 (1)‡ | 9 | C9orf102 | Intergenic | ||||
| 43143496 (1)‡ | 15 | TTBK2 | Against | ||||
| 68171424b (1)‡ | 16 | NFATC3 | Against | ||||
| 3 | K103N | 98261540 (2) | 5 | CHD1 | With | Yes | |
| 203502280 (1) | 2 | FAM117B | Against | ||||
| 151719353 (1) | 4 | LRBA | With | ||||
| 150393718 (1) | 7 | GIMAP2 | Intergenic | ||||
| 1033517 (1) | 19 | CNN2 | Against | ||||
| 34701163 (1) | 19 | LSM14A | Against | ||||
| 247015648 (1)‡ | 1 | AHCTF1 | With | ||||
| 61507403 (1)‡ | 2 | USP34 | With | ||||
| 40498417 (1)‡ | 17 | STAT3 | Against | ||||
| 4 | None | 12501213 (2) | 1 | VPS13D | Against | Yes | |
| 3162 | 5 | I84V, L90M, D67N, K70R, M184V, K219Q | 3124617 (1) | 4 | HTT | Against | No |
| 67837014 (1) | 11 | CHKA | Against | ||||
| 28172853 (1) | 16 | XPO6 | Against | ||||
| 50275074 (1) | 22 | ZBED4 | Against | ||||
| 117733664 (3) | 9 | TNFSF8 | Intergenic | ||||
| 6 | None | 81002803 (1) | 17 | B3GNTL1 | Against | ||
| R-09 | 7 | None | 425766†,¶ (2) | 4 | ABCA11P | Against |
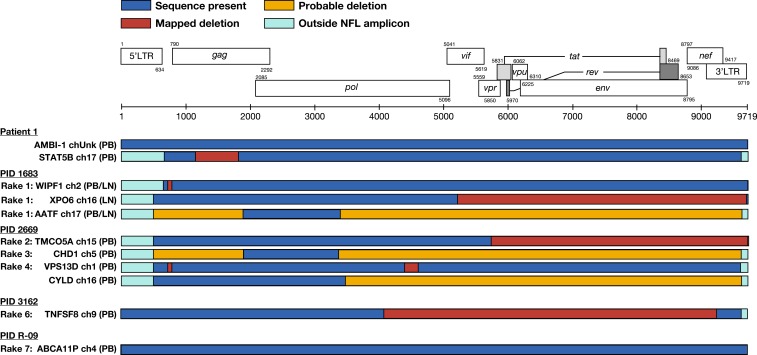
In addition to the AMBI-1 clone, we were also able to use MDA-SGS to characterize the provirus in another expanded T cell clone reported in Maldarelli et al. (1) integrated into the STAT5B gene of chromosome 17 (Fig. 2, black arrow). MDA-SGS revealed that the provirus in the STAT5B clone contained a 663-bp deletion in gag, rendering the capsid protein defective, and, therefore, the virus not replication competent (Fig. 3). Together, performing MDA-SGS on a sample with previously described expanded T cell clones, including one that is known to be replication competent, showed that the MDA-SGS method is robust for characterizing NFL proviral sequences and their sites of integration in host genomes.
Fig. 3.
Near–full-length (NFL) proviral structures. Structures of proviruses within cell clones aligned to the HXB2 reference sequence (adapted from Los Alamos National Laboratory). Corresponding rake ID (Fig. 4), integration site (gene/nearest gene, chromosome), and cell source (PB, LN) are given. A mapped deletion (red box) denotes sequence obtained spanning the deletion. Probable deletion (yellow box) denotes failure at either amplification or sequencing. All proviruses detected were shown to be defective except for the AMBI-1 provirus and the newly identified ABCA11P provirus in R-09. LN, lymph node; PB, peripheral blood.
Identical Subgenomic Sequences Occur More Often than Expected by Chance.
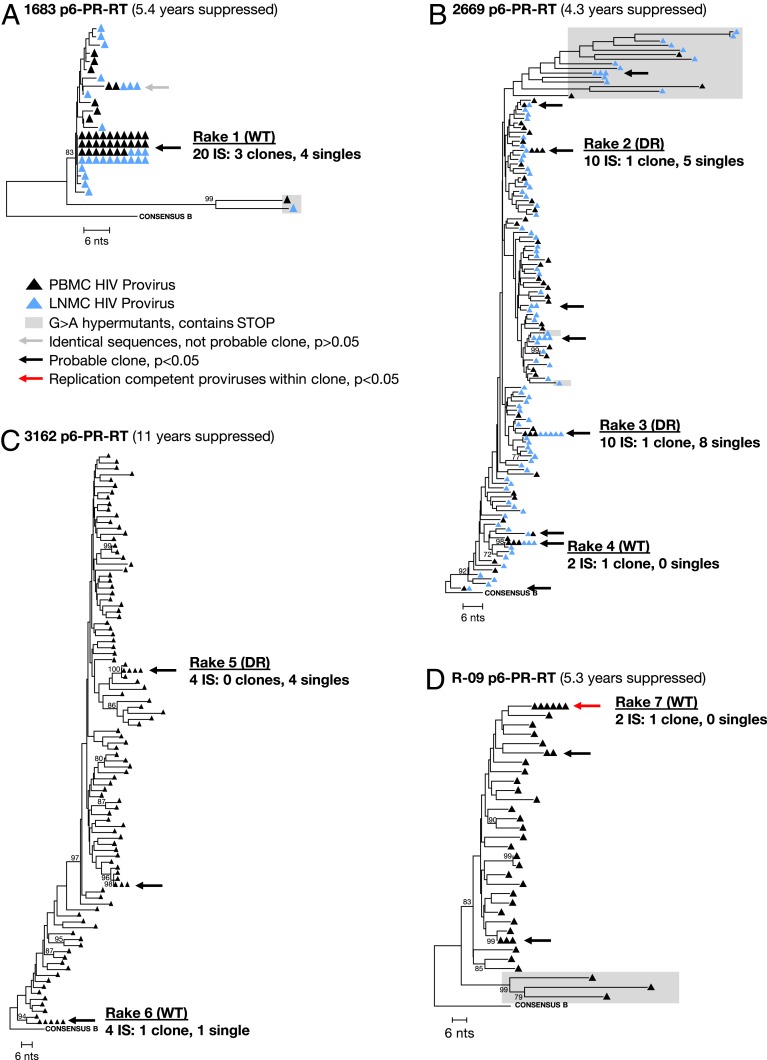
After validating MDA-SGS on ACH-2 cells and on cells donated from patient 1 (1, 10), the method was applied to samples from 3 donors in the SCOPE cohort at the University of California, San Francisco (30), and 1 donor enrolled through the University of Pittsburgh to determine whether proviruses with identical P6–PR–RT sequences are derived from cell clones and to determine the structures of the proviruses in each detected cell clone (Table 2). The intrahost proviral populations in the 4 donors were first analyzed by standard P6–PR–RT SGS, and clusters (rakes) of identical sequences were identified (Fig. 4).
Table 2.
Donor characteristics
| PID | Gender | Maximum pre-ART viral load,* c/mL | CD4+ nadir, cells/µL | Approximate years of infection before initial ART | Current ART regimen | Total years of ART experience | Years of viral suppression on ART at sampling | Plasma viral load on ART, c/mL | CD4+ on ART, cells/µL |
| Patient 1† | Male | 412,033 | 16 | Unknown | TDF, FTC, RTG | 13 | 1.0, not suppressed | <50, 134 | 137, 153 |
| 1683 | Male | 134,406 | 452 | 2.0 | FTC, TDF, RTV, DRV | 5.7 | 5.4 | <40 | 1,348 |
| 2669‡ | Male | Unknown | 205 | ≤22.0 | ABC, 3TC, DTG | 5.5 | 4.3 | <40 | 681 |
| 3162‡ | Male | 171,000 | 147 | 11.0 | RTV, DRV, ABC, DTG, 3TC‡ | 20 | 11.2 | <40 | 558 |
| R-09 | Male | 97,000 | 105 | Unknown | EFV, FTC, TDF | 9.8 | 5.3§ | ≤133 | 380 |
Abbott Diagnostics RealTime HIV-1 PCR Assay.
ART failure due to drug resistance (Fig. 4) prior to suppression on new regimen including dolutegravir.
PID R-09 maintained viral suppression ≤133 copies/mL for 5.3 y and <40 copies/mL for 0.9 y prior to sample time point.
Fig. 4.
Neighbor-joining trees of viral sequences obtained from donors on ART. SGS of P6–PR–RT region of viral DNA from SCOPE donors 1683 (A), 2669 (B), 3162 (C), and Pitt donor R-09 (D). Average pairwise distance: 0.2%, 1.6%, 2.7%, and 1.9%, respectively. The gray arrow indicates a group that had fewer than the number of identical sequences expected by chance (P > 0.05); the black arrows indicate groups with more than the expected number of identical sequences, and the red arrow indicates groups that matched variants that grew out in VOA. Groups that were investigated for clonality are labeled rake #1 to #7. Within each group, the number of total integration sites, confirmed expanded clones, and single integration sites are given. Integration site (IS), wild type (WT), and drug-resistance mutations (DR). Bootstrap values are shown for nodes with greater than 70% confidence.
Because the P6–PR–RT subgenomic region represents only ∼15% of the HIV-1 genome, it is possible that there are proviruses that are identical in this region and are genetically distinct elsewhere. To evaluate whether proviruses with identical P6–PR–RT sequences may have resulted from either clonal expansion of infected cells or from a genetic bottleneck imposed on replicating virus prior to ART, we designed a statistical test to estimate whether the number of identical sequences in each rake was detected more than the number of times expected by chance, given the overall genetic diversity of the proviral population, the number of single genomes sampled, the number of identical single genomes observed, and the length of the fragment sequenced (details in Methods; statistical test available at https://michaelbale.shinyapps.io/prob_identical). The test is a conservative estimate used to eliminate rakes that were unlikely to contain identical integration sites and to identify candidate rakes for further investigation. We found that, for the donor with the lowest proviral diversity (patient identifier [PID] 1683) (n = 64 single genomes obtained; average pairwise p-distance = 0.2%), greater than 5 identical sequences were needed for the identity to be considered statistically significant (Fig. 4A). For the 3 donors with higher proviral diversities (PID 2669, 1362, and R-09) (n = 147, 103, 40 single genomes obtained; average pairwise p-distances = 1.6%, 2.7%, and 1.9%, respectively), only 2 identical sequences were needed to reach significance (Fig. 4 B–D). We used this estimate to select 7 candidate rakes of identical P6–PR–RT variants in the 4 donors for further investigation, including 1 sequence that matched a variant that grew out in a viral outgrowth assay (PID R-09) (31, 32).
MDA-SGS on Wild-Type Proviruses and Proviruses with Drug Resistance Mutations.
MDA-SGS was performed to determine whether the proviruses within phylogenetic clusters (rakes) detected by standard SGS were present in expanded clones of infected cells, verified by the detection of identical sites of integration into the host genome (Table 1). We then characterized the NFL sequences of the proviruses that were confirmed to be in expanded cell clones (Fig. 3). MDA-SGS from PID 1683, 2669, 3162, and R-09 yielded 27, 27, 10, and 2 wells, respectively, from which integration site data were generated (Table 1 and SI Appendix, Table S1). In donor 1683, rake #1 (Fig. 4A) had 41 identical sequences in the PBMCs and the LNMCs, and, therefore, it was statistically unlikely that this rake arose by chance from genetically unlinked sequences. MDA-SGS analysis of 20 wells containing proviruses within rake #1 revealed 7 distinct integration sites, 3 of which were found in multiple MDA wells and/or had matches to bulk integration site data (33), indicating that some of the cells carrying these proviruses had undergone clonal expansion (Table 1). Two of the 3 confirmed clones in rake #1, in the WIPF1 gene on chromosome 2 and in the AATF gene on chromosome 17, were found in both PBMCs and LNMCs. The third clone, integrated in the XPO6 gene on chromosome 16, was found in 2 wells, both from LNMCs. To validate the MDA-SGS results, we confirmed the integration sites obtained for the WIPF1 and XPO6 clones by performing host-provirus PCR amplification and sequencing of the DNA in the MDA wells and obtained the exact sites of integration that were identified by ISA (SI Appendix, Table S2).
Rakes #2–7 (Fig. 4 B–D) were present in the 3 donors with higher proviral diversity (PID 2669, 3162, R-09). Two of these individuals (PID 2669 and 3162) had experienced previous virologic failures due to drug resistance but had subsequently suppressed viremia on a new regimen for more than 4 y at the time of sampling (Table 2). In these donors, we investigated the clonality of rakes with wild-type sequences and those with drug-resistance mutations (Fig. 4 B–D, Table 1, and SI Appendix, Table S1). Rakes #2 and #3 in PID 2669 contained the K103N drug resistance mutation that confers nonnucleoside reverse transcriptase inhibitor resistance. MDA-SGS yielded identical integration sites within both rakes, confirming the clonal expansion of cells containing proviruses with drug resistance mutations. However, MDA-SGS also revealed proviruses with different integration sites in both rakes, suggesting that there was also a genetic bottleneck in the replicating virus population during nonsuppressive ART that resulted in the selection for sequence identity. Rake #4 represented a wild-type (no drug resistance mutations within PR and RT) provirus, and the 2 proviruses that yielded integration site data contained identical integrations into the VSP13D gene on chromosome 1.
In PID 3162, rake #5 proviruses contained multiple drug-resistance mutations both in PR (I84V and L90M) and in RT (D67N, K70R, M184V, and K219Q) (Fig. 4C). After applying MDA-SGS, we found that all 4 of the sampled MDA wells in rake #5 contained unique integration sites, implying a strong genetic bottleneck for the selection for multidrug resistance at the time of their formation and demonstrating that subgenomic sequence identity alone is insufficient to determine clonality, especially when such sequences carry drug-resistance mutations. Like rake #4 in PID 2669, rake #6 in PID 3162 represented a wild-type provirus, of which 3 of the 4 proviruses sampled yielded an identical intergenic integration site on chromosome 9 near the TNFSF8 gene. The discrepancy between the diversity of integration sites in the rakes with wild-type versus drug-resistant proviruses may reflect the different selection pressures on the RT protein prior to ART initiation and upon ART failure. Of note, we detected a second multidrug-resistant variant (in PID 2669 containing K103N, M184V, P225H, and K238T mutations) that was confirmed by MDA-SGS to be in an expanded T cell clone (Table 1) but is not shown in the phylogenetic tree because it was not sampled in the standard SGS (accession nos. MN691965–MN691966). Nonetheless, this finding demonstrates that different stages in the evolution of drug resistance, from wild type to variants with single mutations to those that are resistant to multiple classes of antiretrovirals, can be archived in long-lived and clonally expanded populations of cells. Like rake #4 (PID 2669) and rake #6 (PID 3162), rake #7 (PID R-09) contained a wild-type provirus within an expanded T cell clone. This provirus was integrated into an intron in the ABCA11P gene against the orientation of the gene (Fig. 4D and Table 1) and, upon NFL sequencing, matched a variant that was induced in viral outgrowth assays, implying replication competence. Across the 4 donors, we observed similar representation of defective proviruses in the same orientation and in the opposite orientation of gene, suggesting no orientation bias in defective proviruses, as previously reported (5).
Using MDA-SGS to Characterize Proviral Structure.
In addition to determining the integration sites of HIV-1 proviruses of interest, MDA-SGS can be used to characterize their NFL sequences. Of the 7 rakes of identical P6–PR–RT sequences that were investigated, we found a total of 9 expanded clones, as well as a number of single integrants with identical P6–PR–RT and distinct integration sites. From each, we sequenced the NFL proviruses (as in SI Appendix, Fig. S2) by Sanger and/or PacBio sequencing (Fig. 3). We found one expanded clone (rake #7) to contain a provirus that was inferred to be intact based on our intactness detection pipeline. To inform replication competence of the intact provirus in rake #7, viral outgrowth assays were performed on the same sample. The virus in an outgrowth well with P6–PR–RT RNA matching that of the proviruses in rake #7 was subjected to NFL amplification and sequencing. The resulting 8,821-bp sequence perfectly matched that from the MDA-SGS in rake #7, showing that the ABCA11P clone contains a replication-competent provirus (accession no. MN692189). This finding is the third report (5, 10) of replication-competent proviruses in expanded clones of cells with known site of integration.
Each of the other HIV-1–infected expanded cell clones analyzed by MDA-SGS contained proviruses with deletions rendering them defective, including the drug-resistant proviruses in 3 of the cell clones (Fig. 3, rakes CYLD, #2, and #3). While many of the proviral deletions may be attributed to preintegration errors during reverse transcription, one provirus within an expanded cell clone (rake #2, TMCO5A) may have incurred a deletion postintegration, as it lacks the entire 3′-LTR, the canonical 5-bp host duplication, and 4 additional base pairs of host sequence.
Reconstruction of Intact Proviral Ancestors.
The proviruses in the clone with integration in WIPF1 and the clone with integration in XPO6 have nonoverlapping deletions, but the sequences of the rest of these 2 proviruses are identical to one another (Fig. 3, rake #1). The provirus in WIPF1 has a 52-bp deletion in the 5′-UTR including part of stem loop 1 and stem loop 2, the packaging signal, and the major splice donor site, while the remainder of the provirus is intact. The provirus in XPO6 had an intact 5′-UTR but had a 3′ deletion from Vif through the 3′-LTR. Combining the WIPF1 and XPO6 proviruses should produce the intact, parental virus. Because this individual initiated ART within 2 y of infection and the on-ART proviral diversity is low, we propose that MDA-SGS can be used to reconstruct a viral sequence that is similar, or identical, to the transmitted founder virus responsible for the initial infection or one that arose following a population genetic bottleneck.
Discussion
Characterizing the HIV-1 reservoir during ART is crucial to the design of strategies to eradicate the virus or establish a functional cure. It was first shown, using a laborious patient- and clone-specific method, that a highly expanded clone of cells that carried a replication-competent provirus (AMBI-1) was an important part of the HIV-1 reservoir during ART in one patient sample (10). This patient (1, 10) was diagnosed with a squamous cell carcinoma at the time of sampling, which could have affected the massive expansion of the T cell clone carrying the AMBI-1 provirus. Subsequent studies showed that identical replication-competent proviruses were often present in what appeared to be expanded clones (outgrowth of the same virus was seen in multiple wells in VOA), but clonal expansion was not confirmed in these studies by showing that the proviruses had identical integration sites (2, 4, 6, 11). Recently, using an assay similar to the one we describe here, another study showed that multiple donors carry expanded clones with intact proviruses matching viral outgrowth in culture (5). Although there are minor technical differences between our assays, including the methods for MDA, NFL amplification, and ISA, together, the 2 studies show that, while expanded clones with intact proviruses often constitute a small minority of the total population (34, 35), they can be stably maintained by cellular proliferation and are likely to be the major obstacle preventing a cure for HIV-1 (2, 6). It is therefore crucial to HIV-1 cure efforts to understand the distribution, dynamics, and the mechanisms involved in the origin, persistence, and latency of the T cell clones that carry intact infectious proviruses.
Here, we describe an assay designed to identify expanded clones of infected cells in HIV-1–infected donors and to characterize both the integration sites and the corresponding sequence of individual proviruses. Unlike previous methods for ISA (1, 9, 36), MDA-SGS allows for characterization single proviruses by both ISA and NFL sequencing. After assay validation using the ACH-2 cell line, we applied the MDA-SGS methods to 5 donors with groups of identical P6–PR–RT sequences. The majority of the cell clones contained proviruses with defects, ranging from small deletions in the 5′-UTR to large 3′ deletions including some host sequences. However, the provirus in one clone, integrated in the ABCA11P gene, was determined to be intact and matched the NFL sequence of a virus recovered from the same donor by a viral outgrowth assay. This is the third report of a confirmed expanded clone identified in vivo that harbors an intact, replication-competent HIV-1 provirus. Einkauf et al. (5) observed intact proviruses, some in cell clones, in 3 donors and found the majority to be oriented against the gene in which the provirus was integrated, while defective proviruses had no observed orientation bias. Likewise, we report an intact provirus, persisting in a T cell clone, that was integrated against the gene orientation and we found no such orientation bias for defective proviruses. Additional studies are needed to determine whether replication-competent proviruses in expanded cell clones have other specific characteristics that may be exploited in future curative interventions. The identification of integration sites of intact proviruses may also guide the design of site-specific assays to specifically monitor cells containing these proviruses, determine their anatomical distribution and dynamics during ART, and evaluate their response to therapeutic interventions, such as “shock-and-kill” strategies (37, 38).
When considering identification of clonal cell populations, it is important to bear in mind that we are sampling, at most, a few hundred out of potentially 109 or more infected CD4+ T cells present within the whole donor, or about one 10-millionth of the total population of infected cells. Therefore, a proviral integration site seen in only one cell in the MDA-SGS assay may well be part of a clone of more than a million cells. Indeed, we hypothesize that the majority, if not all, of HIV-infected cells in individuals on suppressive ART are members of such clones. Mathematical modeling has been used to provide support for this contention, but the assumptions regarding the distribution of smaller clones in underlying populations have not been experimentally verified (39).
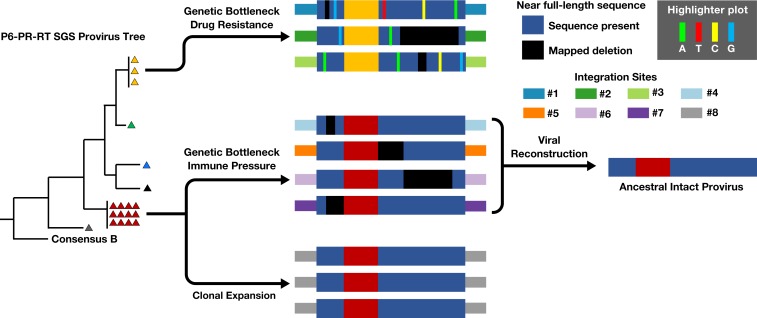
MDA-SGS was successfully used to identify expanded cell clones carrying defective and intact proviruses and expanded clones carrying proviruses with and without drug resistance mutations. The frequent observation of proviruses with identical P6–PR–RT sequences but different sites of integration cautions against the exclusive use of subgenomic SGS to establish cellular clonality and highlights the effect of genetic bottlenecks imposed during transmission/early infection (14), during the selection for drug-resistant virus (15, 16), or, perhaps, during the selection of immune escape variants (17–20). From these observations, we developed a model for the various origins of identical HIV-1 subgenomic proviral sequences that persist on ART (Fig. 5). The model depicts 3 origins (although there could be others): 1) cellular proliferation, 2) selection for drug resistance mutations prior to ART, and 3) immune selection prior to and/or during ART. It should be noted that identical sequences with different integration sites will be more common in patients with low proviral diversity including those initiating ART in acute or early infection; hence, MDA-SGS will prove useful in identifying expanded cell clones with intact proviruses in such individuals.
Fig. 5.
Origin of Identical P6–PR–RT sequences. Identical subgenomic P6–PR–RT sequences observed during ART can be originate from multiple mechanisms including (Top) genetic bottleneck of multiple viruses during selection of drug resistance, (Middle) immune pressure (i.e., CTL/antibody) driving persistence of select proviruses, and (Bottom) clonal expansion of cells containing a provirus with identical integration site and sequence. Hypothetical integration sites and proviral deletions are shown. Reconstruction of the replication-competent ancestor resulting in sequences in red triangles is also shown.
We observed multiple defective proviruses within expanded cell clones across the 5 donors, including 1 aberrantly integrated provirus, an event likely mediated by host-repair mechanisms, similar to integration events observed in vitro in the presence of raltegravir (40). From defective proviruses, we were able to reconstruct a replication-competent ancestral virus in one donor, providing an example for the application of “viral reconstruction” to study the genetic content of intact proviruses that may comprise the HIV-1 reservoir.
The MDA-SGS approach will allow future studies to better characterize the HIV-1 reservoir during ART, determine whether HIV-1–infected cell clones carrying replication-competent proviruses are common among ART-treated individuals, determine whether sequences of intact proviruses in clones closely match founder viruses or other variants of interest, and monitor expanded cell clones harboring replication-competent proviruses after curative interventions. MDA-SGS can also be used to identify proviruses that are likely to be intact by first assaying the MDA wells with the recently described “intact proviral DNA assay” (34). Such detailed characterization of the HIV-1 reservoir should provide insights into the mechanisms by which the reservoir is established and persists and may reveal targets for potential curative interventions.
Methods
Patient Cohorts and Samples.
PBMCs and LNMCs were obtained from the SCOPE cohort, University of California, San Francisco, as described (30), the University of Pittsburgh, and the AVBIO cohort, NIH (1, 10). PID 2669 and 3162 both experienced prior ART failure with drug resistance mutations but sustained full viral suppression for greater than 4 y after the addition of dolutegravir. Paired PBMCs and LNMCs were obtained from PID 1683 and 2669, while PBMCs alone were obtained from PID 3162 and R-09.
Ethics Statement.
All participants signed informed consent in accordance with clinical trial NCT00187512 (SCOPE), NCT00009256 (AVBIO), and PRO14120068 and PRO10070203 (University of Pittsburgh). Protocols were approved by the internal review boards at the University of California, San Francisco, National Institutes of Health, and the University of Pittsburgh.
Phylogenetic Tree Construction.
Phylogenetic trees of P6–PR–RT proviral single-genome sequences were constructed in MEGA7 (https://www.megasoftware.net/) using the neighbor-joining method. Intrapatient proviral diversity is given by average pairwise difference in percent p-distance.
Statistical Test on Groups of Identical Sequences in Standard SGS.
We calculated the probability that a group (or rake) of identical HIV-1 variants contained more than the expected number of identical sequences, making it likely to have arisen by clonal expansion or by selection. A Rshiny web-app of the test is available online at https://michaelbale.shinyapps.io/prob_identical. The test is carried out as follows: We calculated the average pairwise distance of a dataset of S sequences as the average Hamming distance and the sequence length for each pair i, j as . We assume here that and that the probability that any sequence pair has a 0 distance (i.e., is identical) is . Given sequence pairs, the probability of finding or more sequences that have 0 distance is thus . Importantly, we do not define a statistically significant P value at the 95% confidence level to be an arbiter of clonality, but rather that a statistically insignificant P value is more likely to result from a genetic bottleneck. Its use here is the elimination of rakes highly unlikely to contain T cell clones and the identification of candidate rakes for further investigation.
MDA-SGS.
Genomic DNA from donor PBMCs or LNMCs was extracted as previously described (41–43), endpoint diluted in 96-well microtiter plates, and amplified using MDA (details in SI Appendix, Supplemental Methods). Wells were screened for proviruses of interest using P6–PR–RT amplification as described (41–43). Integration site assays were performed on MDA wells exactly as described previously for bulk genomic DNA (1). NFL sequencing was performed by Sanger or PacBio (details in SI Appendix, Supplemental Methods).
Pipeline to Determine Intactness of HIV-1 Proviruses.
To characterize the NFL proviral sequences, we developed an informatic “intactness detection” pipeline (https://psd.cancer.gov/). The pipeline identifies specific genetic elements including the gag, pol, and env ORFs within an NFL sequence, the 5′-LTR, the packaging signal, major splice donor (MSD) site, Rev response element (RRE), and 3′-LTR. It then detects any insertions and/or deletions in these genes/elements and any premature stop codons in the genes. The provirus is considered defective if any one of the following is true: 1) length of the provirus <7,000 nt; 2) deletions in the packaging signal region >7 nt, missing MSD, or MSD is mutated (35); 3) deletions in gag start AUG region (HXB2: 788–798) that pairs with U5 >2 nt; 4) either one of the 2 nt (AG in HXB2) is missing before gag start (AUG); 5) gag deletions >49 nt or insertions >49 nt or at least one premature stop codon; 6) pol deletions >49 nt or insertions ≥50 nt or at least one premature stop codon; 7) env deletions >99 nt, or insertions >149 nt, or at least one premature stop codon; 8) RRE length <235 nt or deletions >8 nt in the tr4–tr5 region of either 5′ or 3′ of tr4 to tr5 regions or deletions >8 nt in the domain II region (44); 9) a frameshift is detected in any of the above genes. If none of the above is true, the provirus is considered intact.
Data Availability Statement.
All sequence data reported in this paper have been deposited publicly in the GenBank database (accession nos. MK147632–MK147695, MK147878–MK148024, and MN691959–MN692189). All integration site data reported in this paper have been deposited in the Retroviral Integration Database (https://rid.ncifcrf.gov/), and all NFL genome sequences reported in this paper have been deposited in the Proviral Sequence Database (https://psd.cancer.gov/) using the PubMed ID of this paper.
Supplementary Material
Acknowledgments
We thank Connie Kinna, Valerie Turnquist, Terri Burdette, and Susan Toms for administrative support; Jack Chen for bioinformatics support; Valerie Boltz, Andrew Musick, and Jennifer Groebner for consultations; and Joseph Meyer for graphical consultation and assistance. This study was supported by National Cancer Institute (NCI) intramural funding (to M.F.K.), by funding from the Office of AIDS Research (to M.F.K.), by federal funds from the NCI under Contract HHSN261200800001E, and by Leidos Contract HHSN261200800001E (to the University of Pittsburgh). J.M.C. was a Research Professor of the American Cancer Society and was supported by Subcontract l3XS110 (to Tufts University).
Footnotes
Competing interest statement: J.W.M. is a consultant to Gilead Sciences, Merck Research Laboratories, Janssen Pharmaceuticals, and AccelevirDx, and a share option holder of Co-Crystal, Inc. B.F.K. and J.A.H. are co-authors on an October 2015 article. The remaining authors have no potential conflicts.
Data deposition: All sequence data reported in this paper have been deposited publicly in the GenBank database (accession nos. MK147632–MK147695, MK147878–MK148024, and MN691959–MN692189). All integration site data reported in this paper have been deposited in the Retroviral Integration Database (https://rid.ncifcrf.gov/), and all NFL genome sequences reported in this paper have been deposited in the Proviral Sequence Database (https://psd.cancer.gov/) using the PubMed ID of this paper.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910334116/-/DCSupplemental.
References
- 1.Maldarelli F., et al. , HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bui J. K., et al. , Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog. 13, e1006283 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boritz E. A., et al. , Multiple origins of virus persistence during natural control of HIV infection. Cell 166, 1004–1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohn L. B., et al. , Clonal CD4+ T cells in the HIV-1 latent reservoir display a distinct gene profile upon reactivation. Nat. Med. 24, 604–609 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Einkauf K. B., et al. , Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J. Clin. Invest. 129, 988–998 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hosmane N. N., et al. , Proliferation of latently infected CD4+ T cells carrying replication-competent HIV-1: Potential role in latent reservoir dynamics. J. Exp. Med. 214, 959–972 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kearney M. F., et al. , Origin of rebound plasma HIV includes cells with identical proviruses that are transcriptionally active before stopping of antiretroviral therapy. J. Virol. 90, 1369–1376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salantes D. B., et al. , HIV-1 latent reservoir size and diversity are stable following brief treatment interruption. J. Clin. Invest. 128, 3102–3115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wagner T. A., et al. , HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Simonetti F. R., et al. , Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc. Natl. Acad. Sci. U.S.A. 113, 1883–1888 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bui J. K., et al. , Ex vivo activation of CD4+ T-cells from donors on suppressive ART can lead to sustained production of infectious HIV-1 from a subset of infected cells. PLoS Pathog. 13, e1006230 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearney M. F., et al. , Lack of detectable HIV-1 molecular evolution during suppressive antiretroviral therapy. PLoS Pathog. 10, e1004010 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey J. R., et al. , Residual human immunodeficiency virus type 1 viremia in some patients on antiretroviral therapy is dominated by a small number of invariant clones rarely found in circulating CD4+ T cells. J. Virol. 80, 6441–6457 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keele B. F., et al. , Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 105, 7552–7557 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kitrinos K. M., Nelson J. A., Resch W., Swanstrom R., Effect of a protease inhibitor-induced genetic bottleneck on human immunodeficiency virus type 1 env gene populations. J. Virol. 79, 10627–10637 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Zyl G., Bale M. J., Kearney M. F., HIV evolution and diversity in ART-treated patients. Retrovirology 15, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deng K., et al. , Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Borrow P., et al. , Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat. Med. 3, 205–211 (1997). [DOI] [PubMed] [Google Scholar]
- 19.Goonetilleke N., et al. ; CHAVI Clinical Core B , The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206, 1253–1272 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sunshine J. E., et al. , Fitness-balanced escape determines resolution of dynamic founder virus escape processes in HIV-1 infection. J. Virol. 89, 10303–10318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes S. H., Coffin J. M., What integration sites tell us about HIV persistence. Cell Host Microbe 19, 588–598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao W., et al. , Retrovirus integration database (RID): A public database for retroviral insertion sites into host genomes. Retrovirology 13, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clouse K. A., et al. , Monokine regulation of human immunodeficiency virus-1 expression in a chronically infected human T cell clone. J. Immunol. 142, 431–438 (1989). [PubMed] [Google Scholar]
- 24.Besson G. J., et al. , HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin. Infect. Dis. 59, 1312–1321 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dean F. B., Nelson J. R., Giesler T. L., Lasken R. S., Rapid amplification of plasmid and phage DNA using Phi 29 DNA polymerase and multiply-primed rolling circle amplification. Genome Res. 11, 1095–1099 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Spits C., et al. , Whole-genome multiple displacement amplification from single cells. Nat. Protoc. 1, 1965–1970 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Symons J., et al. , HIV integration sites in latently infected cell lines: Evidence of ongoing replication. Retrovirology 14, 2 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sunshine S., et al. , HIV integration site analysis of cellular models of HIV latency with a probe-enriched next-generation sequencing assay. J. Virol. 90, 4511–4519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson E. M., Maldarelli F., Quantification of HIV DNA using droplet digital PCR techniques. Curr. Protoc. Microbiol. 51, e62 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Favre D., et al. , HIV disease progression correlates with the generation of dysfunctional naive CD8low T cells. Blood 117, 2189–2199 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siliciano J. D., Siliciano R. F., Enhanced culture assay for detection and quantitation of latently infected, resting CD4+ T-cells carrying replication-competent virus in HIV-1-infected individuals. Methods Mol. Biol. 304, 3–15 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Rosenbloom D. I., et al. , Designing and interpreting limiting dilution assays: General principles and applications to the latent reservoir for human immunodeficiency virus-1. Open Forum Infect. Dis. 2, ofv123 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McManus W. R., et al. , HIV-1 in lymph nodes is maintained by cellular proliferation during antiretroviral therapy. J. Clin. Invest. 130, 4629–4642 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bruner K. M., et al. , A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566, 120–125 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ho Y. C., et al. , Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155, 540–551 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schröder A. R., et al. , HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Martin A. R., Siliciano R. F., Progress toward HIV eradication: Case reports, current efforts, and the challenges associated with cure. Annu. Rev. Med. 67, 215–228 (2016). [DOI] [PubMed] [Google Scholar]
- 38.Archin N. M., Margolis D. M., Emerging strategies to deplete the HIV reservoir. Curr. Opin. Infect. Dis. 27, 29–35 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reeves D. B., et al. , A majority of HIV persistence during antiretroviral therapy is due to infected cell proliferation. Nat. Commun. 9, 4811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Varadarajan J., McWilliams M. J., Hughes S. H., Treatment with suboptimal doses of raltegravir leads to aberrant HIV-1 integrations. Proc. Natl. Acad. Sci. U.S.A. 110, 14747–14752 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kearney M., et al. , Human immunodeficiency virus type 1 population genetics and adaptation in newly infected individuals. J. Virol. 83, 2715–2727 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kearney M., et al. , Frequent polymorphism at drug resistance sites in HIV-1 protease and reverse transcriptase. AIDS 22, 497–501 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Palmer S., et al. , Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 43, 406–413 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Carroll I. P., et al. , Contributions of individual domains to function of the HIV-1 Rev response element. J. Virol. 91, e00746-17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data reported in this paper have been deposited publicly in the GenBank database (accession nos. MK147632–MK147695, MK147878–MK148024, and MN691959–MN692189). All integration site data reported in this paper have been deposited in the Retroviral Integration Database (https://rid.ncifcrf.gov/), and all NFL genome sequences reported in this paper have been deposited in the Proviral Sequence Database (https://psd.cancer.gov/) using the PubMed ID of this paper.