Significance
The 5 subtypes of the muscarinic acetylcholine receptors (mAChRs) are expressed throughout the central and peripheral nervous system where they play a vital role in physiology and pathologies. Recently, the M5 mAChR subtype has emerged as an exciting drug target for the treatment of drug addiction. We have determined the atomic structure of the M5 mAChR bound to the clinically used inverse agonist tiotropium. The M5 mAChR structure now allows for a full comparison of all 5 mAChR subtypes and reveals that small differences in the extracellular loop regions can mediate orthosteric and allosteric ligand selectivity. Together, these findings open the door for future structure-based design of selective drugs that target this therapeutically important class of receptors.
Keywords: muscarinic receptor, G protein-coupled receptor, drug design, kinetics, crystal structure
Abstract
The human M5 muscarinic acetylcholine receptor (mAChR) has recently emerged as an exciting therapeutic target for treating a range of disorders, including drug addiction. However, a lack of structural information for this receptor subtype has limited further drug development and validation. Here we report a high-resolution crystal structure of the human M5 mAChR bound to the clinically used inverse agonist, tiotropium. This structure allowed for a comparison across all 5 mAChR family members that revealed important differences in both orthosteric and allosteric sites that could inform the rational design of selective ligands. These structural studies, together with chimeric swaps between the extracellular regions of the M2 and M5 mAChRs, provided structural insight into kinetic selectivity, where ligands show differential residency times between related family members. Collectively, our study provides important insights into the nature of orthosteric and allosteric ligand interaction across the mAChR family that could be exploited for the design of selective drugs.
The muscarinic acetylcholine (ACh) receptors (mAChRs) are class A G protein-coupled receptors (GPCRs) that together with the nicotinic acetylcholine receptors facilitate the actions of the neurotransmitter, ACh, throughout the body. The mAChR family comprises 5 subtypes in which M1, M3, and M5 are preferentially coupled to the Gq/11 protein-mediated signaling pathways, and M2 and M4 show preference for Gi/o protein-dependent signaling. Localization studies have revealed that the mAChR subtypes are differentially distributed, with M1, M4, and M5 mAChRs found predominantly in the central nervous system (CNS), where they are essential for normal neuronal function, while M2 and M3 mAChRs are expressed more widely, including in the periphery, where they are involved in cardiovascular as well as gut motility and secretory processes (1).
Given the involvement of mAChRs in such a wide range of fundamental physiological processes, they have long been valued as targets for novel therapeutics, in particular the central M1 and M4 mAChRs, which have garnered attention due to their involvement in cognition and memory (2). In contrast, relatively less is known about the M5 mAChR subtype, which represents less than 2% of the total CNS mAChR population (3, 4). Despite its low level of expression, this receptor plays a vital role in the mesolimbic reward pathway due to its presence on dopaminergic neurons of the ventral tegmental area (5–8). Additionally, there is a large population of nonneuronal M5 mAChRs located within the endothelium of the cerebral vasculature, suggesting that the receptor may modulate cerebral vasodilatory processes (9, 10). These observations correlate well with phenotypic data from M5 mAChR knockout mice where the cerebral vasculature is constitutively constricted, resulting in decreased cerebral blood flow (11, 12). Additionally, M5 mAChR knockout mice exhibited attenuated reward-seeking behavior to drugs of addiction, such as cocaine and morphine, in self-administration and conditioned place-preference experiments (13–15). Moreover, in recent studies involving rats (16–18), ethanol-seeking behavior and oxycodone self-administration were attenuated by the selective M5 mAChR negative allosteric modulator (NAM) ML375 (19). From these studies, the M5 mAChR has emerged as a potential target for the treatment of drug addiction.
Despite such promising data, further study of the M5 mAChR has been hindered by a lack of selective small-molecule tool compounds. Designing conventional small-molecule ligands that target the orthosteric ACh-binding site of individual mAChR subtypes has been challenging due to the highly conserved sequence homology of the mAChR orthosteric site residues (1) and in part due to a lack of detailed structural information for all 5 receptor subtypes. While structures of the M1 to M4 mAChRs have been previously determined, there are no available structures for the M5 mAChR. Therefore, to provide a complete structural comparison of all 5 family members, we determined a high-resolution crystal structure of the M5 mAChR, which revealed differences in the extracellular loop (ECL) regions that could mediate orthosteric and allosteric ligand selectivity. Based on these differences and the fact that some medicines are now known to have different binding rates between mAChR subtypes that can result in clinically relevant “kinetic” selectivity, we also made chimeric swaps of the ECL regions between the M2 and M5 mAChRs to investigate the role of these regions in mediating this important mode of drug selectivity.
Results
Crystallization and Determination of the M5 mAChR Structure.
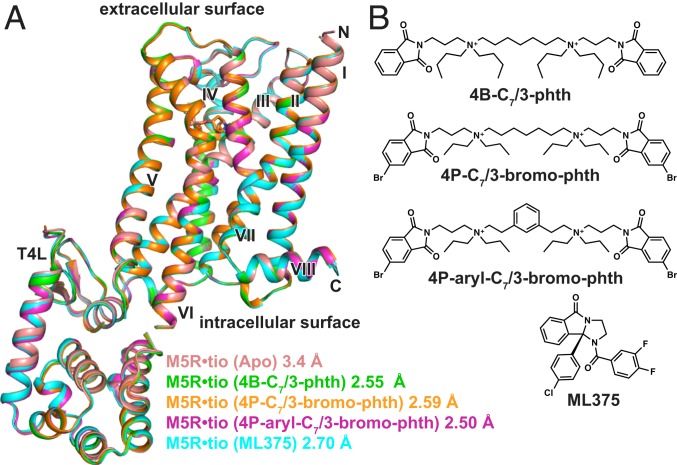
To determine the M5 mAChR structure, we designed a construct in which residues 225 to 430 of intracellular loop 3 were removed and replaced with a T4 lysozyme (T4L) fusion protein. Additionally, to promote crystallization, the first 20 N-terminal amino acids were cleaved by a tobacco etch virus protease site engineered into the receptor (SI Appendix, Fig. S1). The inverse agonist, tiotropium, was used to stabilize the inactive state as it has a slow dissociation rate at the M5 mAChR (20) and was also used in the determination of the M1, M3, and M4 mAChR structures (21, 22). The M5-T4L•tiotropium complex was crystallized in lipidic cubic phase (LCP), and crystals were obtained within 1 to 2 d; however, despite many rounds of optimization, diffraction was limited to 7 Å. To improve the resolution, we built upon a study from Kajiwara et al. (23) that predicted that mutation of the amino acid at position 3.39 [numbered according to Ballesteros-Weinstein (24)] to Arg would create a thermostabilized receptor by promoting an ionic bond between this residue and the highly conserved D2.50 residue. Recently, the same S3.39R mutation was applied to the M2 mAChR, resulting in a series of higher resolution structures (25). Although introduction of the S1173.39R mutation resulted in a construct that binds the antagonists N-methyl scopolamine (NMS) or tiotropium with a slightly reduced affinity relative to the wild-type (WT) M5 mAChR, the effect of the mutation on reducing ACh affinity was substantially more pronounced (SI Appendix, Fig. S1), consistent with the ability of the construct to favor an inactive over an active state. Similar differential effects on antagonist versus agonist affinity were previously observed for S3.39R at the M2 mAChR (25). Notably, introduction of the S1173.39R mutation increased our M5 mAChR yields during purification and resulted in crystals that diffracted to a resolution of 3.4 Å. Data were collected from ∼130 crystals, and the structure was determined by molecular replacement using the M3 structure [Protein Data Bank (PDB) ID 4U15] and an ensemble of T4L structures as templates (Fig. 1A and SI Appendix, Table S1).
Fig. 1.
Structures of M5-T4L bound to tiotropium. (A) Overlay of 5 different M5 mAChR structures determined in the presence of tiotropium and (B) different allosteric modulators. The structure from 4B-C7/3-phth was the most resolved of all of the datasets and is used in all further comparisons.
To investigate the nature of NAM binding to the M5 mAChR, we attempted to obtain cocrystal structures. Given that the bis-ammonium alkane-type ligands tend to have higher affinities for the M5 mAChR than the prototypical modulator, gallamine (26), we tried to obtain a ternary complex structure of the M5 mAChR with tiotropium and several bis-ammonium alkane ligands (Fig. 1B). We initially used the modulator 4B-C7/3-phth, which resulted in crystals that grew to a much larger size and diffracted to a resolution of 2.55 Å (Fig. 1 and SI Appendix, Table S2 and Fig. S2). Based on previous data (27), we predicted that 4B-C7/3-phth would bind in the extracellular vestibule (ECV). While there were regions of strong electron density present in the ECV, we could not unambiguously model 4B-C7/3-phth into the density as a molecule of the precipitant, polyethylene glycol 400 (PEG400), also likely binds in this site (22, 28) and may explain why researchers have had difficulty in obtaining co-NAM–bound structures for the mAChRs.
Subsequently, we designed 2 bis-ammonium alkane analogs using the higher affinity 4P-C7/3-phth scaffold (27) to try to improve modulator affinity (Fig. 1B) and detectability by X-rays. The first modification added 2 bromine (Br) atoms (4P-C7/3-bromo-phth) to increase the size of the pthalamide groups (29), and the second modification rigidified the flexible 7-carbon linker with an aromatic hydrocarbon (4P-aryl-C7/3-bromo-phth). When tested in functional inositol phosphate (IP1) assays and [3H]NMS competition radioligand binding, both ligands had a similar affinity in relation to the parent compound (4P-C7/3-phth) (SI Appendix, Figs. S3A and S4 and Table S3). Like 4B-C7/3-phth, the addition of either 4P-C7/3-bromo-phth or 4P-aryl-C7/3-bromo-phth to purified M5 mAChR and reconstitution into LCP yielded crystals that diffracted to a higher resolution (SI Appendix, Table S1). A full dataset for the 4P-aryl-C7/3-bromo-phth was collected at a wavelength of 0.92 Å to maximize the anomalous Br signal in a single wavelength anomalous diffraction experiment; however, no such signal was detected, suggesting that 4P-aryl-C7/3-bromo-phth was not present in the structure. Since the structure was solved by merging a large number of datasets, there is a possibility that the Br signal for the NAM would be averaged out if NAM occupancy is low. However, inspection of different datasets did not indicate that this was the case.
As an alternate strategy, we attempted to determine a cocrystal structure with the structurally diverse M5 mAChR-selective NAM, ML375 (19). In comparison to the bis-ammonium ligands, the addition of ML375 resulted in a slightly lower resolution structure (2.7 Å, SI Appendix, Table S1), and, as was the case with the bis-ammonium NAMs, we were not able to assign ML375 any electron density. Comparison of all M5 mAChR structures showed that they were nearly identical, with root mean square deviation values of 0.09 to 0.22 Å. The higher-resolution 2.55 Å M5•tiotropium (4B-C7/3-phth) structure was used for further comparison, as this was the best-resolved and modeled structure (SI Appendix, Fig. S5).
Family-Wide Comparison of All mAChR Subtypes.
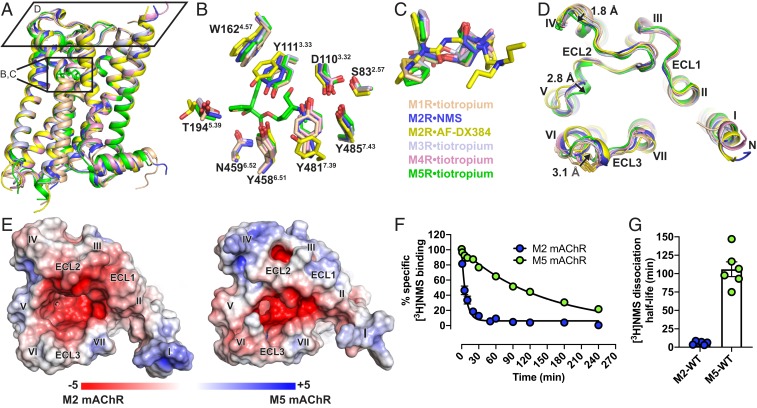
The solution of the M5 mAChR structure allows for a complete subtype-wide comparison of this important GPCR family. The structure of the M5 mAChR is similar to the previously determined structures of the M1 to M4 mAChR subtypes (21, 22, 30) with a root mean squared deviation of 0.5 to 0.8 Å (Fig. 2A) for the 7-transmembrane domain across all subtypes. The 5 mAChR subtypes are most similar in the orthosteric binding site, which is the most conserved region of the receptor. The fact that our M5 mAChR structure was obtained in complex with the same ligand (tiotropium) as the M1, M3, and M4 mAChR structures allowed for a specific, detailed comparison of residues lining this orthosteric binding site (Fig. 2 B and C). This comparison demonstrated that the residues within the orthosteric pocket are absolutely conserved between the receptors. Although there is no tiotropium-bound M2 mAChR structure, there are now 6 different inactive state M2 mAChR structures, which include structures bound with the nonselective ligands 3-quinuclidinyl benzilate (QNB) and NMS and the M2 mAChR selective ligand AF-DX384 (25). The 2.3-Å M2•NMS structure is most similar to the tiotropium-bound mAChR structures, although residues Y3.33 and Y7.39 of the “tyrosine lid” (Y3.33, Y6.51, and Y7.39) are positioned in a distinct conformation in comparison to the tiotropium-bound structures. These differences in the tyrosine lid positions are more pronounced in the M2•AF-DX384 structures, allowing the accommodation of this bulkier ligand into the orthosteric binding pocket (Fig. 2 B and C).
Fig. 2.
Structural comparison of M1 to M5 mAChRs. (A) The overall view of the M1 to M5 mAChR structures aligned with the M5 mAChR and shown as cartoons. M1•tiotropium is colored peach (PDB ID code 5CXV), M2•NMS in dark blue (PDB ID code 5ZKC), M2•AF-DX384 in yellow (PDB ID 5ZKB), M3•tiotropium in light blue (PDB ID 4U15), M4•tiotropium in pink (PDB ID 5DSG) and M5•tiotropium in green (PDB ID 6OL9). (B) Comparison of residues (stick representation) lining the orthosteric site with tiotropium from the M5 mAChR displayed and (C) overlay of the orthosteric ligands. (D) View from the extracellular surface comparing differences in the ECL regions across the M1 to M5 mAChRs. Distances between the backbone of M1 and M5 mAChR residues in ECL2 and ECL3 are shown and indicated by arrows. (E) Electrostatic and surface potential of M2 and M5 mAChR (+5kT/e in blue and −5kT/e in red) mapped on the surface of the receptors calculated at pH 7.0. (F) Comparison of dissociation rate and (G) dissociation half-life of [3H]NMS by the addition of 10 µM atropine at the M2 and M5 mAChRs. Values are significantly different (P value < 0.0001, 2-way ANOVA). Detailed statistical analysis is shown in SI Appendix, Table S5.
Subtle yet notable differences between the mAChR subtypes are observed for ECL2 and ECL3, corresponding to regions that are the least conserved across the receptors (Fig. 2D). At ECL2 there is a 1.8-Å difference across all 5 subtypes beginning at the first nonconserved residue of ECL2. As ECL2 progresses toward TM5, a conserved 310 helix motif moves inward by 2.8 Å in the M5 mAChR when compared to the M1 structure. Similarly, the conserved ECL3 disulphide bond is displaced inward by 3.1 Å for the M3 and M5 mAChRs (Fig. 2D), relative to the other subtypes. These observed differences in the positions of ECL2 and ECL3, along with differences in amino acid composition, contribute to a more constricted entrance to the orthosteric binding site at the M5 (and M3) versus the M2 mAChR (Fig. 2E). Furthermore, this contraction of the entrance in the antagonist-bound structures may contribute to the slower dissociation rate of orthosteric ligands from the M5 and M3 mAChRs, in comparison to other subtypes like the M2 mAChR. For example, despite having similar equilibrium-binding affinities, [3H]NMS dissociates 18-fold more slowly at the M5 than at the M2 mAChR with half-lives of dissociation of 100 ± 11.6 and 5.7 ± 1.2 min, respectively (Fig. 2 F and G).
Structural Differences between the ECVs of the M2 and M5 mAChRs.
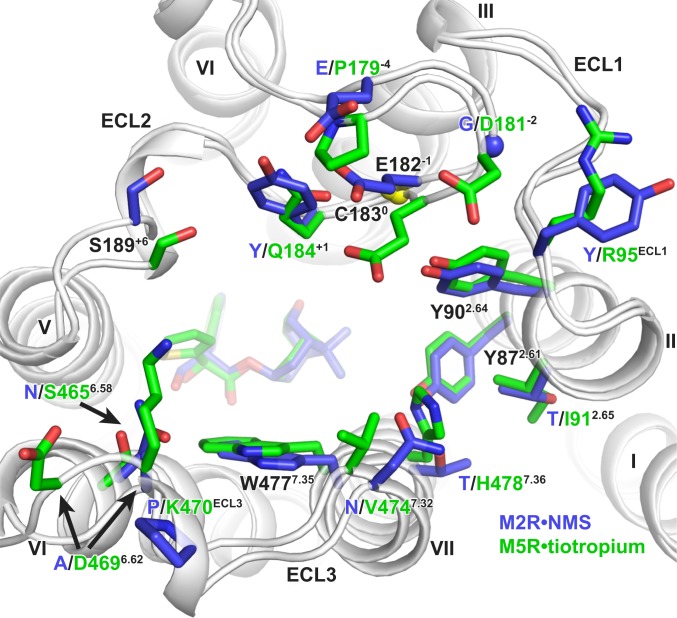
An alternative strategy to generating selective ligands is to target nonconserved allosteric sites (31). This has been extensively explored for the mAChR family in which a palette of both positive and negative allosteric modulators has been identified (32, 33). Structural and mutagenesis studies have established that many of these ligands bind to a “common” allosteric site that is located above the orthosteric site and within an ECV (Fig. 3 and SI Appendix, Fig. S6) (34). In fact, the M5 mAChR has often served as a model system for early research into understanding the binding mode and mechanism of selectivity for prototypical modulators, such as the bis-ammonium alkane ligands (Fig. 1B), that have higher sensitivity for modulating the M2 mAChR and lower sensitivity for the M5 mAChR (26, 35–38). These studies identified nonconserved residues in ECL2 (P179−4, E182−1, and Q184+1; superscript indicates the position of ECL2 residues relative to the conserved Cys in ECL2) and TM7 (V4747.32 and H4847.36) as residues that can account for M2/M5 subtype selectivity. Comparison of the ECV between the M2 and M5 mAChRs confirms differences in the orientations and positions of these residues that could mediate the selectivity. Namely, P179−4 in ECL2 restricts the position of E182−1, forcing the residue into the ECV near Q184+1. Residue Q184+1, which is a F/Y residue for the M1 to M4 mAChRs subtypes, is a key residue for the activity of many allosteric modulators. Other major differences between the M2/M5 ECVs are in the positions of nonconserved residues lining the top of TM6 starting from S4656.58 across ECL3 and down to residue H4787.36 in TM7. At the M5 mAChR, these residues are bulkier and point more inwardly, constricting the overall size of the ECV (Fig. 3).
Fig. 3.
Comparison of residues lining the extracellular vestibule of the M2 and M5 mAChR. M2•NMS is shown in dark blue and M5•tiotropium in green. Conserved residues are labeled black, and nonconserved residues have colored labels based on receptor subtype. Residues are numbered based on the M5 mAChR, with residues in ECL2 numbered relative to the conserved cysteine in ECL2, which is shown as a yellow sphere. Sidechains for D4696.62 and K470ECL3 are truncated to the β-carbon in the deposited model due to a lack of sidechain density and are modeled here as the most probable rotamer.
Role of the M5 and M2 mAChR ECL Regions in Orthosteric and Allosteric Ligand Binding.
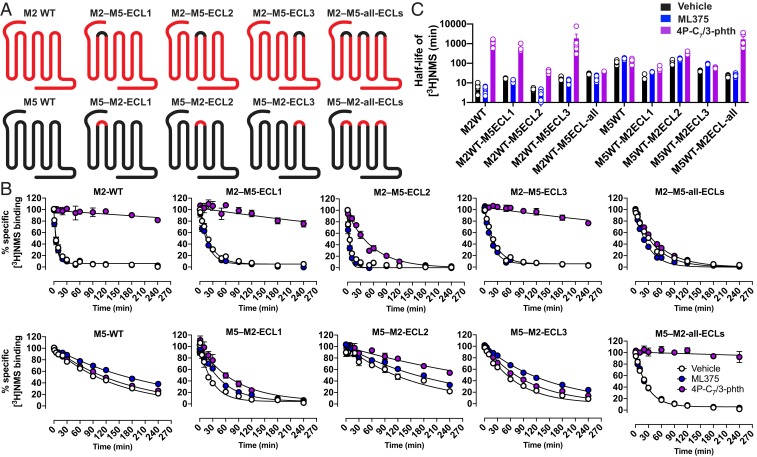
The effect of ECL regions on orthosteric ligand access and egress has significant biological and clinical relevance (39). Therefore, to investigate the role of the ECLs in modulating the slower dissociation kinetics of the M5 mAChR in comparison with the M2 mAChR, we designed full ECL1, ECL2, and/or ECL3 chimeric swaps between the 2 subtypes (Fig. 4 and SI Appendix, Fig. S7). The ECL chimeras had similar levels of expression and binding of [3H]NMS to WT receptors (SI Appendix, Table S4). As previously noted, the M2 mAChR has a shorter half-life for [3H]NMS dissociation in comparison with the M5 mAChR (Fig. 2F). Incorporation of the M2 ECL1 or ECL3 into the M5 mAChR increased [3H]NMS dissociation, while the reciprocal chimeric swap decreased [3H]NMS dissociation at the M2 mAChR. Unexpectedly, it was the ECL1 swaps that had the largest effect on [3H]NMS dissociation between the 2 subtypes, particularly at the M5 mAChR (Fig. 4 and SI Appendix, Table S5). A possible structural explanation for this observation could be that R95ECL1, which is a conserved Tyr residue at the M1 to M4 subtypes, is capable of forming an ionic bond with either the M5 ECL2 residue D181−2 or, in the case of the M2 ECL1 chimera, residue D173−3 (Fig. 3 and SI Appendix, Fig. S6). Such an interaction could tether ECL1 and ECL2, limiting their overall dynamics and thus reduce rates of orthosteric ligand dissociation. It is important to note that R95ECL1 is involved in an ionic interaction mediated through the crystal lattice with a neighboring T4L molecule (SI Appendix, Fig. S2 D–F), and as a result it does not directly interact with D181−2 in the M5 mAChR structure although it is well positioned to do so.
Fig. 4.
[3H]NMS binding dissociation kinetic studies of chimeric swaps between the ECLs of the M2 and M5 mAChRs. (A) Cartoons for the M2 and M5 ECL chimeras used in this study. (B) [3H]NMS reassociation was prevented by the addition of 10 µM atropine, and radioligand dissociation was monitored in the absence (Vehicle) or presence of 10 µM ML375 or 10 µM 4P-C7/3-phth. Data points represent the mean ± SEM of 3 or more independent experiments performed in duplicate. (C) Comparison of the dissociation half-lives for [3H]NMS, showing both individual values and the mean ± SEM (log-scale). Full quantitative parameters derived from this experiment are listed in SI Appendix, Table S5, including a statistical analysis.
A hallmark feature of an allosteric ligand that modulates orthosteric ligand affinity is the ability to either increase or decrease the rate of dissociation of an orthosteric ligand. To examine the effect of allosteric modulators on NMS dissociation across the M5 and M2 ECL chimeras, we used the bis-ammonium alkane ligand 4P-C7/3-phth, which had been previously studied at the M2 mAChR and had high affinity for the M5 mAChR (SI Appendix, Table S3) or the M5 selective modulator ML375 (19, 27). In the presence of ML375, [3H]NMS dissociation was reduced at the M5 mAChR and had no effect at the M2 mAChR, whereas the addition of 4P-C7/3-phth reduced radioligand dissociation at the M2 mAChR but not at the M5 mAChR (Fig. 4 and SI Appendix, Table S5). The ECL1 and ECL3 chimeric swaps had little effect on the activity of ML375 for either receptor subtype and slightly increased the activity of 4P-C7/3-phth at the M5 mAChR. For the ECL2 chimeras, there was no effect on activity of ML375. However, there was a loss of 4P-C7/3-phth activity at the M2 mAChR and a corresponding gain of activity at the M5 mAChR. These results are in line with previous studies and highlight the importance of residues in ECL2, particularly M2-Y177 and M5-E184, on modulating the activity of bis-ammonium alkane ligands. Interestingly, when all 3 ECLs were swapped, the resulting M2 and M5 chimeric constructs functioned more like their swapped receptor counterpart. That is, for the M2–M5-all-ECL construct, 4P-C7/3-phth had little effect, and although ML375 did not retard [3H]NMS dissociation, it slightly increased the rate of [3H]NMS dissociation, suggesting an allosteric mode of action (Fig. 4 and SI Appendix, Table S5). Conversely, for the M5–M2-all-ECL construct, 4P-C7/3-phth retarded radioligand dissociation, and, surprisingly, ML375 had no effect. While none of the chimeric constructs ever fully switched the basal dissociation rate of [3H]NMS or ML375 activity to that observed for the corresponding WT constructs, the data nonetheless suggest that the ECL regions modulate the overall conformation of mAChRs and directly influence the dissociation of ligands from the orthosteric site.
Discussion
Individual mAChR subtypes have long been pursued as drug targets for a range of CNS disorders, and recent studies have begun to validate the M5 mAChR as a target for the treatment of drug addiction (4, 40). In this study, we have determined a high-resolution crystal structure of the M5 mAChR, thus allowing a subtype-wide comparison for any aminergic GPCR subfamily. Introduction of the inactive state stabilizing mutation S1173.39R, which was recently used to stabilize the M2 mAChR (25), was crucial to obtaining well-diffracting crystals and suggests that this mutation could be applied to aid the determination of inactive state structures for other related GPCRs. We further improved the resolution of the M5 mAChR structure by adding allosteric modulators to the purified protein prior to crystallization. Despite the consistent increase in resolution that each of the allosteric modulators provided, we were not able to model any of the modulators into electron density. From a pharmacological perspective, a lack of modulator binding is not surprising, as all of the modulators tested in this study showed strong negative cooperativity with tiotropium (SI Appendix, Fig. S3B). Nevertheless, it is still paradoxical that the addition of an allosteric modulator can clearly improve receptor crystallization and diffraction yet not be visible in any resulting structures. This phenomenon has been noted at other GPCRs, such as the M2 mAChR that was crystallized in the presence of the modulator alcuronium and the CC chemokine receptor 2A that was crystallized in the presence of the modulator AZD-6942, but where neither modulator could be observed in the resulting structures (25, 41).
Comparison of all 5 mAChR structures further confirms the well-conserved transmembrane core and orthosteric binding site that has made the discovery of highly selective drugs for these receptor subtypes incredibly challenging. The most apparent structural differences between the mAChR subtypes are in the ECL regions. Although these differences are generally quite subtle, they are important because they open up the possibility for designing selective molecules in a way that has not previously been possible (SI Appendix, Fig. S6). For example, a recent crystal structure of the M2 mAChR bound to the M2-selective antagonist AF-DX384 revealed that selectivity is mediated by differential interactions between the ligand and residues in ECL2, which lead to an outward displacement in ECL2 and the top of TM5 (Fig. 2D) (25). Likewise, by utilizing knowledge of a single amino acid difference in ECL2 between the M2 and M3 mAChRs, molecular docking and structure-based design led to the discovery of an M3-selective antagonist with 100-fold selectivity over the M2 mAChR (42). These results are similar to the structure-based design of biased ligands targeting the D2 dopamine receptor that were designed by utilizing specific amino acid–ligand contacts in ECL2 and TM5 (43). Taken together, these findings indicate that the differential targeting of ECL residues may be a path forward for creating selective mAChR ligands. This is well supported by the fact that many mAChR-selective allosteric modulators interact with the ECL regions (27, 34) and suggests that designing orthosteric ligands linked to allosteric pharmacophores, known as bitopic ligands, is a potential strategy for future structure-based drug design.
Drug discovery has typically focused on optimizing ligand affinity and selectivity; however, it is now apparent that binding kinetics can play a critical role in these events (39, 44–46). This is illustrated in 2 ways with the drug tiotropium as a pertinent example. First, tiotropium has slow rate of dissociation from the M3 mAChR, which is a key feature of the drug that allows for a once daily dosing for the treatment of chronic obstructive pulmonary disease (47). Second, although tiotropium has the same equilibrium-binding affinity for the M3 and M2 mAChRs, it exhibits kinetic selectivity for the M3 over the M2 mAChR by having substantially different rates of dissociation. This kinetic selectivity over the M2 mAChR is postulated to be due to differences in the electrostatics and dynamics of the ECL region (47). The M5 mAChR is similar to the M3 mAChR with respect to having slow rates of orthosteric ligand dissociation (20), and data from our M2/M5 ECL chimeras support the idea of the ECL regions underpinning kinetic selectivity as [3H]NMS dissociation was switched between the M2 and M5 mAChRs (Fig. 4). Notably, none of the combined ECL chimeras could ever fully switch the dissociation kinetics between subtypes, suggesting that other mechanisms are operative such as the global conformation of the ECLs. Our results also highlight the importance of the ECL regions on conferring sensitivity to allosteric modulators across different subtypes. By swapping out the entire ECL region between the M2 and M5 mAChRs we were able to completely alter the sensitivity of a modulator that is selective for the M2 versus the M5 mAChR and vice versa. These results are in line with previous studies using similar M2/M5 mAChR ECL chimeras (35–38, 48) and, collectively with our findings, highlight the importance of the ECL region for conferring subtype selectivity for different types of mAChR ligands.
In summary, our reported M5 mAChR crystal structure has allowed for the comparison of all 5 mAChR subtypes and has revealed that subtle differences in the ECL regions are a major determinant in ligand selectivity, regardless of the ligand being orthosteric or allosteric. As the M1, M4, and M5 mAChRs continue to emerge as exciting drug targets for the treatment of CNS disorders, it will be important to understand both the structural and the dynamic differences between all 5 mAChR subtypes in order to aid design of safer and more effective small-molecule therapeutics.
Materials and Methods
Detailed information on cloning, receptor purification, synthesis of the bis-ammonium alkane ligands, and molecular pharmacology experiments is provided in SI Appendix, Materials and Methods.
M5 Receptor Expression and Purification.
M5-T4L with the S1173.39R was purified similarly to previous methods (22).
Crystallization and Structure Determination.
Purified M5-T4L S1173.39R bound to tiotropium was crystallized using LCP. For allosteric modulator cocrystallization, the modulator was added to purified protein at a final concentration of 2.5 mM. The sample was incubated on ice for 3 h before it was mixed into 10:1 (wt/wt) monoolein:cholesterol in 1:1.5 wt/wt protein:lipid ratio. LCP crystallization was performed by spotting 25 to 30 nL of samples on a siliconized 96-well glass plate overlaying the samples with 600 nL of precipitant solution using the Gryphon LCP (Art Robbins Instruments). Sealed glass plates were incubated at 20 °C. Crystals appeared in the first 24 h and grew to full size in the following 1 to 2 d. The best diffracting crystals grew in 100 mM DL-Malic acid, pH 6.0; 220 to 280 mM ammonium tartrate dibasic; and 37 to 41% PEG 400. For the data collection, whole drops were harvested using mesh grid loops (Mitegen) and flash-frozen in liquid nitrogen.
X-ray diffraction data were collected at the SPring-8 (Japan) beamline BL32XU (49) and the MX2 beamline at the Australian Synchrotron (50). Diffraction data at SPring-8 was collected using the automatic data-collection system ZOO (51). Diffraction data were processed using KAMO (52) with XDS (53). The structure was solved using molecular replacement with M3-mT4L (4U15) as a search model for the receptor and an ensemble of T4L molecules for T4L. Structure refinement was performed with Phenix (54), and the models were validated with MolProbity (55). Sidechains for residues with no electron density (contoured at 1σ in a 2m|Fo| − D|Fc| map; SI Appendix, Fig. S5) past the β-carbon were truncated. Structure figures were prepared using the program PyMol. Electrostatic and surface potential of M2 and M5 mAChR (+5kT/e in blue and −5kT/e in red) mapped on the surface of the receptors calculated at pH 7.0 were calculated using PDB2PQR and APBS (56).
Data Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.rcsb.org (PDB ID code 6OL9 for M5-T4L). Detailed methods are provided in SI Appendix. Other materials and data are available on request.
Supplementary Material
Acknowledgments
The synchrotron radiation experiments were performed at the BL32XU beamline at SPring-8 with the approval of the Japan Synchrotron Radiation Research Institute (JASRI) (Proposal No. 2017B2731) and the MX2 beamline at the Australian Synchrotron (CAP13670). This work was funded by a Wellcome Trust Collaborative Award (201529/Z/16/Z) and supported by National Health and Medical Research Council of Australia (NHMRC) Project Grant APP1138448 and Program Grants APP1055134 and APP1150083. J.B.B. is a NHMRC Principal Research Fellow; P.M.S. and A.C. are NHMRC Senior Principal Research Fellows; and D.M.T. is an Australian Research Council Discovery Early Career Research Fellow.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Protein Data Bank (PDB), https://www.rcsb.org/ (PDB ID 6OL9).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914446116/-/DCSupplemental.
References
- 1.Caulfield M. P., Birdsall N. J., International Union of Pharmacology. XVII. Classification of muscarinic acetylcholine receptors. Pharmacol. Rev. 50, 279–290 (1998). [PubMed] [Google Scholar]
- 2.Kruse A. C., Hu J., Kobilka B. K., Wess J., Muscarinic acetylcholine receptor X-ray structures: Potential implications for drug development. Curr. Opin. Pharmacol. 16, 24–30 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yasuda R. P., et al. , Development of antisera selective for m4 and m5 muscarinic cholinergic receptors: Distribution of m4 and m5 receptors in rat brain. Mol. Pharmacol. 43, 149–157 (1993). [PubMed] [Google Scholar]
- 4.Bender A. M., Garrison A. T., Lindsley C. W., The muscarinic acetylcholine receptor M5: Therapeutic implications and allosteric modulation. ACS Chem. Neurosci. 10, 1025–1034 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Forster G. L., Blaha C. D., Laterodorsal tegmental stimulation elicits dopamine efflux in the rat nucleus accumbens by activation of acetylcholine and glutamate receptors in the ventral tegmental area. Eur. J. Neurosci. 12, 3596–3604 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Forster G. L., Blaha C. D., Pedunculopontine tegmental stimulation evokes striatal dopamine efflux by activation of acetylcholine and glutamate receptors in the midbrain and pons of the rat. Eur. J. Neurosci. 17, 751–762 (2003). [DOI] [PubMed] [Google Scholar]
- 7.Forster G. L., Yeomans J. S., Takeuchi J., Blaha C. D., M5 muscarinic receptors are required for prolonged accumbal dopamine release after electrical stimulation of the pons in mice. J. Neurosci. 22, RC190 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Steidl S., Miller A. D., Blaha C. D., Yeomans J. S., M5 muscarinic receptors mediate striatal dopamine activation by ventral tegmental morphine and pedunculopontine stimulation in mice. PLoS One 6, e27538 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elhusseiny A., Cohen Z., Olivier A., Stanimirović D. B., Hamel E., Functional acetylcholine muscarinic receptor subtypes in human brain microcirculation: Identification and cellular localization. J. Cereb. Blood Flow Metab. 19, 794–802 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Tayebati S. K., Di Tullio M. A., Tomassoni D., Amenta F., Localization of the m5 muscarinic cholinergic receptor in rat circle of Willis and pial arteries. Neuroscience 122, 205–211 (2003). [DOI] [PubMed] [Google Scholar]
- 11.Araya R., et al. , Loss of M5 muscarinic acetylcholine receptors leads to cerebrovascular and neuronal abnormalities and cognitive deficits in mice. Neurobiol. Dis. 24, 334–344 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Yamada M., et al. , Cholinergic dilation of cerebral blood vessels is abolished in M(5) muscarinic acetylcholine receptor knockout mice. Proc. Natl. Acad. Sci. U.S.A. 98, 14096–14101 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Basile A. S., et al. , Deletion of the M5 muscarinic acetylcholine receptor attenuates morphine reinforcement and withdrawal but not morphine analgesia. Proc. Natl. Acad. Sci. U.S.A. 99, 11452–11457 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fink-Jensen A., et al. , Role for M5 muscarinic acetylcholine receptors in cocaine addiction. J. Neurosci. Res. 74, 91–96 (2003). [DOI] [PubMed] [Google Scholar]
- 15.Thomsen M., et al. , Reduced cocaine self-administration in muscarinic M5 acetylcholine receptor-deficient mice. J. Neurosci. 25, 8141–8149 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berizzi A. E., et al. , Muscarinic M5 receptors modulate ethanol seeking in rats. Neuropsychopharmacology 43, 1510–1517 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gould R. W., et al. , Acutenegative allosteric modulation of M5 muscarinic acetylcholine receptors inhibits oxycodone self-administration and cue-induced reactivity with no effect on antinociception. ACS Chem. Neurosci. 10, 3740–3750 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gunter B. W., et al. , Selective inhibition of M5 muscarinic acetylcholine receptors attenuates cocaine self-administration in rats. Addict. Biol. 23, 1106–1116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gentry P. R., et al. , Discovery of the first M5-selective and CNS penetrant negative allosteric modulator (NAM) of a muscarinic acetylcholine receptor: (S)-9b-(4-chlorophenyl)-1-(3,4-difluorobenzoyl)-2,3-dihydro-1H-imidazo[2,1-a]isoindol-5(9bH)-one (ML375). J. Med. Chem. 56, 9351–9355 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sykes D. A., et al. , The influence of receptor kinetics on the onset and duration of action and the therapeutic index of NVA237 and tiotropium. J. Pharmacol. Exp. Ther. 343, 520–528 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Kruse A. C., et al. , Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature 482, 552–556 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Thal D. M., et al. , Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature 531, 335–340 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kajiwara Y., Yasuda S., Takamuku Y., Murata T., Kinoshita M., Identification of thermostabilizing mutations for a membrane protein whose three-dimensional structure is unknown. J. Comput. Chem. 38, 211–223 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Ballesteros J. A., Weinstein H., “Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors” in Methods in Neurosciences, Sealfon S. C., Ed. (Academic Press, 1995), vol. 25, chap. 19, pp. 366–428. [Google Scholar]
- 25.Suno R., et al. , Structural insights into the subtype-selective antagonist binding to the M2 muscarinic receptor. Nat. Chem. Biol. 14, 1150–1158 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang X. P., Prilla S., Mohr K., Ellis J., Critical amino acid residues of the common allosteric site on the M2 muscarinic acetylcholine receptor: More similarities than differences between the structurally divergent agents gallamine and bis(ammonio)alkane-type hexamethylene-bis-[dimethyl-(3-phthalimidopropyl)ammonium]dibromide. Mol. Pharmacol. 68, 769–778 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Dror R. O., et al. , Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature 503, 295–299 (2013). [DOI] [PubMed] [Google Scholar]
- 28.Thorsen T. S., Matt R., Weis W. I., Kobilka B. K., Modified T4 lysozyme fusion proteins facilitate G protein-coupled receptor crystallogenesis. Structure 22, 1657–1664 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bender W., Staudt M., Tränkle C., Mohr K., Holzgrabe U., Probing the size of a hydrophobic binding pocket within the allosteric site of muscarinic acetylcholine M2-receptors. Life Sci. 66, 1675–1682 (2000). [DOI] [PubMed] [Google Scholar]
- 30.Haga K., et al. , Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature 482, 547–551 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thal D. M., Glukhova A., Sexton P. M., Christopoulos A., Structural insights into G-protein-coupled receptor allostery. Nature 559, 45–53 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Bock A., Schrage R., Mohr K., Allosteric modulators targeting CNS muscarinic receptors. Neuropharmacology 136, 427–437 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Gregory K. J., Sexton P. M., Christopoulos A., Allosteric modulation of muscarinic acetylcholine receptors. Curr. Neuropharmacol. 5, 157–167 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burger W. A. C., Sexton P. M., Christopoulos A., Thal D. M., Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J. Gen. Physiol. 150, 1360–1372 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Buller S., Zlotos D. P., Mohr K., Ellis J., Allosteric site on muscarinic acetylcholine receptors: A single amino acid in transmembrane region 7 is critical to the subtype selectivities of caracurine V derivatives and alkane-bisammonium ligands. Mol. Pharmacol. 61, 160–168 (2002). [DOI] [PubMed] [Google Scholar]
- 36.Gnagey A. L., Seidenberg M., Ellis J., Site-directed mutagenesis reveals two epitopes involved in the subtype selectivity of the allosteric interactions of gallamine at muscarinic acetylcholine receptors. Mol. Pharmacol. 56, 1245–1253 (1999). [DOI] [PubMed] [Google Scholar]
- 37.Prilla S., Schrobang J., Ellis J., Höltje H. D., Mohr K., Allosteric interactions with muscarinic acetylcholine receptors: Complex role of the conserved tryptophan M2422Trp in a critical cluster of amino acids for baseline affinity, subtype selectivity, and cooperativity. Mol. Pharmacol. 70, 181–193 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Voigtländer U., et al. , Allosteric site on muscarinic acetylcholine receptors: Identification of two amino acids in the muscarinic M2 receptor that account entirely for the M2/M5 subtype selectivities of some structurally diverse allosteric ligands in N-methylscopolamine-occupied receptors. Mol. Pharmacol. 64, 21–31 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Lane J. R., May L. T., Parton R. G., Sexton P. M., Christopoulos A., A kinetic view of GPCR allostery and biased agonism. Nat. Chem. Biol. 13, 929–937 (2017). [DOI] [PubMed] [Google Scholar]
- 40.Kruse A. C., et al. , Muscarinic acetylcholine receptors: Novel opportunities for drug development. Nat. Rev. Drug Discov. 13, 549–560 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Apel A. K., et al. , Crystal structure of CC chemokine receptor 2A in complex with an orthosteric antagonist provides insights for the design of selective antagonists. Structure 27, 427–438.e5 (2019). [DOI] [PubMed] [Google Scholar]
- 42.Liu H., et al. , Structure-guided development of selective M3 muscarinic acetylcholine receptor antagonists. Proc. Natl. Acad. Sci. U.S.A. 115, 12046–12050 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McCorvy J. D., et al. , Structure-inspired design of β-arrestin-biased ligands for aminergic GPCRs. Nat. Chem. Biol. 14, 126–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Strasser A., Wittmann H. J., Seifert R., Binding kinetics and pathways of ligands to GPCRs. Trends Pharmacol. Sci. 38, 717–732 (2017). [DOI] [PubMed] [Google Scholar]
- 45.Swinney D. C., Haubrich B. A., Van Liefde I., Vauquelin G., The role of binding kinetics in GPCR drug discovery. Curr. Top. Med. Chem. 15, 2504–2522 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Copeland R. A., The drug-target residence time model: A 10-year retrospective. Nat. Rev. Drug Discov. 15, 87–95 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Tautermann C. S., et al. , Molecular basis for the long duration of action and kinetic selectivity of tiotropium for the muscarinic M3 receptor. J. Med. Chem. 56, 8746–8756 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Wess J., Gdula D., Brann M. R., Structural basis of the subtype selectivity of muscarinic antagonists: A study with chimeric m2/m5 muscarinic receptors. Mol. Pharmacol. 41, 369–374 (1992). [PubMed] [Google Scholar]
- 49.Hirata K., et al. , Achievement of protein micro-crystallography at SPring-8 beamline BL32XU. J. Phys. Conf. Ser. 425, 012002 (2013). [Google Scholar]
- 50.Aragão D., et al. , MX2: A high-flux undulator microfocus beamline serving both the chemical and macromolecular crystallography communities at the Australian synchrotron. J. Synchrotron Radiat. 25, 885–891 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hirata K., et al. , ZOO: An automatic data-collection system for high-throughput structure analysis in protein microcrystallography. Acta Crystallogr. D Struct. Biol. 75, 138–150 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yamashita K., Hirata K., Yamamoto M., KAMO: Towards automated data processing for microcrystals. Acta Crystallogr. D Struct. Biol. 74, 441–449 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kabsch W., Xds. Acta Crystallogr. D Biol. Crystallogr. 66, 125–132 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adams P. D., et al. , PHENIX: A comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen V. B., et al. , MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurrus E., et al. , Improvements to the APBS biomolecular solvation software suite. Protein Sci. 27, 112–128 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank, http://www.rcsb.org (PDB ID code 6OL9 for M5-T4L). Detailed methods are provided in SI Appendix. Other materials and data are available on request.