Significance
There are 20 chemokine receptors that bind their respective chemokines. It is not currently understood how these structurally similar receptors distinguish their ligands; namely, how EC loops and transmembrane domains of these receptors are involved in ligand-binding activities. With the detergent-free GPCRs, we show that it is now possible to design and produce chimeric receptor proteins to study ligand-binding mechanisms. We exchanged the N terminus and 3 EC loops of natural chemokine receptor CXCR4 to append them onto the 7TM α-helices of detergent-free variant CCR5QTY, and systematically studied which ligands it binds. These designer chimeric receptors provide insight into how natural receptors bind their respective ligands. These chimeric receptors with tunable functionality may have applications for bioelectronics sensing devices.
Keywords: CCR5, CXCR4, chimera receptors, GPCR, membrane protein design
Abstract
Chemokine receptors are of great interest as they play a critical role in many immunological and pathological processes. The ability to study chemokine receptors in aqueous solution without detergent would be significant because natural receptors require detergents to become soluble. We previously reported using the QTY code to design detergent-free chemokine receptors. We here report the design of 2 detergent-free chimeric chemokine receptors that were experimentally unattainable in detergent solution. We designed chimeric receptors by switching the N terminus and 3 extracellular (EC) loops between different receptors. Specifically, we replaced the N terminus and 3 EC loops of CCR5QTY with the N terminus and 3 EC loops of CXCR4. The ligand for CXCR4; namely CXCL12, binds to the chimeric receptor CCR5QTY (7TM)-CXCR4 (N terminus+3 EC loops), but with lower affinity compared to CXCR4; the CCL5 ligand of CCR5 binds the chimeric receptor with ∼20× lower affinity. The chimeric design helps to elucidate the mechanism of native receptor-ligand interaction. We also show that all detergent-free QTY-designed chemokine receptors, expressed in Escherichia coli, bind to their respective chemokines with affinities in the nanomolar (nM) range, similar to the affinities of native receptors and SF9-produced QTY variants. These QTY-designed receptors exhibit remarkable thermostability in the presence of arginine and retain ligand-binding activity after heat treatment at 60 °C for 4 h and 24 h, and at 100 °C for 10 min. Our design approach enables affordable scale-up production of detergent-free QTY variant chemokine receptors with tunable functionality for various uses.
G protein coupled receptors (GPCRs), including chemokine receptors, are a family of integral membrane proteins embedded in the lipid bilayer of cell membranes that transduce extracellular stimuli into cellular response (1). As one of the most important classes of proteins, GPCRs regulate a wide range of human body functions including sensory systems, the immune system, brain function, hormone responses, growth, and aging (2). GPCRs are the targets of ∼50% drugs in medicine. Chemokine receptors represent a highly interesting class of GPCRs (3). They detect the gradient of native chemokine and guide migration of immune cells, i.e., chemotaxis. When chemokines are produced under normal or pathological conditions, the chemokine receptors play a critical role in various types of immune responses, tissue repair, embryogenesis, development, and many other functions (4).
CXCR4 and CCR5 are 2 of the most well-studied chemokine receptors. The CXCR4 receptor regulates leukocyte hematopoiesis and trafficking with its native ligand stromal-derived-factor 1 (SDF-1α, renamed as CXCL12). It plays a major role in multiple sclerosis and is involved in the metastasis of 23 types of cancers (4). In addition, CXCR4 signaling serves as axon guidance and prevents aberrant neuron distribution (5–7). CXCR4 and CCR5 are both identified as coreceptors for HIV entry into T cells (8). On the other hand, CXCR7 is an atypical chemokine receptor as it is associated only with β-arrestin and not coupled with G proteins. It forms a hetero-oligomer with CXCR4 and modulates CXCR4 signaling and trafficking due to its higher affinity to SDF-1α (9). CXCR5 with CXCL13 as its native ligand is expressed in mature B cells and Burkitt's lymphoma. It plays a central role in the control of B cell follicles and germinal center reactions (10). CCR10 is expressed primarily in skin cells and regulates many aspects of epithelial immunity. It is also involved in the growth and metastasis of epithelial-homing or epithelial-originated cancers (4). Because of their critical role in various physiological functions related to human health and diseases, these receptors were chosen in this study.
Despite recent significant progress in the study of these chemokine receptors, a thorough understanding of their physiological, functional properties, how these seemingly similar receptors recognize and interact with their distinctive ligands, and the utilization of these receptors for the benefit of mankind still remains a challenge. One of the major reasons is attributed to the hydrophobic residues in the transmembrane (TM) region. Detergent optimization is required for these membrane receptors to be extracted from cell membranes and stabilized in an aqueous environment for an extended time (11, 12). Such detergent-requirement limitations seriously hinder the uses of membrane proteins for practical medical purposes and for bioelectronic technology. In order to use membrane proteins for technological development and medical applications, alternative methods are required.
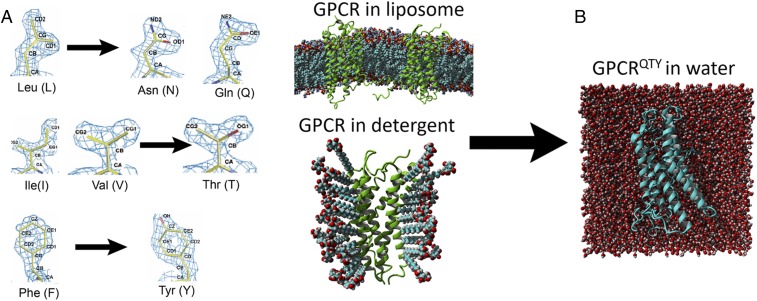
We previously reported a simple QTY code that can make systematic and specific substitutions of amino acids to design the water-soluble, detergent-free, and functional variants of chemokine receptors (13). The QTY code is based on the similarity of chemical structures and electron density maps between amino acids (Fig. 1A). We believe that the QTY code can be used to design water-soluble receptor variants by substituting specific hydrophobic amino acids with hydrophilic ones.
Fig. 1.
QTY code and how it converts transmembrane hydrophobic domains to hydrophilic domains. (A) Crystallographic electron density maps of Leucine (L), Asparagine (N), Glutamine (Q), Isoleucine (I), Valine (V), Threonine (T), Phenylalanine (F), and Tyrosine (Y). QTY code replaces the hydrophobic amino acid residues LIVF to hydrophilic QTY that their side chains can form hydrogen bonds while structurally similar in electron density map. Q is chosen over N due to its higher presence in α-helices and N often exists at turns. The QTY code is reversible if one desires. (B) Graphic illustration of a GPCR before and after applying QTY code. The membrane QTY variant receptor is now stable in aqueous environment without any detergents.
While the QTY code enables the design of water-soluble QTY variant chemokine receptors, protein yields in the SF9 cell are suboptimal. Including a designed CXCR5QTY and a further modified version of CCR5QTY, we describe here the production of these QTY variant chemokine receptors in a conventional Escherichia coli system with yields of ∼5 mg/L in commonly used Luria–Bertani (LB) culture. QTY variant receptors were extracted from E. coli inclusion bodies, affinity and gel filtration purified, and refolded into a stable state in the presence of arginine. Ligand binding of these receptors was confirmed by MicroScale Thermophoresis (MST), and the affinities were similar to the same QTY variant receptors produced in SF9 cells and native counterparts. The yields were sufficient to permit investigations into key questions of function and pathways.
One of the most important questions is how diverse chemokines recognize their receptors at all with similar 7TM. Does a specific ligand only recognize the external parts of the receptor? Or does it also need to interact with the 7TM? In order to address such questions, we ask if we can replace the N terminus and 3 EC loops of 1 receptor with those from another to redesign their functionality, inspired from the concept of chimeric antibodies (14–16). This approach would allow us to understand: 1) how critical is the chemokine CXCL12 interaction with the N terminus and 3 EC loops of the CXCR4 receptor; and, 2) whether or not the interaction involves 7TM α-helical segments that are embedded in the lipid membrane. Therefore, we carried out experiments to design the chimeric receptor. We replaced the N terminus and EC loops of CCR5QTY with those of native CXCR4, or with a negative control; namely, glycine-serine (GS) linkers. The chimera designs permit the fine-tuning of protein solubility and functionality, as well as further investigation into the binding properties of QTY variant receptors. We believe such systematic studies will provide insight into how receptors recognize their ligands.
Results
Sequence Alignments and Bioinformatics of CCR5 vs. CCR5QTY and CXCR5 vs. CXCR5QTY.
The pairwise protein sequences were aligned to compare the amino acid substitutions between the natural receptors and their QTY variants (Fig. 2). This expands on work reported earlier. Alignments for CXCR4 vs. CXCR4QTY, CCR10 vs. CCR10QTY and CXCR7 vs. CXCR7QTY can be found in our previous publication (13). For CXCR5, the corresponding hydrophobic residues in the TM regions were replaced by glutamine (Q), threonine (T) and tyrosine (Y), thus the 7TM α-helices become water soluble. Residues in both intracellular (IC) and EC loops were untouched. However, for CCR5 we started with the same sequence design as published in our last paper, where all of the amino acid exchange occurred in the TM region. The CCR5QTY was stable without detergent in aqueous solution, but the solubility of the protein was still inadequate in E. coli-expressed protein. Since 3 IC loops and C terminus are never exposed to external ligands, we replaced the additional L, I, V, and F in 3 IC loops and C terminus so as to further increase hydrophilicity of the protein for characterization.
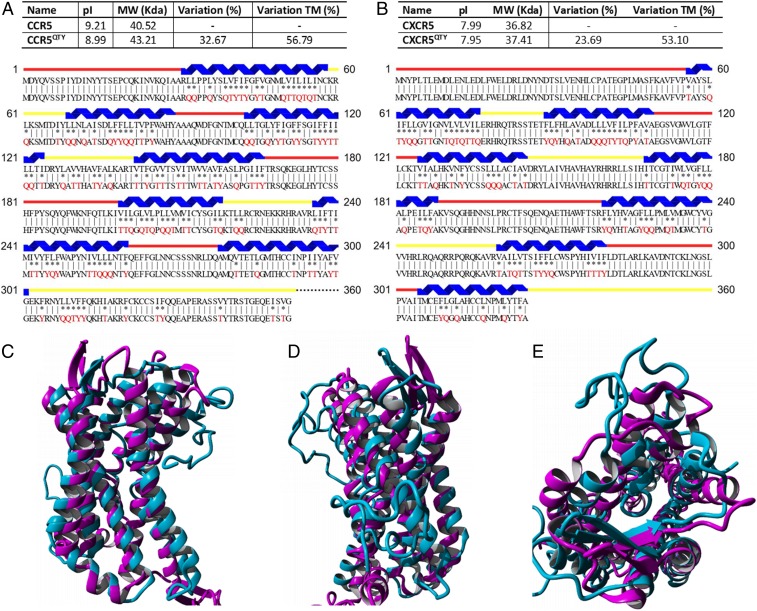
Fig. 2.
Comparison between natural CCR5 vs. CCR5QTY and CXCR5 vs. CXCR5QTY. Protein sequence alignment between natural (Top) (A) CCR5 and CCR5QTY and (B) CXCR5 and CXCR5QTY vs. their QTY variants (Bottom). The substitutions of Q, T, and Y are denoted with “*”, while “|” indicates no change in residues between the 2 sequences. The Q, T, and Y amino acid substitutions are in red. The α-helical segments (blue) are shown above the protein sequences, and the external (red) and internal (yellow) loops of the receptors are indicated. Characteristics of native and QTY proteins’ pI, molecular weight, and overall variation rate and that % changes only transmembrane segments are presented. (C–E) Computer-simulated CCR5QTY (light blue) structure is superimposed with its natural CCR5 (magenta) (PDB ID code 4MBS; for clarity, the IC3 insert was removed from the superimposed images). It has a deviation of ∼2 Å and is shown in 2 different side views in C and D, and in a top view shown in E.
There are some minor differences between native and QTY variant receptors. Despite the total difference of 32.67% (56.79% in 7TM) in primary sequence for CCR5QTY, and 23.69% (53.10% in 7TM) for CXCR5QTY, the change in the isoelectric point (pI) for CCR5 and CXCR5 are only 0.22 and 0.04, respectively. This is attributed to the nonionic nature of glutamine, threonine, and tyrosine. The QTY residues can form numerous hydrogen bonds with surrounding water molecules to solubilize the proteins. The molecular weight of QTY proteins is slightly larger as Q, T, and Y are slightly larger than their hydrophobic analogs L, I, V and F. Following the QTY application, both CCR5QTY and CXCR5QTY contain no TM hydrophobic segments as confirmed by bioinformatics analysis (SI Appendix, Fig. S1).
Computer Simulation of CCR5QTY Variant with Additional Changes.
Crystal structures for QTY-designed chemokine receptors are in progress, but are not available yet. We therefore carried out molecular simulation for the CCR5QTY variant with changes. Fig. 2 C–E compares the simulated molecular structure of the CCR5QTY receptor to the crystal structure of native CCR5 determined in the presence of detergent. The simulation is based on homology using the QTY sequences in an explicit water environment. The simulated CCR5QTY folded at 24.85 °C, pH 7.4 and 0.9% NaCl. These structures formed during the initial 0.3 μs of simulation and did not show further changes for 0.7 μs simulation. Despite the differences in the TM regions and further changes in the IC regions, the structure of CCR5QTY is superimposable with the known crystal structure of its natural counterpart. The structural superimposition shows that CCR5 and CCR5QTY have a deviation of ∼2 Å, similar to that previously reported (13).
Ligand-Binding Measurements in Buffer and 50% Human Serum.
We measured the ligand-binding activity of these E. coli-expressed and -purified QTY variants of chemokine receptors. Fig. 3 shows the ligand-binding measurement using MicroScale Thermophoresis (MST) (17–19). Changes in thermophoretic movement for GPCRQTY and temperature related intensity changes (TRIC) of the protein-attached fluorophore upon ligand-binding were recorded and plotted as a function of ligand concentration (19). No unspecific adhesion or major aggregation of protein was detected during the measurements. For better visualization, the data were replotted as bound fraction vs. concentration. The plot was then used to calculate the Kd value for receptor-ligand interactions using the Kd model, presented in the SI Appendix, Materials and Methods.
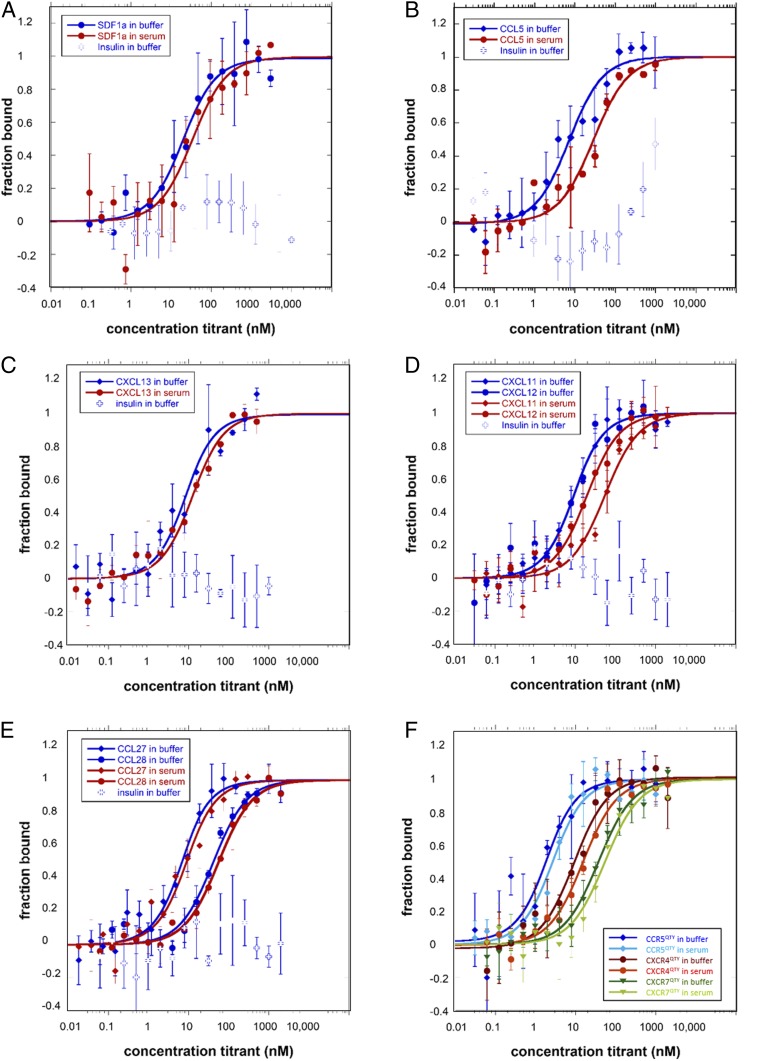
Fig. 3.
Ligand-binding measurements using MST. The receptors were labeled with fluorescent dye since both receptors and ligands contain tryptophans that interfere with the measurement without labeling. The ligands were purchased commercially. They are either dissolved in buffer or in 50% human serum. Human insulin was used as a negative control. Error bars were calculated from 3 independent biological repeats with duplicate measurements of each sample. (A) CXCR4QTY with CXCL12, (B) CCR5QTY with CCL5, (C) CXCR5QTY with CXCL13, (D) CXCR7QTY with CXCL11 and CXCL12, (E) CCR10QTY with CCL27 and CCL28, (F) CCR5QTY, CXCR4QTY, and CXCR7QTY with HIV1-coated glycoprotein gp41–120. The Kd value calculated from the graphs are listed in Table 1. All QTY variants of chemokine receptors did not bind human insulin, suggesting ligand-binding specificity.
The ligand-binding affinity of QTY variants of chemokine receptors was measured both in buffer and 50% human serum since serum conditions more closely resemble in vivo pharmacokinetic conditions. Human insulin was used as a negative control for nonspecific binding. These results demonstrate that CXCR4QTY, CCR5QTY, CXCR5QTY, CXCR7QTY, and CCR10QTY all retain their respective ligand-binding affinities (Fig. 3). The ligand-binding measurements were reproducible over several different expressions and purifications of QTY variant receptors. The affinity values obtained for QTY receptors produced in E. coli are consistent with those from SF9 cells as reported previously, with minor variations (13). The measured Kd for each QTY receptor is also in the same order of magnitude as the highest Kd values of native receptors reported in the literature (Table 1 and Fig. 3). For measurements carried out in 50% human serum, lower Kd values are observed. The reduced ligand-binding affinity is not surprising and it can be attributed to the complex nature of human serum since serum contains numerous substances that can interfere with the ligand binding.
Table 1.
Ligand-binding affinity of QTY code designed chemokine receptor (E. coli)
| CCL5* Kd, nM | CXCL11 Kd, nM | CXCL12* Kd, nM | CXCL13 Kd, nM | CCL27 Kd, nM | CCL28 Kd, nM | gp41-120 Kd, nM | |
| CXCR4 native | ∼5 | ∼200† | |||||
| CXCR4QTY buffer | 17.3 ± 4.2 | 7.0 ± 1.9 | |||||
| CXCR4QTY serum | 31.4 ± 12.2 | 12.7 ± 2.6 | |||||
| CXCR5 native | ∼50.5 | ||||||
| CXCR5QTY buffer | 5.7 ± 2.2 | ||||||
| CXCR5QTY serum | 10.1 ± 2.6 | ||||||
| CXCR7 native | ∼8 | ∼4.5 | N/A | ||||
| CXCR7QTY buffer | 6.4 ± 1.2 | 6.7 ± 2.2 | 34.3 ± 6.1 | ||||
| CXCR7QTY serum | 51.5 ± 11.8 | 16.9 ± 3.4 | 50.0 ± 11.1 | ||||
| CCR5 native | ∼4 | ∼10 | |||||
| CCR5QTY buffer | 6.8 ± 2.0 | 1.3 ± 0.5 | |||||
| CCR5QTY serum | 29.0 ± 8.9 | 2.1 ± 0.6 | |||||
| CCR10 native | ∼5.6 | ∼38 | |||||
| CCR10QTY buffer | 4.2 ± 1.4 | 38.5 ± 8.0 | |||||
| CCR10QTY serum | 6.2 ± 2.4 | 52.7 ± 7.1 |
CCL5 is also called “Rantes,” and CXCL12 is also called “SDF1α” in the literature.
The Kd ∼200 nM was measured by a cell-based assay.
All QTY code designed chemokine receptors were purified from E. coli inclusion bodies and renatured in refolding buffer.
CXCR4 and CCR5 are the 2 well-known natural coreceptors for HIV docking in human cells. CXCR7 was also recently proved to mediate HIV entry (20). We therefore asked if their QTY variants can bind to HIV1 coat glycoprotein gp41–120. The observed measurements showed good affinity (Fig. 3F). QTY variants may be potentially useful for developing decoy therapeutic mechanisms against the HIV virus.
In order to demonstrate the ligand-binding specificity of QTY variant chemokine receptors, human insulin was used as a negative control for the ligand-binding measurements. As shown in Fig. 3, no nonspecific reactions were observed between insulin and QTY variant receptors, suggesting that ligand-binding specificity is maintained.
Design of Chimeric Receptors through Exchanging the EC Loops.
There are 20 chemokine receptors with similar 7TM segments but different N terminus and 3 EC loops that interact with the different ligands. We asked if by exchanging those EC loops between receptors but keeping the same 7TM segments, we can study how ligand-binding activity relates to external and TM parts as well as design variants of chemokine receptors with tunable functionality. CCR5 and CXCR4 were selected for our study since they belong to different chemokine receptor groups and do not share chemokine ligands. The N terminus and 3 EC loops of each receptor were chosen as the graft sites since they are primarily responsible for capturing and interacting with the chemokines, as shown later in this section. The starting and ending sites of the 3 EC loops were chosen based on PDB files of CCR5 (4MBS) and CXCR4 (3ODU) and UniProt entry. For the chimeric receptors design, we kept the 7TM as the stem and 3 IC loops of CCR5QTY. Chimera A was constructed by replacing the EC1, EC2, and EC3 loops of CCR5 with same length of GS linkers, which serves as a negative control for the CXCL12 binding assay. Chimera B was constructed by replacing the N terminus, EC1, EC2, and EC3 loops of CCR5 with the N terminus, EC1, EC2, and EC3 loops of CXCR4. We anticipated that the Chimera B would preferentially bind to CXCR4’s ligand CXCL12, instead of to CCR5′s ligand CCL5. This was supported by our experimental results. The designs are schematically illustrated in Fig. 4A. The amino acid sequence changes for the chimeric proteins are provided in the SI Appendix, Materials and Methods. Multiple cut sites were not explored in our design. We kept the N terminus and EC loops complete and did not explore partial inclusion of specific extracellular components.
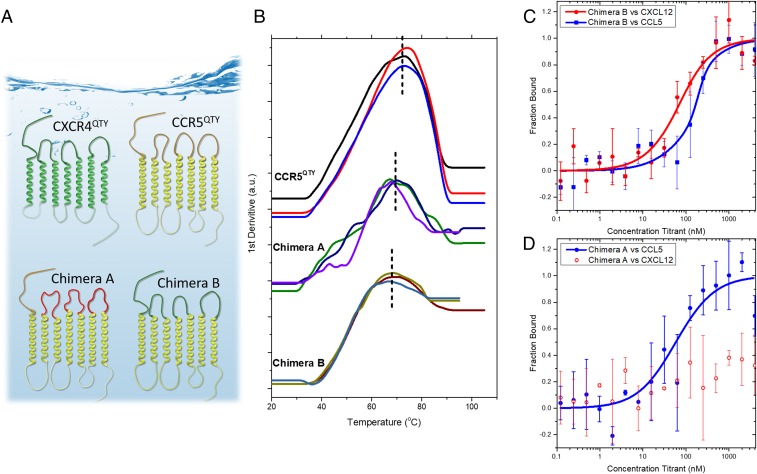
Fig. 4.
Design chimeric QTY receptors. (A) Schematic of the chimera design. CXCR4QTY is green. CCR5QTY’s 7TM regions (yellow) and IC loops were chosen as the backbone of the design (protein sequences are in SI Appendix). In Chimera A (Bottom Left) (CCR5QTY-N-3(GS)n) the 3 EC loops of CCR5QTY were replaced by GS linker sequence (red) with the same length (Upper, yellow loops), but CCR5QTY N terminus (red line) was unchanged. In Chimera B (Bottom Right) (CCR5QTY-7TM: CXCR4QTY-N,3ECL), the N terminus and EC loops of CCR5QTY were replaced by N terminus and 3 EC loops of CXCR4QTY (green N-terminal and 3 green loops). (B) Three independent Tm measurements by NanoDSF to evaluate the thermostability. CCR5QTY: 73.3 ± 0.9 °C, Chimera A: 68.2 ± 1.8 °C, Chimera B: 68.3 ± 1.8 °C. (C and D) MST ligand-binding measurement for Chimera A and Chimera B. (C) Chimera B binds both CXCL12 and CCL5 with affinity of 54.7 ± 19.6 nM and 150 ± 45 nM, respectively. (D) Chimera A binds CCL5 with affinity of 64.4 ± 40.8 nM. These results suggest that the N terminus and 3 EC loops are the most crucial regions for the ligand-binding activities of these chemokine receptors.
Both the Chimera A and Chimera B are detergent free and contain no hydrophobic TM amino acids as confirmed by bioinformatics analysis (SI Appendix, Fig. S2). Production yield of purified chimeric receptors is ∼5 mg/L in LB media, similar to other QTY variant chemokine receptors. Electrophoresis gels of purified proteins are shown in SI Appendix, Fig. S3. Both chimeric receptors exhibited >95% solubility-based refolding yield (soluble yield) in 50 mM Tris⋅HCl pH 9.0 buffer with the addition of arginine content, as shown in SI Appendix, Table S1 (21). These designed chimeric receptors exhibited a typical α-helical structure in far ultraviolet circular dichroism (CD) spectra (SI Appendix, Fig. S4) with signature valleys at 208 nm and 222 nm. The E. coli-synthesized CCR5QTY showed CD spectra similar to the SF9-synthesized variant (13). CCR5QTY and Chimera B contained similar (∼50%) α-helix content due to their similarity in molecular weight. This percentage was higher in Chimera A as the molecular weight of the GS linker is lower compared to the EC loops of both CXCR4 and CCR5.
Fig. 4B shows the measurements from NanoDSF to determine the thermostability of Chimera A and Chimera B as compared to CCR5QTY. The measurements were carried out in protein storage buffer with 100 mM arginine since arginine is required for refolding and important in long-term storage (22, 23). Three independent measurements were carried out for each chimeric receptor. The melting temperatures (Tm) were determined to be 73.3 ± 0.9 °C for CCR5QTY, 68.2 ± 1.8 °C for Chimera A, and 68.3 ± 1.8 °C for Chimera B. Since the 3 variant receptors have the same 7TM segments, the similar Tm suggests that they fold similarly.
The binding affinity for the different ligands of these chimeric receptors was measured with MST as shown in Fig. 4 C and D. Chimera B (CXCR4 N terminus/3 EC loops-CCR5QTY 7TM) binds CXCL12 with a Kd ∼54.7 ± 19.6 nM, which exhibited 3 times less affinity compared to that of CXCR4QTY, but still within the same order of magnitude.
Previously, researchers reported a simulated 3-step binding mechanism between CXCL12 and CXCR4. CXCL12 first forms a binding site with the N terminus of CXCR4, followed by hydrogen bonding and salt bridge interactions with the EC loops (SI Appendix, Fig. S5). After that, the N terminus of CXCL12 interacts with CXCR4’s 7TM region to induce a conformational change and trigger G protein signaling. A full list of sites involved in the interaction has been detailed by other investigators (24, 25).
Chimera B lacks CXCR4’s 7TM region so IC signaling cannot be triggered, yet the N terminus and 3 EC loops that initiate ligand binding stay intact and are able to interact with CXCL12, albeit with a lower affinity. Chimera B also binds to ligand CCL5 with lower Kd values 150 ± 45 nM while the value for Chimera A (CCR5QTY 7TM and N terminus with 3 EC GS linkers) is 64.4 ± 40.8 nM. A computer simulation reported by Tamamis et al. suggests a similar binding mechanism between CCR5 and CCL5, where the 1–6 residues of N terminus of CCL5 inserts into CCR5′s 7TM region; and 7–15 N terminus residues of CCL5 interact with CCR5′s N terminus and EC loops (SI Appendix, Fig. S5 C and D) (26).
Key residues for CCR5 ligand interactions in the 7TM region are still present in both Chimera A and Chimera B including Tyr37, Trp86, Tyr108, Gly163, Tyr251, and Glu28. However, these interaction sites are deeply buried in the α-helical component of the proteins, rendering it difficult for the ligand to access them, resulting in a 20× decrease in affinity between Chimera B and CCL5 as compared to that of CCR5QTY. In Chimera A, the N terminus of CCR5 is still available, which leads to a higher binding affinity compared to that of Chimera B. The binding studies between Chimera A and CXCL12 were carried out as a negative control, and indeed, no detectable interaction was observed. Our systematic results are in agreement with previously simulated native receptor-chemokine interaction mechanisms (20, 21). These studies suggest the feasibility of using the design of interchangeable EC loop systems in water-soluble QTY variants to provide valuable information through which we can further understand native chemokine receptors.
Thermostability of Chemokine Receptor QTY Variants.
As shown in Fig. 4B, we found that CCR5QTY in storage buffer containing 200 mM arginine exhibits a higher stability, with a Tm of ∼73.3 °C, as compared to native CCR5, with a Tm of ∼47.1 °C in detergent (27). We also observed, during experimental handling, that QTY variant receptors show remarkable thermostability. Thus, we evaluated the thermostability and ligand-binding activities of CXCR4QTY and CCR10QTY after treatment at 1) 60 °C for 4 h, 2) 60 °C for 24 h, and 3) 100 °C for 10 min. The 2 temperature conditions are selected because 60 °C is an industrial standard for long-term electronic device performance, whereas 100 °C is the highest attainable temperature for pure water at sea level. Among the QTY variants tested, the CCR10QTY receptor exhibited the highest solubility and thermostability in water. All heat treatments were conducted in 50 mM Tris⋅HCl, 200 mM arginine condition.
After heat treatment, the proteins were centrifuged. No precipitation was visible. Protein concentration was determined using an Implen Nanodrop before and after centrifuge and <10% difference was detected. No extra protein band on gel electrophoresis was observed, comparing the results before and after the heat treatment (SI Appendix, Fig. S6). Protein aggregation was evaluated by MST during ligand-binding measurement and summarized in Fig. 5 A and B. We found that CXCR4QTY showed some minor aggregation, but it was still quite soluble after treatment at 60 °C and did not show visible aggregation after treatment at 100 °C. CCR10QTY also did not show any visible aggregation after treatment at 60 °C and 100 °C.
Fig. 5.
Thermostability of CXCR4QTY and CCR10QTY and extra intracellular hydrogen bond by simulation. (A) Relative affinity of CXCR4QTY toward CXCL12 after heat treatment. (B) Relative affinity of CCR10QTY toward CCL27 after heat treatment. (C–S) Additional internal hydrogen bonds in simulated CXCR4QTY and CCR10QTY. Notation: ‘s’ denotes a side chain bond and ‘b’ denotes a backbone bond. Thus, Q121s-T152s-T148b denotes that the side chain of Q at location 121 forms a hydrogen bond with the side chain of T at location 152, which forms a hydrogen bond with the backbone of T at position 148. In CXCR4QTY: (C) Q260s-S260s-Y256b, (D) T215b-Q216s-Q246s, (E) Y249s-Q253, (F) Q167s-H203b, (G) T169s-Q165b, (H) T204s-Q208s, (I) Q78s-Q69s-Q69b, (J) T112s-Q108b, (K) Q290s-T287b. In CCR10QTY: (L) D35s-R192s-D289s, (M) Y14s-Q172-Q214/Q172-S106b, (N) Q63s-Q82s, (O) Q167s-T163s-H159b, (P) Q54s-Q305s-Y256b-Q252s-Q81s-T308s, (Q) Q259s-Q298s, (R) Y263s-Q211s-S207b, (S) D270s-Q292s.
The ligand-binding affinity reduction of CXCR4QTY and CCR10QTY before and after heat treatment is shown in Fig. 5A. The QTY variant receptors were tested against their native ligands (CXCR4QTY vs. CXCL12 and CCR10QTY vs. CCL27). CXCR4QTY retains 90.5 ± 50.4% of its affinity after treatment at 60 °C for 4 h and 17.1 ± 5.9% of that after 24 h. Only 2.3 ± 0.8% affinity is obtained from CXCR4QTY after being treated at 100 °C for 10 min. The large error bar for the 60 °C test is likely due to soluble aggregation induced by elevated temperatures. At 100 °C it is likely that CXCR4QTY experienced significant structural or biochemical changes, which diminish its ligand-binding activity. However, for the 3 heating conditions for CCR10QTY the ligand affinity retention is 48.2 ± 0.4% (60 °C for 4 h), <1% (60 °C for 24 h), and 48.9 ± 3.1% (100 °C for 10 min). CCR10QTY exhibits better thermostability at higher temperatures but less so under prolonged treatment.
One possible contributing factor for the observed thermostability is the hydrogen bonds from the sidechains of QTY amino acids in the receptors. Close inspection of the simulated protein structures indicates that numerous interhelical and intrahelical hydrogen bonds stabilize the proteins. There are 3 types of hydrogen bonds formed: 1) hydrogen bonds between side chains, 2) hydrogen bonds between side chains and backbones, 3) hydrogen bonds within network of side chain with side chain and with backbones. Fig. 4 C–S shows the potential formation of hydrogen bonds in the simulated CXCR4QTY and CCR10QTY in an explicit water environment. These hydrogen bonds do not exist in natural receptors since L, V, I, and F do not have –OH and H2N-CH-C = O side chains, and thus lack hydrogen bond forming capabilities.
The other contributing factor is likely arginine in storage buffer. Arginine is known to enhance the solubility for aggregation-prone molecules (28). It has been shown that the addition of arginine effectively prevents protein precipitation and aggregation, especially at elevated temperatures, as well as preventing proteolytic degradation (29).
Discussion
The human genome encodes 826 known GPCR genes (12). They regulate a wide range of biological functions, from sight, smell, taste, sensation, immune system, brain function, and growth to hormone responses. The chemokine receptor class is an important subgroup that controls the immune system and accounts for both normal and pathological processes in the human body. Our understanding of how these membrane proteins function in vivo and how they can be utilized in vitro remains inadequate. The QTY code provides a useful tool for further study of these receptors by permitting the study of water-soluble analogs in a detergent-free aqueous environment. It opens the door for the design of chimeric receptors and further analysis of how different ligands bind their respective receptors.
Study of Chimeric Receptors with Alternative N Terminus and 3 EC Loops.
Our study of chimera design with N terminus and EC loops complements previous computational studies. The ligand-binding affinity obtained by exchanging the receptor’s N terminus and EC loops is in good agreement with the ligand-binding mechanism simulations previously reported (24–26). Despite the sequence change, specific bond-forming residues in N terminus, EC loops, and TM regions all contribute to the binding between chemokine receptors and their natural ligands. By comparing the affinity of proteins designed with different binding domains, we are able to elucidate the relative contribution from different parts of the receptors.
Proposed Internal Hydrogen Bonds and Enhanced Thermostability.
The substitution of 7TM hydrophobic residues by nonionic polar amino acids Q, T, and Y substantially increases the possible intramolecular hydrogen bond formations in QTY receptor variants. The results from the present study of thermostability suggest that the additional intrahelix and interhelix hydrogen bonds contribute to structural integrity. It has been found in previous crystallographic studies that natural GPCRs also contain water molecules (30, 31). The existence of hydrogen bonding and water-mediated interactions was suggested for both ligand binding and G protein activation (29). A recent computational study identified a conserved network of mostly mobile water molecules that communicate throughout the protein for signaling in many Class-A GPCRs (32).
Importance of Arginine in Refolding and Stabilizing the QTY Variants.
We observed that in the presence of higher concentrations of arginine, the QTY variant receptor showed good ligand affinity even after treatment in elevated temperatures. The simulated CCR5QTY structure showed a stable folded structure in an explicit water environment that is superimposable with native CCR5 despite substantial QTY changes. Simulations also suggest after the QTY modification, each side chain can form 3 or 4 hydrogen bonds (13). Extensive hydrogen bond formations were observed among intrahelices between the amino acid side chains and peptide backbones, and interhelices between these QTY amino acid side chains of difference α-helices.
Implications and Possible Applications.
Our studies have several implications for membrane protein design and possible uses of these water-soluble chemokine receptors. We can now 1) affordably produce large amounts of the detergent-free QTY receptors in E. coli system, 2) show that the QTY variant receptors’ thermostabilities are better than native receptors, and 3) show that their ligand-binding affinities are similar to native receptors. The ligand binding not only occurs in buffer but also in 50% human serum, which is more realistic for in vivo conditions and pharmacological applications. These QTY variants provide us a robust tool with which we can study how the native counterpart of these receptors work and investigate ligand-binding mechanisms in more detail.
The QTY variant receptors with similar ligand affinities from their native chemokine receptors may be useful for various in vivo and in vitro applications. 1) For example, we showed that CXCR4QTY, CCR5QTY, and CXCR7QTY exhibit a higher affinity for HIV coated gp41–120 glycoprotein without the presence of CD4 coreceptor. This observation has implications in developing decoy therapeutic treatments for HIV infection. 2) The chimera design enables us to design 7TM proteins with fine-tuning ability for physiological properties and functionality. The methodology can potentially be applied to native GPCRs to design variants of TM receptors or other functional membrane proteins in vivo. 3) The thermostability is important in scale-up production and widespread use. The exceptional thermal stability for certain modified TM regions combined with tunable functionality enables the QTY variant receptors to be treated like robust materials instead of delicate and fragile biomolecules. We believe application of the QTY code will be highly beneficial for a broad range of applications.
Materials and Methods
Bioinformatics of the QTY Variants.
The existence of hydrophobic regions within the transmembrane region in the variant protein sequences were determined via the web-based tool TMHMM Server v.2.0: http://www.cbs.dtu.dk/services/TMHMM-2.0/
Computer Simulations of CCR5QTY, CXCR5QTY in an Explicit Water Environment at 24.85 °C, at pH 7.4 and a 0.9% NaCl Ion Concentration.
The published crystal structures of CCR5 (4MBS) were obtained from the Protein Data Bank. Predicted initial structures of the QTY candidates were obtained from the predicted sequence and the GOMoDo modeling server45. The CCR5QTY sequence is 67.33% identical to CCR5. The MD was simulated at a 5 fs timestep using a simulation cell 20 Å larger than each candidate, but with recentering of the candidate at each timestep, and with generation and removal of water at the boundary, to negate the possibility of nonphysical boundary effects. The model was then aligned to its detergent-encapsulated counterparts CCR5 using MUSTANG48 and superimposed. Since there are no available structures of CXCR5QTY, the receptor is not compared with natural CXCR5.
Protein Expression, Purification, and Refolding from E. coli.
Genes of QTY modified chemokine receptor proteins were codon optimized for E. coli expression and obtained from Genscript. The genes were cloned into pET20b expression vector with Carbenicillin resistance. The plasmids were reconstituted and transformed into E. coli BL21(DE3) strain. Transformants were selected on LB medium plates with 100 μg/mL Carbenicillin. E. coli cultures were grown at 37 °C until the OD600 reached 0.4–0.8, after which IPTG (isopropyl-d-thiogalactoside) was added to a final concentration of 1 mM followed by 4 h expression. Cells were lysed by sonication in B-PER protein extraction agent (Thermos-Fisher) and centrifuged (23,000 × g, 40 min, 4 °C) to collect the inclusion body. The biomass was then subsequently washed twice in buffer 1 (50 mM Tris·HCl pH7.4, 50 mM NaCl, 10 mM CaCl2, 0.1%vol/vol Triton X 100, 2 M Urea, 0.2 μm filtered), once in buffer 2 (50 mM Tris·HCl pH7.4, 1 M NaCl, 10 mM CaCl2, 0.1%vol/vol Triton X 100, 2 M Urea, 0.2 μm filtered), and again in buffer 1. Pellets from each washing step were collected by centrifugation (23,000 × g, 25 min, 4 °C).
Washed inclusion bodies were fully solubilized in denaturation buffer (6 M guanidine hydrochloride, 1× phosphate-buffered saline, 10 mM Dithiothreitol, 0.2 μm filtered) at room temperature for 1.5 h with magnetic stirring. The solution was centrifuged at 23,000 × g for 40 min at 4 °C. The supernatant with proteins was then purified by Qiagen Ni-NTA beads (His-tag) followed by size exclusion chromatography using an ÄKTA Purifier system and a GE healthcare Superdex 200 gel-filtration column. Purified protein was collected and dialyzed twice against renaturation buffer (50 mM Tris·HCl pH 9.0, 3 mM reduced glutathione, 1 mM oxidized glutathione, 5 mM Ethylenediaminetetraacetic acid, and 0.5 M l-arginine). Following an overnight refolding process, the renatured protein solution was dialyzed against 50 mM Tris·HCl pH 9.0 with various arginine content, and filtered through a 0.2 µm syringe filter to remove aggregates.
MicroScale Thermophoresis Measurement.
The detailed methods can be found in SI Appendix, Materials and Methods. MST experiments of Chimera A and Chimera B were performed in the Center for Macromolecular Interactions at Harvard Medical School with second generation Monolith NT Protein Labeling Kit RED – NHS. The assay’s buffer was 50 mM Tris⋅HCl pH 9.0, 500 mM arginine for Chimera A and 50 mM Tris⋅HCl pH 9.0, 200 mM arginine for Chimera B.
NanoDSF Measurements of the Thermostability of the QTY Variants.
The detailed methods of NanoDSF can be found in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
We thank Tao Fei for making the Fig. 2 A and B alignment. MST measurements were performed by 2bind GmbH, Germany. This work was primarily funded by OH2 Laboratories and the MIT Center for Bits and Atoms Consortium that includes Bay Valley Innovation Center (Shanghai).
Footnotes
Competing interest statement: This research was in part funded by a MIT startup OH2laboratories. S.Z. fully discloses that he has a minority stake in this startup for the invention.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909026116/-/DCSupplemental.
References
- 1.Fredriksson R., Lagerström M. C., Lundin L. G., Schiöth H. B., The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol. Pharmacol. 63, 1256–1272 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Milić D., Veprintsev D. B., Large-scale production and protein engineering of G protein-coupled receptors for structural studies. Front. Pharmacol. 6, 66 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Charo I. F., Ransohoff R. M., The many roles of chemokines and chemokine receptors in inflammation. N. Engl. J. Med. 354, 610–621 (2006). [DOI] [PubMed] [Google Scholar]
- 4.Wu B., et al. , Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science 330, 1066–1071 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holman D. W., Klein R. S., Ransohoff R. M., The blood-brain barrier, chemokines and multiple sclerosis. Biochim. Biophys. Acta 1812, 220–230 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bagri A., et al. , The chemokine SDF1 regulates migration of dentate granule cells. Development 129, 4249–4260 (2002). [DOI] [PubMed] [Google Scholar]
- 7.Balkwill F., Cancer and the chemokine network. Nat. Rev. Cancer 4, 540–550 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Lopalco L., CCR5: From natural resistance to a new anti-HIV strategy. Viruses 2, 574–600 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keeley E. C., Mehrad B., Strieter R. M., CXC chemokines in cancer angiogenesis and metastases. Adv. Cancer Res. 106, 91–111 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Havenar-Daughton C., et al. ; IAVI Protocol C Principal Investigators , CXCL13 is a plasma biomarker of germinal center activity. Proc. Natl. Acad. Sci. U.S.A. 113, 2702–2707 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vinothkumar K. R., Henderson R., Structures of membrane proteins. Q. Rev. Biophys. 43, 65–158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lv X., et al. , In vitro expression and analysis of the 826 human G protein-coupled receptors. Protein Cell 7, 325–337 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang S., et al. , QTY code enables design of detergent-free chemokine receptors that retain ligand-binding activities. Proc. Natl. Acad. Sci. U.S.A. 115, E8652–E8659 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dall’Acqua W., Carter P., Antibody engineering. Curr. Opin. Struct. Biol. 8, 443–450 (1998). [DOI] [PubMed] [Google Scholar]
- 15.Hudson P. J., Souriau C., Engineered antibodies. Nat. Med. 9, 129–134 (2003). [DOI] [PubMed] [Google Scholar]
- 16.Carter P. J., Potent antibody therapeutics by design. Nat. Rev. Immunol. 6, 343–357 (2006). [DOI] [PubMed] [Google Scholar]
- 17.Duhr S., Braun D., Why molecules move along a temperature gradient. Proc. Natl. Acad. Sci. U.S.A. 103, 19678–19682 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wienken C. J., Baaske P., Rothbauer U., Braun D., Duhr S., Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 100 (2010). [DOI] [PubMed] [Google Scholar]
- 19.Seidel S. A. I., et al. , Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D’huys T., Claes S., Van Loy T., Schols D., CXCR7/ACKR3-targeting ligands interfere with X7 HIV-1 and HIV-2 entry and replication in human host cells. Heliyon 4, e00557 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho J. G. S., Middelberg A. P. J., Estimating the potential refolding yield of recombinant proteins expressed as inclusion bodies. Biotechnol. Bioeng. 87, 584–592 (2004). [DOI] [PubMed] [Google Scholar]
- 22.Arakawa T., Tsumoto K., The effects of arginine on refolding of aggregated proteins: Not facilitate refolding, but suppress aggregation. Biochem. Biophys. Res. Commun. 304, 148–152 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Arakawa T., Uozaki M., Hajime Koyama A., Modulation of small molecule solubility and protein binding by arginine. Mol. Med. Rep. 3, 833–836 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Tamamis P., Floudas C. A., Elucidating a key component of cancer metastasis: CXCL12 (SDF-1α) binding to CXCR4. J. Chem. Inf. Model. 54, 1174–1188 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu L., Li Y., Sun H., Li D., Hou T., Structural basis of the interactions between CXCR4 and CXCL12/SDF-1 revealed by theoretical approaches. Mol. Biosyst. 9, 2107–2117 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Tamamis P., Floudas C. A., Elucidating a key anti-HIV-1 and cancer-associated axis: The structure of CCL5 (Rantes) in complex with CCR5. Sci. Rep. 4, 5447 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knepp A. M., Grunbeck A., Banerjee S., Sakmar T. P., Huber T., Direct measurement of thermal stability of expressed CCR5 and stabilization by small molecule ligands. Biochemistry 50, 502–511 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamada H., Arakawa T., Shiraki K., Effect of additives on protein aggregation. Curr. Pharm. Biotechnol. 10, 400–407 (2009). [DOI] [PubMed] [Google Scholar]
- 29.Golovanov A. P., Hautbergue G. M., Wilson S. A., Lian L. Y., A simple method for improving protein solubility and long-term stability. J. Am. Chem. Soc. 126, 8933–8939 (2004). [DOI] [PubMed] [Google Scholar]
- 30.Pardo L., Deupi X., Dölker N., López-Rodríguez M. L., Campillo M., The role of internal water molecules in the structure and function of the rhodopsin family of G protein-coupled receptors. ChemBioChem 8, 19–24 (2007). [DOI] [PubMed] [Google Scholar]
- 31.Angel T. E., Chance M. R., Palczewski K., Conserved waters mediate structural and functional activation of family A (rhodopsin-like) G protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 106, 8555–8560 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatakrishnan A. J., et al. , Diverse GPCRs exhibit conserved water networks for stabilization and activation. Proc. Natl. Acad. Sci. U.S.A. 116, 3288–3293 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.