Significance
Glycolytic glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is an abundant enzyme whose activity depends on a reactive catalytic cysteine. Here, we determined the effect of 2 cysteine-based redox modifications, namely oxidation and S-glutathionylation, on the functionality and structural stability of GAPDH of Arabidopsis thaliana. Hydrogen peroxide causes the irreversible oxidation of the catalytic cysteine without altering the GAPDH structure. Conversely, S-glutathionylation, consisting of the formation of a glutathionyl-mixed disulfide with its catalytic cysteine, reversibly inactivates GAPDH and protects the enzyme from irreversible oxidation. The persistence, however, of the glutathionylated state alters the native folding of GAPDH, causing the irreversible collapse into insoluble oligomeric aggregates whose growth, but not breakdown, is under the control of physiological reductases such as thioredoxins and glutaredoxins.
Keywords: cysteine, disulfide bond, glyceraldehyde-3-phosphate dehydrogenase, protein aggregation, S-glutathionylation
Abstract
Protein aggregation is a complex physiological process, primarily determined by stress-related factors revealing the hidden aggregation propensity of proteins that otherwise are fully soluble. Here we report a mechanism by which glycolytic glyceraldehyde-3-phosphate dehydrogenase of Arabidopsis thaliana (AtGAPC1) is primed to form insoluble aggregates by the glutathionylation of its catalytic cysteine (Cys149). Following a lag phase, glutathionylated AtGAPC1 initiates a self-aggregation process resulting in the formation of branched chains of globular particles made of partially misfolded and totally inactive proteins. GSH molecules within AtGAPC1 active sites are suggested to provide the initial destabilizing signal. The following removal of glutathione by the formation of an intramolecular disulfide bond between Cys149 and Cys153 reinforces the aggregation process. Physiological reductases, thioredoxins and glutaredoxins, could not dissolve AtGAPC1 aggregates but could efficiently contrast their growth. Besides acting as a protective mechanism against overoxidation, S-glutathionylation of AtGAPC1 triggers an unexpected aggregation pathway with completely different and still unexplored physiological implications.
A single polypeptide chain may adopt a huge number of different conformations among which only one or a few are biologically active. Although native conformations are thermodynamically favored, in some proteins, they are separated from nonnative conformations by small energy barriers (1, 2). Misfolded proteins typically expose hydrophobic residues that are buried in native conformations, and these residues tend to promote protein aggregation in aqueous environments. Aggregation propensity is highly differentiated among different proteins, depending on their amino acid sequence and posttranslational modifications, but still, the capability to aggregate can be considered as an intrinsic property of any type of polypeptide (1, 3).
Protein aggregates may occur in different shapes (1, 4). In some cases, protein aggregation starts with the formation of small oligomers and ends up with amyloid fibrils characterized by a typical cross beta-sheet architecture. Several human disorders collectively known as amyloidoses are associated with amyloid depositions made of disease-specific proteins associated in fibrils (5). Different from amyloid fibrils, protein particulates are insoluble oligomers with a globular shape and no cross beta-sheet architecture as they are formed by proteins that are only partially unfolded (4). Aggregation-prone proteins tend to form particulates at pH close to their isoelectric point and amyloid fibrils at pH values in which they bear a strong net charge (6). Both aggregation products are considered states that virtually any protein can be forced to adopt, even though the process might require harsh treatments such as heat or extreme pH values (6).
The risk of protein aggregation in vivo is exacerbated by the high concentration of proteins in living cells (3). Being at high risk of aggregation, proteins that are particularly abundant tend to adopt conformations that result in higher solubility compared to less abundant ones (7). Moreover, cells of any domain of life possess a large set of molecular chaperones that limit protein aggregation by shielding the exposed hydrophobic patches of misfolded proteins, thereby promoting their refolding (8–10). However, despite the molecular chaperones and the efficiency of the whole machinery that controls proteome homeostasis, hundreds of proteins remain at high risk of aggregation, even under nonpathological conditions. In vivo, protein aggregates have been reported in different model organisms including Escherichia coli (11), yeast (12, 13), Caenorhabditis elegans (14, 15), and tomato and tobacco cell cultures (16, 17), typically as a consequence of aging or heat stress. These protein aggregates are not associated with specific diseases, while they may be associated with oxidative stress conditions shared by different types of stress (13). Oxidative posttranslational modifications (Cys and Met oxidation, carbonylation, etc.) have been shown to favor protein aggregation, presumably by lowering the energy barriers separating native from misfolded conformations (18–21). Although, in some cases, nonamyloid protein aggregation can induce cell death (22, 23), protein aggregation may also be beneficial to cells as long as sequestration of aggregates in specific cell sites prevents toxicity (24). Indeed, nonamyloid protein aggregates may be asymmetrically inherited by daughter cells (25), dissolved by chaperones (12, 26), or digested by proteases (27) or autophagy (28, 29). All these processes limit cell toxicity. In this sense, protein aggregation may be considered as a last line of defense against stress (2).
Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) is a ubiquitous and abundant glycolytic enzyme that was found to aggregate in different types of cells and conditions. In vitro, animal GAPDH forms aggregates under strongly oxidative conditions (21–23, 30–34). The essential catalytic cysteine of GAPDH can be oxidized by hydrogen peroxide (H2O2) to generate a sulfenic acid group that may react with a second H2O2 molecule to form a sulfinic acid. Alternatively, the sulfenic acid of the catalytic cysteine can react with a second thiol, like that of reduced glutathione (GSH), to form a mixed disulfide (S-glutathionylation) (35, 36). While the sulfinic acid cannot be reduced by cell reductants, the glutathionylated cysteine can be reduced back to the thiol group by glutaredoxins or thioredoxins that are active as deglutathionylases (37, 38). S-glutathionylation might also derive from the reaction of protein thiolates with oxidized glutathione (GSSG). The reaction is slow, but can be efficiently catalyzed by specific glutaredoxins like human GRX1 (39).
The sensitivity of GAPDH to reactive oxygen species (ROS) has important consequences. Since the interaction between cytoplasmic GAPDH (GAPC) and autophagy-related protein 3 (ATG3) negatively regulates autophagy, ROS may induce autophagy in plants by impairing GAPC–ATG3 complex formation (40). On the contrary, oxidized GAPC activates phospholipase D at the plasma membrane, creating a connection between ROS signaling and lipid signaling that controls Arabidopsis response to stress (41). In animal cells, GAPDH sensitivity to H2O2 was proposed to have a positive role in oxidative stress conditions because it allows rerouting of the primary metabolism from glycolysis to the oxidative pentose phosphate pathway, the resulting NADPH being essential for the antioxidant response, which in turn allows GAPDH recovery (42). Thanks to its extreme redox sensitivity, both in animals and in plants (43, 44), GAPDH is now regarded as being a hub of controlled redox responses for metabolic regulation (45).
Clearly, all functional interactions, catalytic activity, and regulatory functions of GAPDH are impaired by aggregation. In Arabidopsis plants, GAPC is suggested to form aggregates in leaves infiltrated with flg22, a pathogen-associated molecular pattern that triggers basal immunity, with ROS being implicated in the response (32). Here we show that Arabidopsis GAPC1 specifically aggregates in vitro following oxidation by H2O2 in the presence of GSH at nearly physiological concentrations. These conditions lead to specific glutathionylation of GAPC1 catalytic cysteines with no other amino acids being modified. Although protected from irreversible oxidation, glutathionylated GAPC1 is conformationally destabilized and, surprisingly, slowly induced to aggregate into oligomeric particles. In the next phase, glutathionylated GAPC1 spontaneously releases GSH, and the small oligomeric particles melt into large micrometric clusters made of smaller (pseudo-) globular units. Aggregated GAPC1 is partially unfolded and bears a novel disulfide bond engaging the catalytic Cys149 and the conserved Cys153 of the same subunits. Formation of the disulfide bond speeds up the aggregation process. Although AtGAPC1 aggregation was found irreversible, the aggregation process could be immediately halted by thioredoxin h1 (TRXh1) or, less efficiently, glutaredoxin C1 (GRXC1), which reduced and reactivated the population of not-yet-aggregated GAPC1 proteins. The crystal structure of glutathionylated AtGAPC1 and molecular dynamics (MD) calculations derived thereof provide clues on the mechanism by which a physiological posttranslational modification like S-glutathionylation triggers the collapse of a soluble tetrameric protein into insoluble aggregates in a process that appears to be under strict redox control.
Results
AtGAPC1 Forms Nonamyloid Aggregates.
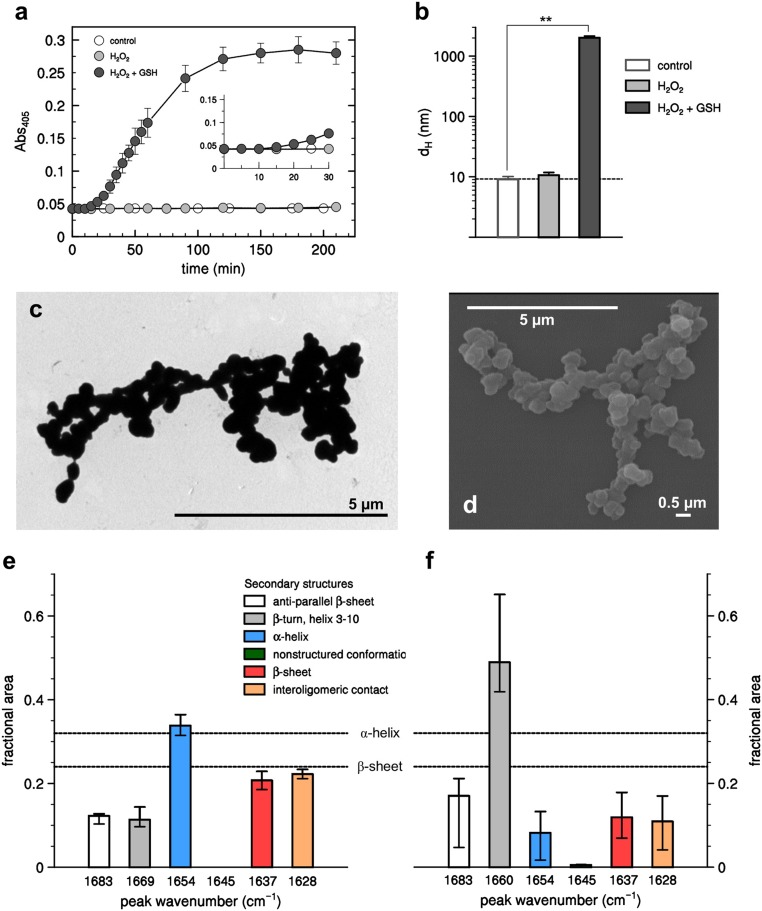
Visible light scattering (turbidity) can be taken as a proxy of protein stability in solution, and turbidity measurements show that native AtGAPC1 in solution (0.2 mg/mL) is stable for hours at room temperature (Fig. 1A). Similar behavior was shown in the presence of H2O2 (125 µM) at a 25:1 ratio with AtGAPC1 subunits (Fig. 1A), whereas addition of GSH together with H2O2 (5:1 ratio) caused a dramatic increase in turbidity over time, indicative of protein aggregation (Fig. 1A). The increase in turbidity followed a lag phase of 15 to 20 min (Fig. 1 A, Inset) and proceeded linearly for more than 1 h, reaching a plateau in about 2 h (Fig. 1A). The kinetics of turbidity were decreased by decreasing AtGAPC1 concentrations, but it was not influenced by the presence of H2O2 and GSH in the solution during the lag phase. Removal of H2O2 and GSH by fast desalting 10 min after the beginning of the experiment did not show any effect on the following aggregation phase (SI Appendix, Fig. S1A).
Fig. 1.
Oxidative treatments alter AtGAPC1 stability, inducing globular aggregation. (A) Turbidity analyses monitored at Abs405 of AtGAPC1 (5 μM) incubated with 0.125 mM H2O2 in the absence (gray closed circles) or presence of 0.625 mM GSH (black closed circles). In the control experiment, change in turbidity was measured following incubation of AtGAPC1 in buffer alone (open circles). The turbidity of control and H2O2-treated AtGAPC1 showed no variation over 210 min, while H2O2/GSH-treated AtGAPC1 has a lag phase (15 to 20 min; Inset) followed by a rapid increase reaching a plateau after 2 h incubation. Data represent mean ± SDs (n = 3 experiments with technical duplicates). When not visible, SDs are within the symbols. (B) Dynamic light scattering (DLS) measurement of AtGAPC1 incubated for 90 min in the presence of buffer alone (white bar), 0.125 mM H2O2 (light gray bar), or 0.125 mM H2O2 supplemented with 0.625 mM GSH (dark gray bar). No appreciable variation of protein diameter (dH) was observed for control and H2O2-treated AtGAPC1, while H2O2/GSH-treated AtGAPC1 formed aggregates with a diameter of ∼2 μm. This value might be underestimated due to technical limitations of the DLS instrument linked to the polydispersity index of samples. Data represent mean ± SDs (n = 3 experiments with technical duplicates; **P < 0.01). Representative TEM (C) and SEM (D) images of aggregated AtGAPC1 obtained after 90 min incubation in the presence of 0.125 mM H2O2 and 0.625 mM GSH. (Scale bars: 5 μm and 0.5 μm.) (E) Relative content in secondary structures of native AtGAPC1 determined by FTIR analysis. FTIR spectra were acquired after H2O-to-D2O substitution achieved through exhaustive concentrating/diluting steps. (F) Relative content in secondary structures of aggregated AtGAPC1 determined by FTIR analysis. The protein sample was incubated for 90 min in the presence of 0.125 mM H2O2 and 0.625 mM GSH, and, after treatment, the sample was centrifuged and the pellet resuspended in D2O. In E and F, the percentages of secondary structures derived from the crystallographic 3D structure of native AtGAPC1 are also indicated as dashed lines.
The formation of products with increasing size was confirmed by dynamic light scattering (DLS) measurements, from which information about the size of the aggregates can be derived from autocorrelation functions. By fitting the autocorrelation function to a spherical model, a hydrodynamic diameter (dH) of 9.2 ± 0.5 nm was calculated for soluble AtGAPC1 (Fig. 1B). This value did not change over time and upon treatment with H2O2 alone (Fig. 1B), and was compatible with the crystal structure of AtGAPC1 tetramers (46). On the contrary, aggregates formed after 90 min incubation with H2O2 and GSH were ∼200-fold larger in terms of hydrodynamic diameter (Fig. 1B), thus roughly corresponding to the volume of 10 million AtGAPC1 tetramers. Protein aggregates could be dissolved by SDS at 95 °C and migrated in reducing polyacrylamide gels as a single band corresponding to AtGAPC1 monomers (SI Appendix, Fig. S1B). Under nonreducing conditions, the protein profile in SDS/PAGE was almost identical, except for a minor band likely corresponding to AtGAPC1 tetramers (SI Appendix, Fig. S1B). Inspection of AtGAPC1 aggregates by transmission and scanning electron microscopy (TEM and SEM, respectively) revealed irregular shapes resulting from the random binding of nearly globular particles of ∼300 to 500 nm (Fig. 1 C and D). No fibrils were observed. Fluorescence spectroscopy demonstrated that AtGAPC1 aggregates could interact with dyes commonly used to stain beta-enriched regions (Thioflavin-T; ThT) and hydrophobic patches (1-anilino-8-naphthalene sulfonate; ANS) in protein aggregates (SI Appendix, Fig. S1 C and D, respectively). Native AtGAPC1, on the contrary, showed minimal interaction with either ThT or ANS dyes, suggesting that the conformation of AtGAPC1 in the aggregates was different from the native one. A detailed analysis of secondary structure elements in both native and aggregated AtGAPC1 was performed by Fourier-transform infrared spectroscopy (FTIR). In order to test the method, the amide I band of native AtGAPC1 was decomposed into 5 Gaussian curves corresponding to different secondary structure motifs according to the literature (47, 48) (SI Appendix, Fig. S2A). The relative content of α-helices (1,654 cm−1) and β-sheets (1,637 cm−1) was in good agreement with crystallographic data (Fig. 1E). Additionally, the absence of a Gaussian component at 1,645 cm−1 was fully consistent with the absence of disordered regions in native AtGAPC1 structure. The same measurements performed on AtGAPC1 aggregates showed an amide I band of different shape (SI Appendix, Fig. S2C). The amide I difference spectrum between the 2 forms (SI Appendix, Fig. S2B) exhibited an increase of absorption at wavenumbers higher than 1,665 cm−1, paralleled by a decrease between 1,665 and 1,625 cm−1. The decomposition of the amide I band showed 2 major effects consisting of a 3-fold decrease of α-helices and a 4-fold increase in short structural motifs (β-turns and 310-helices; 1,669 and 1,660 cm−1 in native and aggregated protein, respectively), suggesting a conversion of the former into the latter ones (Fig. 1F). No clear changes were observed in other spectral components, including unstructured regions (1,645 cm−1) and β-sheets (1,636 cm−1), the latter result suggesting that AtGAPC1 aggregates do not contain the cross-β spine typical of amyloid-like fibrils (49, 50).
Aggregating AtGAPC1 Is Transiently Glutathionylated.
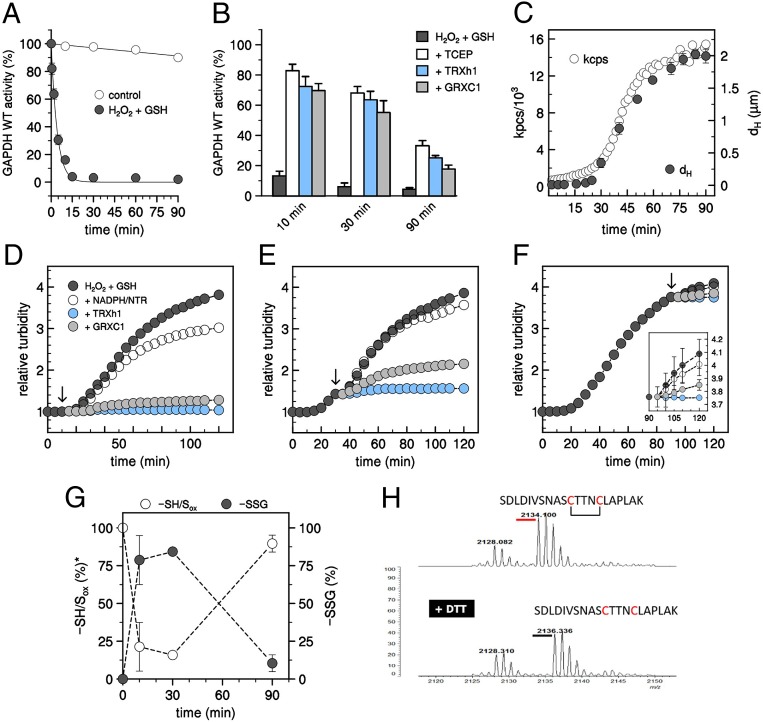
The treatment inducing AtGAPC1 aggregation (i.e., 125 µM H2O2 plus 625 µM GSH) also determined a rapid and complete inactivation of the enzyme in about 15 min (Fig. 2A). Within this time frame, inhibition could be largely reverted by physiological thiol reductants (TRXh1 or GRXC1; Fig. 2B). However, reductants became less and less effective over time, and, after 90 min incubation with H2O2 and GSH, less than 35% of AtGAPC1 activity could be recovered by the reductive treatment, performed with either physiological redoxins (i.e., TRXh1 and GRXC1) or chemically with the strong reductant TCEP (Tris[2-carboxyethyl]phosphine hydrochloride; Fig. 2B).
Fig. 2.
AtGAPC1 aggregation is specifically induced by transient S-glutathionylation. (A) Inactivation kinetics of AtGAPC1 in the presence of H2O2 and GSH. AtGAPC1 was incubated with 0.125 mM H2O2 and 0.625 mM GSH, and, at the indicated time, an aliquot was withdrawn and assayed for GAPDH activity. Data represent mean ± SDs (n = 3 experiments with technical duplicates). (B) Time-course reactivation of inactivated AtGAPC1. At different time points, AtGAPC1 samples treated with H2O2 and GSH (see above) were further incubated for 30 min with 10 mM TCEP (white bars, + TCEP), TRXh1 system (light blue bars, + TRXh1), or GRXC1 system (gray bars, + GRXC1) to assess the inactivation reversibility. Experimental details are provided in SI Appendix, Materials and Methods. Data represent mean ± SDs (n = 3 experiments with technical duplicates). (C) Time-course DLS analysis of AtGAPC1 treated with H2O2 and GSH. Protein diameters (dH, closed black circles) and integrated count rates (kpcs, open circles) were monitored over time and plotted versus time (0 to 90 min). Data correspond to the mean of 3 biological experiments and are represented as mean ± SDs. When not visible, SDs are within the symbols. For the sake of clarity, SDs were omitted for integrated count rates. (D) Turbidity analyses of H2O2/GSH-treated AtGAPC1 following exposure to reducing treatments. AtGAPC1 was treated with H2O2 and GSH as described above, and changes in turbidity were monitored at 405 nm. After 10 min incubation, an aliquot (20 μL) was withdrawn and the corresponding volume was replaced by 0.1 mg/mL catalase supplemented with NADPH/NTR system (open circles, + NADPH/NTR), TRXh1 system (light blue circles, + TRXh1), or GRXC1 system (gray circles, + GRXC1). Control experiments were conducted by replacing the reaction mixture with buffer (dark gray circles). Experimental data were adjusted to the absorbance at 405 nm of control samples measured at 15 min. Experimental details are provided in SI Appendix, Materials and Methods. (E) Turbidity analyses of H2O2/GSH-treated AtGAPC1 following exposure to reducing treatments. AtGAPC1 was treated as described above, and changes in turbidity were monitored at 405 nm. After 30 min incubation, an aliquot (20 μL) was withdrawn, and the corresponding volume was replaced as described in D. Experimental data were adjusted to the absorbance at 405 nm of control samples measured at 35 min. (F) Turbidity analyses of H2O2/GSH-treated AtGAPC1 following exposure to reducing treatments. AtGAPC1 was treated with H2O2 and GSH as described above, and changes in turbidity were monitored at 405 nm. After 90 min incubation, an aliquot (20 μL) was withdrawn, and the corresponding volume was replaced as described in D. Experimental data were adjusted to the absorbance at 405 nm of control samples measured at 95 min. (Inset) Turbidity analysis in the 90- to 120-min time range. For D–F, data represent mean of 3 biological replicates. For the sake of clarity, SDs were omitted. (G) Time-dependent MALDI-TOF signals of AtGAPC1 treated with H2O2 and GSH. AtGAPC1 was incubated as described above. Percentage of glutathionylated (i.e., ∼300 Da shifted, closed black circles) versus native or oxidized other than glutathionylated (SH/Sox, open circles) AtGAPC1 forms were extrapolated from MALDI-TOF spectra (SI Appendix, Fig. S3C) and plotted versus times (0, 10, 30, and 90 min). Data represent mean ± SDs (n = 3). (H) Peptide analysis of aggregated AtGAPC1. Following 90 min treatment with H2O2 and GSH (see above), the protein was centrifuged, and the pellet was resuspended, subjected to tryptic digestion, and analyzed by mass spectrometry analysis before and after treatment with DTT. The peptide containing the catalytic Cys149 and the Cys153 is shown.
A detailed analysis by DLS, with light scattered intensity measured over 100-s intervals during the whole 90-min experiment, provided further hints on the aggregation process that paralleled the progressive, irreversible inactivation of AtGAPC1. The autocorrelation function, averaged over 500-s intervals, clearly demonstrated a slow increase of particle sizes during the first 25 min of the experiment (Fig. 2C and SI Appendix, Fig. S3A) followed by rapid growth in the micrometric range until the end of the experiment (Fig. 2C). The integrated count rate (kpcs), which depends on both the concentration and the size of the aggregates, showed a similar profile (Fig. 2C). Combination of the plots (Fig. 2C) thus suggests that AtGAPC1 is first induced to self-assemble into small nanoparticles that only later start to melt into larger micrometric aggregates.
Although TRXh1 or GRXC1 had limited effect in the recovery of AtGAPC1 activity during the aggregation process, they were extremely effective in arresting particle growth. When reduced TRXh1 was added to the system during the lag phase (10 min after AtGAPC1 treatment with H2O2 plus GSH), further aggregation was fully prevented (Fig. 2D). When reduced TRXh1 was added after aggregation had started, at the beginning (30 min; Fig. 2E) or at the end of the aggregation phase (90 min; Fig. 2F), the turbidity curve suddenly leveled off. Reduced GRXC1 was in general less effective than TRXh1, particularly so during the aggregation phase. In any case, a decrease of turbidity was never observed, indicating that redoxins could not solubilize the aggregated particles.
MALDI-TOF mass spectrometry (MS) analysis showed that AtGAPC1 was transiently glutathionylated during the aggregation process. A large portion of AtGAPC1 (∼75%) became glutathionylated after 10 min of treatment with H2O2 and GSH (1 GSH/monomer; Fig. 2G and SI Appendix, Fig. S3C) and reached ∼85% after 30 min incubation (Fig. 2G and SI Appendix, Fig. S3C). However, after 90 min incubation, glutathionylation decreased to roughly 10% (Fig. 2G and SI Appendix, Fig. S3C) and dropped to zero if the MS analysis was carried out on the insoluble aggregates obtained by centrifugation (SI Appendix, Fig. S3D), indicating that the residual glutathionylated AtGAPC1 was soluble. Aggregation could be prevented by pretreatment of AtGAPC1 with the cysteine-alkylating agent iodoacetamide (IAM; SI Appendix, Fig. S4), known to specifically alkylate Cys149 (37), or by mutating Cys149 into Ser (SI Appendix, Fig. S4), suggesting that the glutathionylation of Cys149 was the trigger of the aggregation process.
With the aim of unraveling how AtGAPC1 may lose its glutathionyl moiety during aggregation, tryptic peptides were obtained from AtGAPC1 aggregates and analyzed by MALDI-TOF MS. The presence of an intramolecular disulfide bond between Cys149 and Cys153 (Fig. 2H) clearly demonstrated that the removal of GSH was the consequence of the nucleophilic attack performed by Cys153 of the same subunit on the proximal sulfur atom of glutathionylated Cys149.
The relevance of the Cys149–Cys153 disulfide bond for aggregation of AtGAPC1 was tested by mutating Cys153 into Ser. The mutation had no effect on the sensitivity to glutathionylation treatment (SI Appendix, Fig. S5 A and B) and glutathionylation of Cys149 that, however, remained constant over time (SI Appendix, Fig. S6 A and B), confirming the role of Cys153 in the deglutathionylation of wild type AtGAPC1 (Fig. 2G). Interestingly, turbidity and DLS measurements showed slower aggregation kinetics of the C153S mutant with respect to the wild type enzyme (SI Appendix, Fig. S7 A–D).
Overall, the plots of Figs. 1 and 2 hence describe a scenario in which AtGAPC1 is rapidly and reversibly inactivated by glutathionylation of Cys149 and slowly induced to self-assemble into small nanoparticles that later rapidly melt into larger micrometric aggregates. During this massive aggregation phase, glutathionylation of Cys149 is progressively substituted by a Cys149–Cys153 disulfide bond, and this thiol-disulfide interchange speeds up the aggregation process. Aggregation can be halted at any time, but not reverted, by redoxins that reactivate redox-modified AtGAPC1 proteins that are still soluble and prevent their late collapse on the surface of the particles.
Structural Snapshots along the Pathway to AtGAPC1 Aggregation.
With the aim of describing the early steps of protein aggregation at the structural level, the crystal structure of AtGAPC1 was determined after treatments with H2O2 alone or H2O2 plus GSH.
Inhibition of AtGAPC1 activity by H2O2 alone is even faster than under glutathionylating conditions (H2O2 plus GSH; Fig. 2A and SI Appendix, Fig. S8A) and is irreversible (SI Appendix, Fig. S8B). These fast and irreversible kinetics indicates that the first oxidation product of catalytic Cys149 (sulfenic acid) is short-lived because it further reacts with H2O2 to generate more oxidized forms (sulfinic or sulfonic acids). Fully oxidized AtGAPC1 remains, however, fully soluble and tetrameric (Fig. 1 A and B and SI Appendix, Fig. S8C).
After soaking AtGAPC1 crystals with H2O2, the effect of the oxidant could be directly evaluated from X-ray diffraction analysis and model structure calculations. The computed electron density map of H2O2-oxidized AtGapC1 showed a clear positive density in the Fo − Fc map around the thiol group of catalytic Cys149. In this additional electron density, a sulfinic acid or sulfinate (-SO2H or -SO2−) was easily built (SI Appendix, Fig. S8D), in agreement with recent quantum-mechanical analyses (46). Interestingly, no additional positive electron densities were observed for other H2O2-sensitive residues, i.e., neither for the second cysteine (Cys153) nor for the 7 methionines of each subunit (SI Appendix, Fig. S9). This observation demonstrates the absolute selectivity of H2O2 for catalytic Cys149 among all other residues of AtGAPC1 in these conditions.
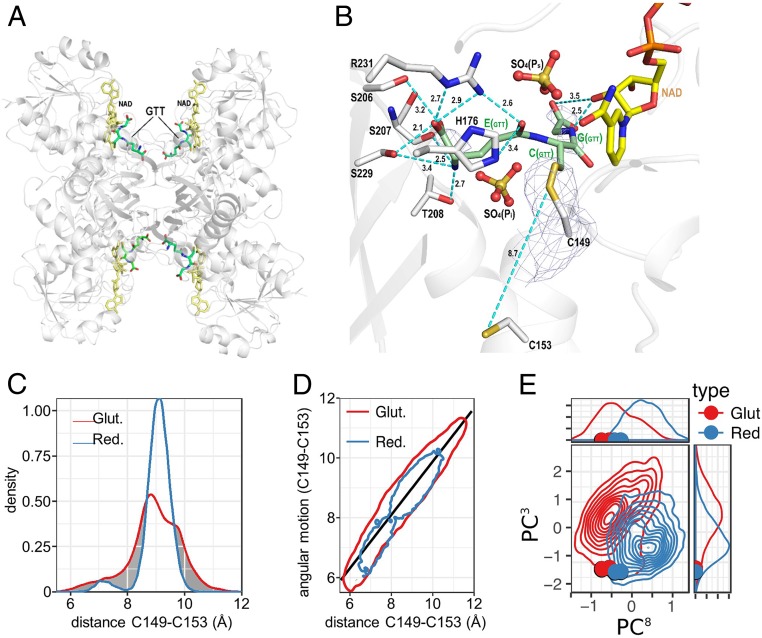
Crystals of AtGAPC1 were also soaked with a solution containing both H2O2 and GSH (1:10 ratio) with the aim of attempting a glutathionylation reaction in crystallo. Again, soaked crystals were subjected to X-ray diffraction analysis, and the 3D structure was solved at 3.0-Å resolution (Fig. 3A). The calculated Fo − Fc electron density map revealed, in both chains O and R of the asymmetric unit, an elongated positive electron density stemming from the thiol group of Cys149, which could be interpreted as a mixed disulfide bond with a glutathione molecule (Fig. 3B). Additional discontinuous electron density regions were ascribed to the carboxylic groups of bound GSH (Fig. 3B). Beyond the mixed disulfide bond, GSH was stabilized by hydrogen bonds with the protein, the NAD+ cofactor, and the sulfate ions bound to the active site (PS and Pi sites) (35) (Fig. 3B). Consistent with the results with H2O2-soaked crystals, no other residues than Cys149 were modified by the treatment with GSH and H2O2. Therefore, even in the crystal, AtGAPC1 may undergo the specific glutathionylation of Cys149 in the presence of H2O2 and GSH, indicating that, under these conditions, sulfenic Cys149 (formed by the reaction of the thiol with H2O2) reacts faster with GSH than with H2O2, thereby escaping the irreversible modification.
Fig. 3.
Structural features and MD simulation of S-glutathionylated AtGAPC1. (A) Cartoon representation of AtGAPC1 tetramer structure with 4 glutathione (GTT in the figure) molecules covalently bound to the catalytic cysteines and 4 cofactor molecules (NAD+) noncovalently interacting with the protein. Glutathione (GTT) and NAD+ molecules are shown as sticks. (B) Representation of the mixed disulfide bond between the catalytic cysteine (Cys149) and glutathione. The hydrogen bonds between glutathione and the protein (cutoff distance 3.5 Å) as well as the 2Fo − Fc electron density map (contoured at 1.5 σ) around the Cys149 and glutathione are shown. The distance between the sulfur atoms of Cys149 and 153 is only slightly shorter in glutathionylated AtGAPC1 with respect to the unmodified protein (SI Appendix, Fig. S11). The interactions between GSH and sulfate ions (Ps and Pi sites) were omitted to mimic the solution environment. (C) MD simulation-based analysis of distance distributions between the 2 sulfur atoms of Cys149 and Cys153 in reduced and glutathionylated forms are shown in blue and red color, respectively. The distributions highlight the impact of glutathionylation on the cysteines 149 to 153 distance. (D) Correlation of a metric combining several key angles with the distance between the 2 sulfur atoms of cysteines 149 and 153. Contour of the lowest density for the glutathionylated and reduced forms are indicated by red and blue color, respectively. (E) Principal component analysis (PCA) applied to the whole MD simulation dataset of GAPC1 in reduced and glutathionylated form. Projection of third (y axis) and eighth (x axis) PCA modes computed on protein secondary structure regions. An overall density contour plot for the whole simulation set consolidating the data on all 4 chains is displayed as blue and red plain lines for reduced and glutathionylated forms, respectively. Crystal structure principal component projections are indicated as colored dots. The probability distribution of both PCA modes is shown above and to the right of the corresponding contour plot axes, respectively. Crystal structure projection on both modes are indicated as colored dots.
Each GAPDH tetramer can bind a maximum of 4 GSH molecules lying at the entrance of the 4 active sites (Fig. 3A). However, GSH molecules did not occupy all of the available sites in AtGAPC1 crystals and were characterized by high thermal parameters. Indeed, the GSH occupancy (q) was 82% for chain O and 67% for chain R, and thermal parameters (B) ranged between 60 and 80 Å2, indicating substantial mobility of the GSH molecules within AtGAPC1 active sites.
Multiple molecular dynamics (MD) simulations, starting from the crystal structure of glutathionylated AtGAPC1, confirmed the high mobility of bound GSH. Six main conformational clusters of the glutathione were observed (SI Appendix, Fig. S10), with only one (cluster 2) corresponding closely to the starting crystal structure. Glutathionylation of Cys149 had no significant effect on the overall conformation of AtGAPC1 tetramers within the crystal (0.45-Å rmsd on Cα atoms between glutathionylated and native AtGAPC1), and the average distance between the S atoms of Cys149 and Cys153 remained prohibitive for the formation of a disulfide (8.7 Å; Fig. 3B). This result is consistent with the fact that the protein was crystallized before undergoing glutathionylation.
MD simulations indicated, however, that glutathionylation had an impact on the intersulfur distance between Cys149 and Cys153. The modification clearly extends the range of intersulfur distances, sampling both shorter and longer distances compared to the 8.8 Å of the native/reduced form (Fig. 3C and SI Appendix, Fig. S3D). A strong correlation between the intercysteine distances and a metric combining key dihedral angles included between Cys149 and Cy153 was found (Fig. 3D), with a marked influence of the redox state. The overall conformational distribution of secondary structure elements analyzed by principal component analysis (PCA) showed that glutathionylation of Cys149 modified the protein conformational landscape and its plasticity by inducing a change in interdomain arrangement (Fig. 3E). These changes observed on a microsecond timescale may represent the early stages of a local unfolding process that selectively prompted the protein to evolve toward an aggregation-prone conformation. Because of the subtle nature of the effects and the restricted timescale that can be sampled, quantitatively linking our observations to the formation of the Cys149–Cys153 disulfide and to the aggregation process on a much longer timescale remains, however, an important challenge.
Discussion
In this work, we describe a mechanism by which an abundant tetrameric protein like GAPC1 from Arabidopsis thaliana is primed to form insoluble oligomeric aggregates (protein particulates) by the specific glutathionylation of its catalytic cysteine.
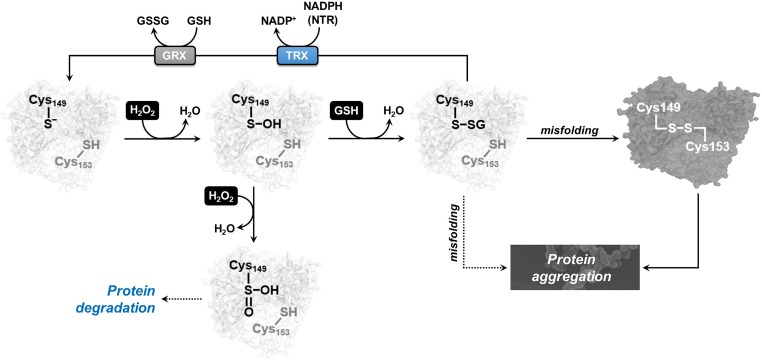
Due to its acidic and nucleophilic catalytic cysteine (Cys149), AtGAPC1 reacts quickly with H2O2. Cys149 thiolate (–S−) is first converted to a sulfenic acid intermediate (–SOH) and then to sulfinic acid (–SO2−; Fig. 4) (46). The modification of Cys149 by H2O2 is specific and irreversibly inhibits enzyme activity. The only other cysteine of AtGAPC1 (Cys153), which is found strictly conserved in GAPDH enzymes from almost all living organisms (42), is less exposed to the solvent and remains reduced even after long incubations with H2O2. This residue, which is 8.8 Å apart from Cys149 in terms of intersulfur distance, does not react with sulfenic Cys149 either. In vitro, the 2-step oxidation of Cys149 thiolate to the sulfinic acid proceeds undisturbed even at AtGAPC1:H2O2 equimolar concentrations (46). In vivo, however, in the presence of millimolar concentrations of glutathione (51), GAPC1 in Arabidopsis (52), and orthologs in other photosynthetic organisms (Synechocystis sp. PCC6803 [53]; Chlamydomonas reinhardtii [54]), are found glutathionylated under oxidative stress conditions. This observation indicates that sulfenic Cys149 may react faster with GSH than with H2O2. Efficient interference of GSH on AtGAPC1 irreversible oxidation apparently relies on the easy accommodation of the GSH molecule within AtGAPC1 active sites, where it can be stabilized by several interactions with the protein, as derived from our crystal structure of glutathionylated AtGAPC1 (Fig. 3 A and B). From its preferential position, GSH may attack sulfenic Cys149, leaving no possibility for H2O2 to compete (Fig. 4). As long as the activity of glutathionylated GAPC can be recovered by cytoplasmic GRXs or TRXs (37), the resulting glutathionylation/deglutathionylation cycle perfectly fits into the metabolic remodeling of aerobic cells under oxidative stress conditions. Observations made on different types of aerobic cells show indeed that (i) H2O2 may inhibit glycolysis by inactivating GAPDH; (ii) the cell antioxidative response consumes NADPH for protective (recycling) functions, including GAPDH deglutathionylation; and (iii) the pentose phosphate pathway and, only in plants, nonphosphorylating GAPDH, both activated by low NADPH levels, provide the NADPH required (35, 42, 45, 55).
Fig. 4.
Oxidative modifications and correlated structural/functional alterations of AtGAPC1. Schematic representation of the proposed sensitivity of AtGAPC1 to oxidation and S-glutathionylation and related conformational and functional changes.
Although proposed as a salvage pathway for redox-sensitive GAPDH, here we show that glutathionylation destabilizes AtGAPC1 conformation, and, in the long run, promotes the formation of insoluble aggregates (Fig. 4).
Aggregation of AtGAPC1 develops as a 3-phase process. Glutathione is quite mobile when it is covalently bound to Cys149 (SI Appendix, Fig. S10), but it interacts with the protein and affects its dynamics, both globally and locally, as clearly shown by MD simulations (Fig. 3 D and E). These changes are indicative of a conformational evolution whose effects need tens of minutes to show up. In our hands, AtGAPC1 could stay glutathionylated for about 10 min without changing significantly its overall native conformation. In this “pre-aggregation phase,” enzyme activity can be fully recovered and late aggregation can be fully prevented upon removal of GSH from AtGAPC1 by TRXh1 or GRXC1 (glutathionylation/deglutathionylation cycle; Fig. 4) (37, 38). The fate of glutathionylated AtGAPC1 is instead fully independent of GSH and H2O2 present in the solution (SI Appendix, Fig. S1A). GSH and H2O2 are required for the glutathionylation of AtGAPC1 (one GSH per monomer), but fully dispensable for the following aggregation process.
True aggregation starts only later, during the “oligomeric phase.” In this phase, the efficiency of reductants (TCEP or redoxins) in recovering the AtGAPC1 activity starts to decline, indicating that oligomerization is associated with a permanently inactivated state of the protein. The oligomeric phase ends up with the completion of the lag phase of the whole aggregation process. At the end of the lag phase, AtGAPC1 aggregates are still rather small (dH of ∼100 nm, corresponding to ∼103 tetramers) and hardly affect the turbidity of the system.
In the third phase, much faster than the previous one, particles abruptly start to grow to reach micrometric dimensions. Final aggregates are made by hundreds of bead-like particles of roughly 500 nm in dH (∼105 tetramers each). Particle units are linked together to form irregular branched chains. During this “particulate phase,” AtGAPC1 loses its glutathionyl moiety in favor of a Cys149–Cys153 disulfide bond. This is a hallmark of an initial conformational shift. Otherwise, the formation of the Cys149–Cys153 disulfide bond would be prevented by the long intersulfur distance and unsuitable orientation of the side chains of the 2 cysteines that, in native AtGAPC1, are fixed into the rigid structure of an α-helix (SI Appendix, Fig. S11). Once formed, the intramolecular disulfide bond must be instrumental for aggregation, as suggested by the slower and limited aggregation of the C153S mutant. The conformational modification undergone by aggregated AtGAPC1 is documented by the (limited) change in secondary structures revealed by FTIR (Fig. 1 E and F and SI Appendix, Fig. S2) and by the increased exposition of hydrophobic regions that bind ANS (SI Appendix, Fig. S1D). The binding of ThT (SI Appendix, Fig. S1C), documented by an increase in fluorescence that is orders of magnitude lower than that shown by amyloid fibrils (4), is in agreement with the absence of a FTIR-positive signal ascribable to interchain β-structures (1,620 cm−1), thus excluding the possibility of a cross–β-sheet architecture typical of amyloids. On the contrary, the lack of unstructured regions (1,645 cm−1) and the maintenance of secondary structures overall seem to exclude the possibility that aggregates are fully disordered and amorphous. Interestingly, the growth of the aggregates during the particulate phase can be halted at any time by TRXh1 (reduced by NADPH and NTR) or can be strongly inhibited by GRXC1 (reduced by GSH). Although thiol reductases could not dissolve the aggregates, they could apparently change the fate of all soluble tetramers that were still at the beginning of their misfolding process. Once the thiolate state of Cys149 is restored, AtGAPC1 tetramers lose their tendency to misfold and feed the particles’ growth.
Shape and features of AtGAPC1 aggregates are reminiscent of animal GAPDH aggregates obtained after nonspecific oxidation by the nitric oxide donor NOR3 (21–23), although more than 15 different residues of GAPDH, including cysteines, methionines, and tyrosines, were found modified by the NOR3 treatment (21). Aggregates contained intermolecular disulfides that bound together a minor portion of GAPDH subunits (21, 22). These aggregates, grown in vitro following a nucleated process under strongly oxidizing conditions, are thought to recapitulate the GAPDH aggregates observed in vivo in different pathological conditions (e.g., Alzheimer, Parkinson, ALS) (34, 56, 57) and under oxidative stress (23). Plant cytoplasmic GAPDH (GAPC) is also extremely sensitive to oxidative stress and a common target of redox posttranslational modifications (35), but, to our knowledge, the only report of GAPC aggregation deals with Arabidopsis plants treated with the flagellin fragment flg22 (32). The flg22 is a pathogen-associated molecular pattern that elicits basal immunity and ROS production and causes AtGAPC1-GFP to coalesce into cytoplasmic fluorescent puncta that are indicative of protein aggregation.
Globular particulates like those formed by glutathionylated AtGAPC1 are also formed by other aggregation-prone proteins under conditions that promote aggregation (4, 6, 58, 59). Often, protein particulates are made of proteins only partially unfolded. As a rule, they do not contain cross β-sheets, but the involvement of β-sheets and hydrophobic interactions in the aggregate formation are suggested by ThT and ANS binding.
In conclusion, here we show that a common thiol-based posttranslational modification that occurs under oxidative stress conditions and consists of the addition of a glutathionyl moiety to a nucleophilic cysteine previously oxidized by H2O2 can trigger the formation of insoluble protein aggregates. In a physiological context, it is also possible that protein glutathionylation might be achieved by other means, including an enzyme-assisted mechanism involving GRXs in the presence of either GSSG or GS• radical (39). Although this reaction has been demonstrated with human GRX1 (39), experimental evidence for protein glutathionylation catalyzed by a plant GRX is still lacking. Whichever the mechanism of glutathionylation, the fate of AtGAPC1, the major glycolytic GAPDH isoform of A. thaliana, depends on 2 cysteines (the catalytic Cys149, and Cys153) that are highly conserved in GAPDHs of any source. The collapse of AtGAPC1 into insoluble particles can be efficiently counteracted by GSH removal, and therefore it is strictly controlled by physiological thiol reductants like TRXh1 and GRXC1. However, if AtGAPC1 remains glutathionylated for too long, aggregation proceeds undisturbed until all of the protein collapses into micrometric globular particles. The whole process primarily depends on catalytic Cys149 that needs to be first oxidized and then glutathionylated. The prompt oxidation of Cys149 by H2O2 is assisted by Cys153 in a mechanism that favors metabolic remodeling of oxidatively stressed yeast cells (42). Interestingly, fully conserved Cys153 is shown here to also assist the aggregation process. Whether AtGAPC1 aggregates are dead-end products of oxidation that cells need to dispose of or whether the native protein might be recovered by chaperone-assisted disassembling of the particulates remains an open question.
Materials and Methods
Oxidation Treatments of Recombinant AtGAPC1.
Recombinant proteins were incubated with 10 mM dithiothreitol (DTT) for 30 min and then desalted using NAP-5 columns (GE Healthcare) preequilibrated with 50 mM Tris⋅HCl, 1 mM EDTA, pH 7.5 (buffer C). For oxidation experiments, prereduced samples (5 µM) were treated with buffer C (control) or with 0.125 mM H2O2 alone or supplemented with 0.625 mM GSH. All treatments were carried out at 25 °C in buffer C supplemented with 0.14 mM NAD+. At the indicated times, an aliquot was withdrawn to assay residual GAPDH activity (further detailed in SI Appendix, Materials and Methods). Activity data expressed as a percentage of maximal control activity were plotted versus time. Interpolation curves were generated by nonlinear regression using CoStat (CoHort Software). For recovery assays, protein samples were incubated at different time points with 0.1 mg/mL catalase supplemented with 10 mM TCEP, NADPH/NTR system (0.2 NADPH and 0.22 μM NTR), TRXh1 system (0.2 NADPH, 0.22 μM NTR, and 10 μM TRXh1), 1 mM GSH, or GRXC1 system (10 μM GRXC1). After 30 min incubation, aliquots were withdrawn for the assay of GAPDH activity.
Aggregation Kinetics Measured by Turbidity.
Kinetics of AtGAPC1 aggregation were assessed by measuring the increase of turbidity at 405 nm. AtGAPC1 samples (WT and Cys variants) were incubated at 25 °C with or without 0.125 mM H2O2 alone or supplemented with 0.625 mM GSH in a low-protein-binding 96-well plate. Reducing treatments were carried by adding 0.1 mg/mL catalase supplemented with NADPH/NTR system (0.2 NADPH and 0.22 μM NTR), TRXh1 system (0.2 NADPH, 0.22 μM NTR, and 10 μM TRXh1), 1 mM GSH, or GRXC1 system (10 μM GRXC1). All treatments were performed in buffer C supplemented with 0.14 mM NAD+. Samples were monitored over time, and turbidity at 405 nm was measured using a plate reader (Victor3 Multilabeling Counter; Perkin-Elmer).
Dynamic Light Scattering.
Size distribution of the species present in samples were obtained from measurements of AtGAPC1 (WT and Cys variants) incubated with or without 0.125 mM H2O2 alone or supplemented with 0.625 mM GSH in buffer C plus 0.14 mM NAD+ at 25 °C. The data were obtained using Zetasizer Nano (Malvern) cuvettes, and 30 spectra were acquired for each DLS analysis, averaged, and used to determine the hydrodynamic diameter and polydispersity using the average autocorrelation function.
Crystallization and Data Collection.
Crystals of NAD+-AtGAPC1 were grown as previously reported (46). Native crystals were soaked in the reservoir solution composed of 3.0 M (NH4)2SO4 and 0.1 M Hepes-NaOH (pH 7.5) plus 0.1 mM H2O2 to obtain oxidized AtGAPC1 (AtGAPC1-Sox) crystals or plus 0.1 mM H2O2 and 1 mM GSH to obtain glutathionylated AtGAPC1 (AtGAPC1-SSG) crystals. The GSH/H2O2 molar ratio used in soaking experiments was doubled with respect to biochemical assays, as the protein molecules were constrained in a crystalline lattice. Preliminary tests showed that the AtGAPC1 crystals were stable in the soaking solutions. The soaking was performed for 1 mo, then the crystals were fished and briefly soaked in a cryo solution containing 3.2 M (NH4)2SO4 and 20% vol/vol glycerol. All details for the structure determination are reported in SI Appendix, Materials and Methods.
Fourier Transform Infrared (FTIR) Analysis.
FTIR absorption measurements of native or aggregated AtGAPC1 were performed at room temperature with a Jasco Fourier transform 6100 spectrometer equipped with a DLATGS detector. The spectra were acquired with 2-cm−1 resolution in the whole mid-IR range (7,000 to 1,000 cm−1) using a standard high-intensity ceramic source and a Ge/KBr beam splitter. All details for the FTIR analysis are reported in SI Appendix, Materials and Methods.
Electron Microscopy.
AtGAPC1 samples were incubated with H2O2 supplemented with GSH as described earlier, then deposited onto carbon-coated copper mesh grids and negatively stained with 2% (wt/vol) uranyl acetate. The excess stain was wicked away, and the sample grids were allowed to air dry. The samples were viewed with an FEI Tecnai 12 BioTwin 85-kV transmission electron microscope (TEM), and digital images were taken with an Advanced Microscopy Techniques camera. The scanning electron microscope (SEM) observations were conducted using a Hitachi S-4000. The samples were deposited on a mica layer and gold-coated (2 nm) before the observations.
MALDI-TOF Mass Spectrometry.
AtGAPC1 samples (WT and C153S mutant; 5 µM) were incubated with H2O2 supplemented with 0.625 as described earlier. At different time points, aliquots were withdrawn for MALDI-TOF mass spectrometry analysis using a Performance Axima MALDI-TOF mass spectrometer (Shimadzu-Kratos) equipped with a 337-nm nitrogen laser. For mass determination of AtGAPC1 samples, spectra were acquired as described previously (60) with a pulse-extraction fixed at 60,000. For peptide analysis, the aggregated AtGAPC1 was prepared as described earlier in a 500-µL reaction mixture. After 90 min incubation, the sample was centrifuged for 5 min (10,000 × g, 20 °C) and the resulting pellet was solubilized in 11 µL of 50 mM ammonium bicarbonate containing 5 mM iodoacetamide and 0.01% of MS-compatible ProteaseMax detergent (Promega). All of the details for protein and peptide mass spectrometry analyses are reported in SI Appendix, Materials and Methods.
Molecular Dynamics.
We set up simulation systems for AtGAPC1 in complex with NAD+ in both reduced (Red.) and glutathionylated (Glut.) form using 3 different treatments of the NAD+ cofactor, leading to 6 independent simulations. The first simulation set was run without any constraints. At the end of the production, for both reduced and glutathionylated forms, 3 out of 4 NAD+ left their binding sites. Although such partial occupation is not unexpected, we wanted to also investigate the scenario of full occupation of all sites. For this purpose, we introduced distance restraints between NAD+ and protein Cα atoms in the 2 other simulation sets. In simulation set 2, distance restraints were applied between the 2 NAD+ ribose O4′ atoms and the closest Gly11 and Ile13 Cα atoms with a force constant of 500 kJ mol−1 nm−2. In simulation set 3, the same 2 distance restraints were applied with a higher force constant of 2,000 kJ mol−1 nm−2 and 2 additional restraints involving the NAD+ adenine N3′ and N315 Cα atoms and the NAD+ nicotinamide C4′ and D34 Cα atoms. All of the details for MD simulations are reported in SI Appendix, Materials and Methods.
Data Availability.
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.wwpdb.org) under PDB ID codes 6QUN (61) and 6QUQ (62) for oxidized AtGAPC1 (AtGAPC1-Sox) and glutathionylated AtGAPC1 (AtGAPC1-SSG), respectively.
Supplementary Material
Acknowledgments
We thank Elettra and the European Synchrotron Radiation Facility for allocation of X-ray diffraction beam time. This work was supported by University of Bologna Alma Idea Grant (to M.Z. and D.G.); CNRS Sorbonne Université, Agence Nationale de la Recherche Grant 17-CE05-0001 CalvinDesign (to C.H.M. and S.D.L.); LABEX DYNAMO ANR-LABX-011 (to C.H.M., M.B., and S.D.L.); and EQUIPEX CACSICE ANR-11-EQPX-0008 (to M.B. and S.D.L.). Computational work was performed using HPC resources from GENCI (grant number 2016-072292 to M.B.). M.Z. and M.B. thank “Cercle project” and Sesame Ile-de-France, respectively.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors of the oxidized AtGAPC1 (AtGAPC1-Sox) and glutathionylated AtGAPC1 (AtGAPC1-SSG) have been deposited in the Protein Data Bank (www.rcsb.org) under accession codes PDB 6QUN and PDB 6QUQ, respectively.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914484116/-/DCSupplemental.
References
- 1.Dobson C. M., Protein folding and misfolding. Nature 426, 884–890 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Tyedmers J., Mogk A., Bukau B., Cellular strategies for controlling protein aggregation. Nat. Rev. Mol. Cell Biol. 11, 777–788 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Vendruscolo M., Proteome folding and aggregation. Curr. Opin. Struct. Biol. 22, 138–143 (2012). [DOI] [PubMed] [Google Scholar]
- 4.Vetri V., Foderà V., The route to protein aggregate superstructures: Particulates and amyloid-like spherulites. FEBS Lett. 589, 2448–2463 (2015). [DOI] [PubMed] [Google Scholar]
- 5.Chiti F., Dobson C. M., Protein misfolding, amyloid formation, and human disease: A summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Krebs M. R., Devlin G. L., Donald A. M., Protein particulates: Another generic form of protein aggregation? Biophys. J. 92, 1336–1342 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldwin A. J., et al. , Metastability of native proteins and the phenomenon of amyloid formation. J. Am. Chem. Soc. 133, 14160–14163 (2011). [DOI] [PubMed] [Google Scholar]
- 8.Hartl F. U., Bracher A., Hayer-Hartl M., Molecular chaperones in protein folding and proteostasis. Nature 475, 324–332 (2011). [DOI] [PubMed] [Google Scholar]
- 9.Finka A., Mattoo R. U., Goloubinoff P., Experimental milestones in the discovery of molecular chaperones as polypeptide unfolding enzymes. Annu. Rev. Biochem. 85, 715–742 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Mannini B., Chiti F., Chaperones as suppressors of protein misfolded oligomer toxicity. Front. Mol. Neurosci. 10, 98 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Winkler J., et al. , Quantitative and spatio-temporal features of protein aggregation in Escherichia coli and consequences on protein quality control and cellular ageing. EMBO J. 29, 910–923 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wallace E. W., et al. , Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell 162, 1286–1298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weids A. J., Ibstedt S., Tamás M. J., Grant C. M., Distinct stress conditions result in aggregation of proteins with similar properties. Sci. Rep. 6, 24554 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.David D. C., et al. , Widespread protein aggregation as an inherent part of aging in C. elegans. PLoS Biol. 8, e1000450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walther D. M., et al. , Widespread proteome remodeling and aggregation in aging C. elegans. Cell 168, 944 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Nover L., Scharf K. D., Neumann D., Formation of cytoplasmic heat shock granules in tomato cell cultures and leaves. Mol. Cell. Biol. 3, 1648–1655 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nakajima Y., Suzuki S., Environmental stresses induce misfolded protein aggregation in plant cells in a microtubule-dependent manner. Int. J. Mol. Sci. 14, 7771–7783 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang L., Colodner K. J., Feany M. B., Protein misfolding and oxidative stress promote glial-mediated neurodegeneration in an Alexander disease model. J. Neurosci. 31, 2868–2877 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulinacci F., Poirier E., Capelle M. A., Gurny R., Arvinte T., Influence of methionine oxidation on the aggregation of recombinant human growth hormone. Eur. J. Pharm. Biopharm. 85, 42–52 (2013). [DOI] [PubMed] [Google Scholar]
- 20.Zhao L., Buxbaum J. N., Reixach N., Age-related oxidative modifications of transthyretin modulate its amyloidogenicity. Biochemistry 52, 1913–1926 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Samson A. L., et al. , Oxidation of an exposed methionine instigates the aggregation of glyceraldehyde-3-phosphate dehydrogenase. J. Biol. Chem. 289, 26922–26936 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakajima H., et al. , The active site cysteine of the proapoptotic protein glyceraldehyde-3-phosphate dehydrogenase is essential in oxidative stress-induced aggregation and cell death. J. Biol. Chem. 282, 26562–26574 (2007). [DOI] [PubMed] [Google Scholar]
- 23.Nakajima H., et al. , Glyceraldehyde-3-phosphate dehydrogenase aggregate formation participates in oxidative stress-induced cell death. J. Biol. Chem. 284, 34331–34341 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carija A., Navarro S., de Groot N. S., Ventura S., Protein aggregation into insoluble deposits protects from oxidative stress. Redox Biol. 12, 699–711 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindner A. B., Madden R., Demarez A., Stewart E. J., Taddei F., Asymmetric segregation of protein aggregates is associated with cellular aging and rejuvenation. Proc. Natl. Acad. Sci. U.S.A. 105, 3076–3081 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover J. R., Lindquist S., Hsp104, Hsp70, and Hsp40: A novel chaperone system that rescues previously aggregated proteins. Cell 94, 73–82 (1998). [DOI] [PubMed] [Google Scholar]
- 27.Ruan L., et al. , Cytosolic proteostasis through importing of misfolded proteins into mitochondria. Nature 543, 443–446 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hara T., et al. , Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 441, 885–889 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Komatsu M., et al. , Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 441, 880–884 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Torres-Bugeau C. M., et al. , Characterization of heparin-induced glyceraldehyde-3-phosphate dehydrogenase early amyloid-like oligomers and their implication in α-synuclein aggregation. J. Biol. Chem. 287, 2398–2409 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ávila C. L., et al. , Structural characterization of heparin-induced glyceraldehyde-3-phosphate dehydrogenase protofibrils preventing α-synuclein oligomeric species toxicity. J. Biol. Chem. 289, 13838–13850 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Henry E., Fung N., Liu J., Drakakaki G., Coaker G., Beyond glycolysis: GAPDHs are multi-functional enzymes involved in regulation of ROS, autophagy, and plant immune responses. PLoS Genet. 11, e1005199 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Itakura M., et al. , Glyceraldehyde-3-phosphate dehydrogenase aggregates accelerate amyloid-β amyloidogenesis in Alzheimer disease. J. Biol. Chem. 290, 26072–26087 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Muronetz V. I., Barinova K. V., Stroylova Y. Y., Semenyuk P. I., Schmalhausen E. V., Glyceraldehyde-3-phosphate dehydrogenase: Aggregation mechanisms and impact on amyloid neurodegenerative diseases. Int. J. Biol. Macromol. 100, 55–66 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Zaffagnini M., Fermani S., Costa A., Lemaire S. D., Trost P., Plant cytoplasmic GAPDH: Redox post-translational modifications and moonlighting properties. Front. Plant Sci. 4, 450 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trost P., Fermani S., Calvaresi M., Zaffagnini M., Biochemical basis of sulphenomics: How protein sulphenic acids may be stabilized by the protein microenvironment. Plant Cell Environ. 40, 483–490 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Bedhomme M., et al. , Glutathionylation of cytosolic glyceraldehyde-3-phosphate dehydrogenase from the model plant Arabidopsis thaliana is reversed by both glutaredoxins and thioredoxins in vitro. Biochem. J. 445, 337–347 (2012). [DOI] [PubMed] [Google Scholar]
- 38.Zaffagnini M., et al. , Redox regulation in photosynthetic organisms: Focus on glutathionylation. Antioxid. Redox Signal. 16, 567–586 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Zaffagnini M., et al. , Redox homeostasis in photosynthetic organisms: Novel and established thiol-based molecular mechanisms. Antioxid. Redox Signal. 31, 155–210 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Han S., et al. , Cytoplastic glyceraldehyde-3-phosphate dehydrogenases interact with ATG3 to negatively regulate autophagy and immunity in nicotiana benthamiana. Plant Cell 27, 1316–1331 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guo L., et al. , Cytosolic glyceraldehyde-3-phosphate dehydrogenases interact with phospholipase Dδ to transduce hydrogen peroxide signals in the Arabidopsis response to stress. Plant Cell 24, 2200–2212 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peralta D., et al. , A proton relay enhances H2O2 sensitivity of GAPDH to facilitate metabolic adaptation. Nat. Chem. Biol. 11, 156–163 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Wang H., et al. , Proteomic analysis of early-responsive redox-sensitive proteins in Arabidopsis. J. Proteome Res. 11, 412–424 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Araki K., et al. , Redox sensitivities of global cellular cysteine residues under reductive and oxidative stress. J. Proteome Res. 15, 2548–2559 (2016). [DOI] [PubMed] [Google Scholar]
- 45.Hildebrandt T., Knuesting J., Berndt C., Morgan B., Scheibe R., Cytosolic thiol switches regulating basic cellular functions: GAPDH as an information hub? Biol. Chem. 396, 523–537 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Zaffagnini M., et al. , Tuning cysteine reactivity and sulfenic acid stability by protein microenvironment in glyceraldehyde-3-phosphate dehydrogenases of Arabidopsis thaliana. Antioxid. Redox Signal. 24, 502–517 (2016). [DOI] [PubMed] [Google Scholar]
- 47.Yang H., Yang S., Kong J., Dong A., Yu S., Obtaining information about protein secondary structures in aqueous solution using Fourier transform IR spectroscopy. Nat. Protoc. 10, 382–396 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Jackson M., Mantsch H. H., The use and misuse of FTIR spectroscopy in the determination of protein structure. Crit. Rev. Biochem. Mol. Biol. 30, 95–120 (1995). [DOI] [PubMed] [Google Scholar]
- 49.Barth A., Infrared spectroscopy of proteins. Biochim. Biophys. Acta 1767, 1073–1101 (2007). [DOI] [PubMed] [Google Scholar]
- 50.Seo J., et al. , An infrared spectroscopy approach to follow β-sheet formation in peptide amyloid assemblies. Nat. Chem. 9, 39–44 (2017). [DOI] [PubMed] [Google Scholar]
- 51.Rouhier N., Lemaire S. D., Jacquot J. P., The role of glutathione in photosynthetic organisms: Emerging functions for glutaredoxins and glutathionylation. Annu. Rev. Plant Biol. 59, 143–166 (2008). [DOI] [PubMed] [Google Scholar]
- 52.Dixon D. P., Skipsey M., Grundy N. M., Edwards R., Stress-induced protein S-glutathionylation in Arabidopsis. Plant Physiol. 138, 2233–2244 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chardonnet S., et al. , First proteomic study of S-glutathionylation in cyanobacteria. J. Proteome Res. 14, 59–71 (2015). [DOI] [PubMed] [Google Scholar]
- 54.Zaffagnini M., et al. , Glutathionylation in the photosynthetic model organism Chlamydomonas reinhardtii: A proteomic survey. Mol. Cell. Proteomics 11, M111.014142 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuehne A., et al. , Acute activation of oxidative pentose phosphate pathway as first-line response to oxidative stress in human skin cells. Mol. Cell 59, 359–371 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Chuang D. M., Hough C., Senatorov V. V., Glyceraldehyde-3-phosphate dehydrogenase, apoptosis, and neurodegenerative diseases. Annu. Rev. Pharmacol. Toxicol. 45, 269–290 (2005). [DOI] [PubMed] [Google Scholar]
- 57.Butterfield D. A., Hardas S. S., Lange M. L., Oxidatively modified glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and Alzheimer’s disease: Many pathways to neurodegeneration. J. Alzheimers Dis. 20, 369–393 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mukai H., et al. , Formation of morphologically similar globular aggregates from diverse aggregation-prone proteins in mammalian cells. Proc. Natl. Acad. Sci. U.S.A. 102, 10887–10892 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.El Moustaine D., Perrier V., Smeller L., Lange R., Torrent J., Full-length prion protein aggregates to amyloid fibrils and spherical particles by distinct pathways. FEBS J. 275, 2021–2031 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Berger H., et al. , A light switch based on protein S-nitrosylation fine-tunes photosynthetic light harvesting in Chlamydomonas. Plant Physiol. 171, 821–832 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Fermani S., Zaffagnini M., Falini G., Trost P., Crystal structure of AtGapC1 with the catalytic Cys149 irreversibly oxidized by H2O2 treatment. Protein Data Bank. https://www.rcsb.org/structure/6QUN. Deposited 28 February 2019.
- 62.Fermani S., Zaffagnini M., Falini G., Trost P., Crystal structure of glutathionylated glycolytic glyceraldehyde-3-phosphate dehydrogenase from Arabidopsis thaliana (AtGAPC1). Protein Data Bank. https://www.rcsb.org/structure/6QUQ. Deposited 28 February, 2019.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Atomic coordinates and structure factors have been deposited in the Protein Data Bank (www.wwpdb.org) under PDB ID codes 6QUN (61) and 6QUQ (62) for oxidized AtGAPC1 (AtGAPC1-Sox) and glutathionylated AtGAPC1 (AtGAPC1-SSG), respectively.