Significance
It is generally accepted that ethanol and its metabolite acetaldehyde are primarily metabolized in the liver via the alcohol dehydrogenase and the aldehyde dehydrogenase-2 (ALDH2), respectively. However, by using tissue-specific Aldh2 knockout mice, we demonstrated that the liver ALDH2 is only responsible for approximately half of circulating acetaldehyde clearance after acute alcohol intake. Thus, cumulative ALDH2 activity from multiple organs may contribute to circulating acetaldehyde clearance. The present study shows that, although the liver ALDH2 only partially contributes to acetaldehyde clearance, genetic deletion or knockdown of the liver Aldh2 decreases excessive but not light to moderate alcohol drinking. Our results suggest that liver-specific ALDH2 inhibition may be an effective strategy for the treatment of alcohol user disorder with excessive drinking.
Keywords: alcohol metabolism, acetaldehyde, alcohol use disorder, neuron, shAldh2
Abstract
Aldehyde dehydrogenase 2 (ALDH2), a key enzyme for detoxification the ethanol metabolite acetaldehyde, is recognized as a promising therapeutic target to treat alcohol use disorders (AUDs). Disulfiram, a potent ALDH2 inhibitor, is an approved drug for the treatment of AUD but has clinical limitations due to its side effects. This study aims to elucidate the relative contribution of different organs in acetaldehyde clearance through ALDH2 by using global- (Aldh2−/−) and tissue-specific Aldh2-deficient mice, and to examine whether liver-specific ALDH2 inhibition can prevent alcohol-seeking behavior. Aldh2−/− mice showed markedly higher acetaldehyde concentrations than wild-type (WT) mice after acute ethanol gavage. Acetaldehyde levels in hepatocyte-specific Aldh2 knockout (Aldh2Hep−/−) mice were significantly higher than those in WT mice post gavage, but did not reach the levels observed in Aldh2−/− mice. Energy expenditure and motility were dramatically dampened in Aldh2−/− mice, but moderately decreased in Aldh2Hep−/− mice compared to controls. In the 2-bottle paradigm and the drinking-in-the-dark model, Aldh2−/− mice drank negligible volumes from ethanol-containing bottles, whereas Aldh2Hep−/− mice showed reduced alcohol preference at high but not low alcohol concentrations. Glial cell- or neuron-specific Aldh2 deficiency did not affect voluntary alcohol consumption. Finally, specific liver Aldh2 knockdown via injection of shAldh2 markedly decreased alcohol preference. In conclusion, although the liver is the major organ responsible for acetaldehyde metabolism, a cumulative effect of ALDH2 from other organs likely also contributes to systemic acetaldehyde clearance. Liver-targeted ALDH2 inhibition can decrease heavy drinking without affecting moderate drinking, providing molecular basis for hepatic ALDH2 targeting/editing for the treatment of AUD.
Excessive alcohol consumption is a leading risk factor for global disease burden (1). Alarmingly, deaths due to alcoholic cirrhosis have increased in recent years (2–4), thus highlighting the need to prevent or treat alcohol use disorder (AUD) (5, 6). Pharmaceutical approaches to treat AUD include naltrexone (7), acamprosate (8), and disulfiram, an aldehyde dehydrogenase-2 (ALDH2) inhibitor (9–12). ALDH2 is a conserved detoxifying mitochondrial enzyme, notably implicated in the metabolism of aldehydes. Ingested alcohol is first metabolized into acetaldehyde by alcohol dehydrogenase (ADH), then into acetate by ALDH2 (13, 14). Additionally, ALDH2 plays a key role in oxidizing lipid peroxidation products generated under oxidative stress, such as 4-hydroxy-2-nonenal and malondialdehyde (15). An estimated 8% of the world population, mainly of East Asian descent, harbor the ALDH2*2 allele which encodes for a nonfunctioning ALDH2 enzyme, resulting in acetaldehyde buildup in the blood and organs, such as liver and brain, after alcohol consumption (14, 16–20). In these individuals, acetaldehyde accumulation causes facial flushing, and unpleasant feelings such as nausea, headaches, cardiac palpitations, and overall discomfort (13, 14, 16). Therefore, ALDH2-deficient individuals are at lower risks of developing AUD (13, 14). For these reasons, approaches that aim to specifically and reversibly inhibit ALDH2 activity are of great interest in the treatment of AUD.
Therapies targeting ALDH2 have been studied for decades. Disulfiram (Antabuse), a potent ALDH2 inhibitor, has been approved by the Food and Drug Administration for the treatment of AUD; however, its use has been modest largely due to poor efficacy because of compliance (9–12, 21). Disulfiram induces general physical discomfort similar to that observed in ALDH2-deficient individuals upon alcohol consumption. Thus, new methods are being developed to verify patients’ compliance to the treatment (22). Disulfiram is also not specific to ALDH2, since it has been shown to also inhibit cytosolic ALDH1 (23). Thus, development of more-specific ALDH2 inhibitors may be a better strategy for the treatment of AUD (24–26).
The liver is regarded as the main organ responsible for acetaldehyde clearance via its high levels of ALDH2 in hepatocytes (27). Liver drug targeting is not challenging, and numerous technologies have proven their efficacy in delivering drugs specifically to hepatocytes, notably, by targeting the asialoglycoprotein receptor, by using viral systems or apolipoproteins (28). Cytochrome P450 enzymes, despite generating toxic drug metabolites in some instances, may also metabolize drug precursors, thus releasing the effective drug locally in the liver (29). Taken together, it is reasonable to speculate that a therapeutic agent aimed at specifically targeting liver ALDH2 could be used as a treatment for AUD while having limited systemic side effects. However, the ALDH2 enzyme is also expressed by numerous organs other than the liver, but whether ALDH2 in these organs also contributes to acetaldehyde clearance and whether specific blockade of ALDH2 in the liver can efficiently decrease alcohol drinking remain unknown.
In this study, we generated several strains of cell-specific Aldh2-deficient mice and aimed to determine the contribution of ALDH2 from different organs to acetaldehyde metabolism. We also evaluated the relevance of a liver-targeted approach for Aldh2 knockdown by using an adenoviral vector. By using these mouse models in the 2-bottle paradigm and the drinking in the dark (DID) binge-like model, we were able to conclude that the liver is the major but not the sole organ responsible for acetaldehyde metabolism. In addition, our data also reveal that an approach aiming to target liver ALDH2 expression can decrease heavy alcohol drinking.
Results
The Liver Is the Major but Not Sole Organ Responsible for Acetaldehyde Metabolism.
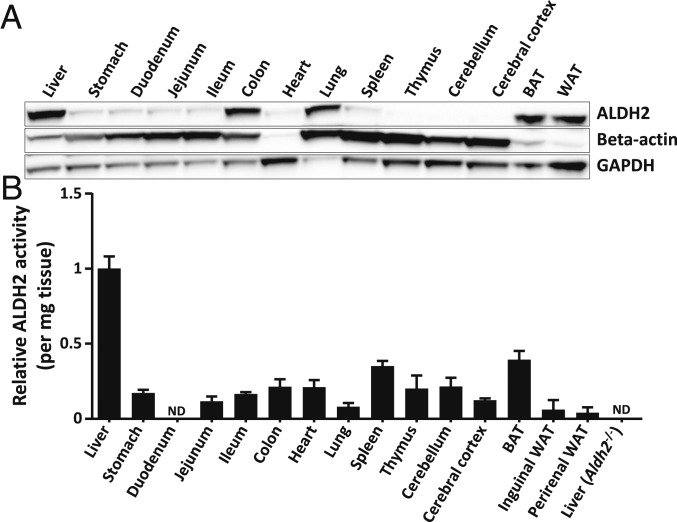
It is known that ALDH2 is expressed in numerous organs (14); however, the relative importance of organ-specific ALDH2 activity in acetaldehyde metabolism has not been investigated. Numerous organs from wild-type (WT) C57BL/6N mice were assessed for ALDH2 expression by Western blot. While the liver exhibited the strongest ALDH2 expression, several other organs also expressed significant ALDH2 protein levels, including intestine, lungs, white and brown adipose tissues, brain, and spleen (Fig. 1A). This widespread ALDH2 expression was further confirmed by measuring ALDH2 enzymatic activity in these organs, with the highest activity in the liver (Aldh2 global knockout [Aldh2−/−] livers were used as a negative control) (Fig. 1B). These data indicate that ALDH2 activity is not limited to the liver, and that other organs harbor a significant ALDH2 activity and might also be of importance in clearing acetaldehyde in the body.
Fig. 1.
Mouse ALDH2 protein expression and enzyme activity are widespread in the organism. To evaluate ALDH2 expression distribution in the organism, major organs from naïve C57BL/6N mice were collected. (A) ALDH2 protein expression was evaluated by Western blot, and beta-actin and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as loading controls. (B) ALDH2 enzymatic activity was measured on fresh tissue homogenates and is represented as the relative mean ± SEM. Relative ALDH2 activity was compared to liver (n = 3 to 6 per organ). Liver samples from Aldh2−/− mice were used as blank negative controls. BAT, brown adipose tissue; WAT, white adipose tissue; ND, not detectable.
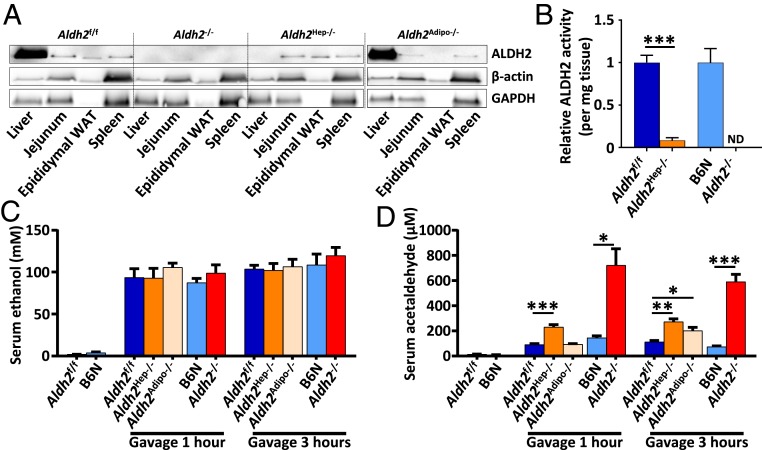
To dissect the relative role of major ALDH2-expressing organs, we generated tissue-specific Aldh2-deficient mice, as depicted in SI Appendix, Fig. S1. Tissue-specific ALDH2 protein deficiency was verified by Western blot and revealed successful tissue-targeted Aldh2 ablation as shown in Fig. 2A. For example, ALDH2 protein was detected at high levels in the liver from Aldh2-expressing WT mice but was barely detected in the liver from hepatocyte-specific Aldh2 knockout (Aldh2Hep−/−) mice, suggesting that Aldh2 was completely deleted in the liver of Aldh2Hep−/− mice (Fig. 2A). Hepatocyte Aldh2 deletion was further demonstrated by an almost complete loss of hepatic ALDH2 enzymatic activity in Aldh2Hep−/− mice compared to their Aldh2-floxed control (Aldh2f/f) littermates (Fig. 2B). Note that the residual ALDH2 activity that was still detected in the liver of Aldh2Hep−/− mice (Fig. 2B) was probably derived from nonparenchymal cells in the liver.
Fig. 2.
Organ-specific ALDH2 deficiency reveals a potent but not exclusive role of the liver in acetaldehyde clearance. (A) Tissue-specific ALDH2 protein deletion was confirmed at the protein level by Western blot analysis. Beta-actin and GAPDH were used as loading controls. (B) Relative liver ALDH2 enzymatic activity was measured in Aldh2−/−, Aldh2Hep−/−, and their WT control mice. Liver samples from Aldh2−/− mice were used as blank negative controls. Mice were given a single oral ethanol gavage (5 g/kg), then (C) ethanol and (D) acetaldehyde serum concentrations were measured by GC-MS at the indicated time points. ND, not detectable. Data are represented as mean ± SEM (n = 3 to 12). *P < 0.05, **P < 0.01, ***P < 0.005, unpaired Student’s t test (in B) and 1-way ANOVA (in C and D).
Previous studies reported that Aldh2−/− mice have a dampened acetaldehyde clearance after acetaldehyde injection (20). To test whether this acetaldehyde accumulation is mainly caused by an impaired liver ALDH2 activity, Aldh2−/− and tissue-specific Aldh2-deficient mice were euthanized 1 h or 3 h after a single dose of 5 g/kg ethanol gavage. Blood was collected for ethanol and acetaldehyde measurement by gas chromatography followed by mass spectrometry (GC-MS). All mice that received an ethanol gavage had comparable ethanol levels in the serum; thus ethanol levels were not impacted by ALDH2 activity or strain differences (Fig. 2C). In addition, serum levels of acetaldehyde were comparable between C57BL/6N (as WT control for Aldh2−/− mice) and Aldhf/f mice (as WT littermate control for tissue-specific Aldh2-specific mice) (Fig. 2D). Aldh2−/− mice showed markedly higher acetaldehyde concentrations than WT mice one and 3 h after gavage. Note that acetaldehyde levels in Aldh2Hep−/− mice were significantly higher than those in WT mice 1 and 3 h post gavage but did not elevate to the concentrations observed in Aldh2−/− mice, reaching an intermediate level between WT and Aldh2−/− mice instead (Fig. 2D). At 3 h postgavage, acetaldehyde levels in Aldh2Hep−/− mice were approximately half of the levels in Aldh2−/− mice (Fig. 2D). These data suggest that the liver is only partially responsible for acetaldehyde metabolism and that other organs play a role in circulating acetaldehyde clearance. Indeed, adipose tissue-specific Aldh2KO (Aldh2Adipo−/−) mice had slightly but significantly higher circulating levels of acetaldehyde compared to WT mice, at 3 h postgavage but not 1 h postgavage (Fig. 2D), suggesting that ALDH2 activity in the adipose tissue also partially contributes to acetaldehyde clearance in vivo. In contrast, intestinal epithelial cell-specific Aldh2KO (Aldh2IEC−/−) and myeloid-specific Aldh2KO mice (Aldh2Mye−/−) did not show any significantly increased acetaldehyde concentrations compared to the WT control group (SI Appendix, Fig. S2 A–C).
Aldh2Hep−/− Mice Have Lower Serum Alanine Aminotransferase Levels but Slightly Higher Levels of Hepatic Ccl2 and Ifng Messenger RNAs after Chronic-Plus-Binge Ethanol Challenge Compared to Their Littermate Controls.
We have previously demonstrated that acetaldehyde accumulation in Aldh2−/− mice was associated with lower serum alanine aminotransferase (ALT) activity but stronger liver inflammation in the chronic-plus-binge model (30). To evaluate whether liver ALDH2 activity was responsible for this phenotype, Aldh2Hep−/− mice were subjected to the chronic-plus-binge model, and liver inflammation was assessed. As shown in SI Appendix, Fig. S3A, no differences were observed in body weight gain. However, serum ALT activity was lower in Aldh2Hep−/− compared to Aldh2f/f littermates (SI Appendix, Fig. S3B), suggesting that liver-specific Aldh2 deficiency ameliorates alcohol-induced liver injury. Circulating neutrophil numbers were also lower in Aldh2Hep−/− mice (SI Appendix, Fig. S3C). Furthermore, F4/80 and myeloperoxidase staining analyses did not show significant differences in macrophage and neutrophil accumulation, respectively, between Aldh2f/f and Aldh2Hep−/− mice (SI Appendix, Fig. S3D), whereas qRT-PCR analyses revealed that expression of some inflammation-associated genes (such as Ccl2, Ifng) was higher in Aldh2Hep−/− than in Aldh2f/f mice (SI Appendix, Fig. S3E). Collectively, compared to Aldh2f/f littermates, Aldh2Hep−/− mice had reduced liver injury (as shown by serum ALT levels) but slightly increased liver inflammation, which is similar to the phenotypes observed in Aldh2−/− mice, but to a much lesser extent (30).
Aldh2Hep−/− and Aldh2−/− Mice Have Comparable Levels of Hepatic and Systemic Inflammatory Responses after Acute Ethanol Gavage.
Next, we evaluated the effects of acute ethanol gavage on liver and systemic inflammation. Our results revealed that acute ethanol gavage slightly up-regulated hepatic expression of some inflammation-associated genes, but no statistical differences were found between Aldh2Hep−/− and Aldh2−/− mice (SI Appendix, Fig. S4A). Hepatic expression of Tnfa and Ifng trended lower for in Aldh2−/− mice than in Aldh2Hep−/− mice, but it did not reach statistical difference. Similarly, acute ethanol gavage slightly elevated serum levels of several proinflammatory cytokines with similar elevation between Aldh2Hep−/− and Aldh2−/− mice (SI Appendix, Fig. S4B). These data suggest that the differences in drinking behavior between Aldh2Hep−/− and Aldh2−/− mice (see below) were likely due to the differences in acetaldehyde levels rather than due to the differences in inflammation, although inflammation is known to modulate alcohol drinking behavior (31).
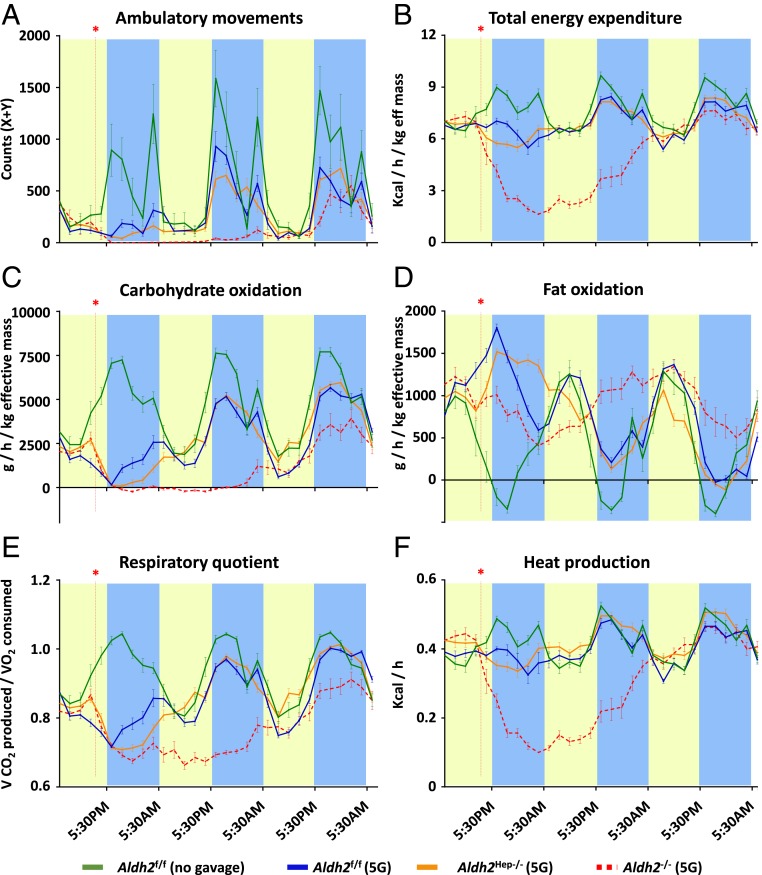
Global Deletion of the Aldh2 Gene Severely Impairs Energy Expenditure and Motility in Mice after Alcohol Binging, whereas Hepatocyte-Specific Deletion of the Aldh2 Gene only Has Modest Effects on Metabolic Parameters.
Since the liver remained the organ with the most prominent role in acetaldehyde clearance, we used Aldh2f/f (used as WT control), Aldh2Hep−/−, and Aldh2−/− mice for the following experiments. These mice received an acute ethanol gavage and were individually housed for indirect metabolic rate measurements. In this experimental approach, ethanol gavage induced a marked reduction in ambulatory activity, energy expenditure, respiratory quotient, heat production, and carbohydrate oxidation but, at the same time, an increased fat oxidation for the first night compared to vehicle control in Aldh2f/f mice (Fig. 3). These results highlight the expected consequences of an alcohol binge, such as limited physical activity and reduced food intake. These parameters progressively reached normal trends within the second night. In contrast, Aldh2−/− mice exhibited dramatically altered metabolic rates and ambulatory movements until the third night following ethanol intake, most likely due to the accumulation of acetaldehyde in the body. Notably, after receiving an oral gavage of ethanol, Aldh2Hep−/− mice also showed significantly reduced metabolic rates and activity for the first night postgavage compared to Aldh2f/f mice, but these effects remained moderate (Fig. 3). These results are consistent with the differences we observed above in terms of acetaldehyde levels in the respective mouse strains. Of note is that, despite differences in circulating acetaldehyde levels, Aldh2Adipo−/− mice did not show any difference in terms of metabolic or ambulatory parameters, compared to Aldh2f/f mice (SI Appendix, Fig. S5 A–F).
Fig. 3.
Global and hepatocyte-specific Aldh2 deficiency leads to severe and moderate inhibition, respectively, of metabolic rates. WT control or liver or global Aldh2-deficient mice received a single dose of 5 g/kg ethanol gavage and were placed in metabolic cages for indirect and noninvasive measurement of metabolic rates. The following parameters were evaluated: (A) ambulatory movements, (B) total energy expenditure, (C) carbohydrate oxidation, (D) fat oxidation, (E) respiratory quotient, and (F) heat production. In each panel, the red asterisk and line indicate the time at which oral gavage was performed. Yellow and blue bars represent the 12-h periods when the lights were on or off, respectively, in the animal holding room. Data are represented as mean ± SEM (n = 3 to 4 mice per group).
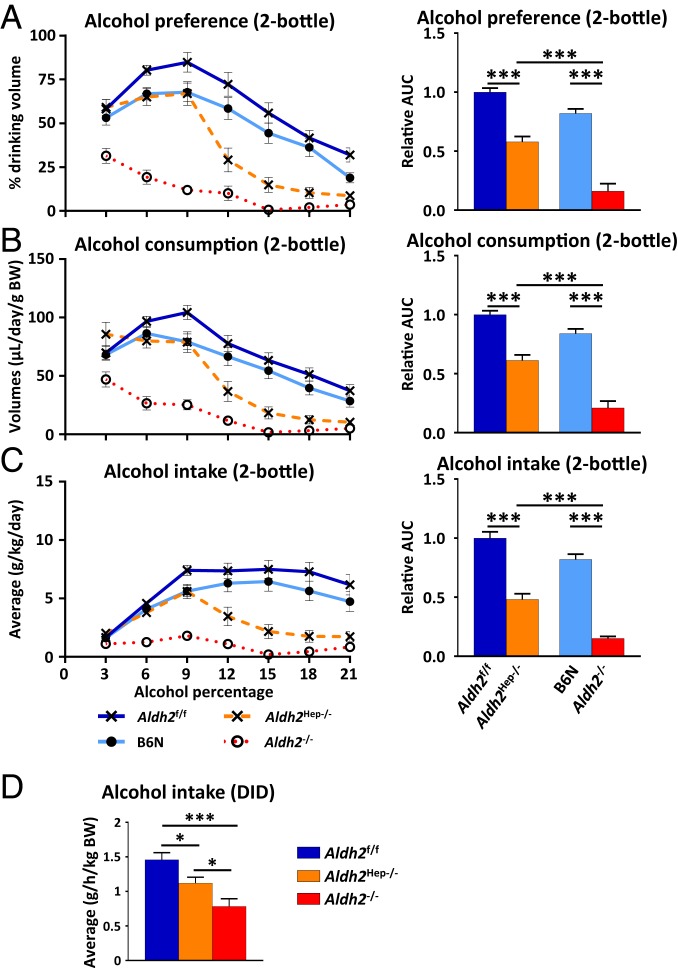
Hepatocyte-Specific Deletion of the Aldh2 Gene Decreases Excessive but Not Moderate Alcohol Drinking Preference.
Acetaldehyde buildup is classically designated as the cause for general physical discomfort in ALDH2-deficient individuals, often discouraging them from consuming alcoholic beverages. Based on the differences we observed in acetaldehyde concentrations and metabolic parameters in Aldh2Hep−/− mice, we next asked whether targeting hepatocyte ALDH2 enzyme would be sufficient to decrease voluntary alcohol drinking. For this purpose, mice were subjected to a 2-bottle choice paradigm in which they were simultaneously offered 2 bottles, one of which contained regular drinking water while the other one contained increasing concentrations of alcohol. Body weights and total daily drinking volumes remained constant in all groups (SI Appendix, Fig. S6 A and B, respectively). Alcohol preference and consumption in Aldh2f/f (used as WT controls for Aldh2Hep−/− mice) progressively increased as the concentration of alcohol increased in the bottle up to 9% vol/vol, then decreased as the ethanol concentration increased (Fig. 4 A–C). C57BL/6N mice (used as WT controls for Aldh2−/− mice) showed similar results to WT Aldh2f/f mice as described in Fig. 4. In contrast, alcohol preference was much lower in the Aldh2−/− mice, starting with the lowest concentrations (3%) used in this experiment. Notably, Aldh2Hep−/− mice trended lower for alcohol intake than their Aldh2f/f control mice when exposed to ethanol concentrations of 6% and 9%, although it did not reach statistical difference. When exposed to higher concentrations (12 to 21% ethanol), Aldh2Hep−/− mice decreased their ethanol preference more rapidly than Aldh2f/f littermates but less obviously than Aldh2−/− mice. Fig. 4C shows that absolute alcohol intake by WT mice reached a maximum of 6.4 ± 0.8 g⋅d−1⋅/kg−1 and remained steady until the end of the experiment, while Aldh2−/− had a much lower alcohol intake. Consistent with ethanol preference, the alcohol intake tended to be lower in Aldh2Hep−/− mice than in Aldh2f/f mice up to 9% ethanol in drinking water, but it did not reach statistical difference, and then was progressively reduced to the levels of Aldh2−/− mice, at 15%, 18%, and 21% concentrations. This result is also highlighted by the cumulated area under the ethanol intake curve in Fig. 4C, and further argues in favor of an intermediate ethanol preference in Aldh2Hep−/− mice. The difference in ethanol preference between WT and Aldh2−/− mice was not attributable to different sweet or bitter taste preference (SI Appendix, Fig. S7). Moreover, despite higher serum acetaldehyde levels after ethanol gavage, ethanol preference and intake in Aldh2Adipo−/− mice was similar to that of Aldh2f/f mice (SI Appendix, Fig. S8 A–F).
Fig. 4.
Hepatocyte-specific Aldh2 deficiency decreases excessive, but not light-to-moderate, alcohol drinking preference and binge-like drinking behavior in mice. (A–C) (Left) Mice were subjected to a 2-bottle choice paradigm, and consumption of regular drinking water and ethanol-containing water were measured daily. (A) Alcohol preference is represented as the percentage drank from the alcohol-containing bottle relative to cumulated water and alcohol drinking volumes. (B) Volumes drank from the alcohol-containing bottle per day per gram body weight. (C) Absolute alcohol intake per day per gram body weight. (Right) For all measurements in A−C, the relative area under the curve. (D) A DID was used to study binge-like drinking behavior of Aldh2f/f, Aldh2Hep−/−, B6N, and Aldh2−/− mice. Data are represented as mean ± SEM (n = 5 to 7 mice per group). *P < 0.05, ***P < 0.005, 1-way ANOVA.
Global or Hepatocyte-Specific Deletion of the Aldh2 Gene Decreases Binge Drinking Behavior but to a Lesser Extent in Mice with Hepatic Aldh2 Deletion.
The above data suggest that dampening liver ALDH2 expression is efficient to prevent excessive alcohol consumption, but not chronic low-dose alcohol drinking. In order to evaluate the consequences of liver ALDH2 inhibition in binge-like drinking behavior, mice were then subjected to the DID model. In this model, Aldh2−/− mice had an alcohol intake of about half that of WT mice, while Aldh2Hep−/− mice showed an intermediate value of alcohol intake as illustrated in Fig. 4D. Notably, WT control, Aldh2−/−, and Aldh2Hep−/− mice drank higher quantities of ethanol in the DID model than in the 2-bottle choice paradigm. Taken together, these findings further demonstrate that targeting liver ALDH2 enzyme might lead to a reduction in the high-dose alcohol drinking behavior.
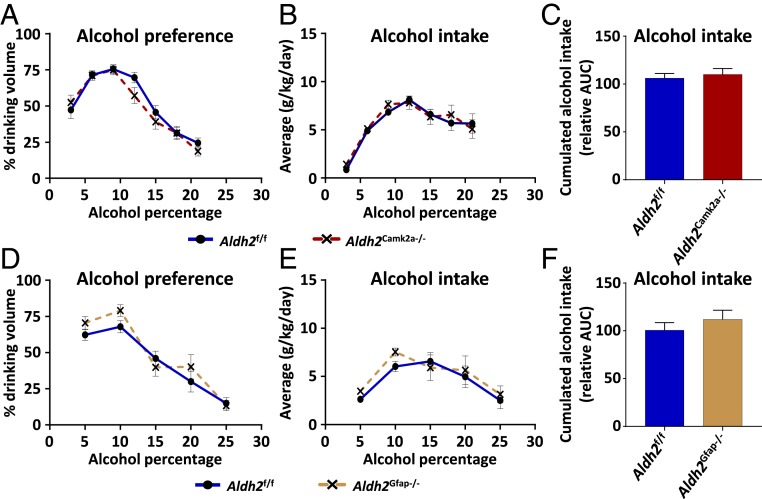
Glial Cell- or Neuron-Specific Deletion of the Aldh2 Gene Does Not Affect Alcohol Drinking Preference.
Several studies reported that ALDH2 expression was detected in the brain (32). To determine whether brain ALDH2 affects voluntary alcohol consumption, we generated glial cell- (Aldh2Gfap−/−) and neuron-specific (Aldh2Camk2a−/−) Aldh2-deficient mice. Our data reveal that both strains of Aldh2Gfap−/− and Aldh2Camk2a−/− mice did not show any differences with WT mice in voluntary alcohol consumption in the 2-bottle paradigm (Fig. 5).
Fig. 5.
Forebrain neuron or glial cell targeted Aldh2 deficiency does not affect alcohol drinking preference. Forebrain neuron- (Aldh2Camk2a−/−) and glial cell-specific (Aldh2Gfap−/−) Aldh2-deficient mice were subjected to the 2-bottle paradigm. (A and D) Alcohol preference and (B, C, E, and F) alcohol intake were evaluated in Aldh2Camk2a−/− and Aldh2Gfap−/− mice, respectively. Data are represented as mean ± SEM (n = 5 mice per group), unpaired Student’s t test (in C and F).
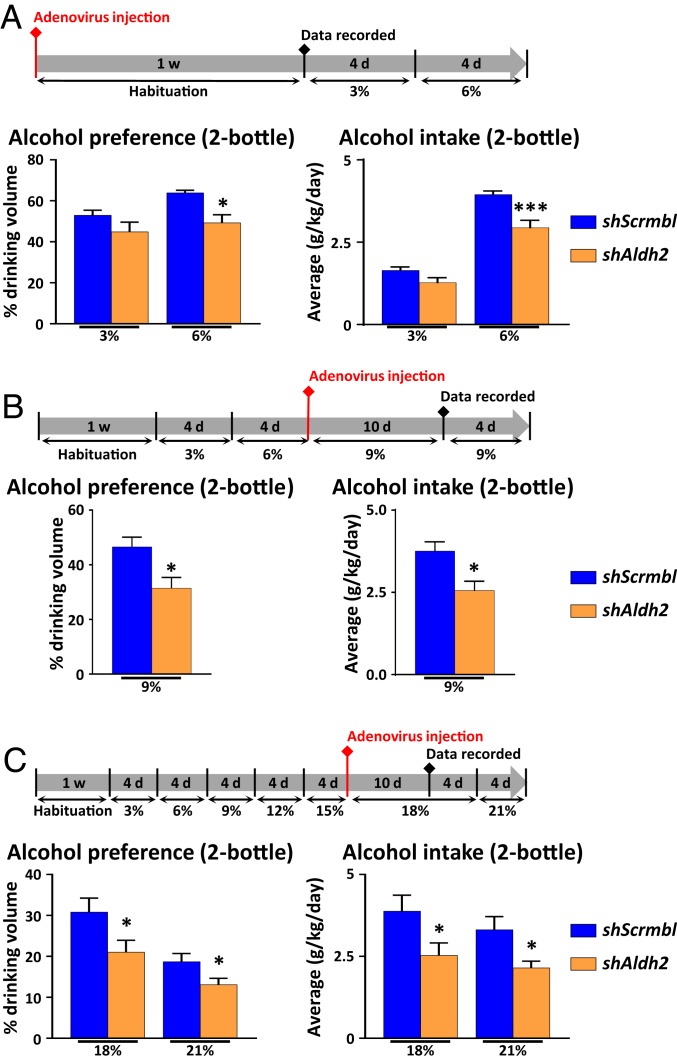
Knockdown of Hepatic Aldh2 Gene by Short Hairpin RNA Decreases Excessive Drinking Behavior.
To evaluate the therapeutic potential of liver ALDH2 inhibition for the treatment of AUD, we tested whether knockdown of liver Aldh2 gene is also effective in decreasing voluntary alcohol drinking. For this purpose, we knocked down hepatic Aldh2 expression by infecting mice with an adenovirus encoding a short hairpin sequence directed against Aldh2 and subsequently performed the 2-bottle choice test on these mice. An adenovirus encoding for a scrambled short hairpin RNA (shRNA) was used as control. For each experiment, Western blot analysis on liver homogenates revealed that the shRNA designed to silence Aldh2 gene expression via RNA interference (shAldh2) adenovirus repressed liver ALDH2 expression by 40 to 70% (SI Appendix, Fig. S9 A–C). We successfully infected a majority of hepatocytes, as shown by a widespread green fluorescent protein (GFP) staining (GFP was included in the expression vector) in scramble and shAldh2 adenovirus-injected mice (SI Appendix, Fig. S9D). Firstly, we evaluated whether liver-targeted Aldh2 knockdown could alter mouse alcohol preference from low concentration. For this purpose, naïve mice with shAldh2 injection were subjected to the 2-bottle preference experiment. Alcohol preference was identical at 3% ethanol in the drinking water, but liver ALDH2 impairment reduced voluntary alcohol intake when alcohol concentration reached 6% (Fig. 6A). In a separate group of mice, shAldh2 adenovirus was injected when the mice were exposed to 9% ethanol in the drinking water (Fig. 6B). The reduction in ALDH2 expression was sufficient to partially decrease ethanol preference and ethanol intake in shAldh2 adenovirus-treated mice compared to scrambled shRNA adenovirus-treated mice (Fig. 6B). Lastly, in a third group of mice, Aldh2 shRNA adenoviruses also markedly repressed alcohol drinking with 18% or 21% ethanol in the drinking water (Fig. 6C).
Fig. 6.
Hepatocyte-specific knockdown of Aldh2 decreases excessive alcohol preference in mice. (A) Naïve mice were injected with scrambled shRNA or shAldh2 adenovirus prior to being subjected to the 2-bottle paradigm. Briefly, mice were injected with adenovirus for 7 d and subsequently presented with 2 bottles, one containing regular drinking water, and the other one containing 3% or 6% ethanol in drinking water. Ethanol preference as well as alcohol intake were calculated. (B and C) (Lower) Mice were subjected to the 2-bottle paradigm with increasing ethanol concentration up to (B) 9% or (C) 18%, before being injected with scramble shRNA or shAldh2 adenovirus. Briefly, mice were given free choice between water or ethanol-containing water at the indicated concentrations (for 4 d each). Adenoviruses were then injected when ethanol concentration reached (B) 9% or (C) 18% in their drinking water. Mice were continuously kept on the 2-bottle choice test. (Upper) For each experiment, the experimental outline indicating the percentage of alcohol (vol/vol) in the drinking water; 1 w, 1 wk. Data are represented as mean ± SEM (n = 5 to 8 mice per group). *P < 0.05, ***P < 0.005, unpaired Student’s t test.
Discussion
An unexpected finding reported in this study is that the liver (hepatocytes) is responsible for only approximately half of circulating acetaldehyde clearance after alcohol intake. It is generally accepted that ethanol metabolism via ADH and acetaldehyde metabolism via ALDH2 mainly occur in the liver (hepatocytes) (27). However, ALDH2 expression was also detected in multiple organs other than the liver (32, 33), and whether ALDH2 in these organs also contributes to circulating acetaldehyde clearance had not been investigated. In the current study, we found that ALDH2 protein expression and activity were detected in a variety of organs with the highest levels in the liver. By measuring acetaldehyde levels in organ-specific Aldh2-deficient mice, we demonstrate that the liver is accountable for only approximately half of circulating acetaldehyde clearance in mice that received a single dose of ethanol by gavage. This partial contribution in Aldh2Hep−/− mice was not due to incomplete Aldh2 deletion, because ALDH2 protein levels and activity were almost completely depleted in the liver of Aldh2Hep−/− mice. In agreement with the partial contribution of the liver ALDH2 to acetaldehyde clearance, liver-specific Aldh2 deletion only modestly attenuated energy expenditure and motility in mice after acute alcohol drinking compared to the dramatic attenuation observed in global Aldh2−/− mice. This was a surprising result that contrasts with the classical view that the liver is the major if not sole organ responsible for ethanol and its metabolite acetaldehyde metabolism (27). This unexpected result could be explained by widespread ALDH2 expression in multiple organs other than the liver. However, which organ or organs are responsible for the remnant acetaldehyde clearance activity observed in Aldh2Hep−/− mice remains unclear, and we believe that cumulative ALDH2 activity in multiple organs is likely involved. First, ALDH2 protein expression and activity were detected in many organs other than the liver. Second, by testing 3 additional strains of tissue-specific Aldh2 knockout mice, we found that Aldh2Adipo−/− mice had ∼1.7-fold higher levels of acetaldehyde than their WT mice 3 h, but not 1 h, after acute ethanol gavage. These data suggest that adipose tissue partially contributes to acetaldehyde metabolism, but this contribution does not explain the remaining acetaldehyde clearance activity observed in Aldh2Hep−/− mice. Although Aldh2Adipo−/− mice had higher serum acetaldehyde levels than WT mice after acute ethanol gavage, both groups had comparable levels of energy expenditure and motility, and alcohol drinking preference. These results may suggest that adipocyte ALDH2 contributes to the metabolism of high levels of acetaldehyde occurring after acute ethanol gavage, but not low levels of acetaldehyde occurring during alcohol preference or DID experiments. Finally, it is also possible that many other organs, such as heart and lung, may also play a role in acetaldehyde clearance, contributing to the cumulative effect of ALDH2 from multiple organs in metabolizing acetaldehyde. This hypothesis will require further studies.
The second important finding from this study is that, although the liver only contributes to approximately half of acetaldehyde clearance, knockdown of Aldh2 in the liver may still have therapeutic potential for the treatment of AUD, because such an approach is sufficient to prevent excessive drinking preference and binge-like drinking behavior, as demonstrated in the current study. Systemic ALDH2 inhibition by disulfiram presents several drawbacks that must be considered in a clinical setting, especially in patients suffering from liver diseases and multiorgan pathology (9–12). Indeed, ALDH2 has been shown to play a protective role in ameliorating oxidative stress and tissue injury in multiple organs. For example, ALDH2 in the brain was shown to be involved in dopamine metabolism by reducing aldehyde accumulation and neurodegeneration induced by oxidative stress (34–36). Alda-1, a potent ALDH2 agonist that can restore ALDH2 activity in ALDH2*2 individuals, was shown to protect intestine, lung, and heart from oxidative stress and ischemia/reperfusion injury (37–41). All these data suggest that global inhibition of ALDH2 may not only decrease alcohol consumption but also have off-target effects. Indeed, investigators have been searching for ALDH2 inhibitors that may have fewer side effects compared to disulfiram. For example, CVT-10216, a highly selective reversible ALDH2 inhibitor, was synthesized based on the cocrystal structure of ALDH2 and daidzin (26). Intraperitoneal injection of CVT-10216 dose-dependently increased circulating acetaldehyde levels, reduced alcohol intake in the 2-bottle paradigm, and reduced alcohol self-administration, without affecting locomotor activity or food intake (26). Alternatively, specific inhibition of liver ALDH2 may be another promising strategy for the treatment of AUD, because deletion of the Aldh2 in the liver produced much less inhibition on ambulatory activity and metabolic rates than global deletion of the Aldh2 gene, while still effectively preventing excessive alcohol seeking behavior. These data indicate that targeting liver ALDH2 may generate fewer off-target effects (e.g., discomfort) than a nonspecific approach such as global deletion of ALDH2 or using disulfiram. Importantly, liver-specific Aldh2 deletion moderately reduced alcohol-induced liver injury but slightly increased liver inflammation, which is similar to the phenotypes observed in Aldh2−/− mice, but to a much lesser extent (30). These data suggest that a liver-targeted ALDH2 therapy likely has fewer detrimental effects for the liver than the global inhibition of ALDH2.
Generally, alcohol abstinence is the recommended treatment for AUD. However, because of the sociocultural factors encouraging alcohol consumption, total abstinence may not always be an attainable goal. Thus, it appears that limiting excessive but not low to moderate alcohol drinking might represent an alternative approach to limiting serious adverse consequences of AUD. Thus, targeting liver ALDH2, rather than globally inhibiting its activity, may have reduced side effects but still prevent heavy drinking. In addition, due to the liver's unique architecture, it is feasible to do gene targeting/editing hepatic ALDH2, which may open promising avenues for AUD treatment.
Materials and Methods
Mice.
The study was approved by the National Institute on Alcohol Abuse and Alcoholism’s Animal Care and Use Committee. Mice had free access to food and water unless otherwise specified and were housed in a 12-h light/dark cycle. Aldh2−/− mice on a C57BL/6N background were described previously (30, 42). The Aldh2f/f mice were generated by activating the Aldh2 gene in Aldh2tm1a(EUCOMM)Wtsi mice, by crossing of Aldh2tm1a(EUCOMM)Wtsi mice (kindly provided by K. J. Patel, Molecular Research Council [MRC] Laboratory of Molecular Biology, Cambridge, United Kingdom) (43) with homozygous FLPcR mice (Jax Laboratory, Bar Harbor, ME), which express Flippase (flip) in the germline cells as described previously (44). Tissue-specific Aldh2 knockout mice were generated via crossing Aldh2f/f mice with several strains of Cre transgenic mice from the Jackson Laboratory, including B6.Cg-Tg(Alb-cre)21Mgn/J (stock #003574), B6;FVB-Tg(Adipoq-cre)1Evdr/J (stock #010803), B6.Cg-Tg(Vil1-cre)997Gum/J (stock #004586), B6.129P2-Lyz2tm1(cre)Ifo/J (stock #004781), B6.Cg-Tg(Gfap-cre)73.12Mvs/J (stock #012886), and B6.Cg-Tg(Camk2a-cre) T29-1Stl/J (stock #005359). The newly generated liver (Aldh2Hep−/−), adipose tissue (Aldh2Adipo−/−), intestinal epithelial cell (Aldh2IEC−/−), myeloid cell (Aldh2Mye−/−), and glial cell- (Aldh2Gfap−/−) and neuron-specific (Aldh2Cam2a−/−) Aldh2-deficient mice were then used in this study.
Assessment of Tissue-Specific Aldh2 Deletion.
Tissue-specific Aldh2 deletion was confirmed at the protein levels by Western blot as described in SI Appendix.
ALDH2 Enzymatic Activity Assay.
Fresh tissue lysates were prepared with a Dounce homogenizer in cold phosphate-buffered saline, and ALDH2 enzymatic activity was measured using the Mitochondrial Aldehyde Dehydrogenase (ALDH2) Activity Assay Kit (Abcam) following manufacturer’s instructions.
Acute Ethanol Gavage.
Mice received an oral gavage of 5 g/kg ethanol prepared in mouse usual drinking water. Mice were kept on a heating pad throughout the experiment to prevent hypothermia.
Serum Sample Extraction Procedure and GC-MS Analysis.
Serum ethanol and acetaldehyde levels were measured 1 h or 3 h after 5 g/kg ethanol gavage by GC-MS. Further details are available in SI Appendix.
Indirect Measurement of Metabolic Parameters.
Indirect metabolic rate measurements were performed on an Oxymax Metabolic Cage System (Columbus Instruments) in a dedicated animal holding room to limit noise and disturbance in the surroundings. The Oxymax System is a noninvasive setup that directly records respiratory exchange ratio (O2 and CO2), as well as mouse physical activity measured by infrared beams and detectors. Mice were individually housed and, after habituation, received a single 5 g/kg ethanol gavage, 2.5 h before lights were turned off in the procedure room. Cages remained unopened during the measurements.
The 2-Bottle Choice Paradigm.
Mice were individually housed and given free access to 2 water bottles. Following 1 wk of habituation, mice were given free choice between 2 bottles, one of which contained regular drinking water while the other one contained drinking water with increasing alcohol concentrations (3%, 6%, 9%, 12%, 15%, 18%, and 21% vol/vol) for 4 d each. Drinking volumes were measured daily, and bottle positions were interchanged to prevent learned preference.
Liver-Specific Aldh2 Knockdown.
Type 5 replication-deficient adenovirus encoding for GFP and a short hairpin sequence designed to target mouse Aldh2 (Ad-GFP-U6-mALDH2-shRNA; Vector Biolabs) was injected in B6N mice via tail vein, at a dose of 109 plaque-forming unit (PFU) at the indicated times. Control adenovirus containing a scramble sequence was similarly injected in control mice (Ad-GFP-U6-scrmb-shRNA; Vector Biolabs). Drinking volumes have been recorded various days after adenovirus injections to allow for efficient gene knockdown.
Statistical Analysis.
The data are expressed as means ± SEM. Statistical significance was determined by a 2‐tailed Student t test, or 1‐way ANOVA followed by a Tukey’s multiple comparison test, as indicated (GraphPad Prism; GraphPad Software Inc.). Results were considered significantly different for p < 0.05.
Additional methods are available in SI Appendix.
Data Availability.
All data and protocols are described in the manuscript or SI Appendix.
Supplementary Material
Acknowledgments
We greatly appreciate Dr. Ketan J. Patel (MRC Laboratory of Molecular Biology, Cambridge, United Kingdom) for providing Aldh2tm1a(EUCOMM)Wtsi mice. We also thank Dr. Bryan Mackowiak and other laboratory members for critically reading the manuscripts.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.A.B. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1908137116/-/DCSupplemental.
References
- 1.Collaborators G. B. D. A.; GBD 2016 Alcohol Collaborators , Alcohol use and burden for 195 countries and territories, 1990-2016: A systematic analysis for the global burden of disease study 2016. Lancet 392, 1015–1035 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fuster D., Samet J. H., Alcohol use in patients with chronic liver disease. N. Engl. J. Med. 379, 1251–1261 (2018). [DOI] [PubMed] [Google Scholar]
- 3.Tapper E. B., Parikh N. D., Mortality due to cirrhosis and liver cancer in the United States, 1999-2016: Observational study. BMJ 362, k2817 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gonzales K., et al. ; Centers for Disease Control and Prevention (CDC) , Alcohol-attributable deaths and years of potential life lost–11 States, 2006-2010. MMWR Morb. Mortal. Wkly. Rep. 63, 213–216 (2014). [PMC free article] [PubMed] [Google Scholar]
- 5.Caputo F., Domenicali M., Bernardi M., Diagnosis and treatment of alcohol use disorder in patients with end-stage alcoholic liver disease. Hepatology 70, 410–417 (2019). [DOI] [PubMed] [Google Scholar]
- 6.Szabo G., Kamath P. S., Shah V. H., Thursz M., Mathurin P.; EASL-AASLD Joint Meeting , Alcohol-related liver disease: Areas of consensus, unmet needs and opportunities for further study. Hepatology 69, 2271–2283 (2019). [DOI] [PubMed] [Google Scholar]
- 7.Nutt D. J., The role of the opioid system in alcohol dependence. J. Psychopharmacol. 28, 8–22 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Plosker G. L., Acamprosate: A review of its use in alcohol dependence. Drugs 75, 1255–1268 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Suh J. J., Pettinati H. M., Kampman K. M., O’Brien C. P., The status of disulfiram: A half of a century later. J. Clin. Psychopharmacol. 26, 290–302 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Jørgensen C. H., Pedersen B., Tønnesen H., The efficacy of disulfiram for the treatment of alcohol use disorder. Alcohol. Clin. Exp. Res. 35, 1749–1758 (2011). [DOI] [PubMed] [Google Scholar]
- 11.Blanc M., Daeppen J. B., Does disulfiram still have a role in alcoholism treatment? Rev. Med. Suisse 1, 1728–1730, 1732–1733 (2005). [PubMed] [Google Scholar]
- 12.Malcolm R., Olive M. F., Lechner W., The safety of disulfiram for the treatment of alcohol and cocaine dependence in randomized clinical trials: Guidance for clinical practice. Expert Opin. Drug Saf. 7, 459–472 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Chen C. H., Ferreira J. C., Gross E. R., Mochly-Rosen D., Targeting aldehyde dehydrogenase 2: New therapeutic opportunities. Physiol. Rev. 94, 1–34 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Edenberg H. J., McClintick J. N., Alcohol dehydrogenases, aldehyde dehydrogenases, and alcohol use disorders: A critical review. Alcohol. Clin. Exp. Res. 42, 2281–2297 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoval-Sánchez B., Rodríguez-Zavala J. S., Differences in susceptibility to inactivation of human aldehyde dehydrogenases by lipid peroxidation byproducts. Chem. Res. Toxicol. 25, 722–729 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Brooks P. J., Enoch M. A., Goldman D., Li T. K., Yokoyama A., The alcohol flushing response: An unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Med. 6, e50 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eng M. Y., Luczak S. E., Wall T. L., ALDH2, ADH1B, and ADH1C genotypes in Asians: A literature review. Alcohol Res. Health 30, 22–27 (2007). [PMC free article] [PubMed] [Google Scholar]
- 18.Chen Y. C., et al. , Pharmacokinetic and pharmacodynamic basis for overcoming acetaldehyde-induced adverse reaction in Asian alcoholics, heterozygous for the variant ALDH2*2 gene allele. Pharmacogenet. Genomics 19, 588–599 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Isse T., Matsuno K., Oyama T., Kitagawa K., Kawamoto T., Aldehyde dehydrogenase 2 gene targeting mouse lacking enzyme activity shows high acetaldehyde level in blood, brain, and liver after ethanol gavages. Alcohol. Clin. Exp. Res. 29, 1959–1964 (2005). [DOI] [PubMed] [Google Scholar]
- 20.Jamal M., et al. , Ethanol and acetaldehyde after intraperitoneal administration to aldh2-knockout mice-Reflection in blood and brain levels. Neurochem. Res. 41, 1029–1034 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Skinner M. D., Lahmek P., Pham H., Aubin H. J., Disulfiram efficacy in the treatment of alcohol dependence: A meta-analysis. PLoS One 9, e87366 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fletcher K., et al. , A breath test to assess compliance with disulfiram. Addiction 101, 1705–1710 (2006). [DOI] [PubMed] [Google Scholar]
- 23.MacDonagh L., et al. , Targeting the cancer stem cell marker, aldehyde dehydrogenase 1, to circumvent cisplatin resistance in NSCLC. Oncotarget 8, 72544–72563 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cortínez G., Sapag A., Israel Y., RNA interference against aldehyde dehydrogenase-2: Development of tools for alcohol research. Alcohol 43, 97–104 (2009). [DOI] [PubMed] [Google Scholar]
- 25.Sanchez A. C., Li C., Andrews B., Asenjo J. A., Samulski R. J., AAV gene therapy for alcoholism: Inhibition of mitochondrial aldehyde dehydrogenase enzyme expression in hepatoma cells. Hum. Gene Ther. 28, 717–725 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arolfo M. P., et al. , Suppression of heavy drinking and alcohol seeking by a selective ALDH-2 inhibitor. Alcohol. Clin. Exp. Res. 33, 1935–1944 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zakhari S., Li T. K., Determinants of alcohol use and abuse: Impact of quantity and frequency patterns on liver disease. Hepatology 46, 2032–2039 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Poelstra K., Prakash J., Beljaars L., Drug targeting to the diseased liver. J. Control. Release 161, 188–197 (2012). [DOI] [PubMed] [Google Scholar]
- 29.Ortiz de Montellano P. R., Cytochrome P450-activated prodrugs. Future Med. Chem. 5, 213–228 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kwon H. J., et al. , Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty liver but worsens liver inflammation and fibrosis in mice. Hepatology 60, 146–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Crews F. T., Lawrimore C. J., Walter T. J., Coleman L. G. Jr, The role of neuroimmune signaling in alcoholism. Neuropharmacology 122, 56–73 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alnouti Y., Klaassen C. D., Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol. Sci. 101, 51–64 (2008). [DOI] [PubMed] [Google Scholar]
- 33.Oyama T., et al. , Tissue-distribution of aldehyde dehydrogenase 2 and effects of the ALDH2 gene-disruption on the expression of enzymes involved in alcohol metabolism. Front. Biosci. 10, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Deza-Ponzio R., Herrera M. L., Bellini M. J., Virgolini M. B., Hereñú C. B., Aldehyde dehydrogenase 2 in the spotlight: The link between mitochondria and neurodegeneration. Neurotoxicology 68, 19–24 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Doorn J. A., Florang V. R., Schamp J. H., Vanle B. C., Aldehyde dehydrogenase inhibition generates a reactive dopamine metabolite autotoxic to dopamine neurons. Parkinsonism Relat. Disord. 20 (suppl. 1), S73–S75 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Florang V. R., et al. , Inhibition of the oxidative metabolism of 3,4-dihydroxyphenylacetaldehyde, a reactive intermediate of dopamine metabolism, by 4-hydroxy-2-nonenal. Neurotoxicology 28, 76–82 (2007). [DOI] [PubMed] [Google Scholar]
- 37.Fu S. H., et al. , Alda-1 reduces cerebral ischemia/reperfusion injury in rat through clearance of reactive aldehydes. Naunyn Schmiedebergs Arch. Pharmacol. 387, 87–94 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Ding J., et al. , Alda-1 attenuates lung ischemia-reperfusion injury by reducing 4-hydroxy-2-nonenal in alveolar epithelial cells. Crit. Care Med. 44, e544–e552 (2016). [DOI] [PubMed] [Google Scholar]
- 39.Zhu Q., He G., Wang J., Wang Y., Chen W., Pretreatment with the ALDH2 agonist Alda-1 reduces intestinal injury induced by ischaemia and reperfusion in mice. Clin. Sci. (Lond.) 131, 1123–1136 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Panisello-Roselló A., et al. , Role of aldehyde dehydrogenase 2 in ischemia reperfusion injury: An update. World J. Gastroenterol. 24, 2984–2994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Perez-Miller S., et al. , Alda-1 is an agonist and chemical chaperone for the common human aldehyde dehydrogenase 2 variant. Nat. Struct. Mol. Biol. 17, 159–164 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao Y., et al. , Alcohol inhibits T-cell glucose metabolism and hepatitis in ALDH2-deficient mice and humans: Roles of acetaldehyde and glucocorticoids. Gut 68, 1311–1322 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langevin F., Crossan G. P., Rosado I. V., Arends M. J., Patel K. J., Fancd2 counteracts the toxic effects of naturally produced aldehydes in mice. Nature 475, 53–58 (2011). [DOI] [PubMed] [Google Scholar]
- 44.Seo W., et al. , ALDH2 deficiency promotes alcohol-associated liver cancer by activating oncogenic pathways via oxidized DNA-enriched extracellular vesicles. J. Hepatol. 71, 1000–1011 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data and protocols are described in the manuscript or SI Appendix.