Significance
The human population contains thousands of MHC-I alleles, showing a range of dependencies on molecular chaperones for loading of their peptide cargo, which are then displayed on the cell surface for T cell surveillance. Using the chaperone TAPBPR as a model, we combine deep mutagenesis with functional and biophysical data, especially solution NMR, to provide a complete view of the molecular determinants of chaperone recognition. Our data provide significant evidence that localized protein motions define the intrinsic ability of MHC-I molecules to interact with chaperones. The importance of MHC-I dynamics unifies our findings, with broad recognition of conformationally unstable, nascent MHC-I molecules becoming restricted to a smaller set of MHC-I alleles that retain relevant dynamic motions in their folded state.
Keywords: major histocompatibility complex, molecular chaperone, peptide repertoire, NMR spectroscopy, peptide editing
Abstract
The interplay between a highly polymorphic set of MHC-I alleles and molecular chaperones shapes the repertoire of peptide antigens displayed on the cell surface for T cell surveillance. Here, we demonstrate that the molecular chaperone TAP-binding protein related (TAPBPR) associates with a broad range of partially folded MHC-I species inside the cell. Bimolecular fluorescence complementation and deep mutational scanning reveal that TAPBPR recognition is polarized toward the α2 domain of the peptide-binding groove, and depends on the formation of a conserved MHC-I disulfide epitope in the α2 domain. Conversely, thermodynamic measurements of TAPBPR binding for a representative set of properly conformed, peptide-loaded molecules suggest a narrower MHC-I specificity range. Using solution NMR, we find that the extent of dynamics at “hotspot” surfaces confers TAPBPR recognition of a sparsely populated MHC-I state attained through a global conformational change. Consistently, restriction of MHC-I groove plasticity through the introduction of a disulfide bond between the α1/α2 helices abrogates TAPBPR binding, both in solution and on a cellular membrane, while intracellular binding is tolerant of many destabilizing MHC-I substitutions. Our data support parallel TAPBPR functions of 1) chaperoning unstable MHC-I molecules with broad allele-specificity at early stages of their folding process, and 2) editing the peptide cargo of properly conformed MHC-I molecules en route to the surface, which demonstrates a narrower specificity. Our results suggest that TAPBPR exploits localized structural adaptations, both near and distant to the peptide-binding groove, to selectively recognize discrete conformational states sampled by MHC-I alleles, toward editing the repertoire of displayed antigens.
Class I major histocompatibility complex (MHC-I) molecules display a diverse set of 8 to 14 residue peptide antigens to CD8+ cytotoxic T lymphocytes (1). This process provides a means for immune surveillance of the endogenous proteome to detect invading pathogens or developing tumors. MHC-I molecules are extremely polymorphic, with thousands of known human alleles, categorized in the HLA-A, -B, and -C classes. Specific interactions with highly polymorphic pockets along the peptide binding groove (termed A- to F-pockets) define a repertoire of up to 104 peptide antigens that can bind to each HLA protein (1). Proper folding of nascent MHC-I molecules and loading with high-affinity peptides requires association with an invariant light-chain, β2-microglobulin (β2m), and is facilitated by dedicated molecular chaperones, tapasin, which is restricted within the peptide-loading complex (PLC), and the homologous, PLC-independent TAPBPR (TAP-binding protein related) (2, 3). Furthermore, through the catalytic enhancement of peptide association and dissociation within the MHC-I groove, chaperones can influence the selection of immunodominant antigens by promoting the exchange of low- and intermediate-affinity for high-affinity peptides (termed “peptide editing”) (2, 3). Further quality control and narrowing of the displayed antigen repertoire along the trafficking pathway is accomplished through the combined functions of TAPBPR and UDP-glucose:glycoprotein glucosyltransferase (UGGT) (3–5). The discovery that TAPBPR can function as a peptide-exchange catalyst outside the peptide-loading complex and can maintain empty MHC-I molecules in a peptide-receptive conformation has opened a new window to study the peptide-loading process in a range of detailed functional and mechanistic studies (4, 6, 7).
Improper function of the antigen processing and presentation pathway confers susceptibility to diseases in a manner that is highly dependent on the individual’s MHC-I haplotype and the disease-relevant immunodominant peptides (8). Several studies have provided key insights into the allelic preferences of chaperone interactions. It has been long established that some MHC-I alleles require tapasin for proper peptide loading, trafficking, and cell surface display, while others can intrinsically load peptides in tapasin knockouts (4, 6–10). In an extreme case demonstrated by the HLA-B*44 alleles, a single amino acid polymorphism at the F-pocket is sufficient to switch between chaperone-dependent and -independent peptide loading (11–13). Similarly, TAPBPR expression exhibits a marked effect on the displayed repertoire of specific alleles (4). While structure-based modeling using the available cocrystal structures as templates can provide some clues for the molecular basis of MHC-I allelic dependence of chaperone interactions, molecular dynamics (MD) simulations, and hydrogen deuterium exchange experiments have further shown that chaperone-dependent and -independent alleles exhibit widespread differences in their dynamic profiles (14, 15). In particular, these experiments have suggested the rate of sampling of an “open” conformation confers chaperone recognition of different MHC-I alleles (16, 17).
Here, using the chaperone TAPBPR as a model, we apply a range of complementary functional and biophysical experiments using fusion and recombinant proteins in a native cellular context and in solution, respectively. We find that TAPBPR interacts broadly with nascent MHC-I of varying sequences within cellular compartments, with a binding mode that is highly polarized toward the α2 side of the peptide-binding groove, and chaperones misfolded molecules. However, interactions with properly conformed pMHC-I molecules toward editing of the peptide cargo are restricted to a limited set of alleles, where the dynamic sampling of a sparse minor-state conformation in solution is important. Our results provide a molecular blueprint for the design of MHC-I molecules with improved antigen processing and presentation properties, through the engineering of conformational states across the entire folding landscape.
Results
TAPBPR Associates with a Broad Range of MHC-I Alleles inside the Cell.
TAPBPR has been reported to exhibit a narrow MHC-I allele specificity (4, 7). However, it remains undetermined whether the allelic preferences of TAPBPR manifest in a similar manner for nascent, empty MHC-I, or properly conformed, peptide-loaded MHC-I molecules. To explore these concepts further, we first investigated the propensity for TAPBPR to interact with MHC-I molecules composed of different heavy-chain sequences (mouse H2 or human HLA) in a native cellular environment. We adapted established bimolecular fluorescence complementation (BiFC) assays in human Expi293F cells transfected using gene constructs containing TAPBPR fused to the C-terminal half (VC) of split fluorescent Venus, and either of a panel of heavy-chain MHC-I fused to the N-terminal half (VN) (18). Here, when TAPBPR-VC comes into proximity with MHC-I–VN as the result of protein–protein interactions inside the cell, an active yellow fluorescent protein can fold resulting in high BiFC signal, which can be readily detected by flow cytometry. As a negative control, we used the unrelated protein CXCR4-VN (C-X-C chemokine receptor type 4), which is not expected to interact with TAPBPR-VC. After gating for cells with similar expression levels based on intracellular staining of N-terminal epitope tags (SI Appendix, Fig. S1 A–C), we observed significant BiFC signal across all H2 and HLA alleles tested, suggesting that a broad range of MHC-I alleles with diverse sequences have the potential to interact with TAPBPR in a native cellular environment (SI Appendix, Fig. S1D). Background BiFC signal for CXCR4 from nonspecific protein associations is substantially lower. We note that gating based on expression levels did not change observed trends, but rather highlighted differences by excluding high-expressing cells with saturated BiFC signal (SI Appendix, Fig. S1 A and B). Overexpression of TAPBPR in this assay also promotes intracellular retention of MHC-I, consistent with previous reports (6). Unlike the negative control CXCR4, the MHC-I molecules tested here exhibit severely reduced cell surface expression in the presence of TAPBPR (SI Appendix, Fig. S1E).
Spontaneous and nonspecific self-assembly of the Venus halves due to protein overexpression can result in high background and low signal-to-noise, leading to false-positive BiFC signals for some protein–protein interactions (18). Thus, we coexpressed and coimmunoprecipitated TAPBPR with MHC-I in the absence of VN and VC fusions. Unlike the CXCR4 control, all of the MHC-I alleles bound TAPBPR, thereby independently confirming the BiFC results using a different experimental method (SI Appendix, Fig. S2). Together, these data suggest that TAPBPR associates with a broad range of MHC-I alleles inside the cell, potentially through partially folded, empty MHC-I conformations within the endoplasmic reticulum or Golgi.
TAPBPR Recognizes a Partially Folded Epitope on MHC-I Molecules with Broad Specificity.
Deep mutagenesis, in which directed evolution of a diverse library of sequences is tracked by next-generation sequencing, has emerged as a powerful tool for interrogating how protein amino acid sequence dictates structure and function in living cells (19). The relative activities of thousands of mutations can be assessed simultaneously to reveal critical features for protein stability or ligand binding. To identify important features of the human MHC-I heavy chain required for TAPBPR association within cells, we generated a single-site saturation mutagenesis (SSM) library on HLA-A*02:01. We chose to focus on HLA-A*02:01, as it is a common human allele that associates with TAPBPR in vitro with high affinity and presents several important autoimmune, viral, and cancer epitopes (4, 7). To improve sampling of individual mutations and thereby increase data quality, library diversity was limited by restricting mutational scanning to the MHC-I α1/α2 “platform” domain (residues 2 to 181), which mediates both peptide and TAPBPR binding (20, 21). Using 2 complementary fluorescence-based selections, we investigated the effects of single-site mutations throughout the groove on 1) HLA-A*02:01 cell surface expression, to reveal how particular mutations influence MHC-I stability, and 2) HLA-A*02:01/TAPBPR recognition, to identify structural elements that are important for chaperone recognition. There were concerns that high BiFC signal may simply report on assembly with MHC-I–VN mutants that are retained in the same intracellular compartments as TAPBPR-VC (see above). To resolve this potential issue, we used cells stably expressing a variant called TAPBPR-TM, in which the native transmembrane (TM) helix and cytosolic tail are replaced with a generic TM domain. Confocal microscopy (SI Appendix, Fig. S3A) and flow cytometry (SI Appendix, Fig. S3B) reveal that TAPBPR-TM localizes intracellularly and at the plasma membrane, and therefore reports on interactions regardless of whether MHC-I mutants traffic to the surface or not. Following transfection of the HLA-A*02:01 SSM library and FACS for MHC-I surface expression in wild-type cells (SI Appendix, Fig. S3C) or MHC-I/TAPBPR BiFC in TAPBPR-TM-VC–expressing cells (SI Appendix, Fig. S3D), the relative effects of nearly all single amino acid substitutions in the MHC-I groove are determined based on their enrichment or depletion (SI Appendix, Fig. S4 A and B). Enrichment ratios of specific mutations and conservation scores for residue positions have excellent agreement between independent replicates, providing high confidence to the mutational landscapes (SI Appendix, Fig. S4 C–G). Processed and raw data are available online (https://www.ncbi.nlm.nih.gov/geo, GEO accession no. GSE128957; ref. 22).
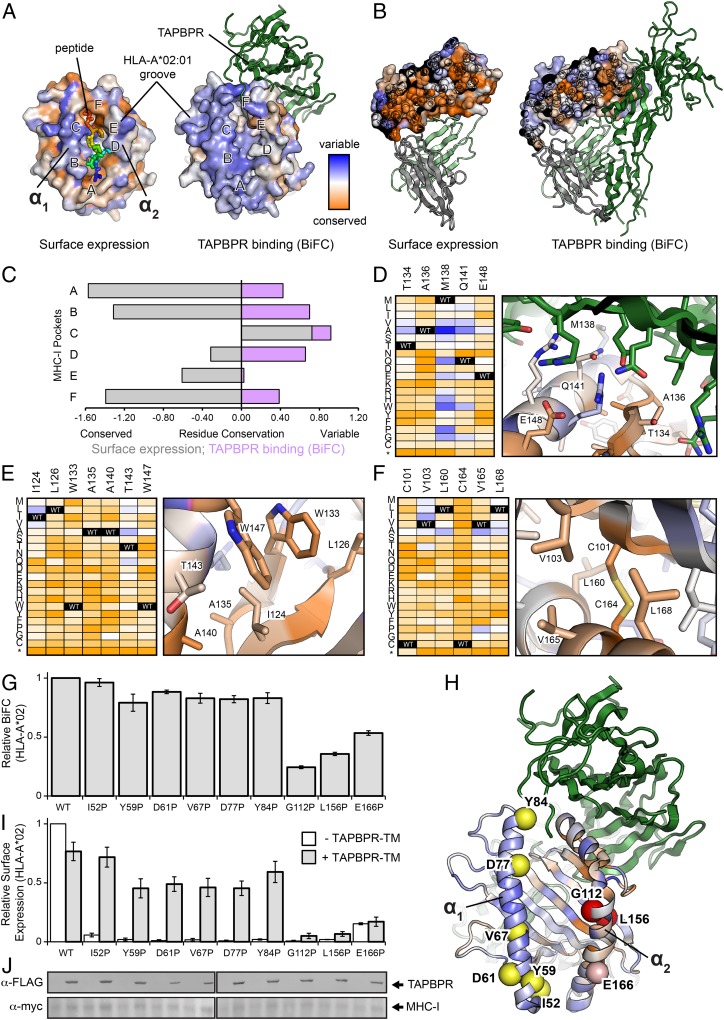
We find that surface expression of HLA-A*02:01 imposes strict sequence constraints throughout the peptide binding groove, consistent with the preferential trafficking of stable, peptide-loaded MHC-I to the plasma membrane (Fig. 1 A and B and SI Appendix, Fig. S4A) (23, 24). Not only are the residues comprising the MHC-I pockets that anchor the peptide tightly conserved for surface expression (residues 5, 7, 59, 63, 66, 99, 159, 163, 167, 171 for the A-pocket and 77, 80, 81, 84, 116, 123, 143, 146, 147 for the F-pocket), but so are residues within the core of the MHC-I groove, and the underlying surface contacting β2m and the α3 domain (Fig. 1 A–C and SI Appendix, Fig. S4A). Proper pMHC-I folding and stability in human cells, which express endogenous tapasin, is therefore intolerant of most single amino acid substitutions. In an interesting exception to this trend, sites on the α1 helix (MHC-I residues 67 to 75) that do not participate in direct interactions with either the bound peptide or TAPBPR, but would be engaged by T cell receptors, are more amenable to mutation (Fig. 1A and SI Appendix, Fig. S4A). Taken together, these data reveal that conserved MHC-I sites important for surface expression are dispersed throughout the groove, and that the most stringent sequence constraints are contained within the peptide anchoring pockets, consistent with the established trend that MHC-I surface expression is tightly coupled to proper peptide loading (23, 24).
Fig. 1.
TAPBPR recognizes a local conformation of the HLA-A*02:01 α2 domain. (A) Sequence conservation from deep mutagenesis mapped to the surface of HLA-A*02:01. Conserved residues are colored in dark orange, while residues exhibiting mutational tolerance in pale and blue. Residue conservation for HLA-A*02:01 surface expression is shown on the structure of the free pMHC-I molecule (PDB ID code 1HHJ), with bound nonamer peptide colored from blue (residue 1) to red (residue 9). HLA-A*02:01 conservation for TAPBPR (dark green) binding is plotted on the structure of the empty MHC-I, in complex with TAPBPR (PDB ID code 5WER). The location of the A- to F-pockets of the MHC-I groove are noted. (B) Cross sections through the core of the α2 domain, colored by conservation as in A. TAPBPR is dark green, β2m is pale green, and the α3 domain is gray. (C) Conservation and variation of residues in the MHC-I pockets for surface expression (gray) or TAPBPR binding by BiFC (purple). (D–F) Heat maps of mutation log2 enrichment ratios for HLA-A*02:01/TAPBPR BiFC (depleted mutations are orange, enriched mutations are dark blue) shown alongside modeled structures of the (D) α2 domain/TAPBPR interface, (E) the hydrophobic core between the α2–1 helix and β-sheet, and (F) the core between the α2–2 helix and β-sheet. (G) Relative BiFC signal between TAPBPR-TM-VC and MHC-I–VN for different proline substitutions in either the α1 or α2 helices, or β-sheet underlying the α2 helix. (H) Location of proline substitutions on the MHC-I groove mapped onto the TAPBPR complex structure (PDB ID code 5WER). (I) Relative surface expression for the different MHC-I proline substitutions in the presence or absence of TAPBPR-TM. (J) Immunoblots comparing total expression levels for TAPBPR-TM (α-FLAG) and HLA-A*02:01 (α-myc) constructs. Lanes are aligned with graphs above.
While surface expression imposes strict sequence constraints throughout the MHC-I groove, we find that TAPBPR association persists following many mutations, and is especially tolerant of mutations to the 310 and α1 helices that demarcate the α1 domain of the peptide binding groove (Fig. 1 A–C). This also includes tolerance to many mutations at positions for which crystallographically defined intermolecular contacts are observed in the homologous mouse H2-Dd/TAPBPR complex (21), such as HLA-A*02:01 residues 83, 84, 127, and 144 (Fig. 1 A and B and SI Appendix, Fig. S4B). Other HLA-A*02:01 residues that are buried in the homologous H2-Dd/TAPBPR interface—such as 134, 135, 136, and 148—are under tight sequence conservation (Fig. 1D), revealing that only a subset of contacts at the MHC-I/TAPBPR interface are amenable to mutation. While HLA-A*02:01 residues comprising the A-, B-, and F-pockets of the groove are conserved for surface expression, they tolerate variability for TAPBPR binding (Fig. 1C), consistent with TAPBPR engaging HLA-A*02:01 mutants that are defective for peptide loading. There are 2 prominent buried clusters of conserved MHC-I residues in the mutational landscape. The first cluster contains HLA-A*02:01 residues from the region 124 to 147, located within and adjacent to the E- and F-pockets (Fig. 1 D and E). A second, smaller cluster of MHC-I residues conserved for TAPBPR binding mediates contacts between the α2–2 helix and β-sheet around the conserved C101-C164 disulfide within the A-pocket (Fig. 1F). We find that mutation of the C101-C164 disulfide abrogates interaction with TAPBPR, likely because disulfide bond formation contributes to stabilization of the α2 domain (25). Elsewhere, HLA-A*02:01/TAPBPR BiFC shows intermediate mutational tolerance at sites of the MHC-I groove contacting the β2m/α3 domains, while the core between the α1 helix and β-sheet has very high mutational tolerance (SI Appendix, Fig. S4B). Thus, in contrast to the strict sequence conservation of MHC-I sites dispersed throughout the groove for surface expression, sites conserved for TAPBPR association are highly polarized toward the α2 domain of the groove, suggesting that TAPBPR has the potential to recognize nascent MHC-I molecules with broad sequence specificity, at least when single amino acid substitutions are considered.
TAPBPR Recognition of Nascent Species Is Polarized toward the α2 Domain of the MHC-I Groove.
We hypothesized that TAPBPR can associate with partially folded or misfolded MHC-I molecules, with an interaction mode that is dependent upon the formation of a local conformational epitope adjacent to the α2 helix. Such MHC-I species likely form intracellularly during the processing of nascent, peptide-deficient molecules. To investigate this, we created a panel of proline substitutions along either the α1 or α2 helices of HLA-A*02:01 groove. Incorporation of a proline is expected to destabilize secondary structure, especially helices (26, 27). Destabilizing proline substitutions within the α1 helix (I52P, Y59P, D61P, V67P, D77P, Y84P) do not abrogate BiFC signal with TAPBPR-TM compared to wild-type HLA-A*02:01, while those within or adjacent to the α2 helix (G112P, L156P, E166P) severely reduce BiFC signal with TAPBPR-TM (Fig. 1 G and H). We simultaneously examined MHC-I surface localization in the transfected cells. Ordinarily, destabilized MHC-I proline mutants would not escape intracellular quality control to reach the plasma membrane, but stable association with TAPBPR-TM, which traffics to the cell surface, is expected to rescue MHC-I surface localization. Indeed, we find that proline substitutions within the α1 helix of the groove are able to reach the cell surface only in the presence of TAPBPR-TM, while those within the α2 helix are retained intracellularly in the absence and presence of TAPBPR-TM (Fig. 1I). Importantly, expression of all of the mutants remain at near wild-type levels, and hence observed differences cannot be explained by a simple loss of expression (Fig. 1J). Thus, while destabilization of the HLA-A*02:01 α1 helix induces a misfolded, peptide-deficient state, these conformations do not compromise the TAPBPR recognition epitope and a stable complex can form (Fig. 1H). This is in contrast with proline substitutions within or near to the α2 helix, which disrupt the local MHC-I structure recognized by TAPBPR (Fig. 1H).
TAPBPR Recognizes Folded pMHC-I Using a Conserved Binding Mode of Narrow Specificity.
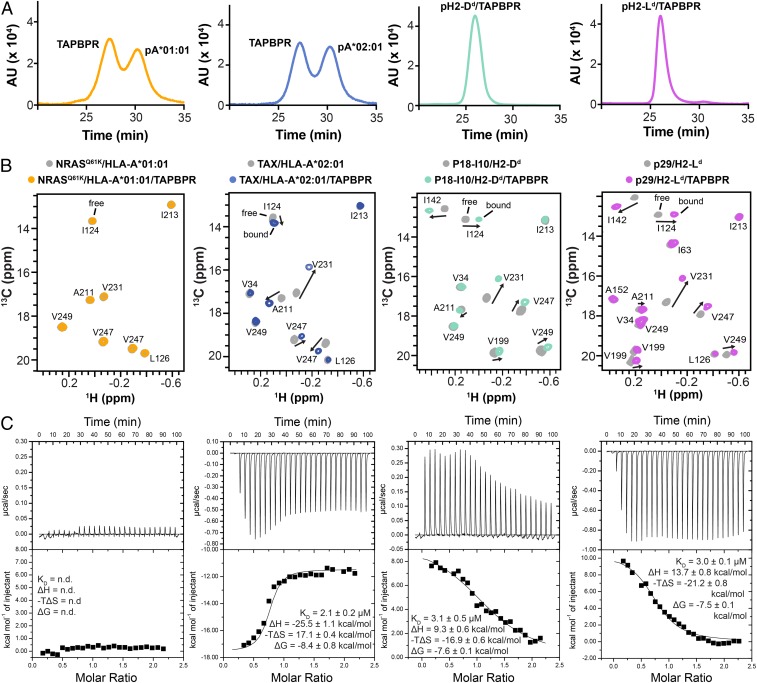
To characterize TAPBPR recognition of purified pMHC-I molecules in solution, we focused on a representative set corresponding to 2 mouse (H2-Dd and H2-Ld) and 2 human (HLA-A*01:01 and HLA-A*02:01) alleles, displaying between 70% and 85% similarity at the sequence level (SI Appendix, Fig. S5). Each MHC-I heavy chain was expressed in Escherichia coli and refolded together with a high-affinity peptide and human β2m (hβ2m) (7, 28). We utilized well-characterized epitopes: The p29 peptide (YPNVNIHNF) for H2-Ld (29), the HIV gp120 P18-I10 peptide (RGPGRAFVTI) for H2-Dd (30), the neuroblastoma related NRASQ61K peptide (ILDTAGKEEY) for HLA-A*01:01 (31), and the HTLV-1 TAX peptide (LLFGYPVYV) for HLA-A*02:01 (32). Using a differential scanning fluorimetry assay (33), we further confirmed that all purified samples displayed thermal stabilities that were characteristic of properly conformed, peptide-bound molecular species with Tm values ranging from 51 to 63 °C (SI Appendix, Table S1). Using a size-exclusion chromatography (SEC) assay, we observed formation of pMHC-I/TAPBPR complexes for both mouse pMHC-I (P18-I10/H2-Dd/hβ2m and NIH/H2-Ld/hβ2m), but neither of the human pMHC-I (NRASQ61K/HLA-A*01:01/hβ2m and TAX/HLA-A*02:01/hβ2m) (Fig. 2A). A potential drawback of SEC is that measurements carried out under nonequilibrium conditions may favor complex or peptide dissociation, depending on kinetic off-rates (34). Thus, we also monitored TAPBPR binding under equilibrium conditions using solution NMR. Here, we used isotopically AILV-methyl labeled (Ala 13Cβ; Ile 13Cδ1; Leu 13Cδ1/13Cδ2; Val 13Cγ1/13Cγ2) pMHC-I (where the hβ2m subunit and peptide are at natural isotopic abundance), which allows us to quantify specific chemical-shift changes upon formation of the 87 kDa pMHC-I/TAPBPR complex (28). NMR can also detect the presence of peptide-deficient complex in the sample, through the observation of unique chemical shifts of MHC-I methyl groups of residues in the peptide-binding groove (28). Toward this goal, we obtained complete stereospecific assignment for the heavy-chain AILV methyl groups of each pMHC-I system in our set (28, 35–39). Simultaneously, to obtain precise thermodynamic parameters describing the binding process, we performed isothermal calorimetry titration (ITC) experiments (40). To ensure that the complex formed with TAPBPR contained exclusively peptide-bound MHC-I (rather than empty molecules), all ITC experiments were performed in the presence of 10-fold molar excess of free peptide relative to the pMHC-I (Materials and Methods).
Fig. 2.
TAPBPR interactions with peptide-loaded MHC-I molecules are allele-dependent. (A) SEC traces of 1:1 molar ratio of TAPBPR with different pMHC-I heavy chains, each refolded with human β2m and a high-affinity peptide. (B) Representative 2D 1H-13C HMQC spectra of heavy-chain 13C AILV methyl labeled pMHC-I at 105 μM without TAPBPR (gray) and in the presence of 3-fold molar excess TAPBPR. From left to right: NRASQ61K/HLA-A*01:01/hβ2m (orange), TAX/HLA-A*02:01/hβ2m (blue), P18-I10/H2-Dd/hβ2m (green), and NIH/H2-Ld/hβ2m (purple). Experiments were performed at a 1H NMR field of 800 MHz at 25 °C. Arrows indicate significant chemical-shift changes in MHC-I heavy-chain NMR resonances upon TAPBPR binding. (C) Representative ITC data titrating ∼100 to 150 μM pMHC-I (from Left to Right: NRASQ61K/HLA-A*01:01, TAX/HLA-A*02:01, P18-I10/H2-Dd, and p29/H2-Ld) into a sample containing 12 μM TAPBPR and 1 mM peptide. Black lines are the fits of the isotherm. Fitted values for KD, ΔH, -TΔS, and ΔG were determined using a 1-site binding model. n.d., not determined. Errors were determined from experimental replicates (n = 2).
Both NMR and ITC experiments confirm that peptide-bound TAX/HLA-A*02:01/hβ2m, P18-I10/H2-Dd/hβ2m, and NIH/H2-Ld/hβ2m form high-affinity complexes with TAPBPR in vitro (Fig. 2 B and C and SI Appendix, Fig. S6). In NMR experiments, tight complex formation is highlighted by the slow-exchange chemical-shift changes of resonances corresponding to methyl groups of conserved heavy-chain residues, such as I124, A211, V231, and V247 (Fig. 2B and SI Appendix, Fig. S6), suggesting a similar TAPBPR binding mode. Peptide release from the pMHC-I/TAPBPR complex to form an empty complex was not detected under the NMR sample conditions (Fig. 2B). Whereas the binding footprint of TAPBPR probed by methyl NMR is highly conserved among the 3 interacting pMHC-I molecules, and is further consistent with the published X-ray structures (20, 21), our ITC data reveal that the same complex structure can be attained via distinct thermodynamic processes. Specifically, HLA-A*02 exhibits exothermic binding behavior, in contrast to the endothermic association observed with H2-Dd and H2-Ld. While the enthalpic (ΔH) and entropic (-T*ΔS) contributions may differ between HLA-A*02:01 and H2-Dd/H2-Ld, the resulting binding free energy (ΔG) values are very similar (Fig. 2C). Fitting of the sigmoidal isotherms using a 1-site interaction model yields very similar apparent dissociation constants of 2.1 μM (TAX/HLA*A*02:01/ hβ2m), 3.1 μM (P18-I10/H2-Dd/hβ2m), and 3.0 μM (NIH/H2-Ld/hβ2m) (Fig. 2C). Notably, NRASQ61K/HLA-A*01:01/hβ2m does not interact with TAPBPR by either SEC, NMR, or ITC, up to millimolar-range concentrations (Fig. 2 and SI Appendix, Fig. S6). Together, our biophysical data reveal that relative to the broad specificity exhibited toward MHC-I molecules of different sequences inside the cell (SI Appendix, Figs. S1 and S2), TAPBPR recognizes properly conformed, peptide-loaded MHC-I molecules with a narrow specificity in solution, even when comparing pMHC-I complexes that are very similar in terms of overall fold, amino acid composition, and thermal stability.
pMHC-I Molecules Exhibit Allele-Specific Conformational Dynamics.
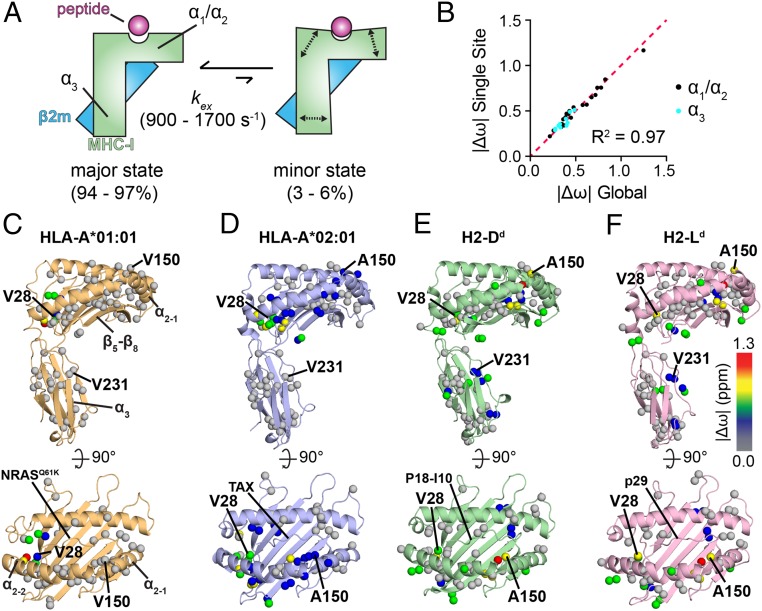
Conformational dynamics have been implicated in several aspects of MHC-I function, including peptide loading, T cell receptor triggering, and chaperone recognition (2, 41, 42). Solution NMR allows the quantitative measurement of dynamics across biologically relevant timescales ranging from picosecond to nanosecond (sidechain and loop motions), microsecond to millisecond (minor domain movements), to seconds (major domain reorientation) (43). Previous NMR measurements of picosecond to nanosecond dynamics in pMHC-I molecules did not uncover molecular flexibility other than in flexible loop regions (28, 44). In contrast, MHC-I regions involved in peptide and chaperone recognition exhibit microsecond to millisecond exchange between major and minor conformational states (28, 45). We have previously established the use of methyl groups as highly sensitive NMR probes with widespread coverage of the entire MHC-I structure to characterize unchaperoned and chaperoned MHC-I complexes (28).
Here, we have adopted a similar approach to quantitatively compare and contrast conformational dynamics in our representative set of pMHC-I molecules of different allelic composition, toward uncovering differences that could explain variation trends in TAPBPR recognition. We examined dynamics of AILV methyl groups of unchaperoned pMHC-I (where the heavy chain is isotopically labeled, while hβ2m and the peptide are at natural isotopic abundance) using 13C single-quantum Carr-Purcell-Meiboom-Gill (13C-SQ CPMG) relaxation dispersion NMR experiments (45, 46). In these experiments, methyl groups undergoing conformational exchange at the microsecond to millisecond timescale exhibit marked changes in the measured effective relaxation rate (R2eff) as a function of the frequency of the refocusing pulses (νCPMG) (43, 46). For each pMHC-I, we acquired 13C-SQ CPMG relaxation dispersion data at 25 °C at 2 NMR field strengths (600 MHz/14.1 Tesla and 800 MHz/18.8 Tesla). All AILV methyl groups with observable dynamics (Rex > 1 s−1) were fit to a 2-state exchange model (Fig. 3A). This procedure allowed us to extract quantitative NMR parameters that report on conformational exchange, including the exchange rate constant (kex = kAB + kBA), populations of the major and minor states (pA and pB) and absolute values of the 13C chemical-shift differences between the minor and the major state (|Δω|) (43, 46). CPMG experiments were performed in the presence of 3-fold molar excess of peptide to minimize the population of aggregation-prone empty MHC-I, as the half-lives of moderate to high-affinity peptides have been reported in the 0.3- to 28-h range (47–49). Specifically, we found no significant difference in the CPMG profiles and derived parameters in experiments performed for HLA-A*02:01 in the absence or presence of excess TAX peptide (SI Appendix, Fig. S7), suggesting that the excited state observed in our CPMG experiments does not correspond to a transition between the peptide-bound and peptide-free form, but rather to a conformational change occurring while the peptide remains bound in the MHC-I groove.
Fig. 3.
Dynamic profiles of different pMHC-I molecules probed by methyl NMR. (A) Summary of microsecond to millisecond timescale conformational exchange of peptide-bound MHC-I molecules between a major, ground state and a minor, excited state. Experimentally determined populations of the major and minor states and the exchange rate (kex) are noted in SI Appendix, Table S2. (B) Comparison of |Δω| values obtained from a global fit of CPMG data for all sites together (x axis) or independent fits of each site (y axis), shown for methyl groups in the groove (black) or α3 domain (cyan). (C–F) The sites that participate in the global conformational exchange process are represented as spheres on the structure of each pMHC-I viewed from the side (Upper) or above the groove (Lower). hβ2m is omitted for clarity. (C) NRASQ61K/HLA-A*01:01/hβ2m (PDB ID 6MPP), orange; (D) TAX/HLA-A*02:01/hβ2m (PDB ID code 1DUZ) (32), blue; (E) P18-I10/H2-Dd/hβ2m (PDB ID code 3ECB) (30), green; and (F) NIH/H2-Ld/hβ2m (PDB ID code 1LD9) (29), purple. Methyl groups undergoing dispersion are shown as spheres and color-coded based on |Δω| values obtained from a global fit of each pMHC-I. Methyl sites of increased dynamics are shown with warmer colors.
We observed conformational exchange at pMHC-I sites important for function, including the α2–1 helix, the β5-β8 sheet forming the floor of the groove, the A-pocket, and α3 domain. Notably, all CPMG profiles of methyl resonances showing conformational exchange could be individually fit to a model yielding the same population and kinetic parameters (Fig. 3B), suggesting the presence of a global conformational change connecting disparate sites on the MHC-I structure. However, the extent of dynamics within the different MHC-I domains was allele-dependent. While all pMHC-I molecules displayed A-pocket dynamics (Fig. 3 C–F, residue 28, and SI Appendix, Fig. S8A), α2–1 helix dynamics were only observed in the 3 pMHC-I molecules recognized by TAPBPR (Fig. 3 C–F, residue 150, and SI Appendix, Fig. S8B). In addition, microsecond to millisecond conformational exchange in the α3 domain was only observed for pMHC-I molecules prepared with mouse heavy chains H2-Dd or H2-Ld (Fig. 3 C–F, residue 231, and SI Appendix, Fig. S8C). In all molecules, methyl groups span the entire length of the MHC-I groove and α3 domain, thus a lack of observed dynamics is not due to incomplete probe coverage (SI Appendix, Fig. S9). Following a global fitting procedure of all methyl sites in each molecule, we obtained kex values ranging from 976 to 1,700 s−1 and pB values between 3.2% and 5.4% (SI Appendix, Table S2). Fitted |Δω| values range between 0.2 and 1.3 ppm (Fig. 3 D–F and SI Appendix, Table S2). In summary, our CPMG data revealed unique allele-specific, dynamic profiles of unchaperoned pMHC-I molecules, for which differences were more pronounced at functionally relevant sites located within the α2 helix and α3 domain.
A Minor Conformational State Underpins TAPBPR Recognition of Different MHC-I Alleles.
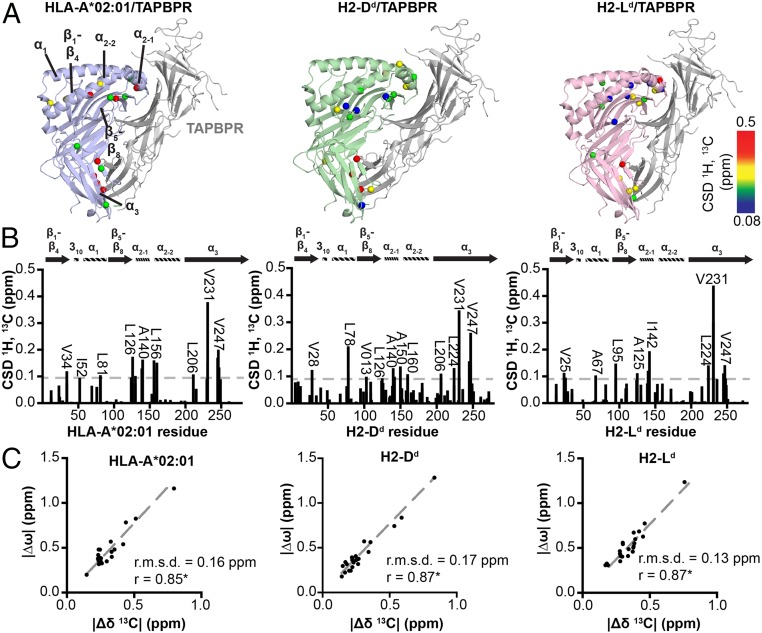
To confirm that TAPBPR utilizes a conserved overall binding mode to recognize pMHC-I molecules of different allelic compositions, we performed a chemical-shift deviation (CSD) analysis for complexes prepared using labeled HLA-A*02:01 and H2-Ld heavy chains, in addition to our previous characterization of H2-Dd (28). Here, the directly observed chemical shifts of the TAPBPR-bound state serve as unique identifiers of the induced local conformational changes, relative to the free pMHC-I state, for each residue. Using our established AILV methyl assignments for each system, the measured CSDs reveal that the same surfaces spanning multiple MHC-I domains on peptide-loaded H2-Dd, H2-Ld, and HLA-A*02:01 are engaged to form tight complexes with TAPBPR, consistent with the peptide-deficient MHC-I/TAPBPR crystal structures (20, 21, 28) (Fig. 4 A and B). Specifically, the NMR resonances corresponding to equivalent methyl probes in the α1 helix (residues 28 to 76), the β5-β8 strands (residues 104 to 130), the α2–1 helix (residues 139 to 158), and α3 domain (residues 230 to 251) of the heavy chain show significant CSDs upon TAPBPR binding across the 3 pMHC-I molecules analyzed here (Fig. 4 A and B).
Fig. 4.
Dynamic surfaces of pMHC-I structures correlate with TAPBPR recognition sites. (A) Methyl CSDs upon chaperone binding plotted onto the structure of MHC-I/TAPBPR complexes: TAX/HLA-A*02:01/hβ2m (blue), P18-I10/H2-Dd/hβ2m (green), and NIH/H2-Ld/hβ2m (pink) with TAPBPR shown in gray. The H2-Dd/TAPBPR crystal structure was obtained from PDB ID 5WER. The other structures are Rosetta homology models using PDB ID 5WER as a template (70). Affected regions are labeled. ΔCSDs are colored based on the scale shown at right. (B) Methyl CSDs measured from NMR titration experiments upon TAPBPR binding are shown as a function of heavy-chain methyl residue number. Select methyl groups affected are noted. The gray dotted line represents the average CSD + 1 SD. (C) The absolute value of the difference in the 13C chemical shift of the major and minor states of unchaperoned pMHC-I obtained from CPMG relaxation dispersion data (|Δω|, ppm) is shown as a function of the absolute value of the difference in the 13C chemical shift between the free and TAPBPR bound pMHC-I states determined from NMR titrations (|Δδ13C|, ppm). The slope (dotted gray line), the Pearson correlation coefficient (r), and the root mean square deviation (r.m.s.d.) are given for each correlation graph. The correlations are statistically significant with a P < 0.0001. HLA-A*01:01 is not included because there is no detectable TAPBPR binding under the NMR sample conditions (see Fig. 2).
Our previous NMR study of the H2-Dd system suggested that MHC-I surfaces exhibiting microsecond to millisecond conformational dynamics in solution correlate with the TAPBPR recognition sites (28). To investigate whether this is a general trend, we compared heavy-chain methyl groups displaying significant chemical shift changes upon complex formation (determined from NMR titrations) with those exhibiting microsecond to millisecond conformational exchange in the unchaperoned pMHC-I (determined from CPMG experiments). A comparison of the absolute value of the difference between the 13C chemical shifts (|Δ13Cδ|) of the unchaperoned and TAPBPR-bound pMHC-I states versus |Δω| values, obtained from fits of CPMG dispersion profiles of free pMHC-I, uncovers a strong positive correlation (R2 0.85 to 0.87) (Fig. 4C). Any conformational change in the pMHC-I induced by TAPBPR binding therefore occurs on pMHC-I surfaces of considerable structural plasticity in the free pMHC-I state.
Moreover, given that the |Δω| values directly report on conformational changes upon formation of a minor (3 to 6% population) state sampled by free pMHC-I molecules in an aqueous solution, our observed quantitative correlation of |Δ13Cδ| with |Δω| supports a plausible model where the minor state is directly recognized by TAPBPR to form an initial encounter complex. Further structural adaptations, induced by direct interactions between the 2 proteins, then lead to the formation of the final, tight complex with each pMHC-I molecule. Since the extent of MHC-I regions participating in the formation of the minor state vary between MHC-I molecules of different heavy-chain composition (Fig. 3 C–F), with the noninteracting heavy-chain HLA-A*01:01 exhibiting the most restricted dynamics, this model can explain allelic dependencies by differences in the minor state conformation.
Restriction of the α2–1 Helix Abrogates Sampling of the Minor State and TAPBPR Recognition.
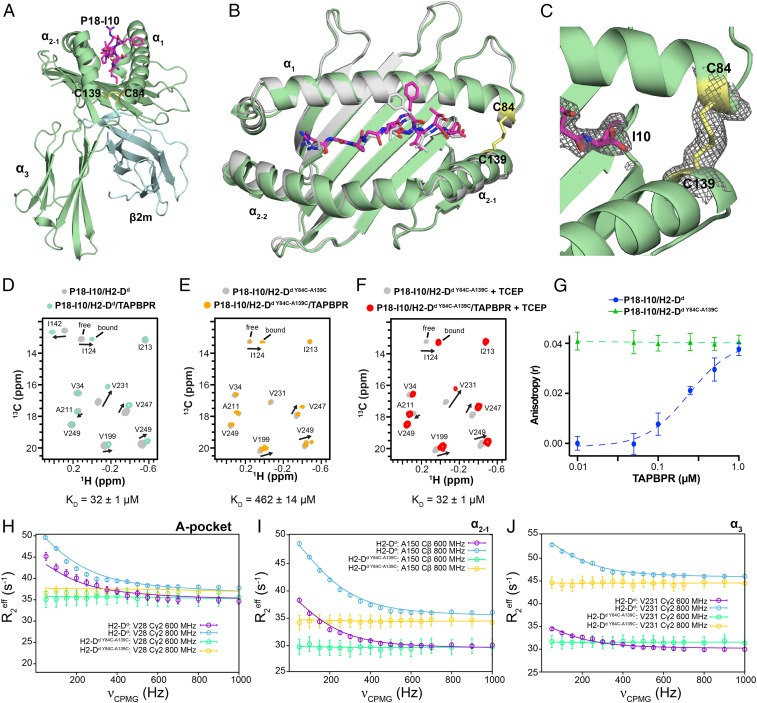
Our data, in conjunction with previous reports (42), provide strong evidence that the conformational plasticity of MHC-I molecules plays a key role in chaperone recognition. TAPBPR binding persists within the cell even in the presence of many disruptive mutations to the α1 domain, demonstrating that nascent MHC-I substrates for TAPBPR may be partially folded, with a high degree of conformational entropy (SI Appendix, Fig. S4 A and B). To further test this model in a controlled manner, we attempted to engineer an MHC-I molecule of restricted groove mobility. Such an engineered variant should also be stable for purification and detailed biophysical characterization in solution. We borrowed a design strategy previously applied to mouse H2-Kb to demonstrate that stabilization of the groove can rescue cell surface expression in a functional tapasin knockout (50, 51). A mutant MHC-I molecule, H2-Dd Y84C-A139C, was prepared with an engineered disulfide between the sidechains of residues 84 and 139 across the F-pocket, which restricts the accessible conformational space for the MHC-I groove. Mouse H2-Dd was chosen as it is closely related to H2-Kb, displays tight TAPBPR affinity in biophysical experiments, and finally because H2-Dd can still traffic to the plasma membrane when overexpressed together with suitable TAPBPR constructs. This property further allows us to distinguish between interactions with nascent intracellular MHC-I versus folded pMHC-I that escapes to the cell surface. By comparison, HLA-A*02:01 is retained internally (possibly in a nascent form) when coexpressed with TAPBPR (SI Appendix, Fig. S3 A and B). Soluble H2-Dd Y84C-A139C, prepared as a complex with the P18-I10 peptide and human β2m, has comparable thermal stability to the wild-type molecule, suggesting that the disulfide does not compromise the pMHC-I structure (SI Appendix, Table S1). Finally, we solved the crystal structure of the P18-I10/H2-Dd Y84C-A139C/hβ2m complex at 2.37-Å resolution (SI Appendix, Table S3, PDB ID 6NPR; ref. 52), which confirmed the presence of the oxidized C84-C139 disulfide bond, bound P18-I10 peptide, and that the structure is very similar to the wild-type molecule (Fig. 5 A–C).
Fig. 5.
Restriction of dynamics in the pMHC-I groove abrogates binding to TAPBPR. (A) View of the P18-I10/H2-Dd Y84C-A139C/hβ2m complex solved at 2.4-Å resolution (PDB ID code 6NPR). The H2-Dd heavy chain is colored green, hβ2m cyan and P18-I10 magenta. The oxidized disulfide bond between C84 and C139 of H2-Dd is shown in yellow. (B) Overlay of the MHC-I groove (residues 1 to 180) and bound P18-I10 peptide for wild-type H2-Dd (PDB ID code 3ECB, gray) and H2-Dd Y84C-A139C (colored as in A). The α3 domain and hβ2m are omitted for clarity. Backbone r.m.s.d. over the entire pMHC-I is 1.4/1.7 Å; (backbone/all-atom). (C) View of the P18-I10/H2-Dd Y84C-A139C structure showing the 2Fo − Fc electron density map at 1.0 σ (gray mesh) around the P18-I10 peptide (magenta) and C84-C139 disulfide bond (yellow). (D–F) Representative 2D 1H-13C HMQC spectra from NMR titrations between TAPBPR and isotopically labeled (at the heavy chain) (D) wild-type P18-I10/H2-Dd/hβ2m, (E) P18-I10/H2-Dd Y84C-A139C/hβ2m, and (F) P18-I10/H2-Dd Y84C-A139C/hβ2m in the presence of 1 mM TCEP. The NMR spectra shown were performed with 3-fold molar excess TAPBPR. Dissociation constants obtained from NMR line shape fitting in TITAN are noted. (G) Fluorescence anisotropy experiments comparing exchange of TAMRA-labeled P18-I10 peptide with wild-type H2-Dd and H2-Dd Y84C-A139C as a function of TAPBPR concentration. (H–J) Comparison of representative 13C-SQ CPMG relaxation dispersion profiles for methyl groups of the heavy chain between wild-type P18-I10/H2-Dd/hβ2m (800 MHz, blue; 600 MHz, purple) and P18-I10/H2-Dd Y84C-A139C/hβ2m (800 MHz, yellow; 600 MHz, green) performed at 25 °C. Both experiments were performed in the presence of 3-fold molar excess P18-I10 peptide.
As predicted by our model, H2-Dd Y84C-A139C exhibits 15-fold lower affinity for TAPBPR relative to wild-type P18-I10/H2-Dd/hβ2m by line-shape analysis of NMR titrations performed using the program TITAN (Fig. 5 D and E). The impaired interaction with TAPBPR is due to the presence of the C84-C139 disulfide, since specific reduction of this exposed disulfide with 1 mM of the mild reducing agent Tris(2-carboxyethyl)phosphine (TCEP), further confirmed by NMR, rescues TAPBPR binding (Fig. 5F). As expected, the methyl chemical shifts of reduced H2-Dd Y84C-A139C/TAPBPR complexes show no observable differences relative to the wild-type sample, suggesting that addition of small amounts of mild reducing agent does not affect the conserved MHC-I disulfide bonds between C101-C164 and C203-C259, located at the buried interface of the α2 helix with the MHC platform and core of the α3 domain, respectively (Fig. 5 D–F). In contrast to wild-type H2-Dd, the disulfide-linked molecule demonstrates high levels of peptide exchange in fluorescence anisotropy experiments performed using a TAMRA-linked version of the P18-I10 peptide, even in the absence of TAPBPR (Fig. 5G), in agreement with recent reports that the same disulfide design approach can stabilize empty MHC-I molecules for a range of allelic compositions (50, 51). Notably, 13C-SQ CPMG relaxation dispersion experiments of P18-I10/H2-Dd Y84C-A139C/hβ2m under the identical NMR conditions to experiments performed using wild-type H2-Dd clearly show that microsecond to millisecond conformational dynamics are quenched throughout the entire MHC-I structure, including the A-pocket, α2–1 helix, and α3 domain regions, which exhibit clear dispersion profiles in the wild-type molecule (Fig. 5 H–J). Together, these results further support our model of TAPBPR recognition through a network of dynamically coupled surfaces spanning multiple domains.
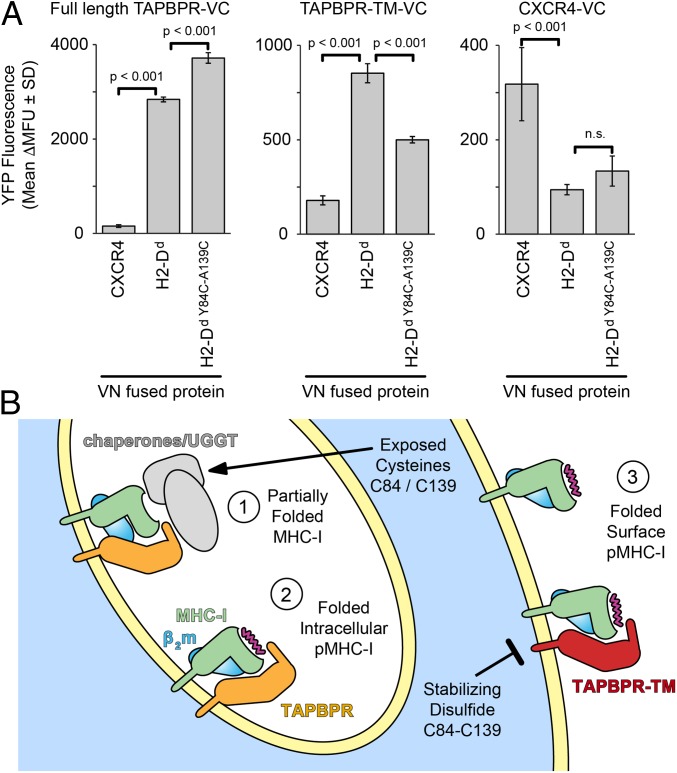
To complement these conclusions with experiments in the native cellular environment, tagged TAPBPR and H2-Dd constructs were coexpressed in human Expi293F cells. H2-Dd Y84C-A139C coimmunoprecipitated with TAPBPR similarly to wild-type H2-Dd (SI Appendix, Fig. S10), despite showing reduced binding in the NMR experiments described above. Whereas binding interactions measured by NMR utilized purified, properly conformed H2-Dd Y84C-A139C in complex with the high-affinity P18-I10 peptide, within the cell nascent, empty MHC-I progressing through the secretory pathway are likely partially folded and exposing reduced cysteines. This effect is further pronounced under our overexpression conditions, leading to increased protein concentrations in the endoplasmic reticulum. To resolve TAPBPR interactions with nascent/misfolded versus properly conformed MHC-I species, we compared wild-type TAPBPR to the TAPBPR-TM variant that can traffic to the plasma membrane, and measured interactions by BiFC. In agreement with the coimmunoprecipitation experiments, H2-Dd Y84C-A139C shows slightly enhanced BiFC with intracellular TAPBPR (Fig. 6 A, Left), presumably due to associations with nascent, partially reduced H2-Dd Y84C-A139C (Fig. 6B) (5). However, when folded H2-Dd Y84C-A139C molecules harboring a stabilizing disulfide are localized to the plasma membrane, association with TAPBPR-TM is substantially reduced, in agreement with our solution NMR results (Fig. 6 A, Center).
Fig. 6.
Restriction of MHC-I dynamics diminishes TAPBPR association on a cell membrane. (A) Cells were cotransfected with VC-fused FLAG-TAPBPR (Left), FLAG-TAPBPR-TM (Center), or FLAG-CXCR4 (Right), and with VN-fused myc-H2-Dd or myc-CXCR4. Yellow fluorescence (shown as average Δmean fluorescence units ± SD, n = 4) was measured after gating for equivalent expression levels. P values are calculated from 2-tailed Student’s t test. n.s., not significant. (B) Nascent MHC-I (step 1) within intracellular compartments associates with quality control machinery. The presence of additional, partially reduced cysteines is hypothesized to enhance chaperone associations, including direct or indirect recruitment of TAPBPR. As MHC-I completes its folding, it may transiently interact with TAPBPR (step 2) before mature pMHC-I traffics to the plasma membrane (step 3). The stabilizing disulfide diminishes direct interactions between folded pMHC-I molecules and TAPBPR.
Discussion
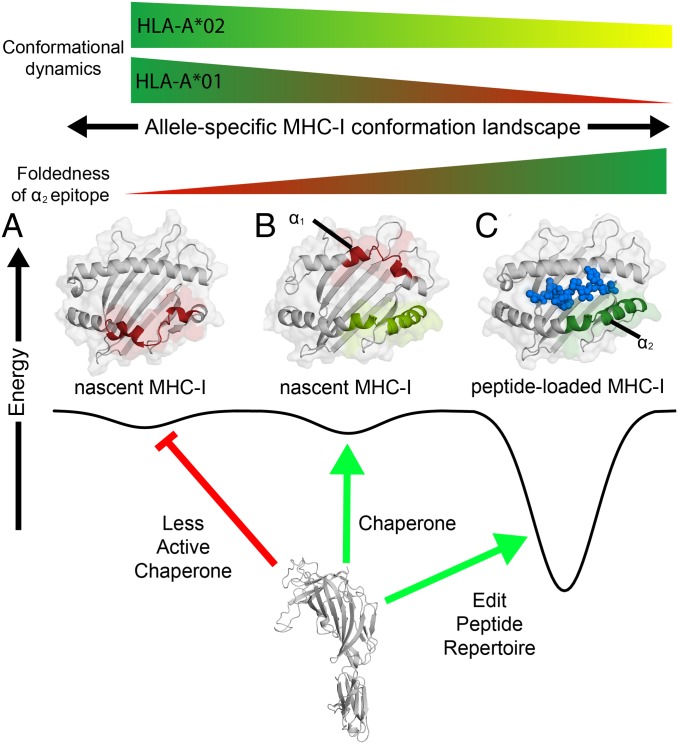
Our combined results suggest that TAPBPR has the capacity for multiple cellular functions: 1) A molecular chaperone of destabilized, empty MHC-I molecules at early stages of their folding process with broad substrate sequence specificity, and 2) a peptide editor acting on properly conformed, peptide-loaded MHC-I molecules with a more exclusive specificity range (Fig. 7). The chaperone function may provide an important protein homeostasis mechanism, by stabilizing misfolded, aggregation-prone MHC-I species that form inside the cell (5, 53, 54) and are known to contribute to the onset of disease (55). This cellular function is often attributed to a class of generic chaperones known as “holdases” that includes essential heat-shock proteins (56). Our deep mutagenesis data suggest that TAPBPR has the capacity to function as a holdase by chaperoning the minimally folded epitope on the MHC α2 domain with a conserved disulfide (Fig. 7), which is further consistent with the observed TAPBPR associations of a large panel of diverse MHC-I sequences and destabilizing mutants tested here. Such a putative holdase function of TAPBPR can be further addressed in protein trafficking studies, aiming to reveal a positive effect on the maturation of MHC-I allotypes whose bound peptides are generally not edited by TAPBPR, such as HLA-A*01:01. The role of the “editing” mode is likely to further scrutinize the peptide cargo of properly conformed MHC-I molecules during their egress to the cell surface. In this manner, TAPBPR helps to “focus” the antigen repertoire toward higher-affinity peptides, such that pMHC-I molecules displayed at the cell surface exhibit sufficient stability and lifetime for immune recognition (4, 5, 57, 58). In contrast to chaperoning activity, the editing function of TAPBPR exhibits a narrow specificity range, where recognition of pMHC-I molecules is mediated by an excited state of 3 to 6% population in solution. This state is attained via a global conformational transition of the pMHC-I molecule as a whole through a network of allosterically coupled sites spanning both ends of the MHC groove and on the α3 domain. While the exact nature of the MHC-I conformations recognized by the chaperone and editing functions of TAPBPR remains to be fully described, our studies provide evidence that dynamics of the α2 domain are important for both recognition events. We propose that the chaperone and editing functions of TAPBPR have the same underlying structural basis. Based on crystal structures, TAPBPR binds to a conformation of the MHC-I α2 domain encompassing a widened groove. Nascent MHC-I molecules will readily sample multiple conformations, including those recognized by TAPBPR. However, as the MHC-I folds and binds to peptides, only MHC-I molecules that retain dynamic motions will sufficiently sample conformations selectively recognized by TAPBPR (Fig. 7). This property of pMHC-I molecules is defined both by the allelic composition of the heavy chain and the bound peptide.
Fig. 7.
Chaperone recognition of a dynamic MHC-I conformational landscape. Conceptual example of the interaction between the chaperone TAPBPR and different MHC-I conformations of varying energetic and structural features. The vertical axis is free energy and the horizontal axis represents the conformational landscape of the MHC-I, which is influenced by specific polymorphisms in the MHC-I groove (α1/α2) and α3 domains. (A) TAPBPR does not associate with an MHC-I state comprising a misfolded α2 domain (red). (B) In the chaperoning function, TAPBPR interacts with nascent MHC-I conformations consisting of a folded α2 domain (light green) with an oxidized disulfide bond between the conserved Cys-101/164, even if the α1 domain remains in a misfolded state (red). (C) As a peptide editor, TAPBPR recognizes properly conformed molecules loaded with peptides toward exchange of the bound peptide cargo, for those alleles that exhibit μs-ms time scale conformational dynamics at the α2–1 helix (green). The peptide in the properly conformed pMHC-I state is shown as blue spheres.
Dynamic sampling of different MHC-I states has been previously suggested to be affected by both the bound peptide and the identity of amino acid sidechains in the D-, E-, and F-pockets (13, 14, 59). Polymorphisms at specific positions—such as 114, 116, 147, and 156—have been shown to confer chaperone recognition of different alleles, due to these sites modulating the conformational flexibility and stability of peptide-deficient MHC-I (9, 59, 60). A sequence alignment of MHC-I molecules that recognize TAPBPR (H2-Dd, H2-Ld, HLA-A*02:01) versus 1 that does not (HLA-A*01:01) leads to no obvious clues to chaperone recognition, as the sites forming D-, E-, and F-pockets for HLA-A*02:01 and HLA-A*01:01 are highly conserved (SI Appendix, Fig. S5). Thus, future MD simulation experiments validated through measurements of dynamics by NMR and other biophysical techniques will be invaluable in understanding the exact molecular determinants that regulate MHC-I dynamics, given that protein dynamics involve transitions between multiple conformational basins with relatively low energy barriers, which can shift drastically in the presence of mutations due to allosteric effects (61).
A limitation of the present study is that the panel of heavy-chain molecules tested has a very limited coverage of the more than 8,000 unique HLA sequences reported in the Immuno Polymorphism Database-International IMmunoGeneTcs Database (IPD-IMGT)/HLA database (62). Additional experiments are needed to establish functionally important trends for TAPBPR across the human HLA allelic landscape, including the HLA-B and HLA-C classes (63). Equally important features of the MHC-I that contribute to the sequence-specificity of chaperone recognition remain to be characterized. First, with respect to the editing function of TAPBPR, bound peptides are known to contribute to the dynamic network on MHC-I structure (28, 64, 65). Second, the β2m light chain contributes to peptide binding, stabilization of the MHC-I groove, and forms direct contacts with TAPBPR (20, 21, 28). Although this is not addressed here, TAPBPR might also chaperone MHC-I molecules prior to their association with β2m, in a similar manner to tapasin (66). In our biophysical characterization of MHC-I/TAPBPR association, both human and mouse MHC-I molecules were prepared with human β2m. It remains unclear how heavy-chain interactions with human versus mouse β2m, known to exhibit differences in affinity for the heavy chain, may modulate activity of the editing mode of TAPBPR (67). Furthermore, MHC-I molecules that are prepared in E. coli lack important glycan modifications present in vivo, which could further contribute to chaperone recognition, either via direct interactions or due to changes in the dynamic landscape of the MHC-I (68, 69). Finally, we have interpreted our cellular data on the basis of direct interactions between TAPBPR and MHC-I, due to their known physical association and because the HLA-A*02:01 mutational landscape is consistent with crystal structures. However, we cannot yet definitively rule out that allele specificity is broadened within the cell due to indirect associations mediated by higher-order chaperone assemblies.
In summary, our data provide insights into the conformational plasticity of different MHC-I molecules by a range of techniques, and suggest a plausible role of these coupled motions in shaping the allelic specificity of TAPBPR recognition. The conserved MHC-I recognition mechanism shared between TAPBPR and tapasin implies that a similar paradigm of dynamically driven recognition may also influence MHC-I allelic dependencies for tapasin in the peptide-loading complex. These findings are a first step in elucidating how molecular chaperones selectively edit the displayed antigen repertoire for different alleles, toward understanding their role in the highly complex associations between HLA polymorphisms and human diseases.
Materials and Methods
Specific details about protein expression and purification, ITC experiments, NMR assignments, chemical-shift mapping and CPMG relaxation dispersion experiments, X-ray crystallography, fluorescence anisotropy, coimmunoprecipitation/immunoblots, and BiFC/deep-sequencing assays are outlined in detail in the SI Appendix.
Data and Materials Availability.
Raw and processed Illumina sequencing data are deposited with National Center for Biotechnology Informaton’s Gene Expression Omnibus under series accession number GSE128957. NMR assignments have been deposited into the Biological Magnetic Resonance Data Bank (http://www.bmrb.wisc.edu) under accession numbers 27249 [H2-Dd], 27682 [H2-Ld], 27632 [HLA-A*01:01] and 27631 [HLA-A*02:01]. The atomic coordinates and structure factors for the P18-I10/H2-Dd Y84C-A139C/β2m complex have been deposited in the Protein Data Bank (https://www.rcsb.org/) under the PDB ID code 6NPR. Plasmids are available from Addgene (https://www.addgene.org).
Supplementary Material
Acknowledgments
This research was supported by NIH Grants 1R01AI129719 (to E.P.) and 1R01AI143997 and 5R35GM125034 (to N.G.S.); and by High End Instrumentation Grant S10OD018455, which funded the 800 MHz NMR spectrometer at the University of California, Santa Cruz.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Raw and processed Illumina sequencing data in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE128957). NMR assignments have been deposited into the Biological Magnetic Resonance Data Bank, http://www.bmrb.wisc.edu (accession nos. 27682 [H2-Ld], 27632 [HLA-A*01:01], and 27631 [HLA-A*02:01]). The atomic coordinates and structure factors for the P18-I10/H2-Dd Y84C-A139C/β2m complex have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 6NPR).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915562116/-/DCSupplemental.
References
- 1.Rock K. L., Reits E., Neefjes J., Present yourself! By MHC class I and MHC class II molecules. Trends Immunol. 37, 724–737 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thomas C., Tampé R., Proofreading of peptide-MHC complexes through dynamic multivalent interactions. Front. Immunol. 8, 65 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neerincx A., Boyle L. H., Properties of the tapasin homologue TAPBPR. Curr. Opin. Immunol. 46, 97–102 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Hermann C., et al. , TAPBPR alters MHC class I peptide presentation by functioning as a peptide exchange catalyst. eLife 4, e09617 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neerincx A., et al. , TAPBPR bridges UDP-glucose:glycoprotein glucosyltransferase 1 onto MHC class I to provide quality control in the antigen presentation pathway. eLife 6, e23049 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle L. H., et al. , Tapasin-related protein TAPBPR is an additional component of the MHC class I presentation pathway. Proc. Natl. Acad. Sci. U.S.A. 110, 3465–3470 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morozov G. I., et al. , Interaction of TAPBPR, a tapasin homolog, with MHC-I molecules promotes peptide editing. Proc. Natl. Acad. Sci. U.S.A. 113, E1006–E1015 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Geironson L., et al. , Tapasin facilitation of natural HLA-A and -B allomorphs is strongly influenced by peptide length, depends on stability, and separates closely related allomorphs. J. Immunol. 191, 3939–3947 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Park B., Lee S., Kim E., Ahn K., A single polymorphic residue within the peptide-binding cleft of MHC class I molecules determines spectrum of tapasin dependence. J. Immunol. 170, 961–968 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Peh C. A., et al. , HLA-B27-restricted antigen presentation in the absence of tapasin reveals polymorphism in mechanisms of HLA class I peptide loading. Immunity 8, 531–542 (1998). [DOI] [PubMed] [Google Scholar]
- 11.Williams A. P., Peh C. A., Purcell A. W., McCluskey J., Elliott T., Optimization of the MHC class I peptide cargo is dependent on tapasin. Immunity 16, 509–520 (2002). [DOI] [PubMed] [Google Scholar]
- 12.Zernich D., et al. , Natural HLA class I polymorphism controls the pathway of antigen presentation and susceptibility to viral evasion. J. Exp. Med. 200, 13–24 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rizvi S. M., et al. , Distinct assembly profiles of HLA-B molecules. J. Immunol. 192, 4967–4976 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sieker F., Springer S., Zacharias M., Comparative molecular dynamics analysis of tapasin-dependent and -independent MHC class I alleles. Protein Sci. 16, 299–308 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abualrous E. T., et al. , F pocket flexibility influences the tapasin dependence of two differentially disease-associated MHC Class I proteins. Eur. J. Immunol. 45, 1248–1257 (2015). [DOI] [PubMed] [Google Scholar]
- 16.Serçinoğlu O., Ozbek P., Computational characterization of residue couplings and micropolymorphism-induced changes in the dynamics of two differentially disease-associated human MHC class-I alleles. J. Biomol. Struct. Dyn. 36, 724–740 (2018). [DOI] [PubMed] [Google Scholar]
- 17.van Hateren A., et al. , Direct evidence for conformational dynamics in major histocompatibility complex class I molecules. J. Biol. Chem. 292, 20255–20269 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kerppola T. K., Design and implementation of bimolecular fluorescence complementation (BiFC) assays for the visualization of protein interactions in living cells. Nat. Protoc. 1, 1278–1286 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler D. M., Fields S., Deep mutational scanning: A new style of protein science. Nat. Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thomas C., Tampé R., Structure of the TAPBPR–MHC I complex defines the mechanism of peptide loading and editing. Science 358, 1060–1064 (2017). [DOI] [PubMed] [Google Scholar]
- 21.Jiang J., Natarajan K., Boyd L. F., Morozov G. I., Mage M. G., Margulies D. H., Crystal structure of a TAPBPR–MHC-I complex reveals the mechanism of peptide editing in antigen presentation. Science, eaao5154 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Procko E., Devlin C. A., Deep mutagenesis of HLA-A*02:01 reveals elements of folded structure necessary for MHC-I-specific chaperone interactions and plasma membrane trafficking. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse128957. Deposited 27 March 2019.
- 23.Hsu V. W., et al. , A recycling pathway between the endoplasmic reticulum and the Golgi apparatus for retention of unassembled MHC class I molecules. Nature 352, 441–444 (1991). [DOI] [PubMed] [Google Scholar]
- 24.Baas E. J., et al. , Peptide-induced stabilization and intracellular localization of empty HLA class I complexes. J. Exp. Med. 176, 147–156 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warburton R. J., et al. , Mutation of the alpha 2 domain disulfide bridge of the class I molecule HLA-A*0201. Effect on maturation and peptide presentation. Hum. Immunol. 39, 261–271 (1994). [DOI] [PubMed] [Google Scholar]
- 26.Kim M. K., Kang Y. K., Positional preference of proline in alpha-helices. Protein Sci. 8, 1492–1499 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bhattacharjee N., Biswas P., Position-specific propensities of amino acids in the β-strand. BMC Struct. Biol. 10, 29 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McShan A. C., et al. , Peptide exchange on MHC-I by TAPBPR is driven by a negative allostery release cycle. Nat. Chem. Biol. 14, 811–820 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balendiran G. K., et al. , The three-dimensional structure of an H-2Ld-peptide complex explains the unique interaction of Ld with beta-2 microglobulin and peptide. Proc. Natl. Acad. Sci. U.S.A. 94, 6880–6885 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang R., Natarajan K., Margulies D. H., Structural basis of the CD8 α β/MHC class I interaction: Focused recognition orients CD8 β to a T cell proximal position. J. Immunol. 183, 2554–2564 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Toor J. S., et al. , A recurrent mutation in anaplastic lymphoma kinase with distinct neoepitope conformations. Front. Immunol. 9, 99 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Khan A. R., Baker B. M., Ghosh P., Biddison W. E., Wiley D. C., The structure and stability of an HLA-A*0201/octameric tax peptide complex with an empty conserved peptide-N-terminal binding site. J. Immunol. 164, 6398–6405 (2000). [DOI] [PubMed] [Google Scholar]
- 33.Hellman L. M., et al. , Differential scanning fluorimetry based assessments of the thermal and kinetic stability of peptide-MHC complexes. J. Immunol. Methods 432, 95–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blom K. F., Larsen B. S., McEwen C. N., Determining affinity-selected ligands and estimating binding affinities by online size exclusion chromatography/liquid chromatography-mass spectrometry. J. Comb. Chem. 1, 82–90 (1999). [DOI] [PubMed] [Google Scholar]
- 35.Natarajan K., et al. , An allosteric site in the T-cell receptor Cβ domain plays a critical signalling role. Nat. Commun. 8, 15260 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McShan A. C., Sgourakis N. G., Kumirov V., Data from “Backbone amide and AILV methyl chemical shift assignments for H2-Dd, a murine class I major histocompatibility molecule heavy chain.” Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/?bmrbId=27249. Accessed 5 November 2018.
- 37.Overall S. A., McShan A. C., Sgourakis N. G., Backbone amide and AILV methyl chemical shift assignments for H2-Ld, a mouse class I major histocompatibility molecule heavy chain. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/?bmrbId=27682. Deposited 5 November 2018.
- 38.Floris-Solid D., McShan A. C., Sgourakis N. G., Backbone amide and AILV methyl chemical shift assignments for HLA-A*01:01, a human class I major histocompatibility molecule heavy chain. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/?bmrbId=27632. Deposited 26 September 2018.
- 39.McShan A., Sgourakis N., Backbone amide and AILV methyl chemical shift assignments for HLA-A*02:01, a human class I major histocompatibility molecule heavy chain. Biological Magnetic Resonance Data Bank. http://www.bmrb.wisc.edu/data_library/summary/?bmrbId=27631. Deposited 26 September 2018.
- 40.Velazquez-Campoy A., Leavitt S. A., Freire E., Characterization of protein-protein interactions by isothermal titration calorimetry. Methods Mol. Biol. 261, 35–54 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Natarajan K., et al. , The role of molecular flexibility in antigen presentation and T cell receptor-mediated signaling. Front. Immunol. 9, 1657 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wieczorek M., et al. , Major histocompatibility complex (MHC) class I and MHC class II proteins: Conformational plasticity in antigen presentation. Front. Immunol. 8, 292 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kleckner I. R., Foster M. P., An introduction to NMR-based approaches for measuring protein dynamics. Biochim. Biophys. Acta 1814, 942–968 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sgourakis N. G., et al. , A novel MHC-I surface targeted for binding by the MCMV m06 immunoevasin revealed by solution NMR. J. Biol. Chem. 290, 28857–28868 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yanaka S., et al. , Peptide-dependent conformational fluctuation determines the stability of the human leukocyte antigen class I complex. J. Biol. Chem. 289, 24680–24690 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lundström P., Vallurupalli P., Religa T. L., Dahlquist F. W., Kay L. E., A single-quantum methyl 13C-relaxation dispersion experiment with improved sensitivity. J. Biomol. NMR 38, 79–88 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Binz A.-K., Rodriguez R. C., Biddison W. E., Baker B. M., Thermodynamic and kinetic analysis of a peptide-class I MHC interaction highlights the noncovalent nature and conformational dynamics of the class I heterotrimer. Biochemistry 42, 4954–4961 (2003). [DOI] [PubMed] [Google Scholar]
- 48.Harndahl M., et al. , Peptide-MHC class I stability is a better predictor than peptide affinity of CTL immunogenicity. Eur. J. Immunol. 42, 1405–1416 (2012). [DOI] [PubMed] [Google Scholar]
- 49.Axelsson-Robertson R., et al. , Extensive major histocompatibility complex class I binding promiscuity for Mycobacterium tuberculosis TB10.4 peptides and immune dominance of human leucocyte antigen (HLA)-B*0702 and HLA-B*0801 alleles in TB10.4 CD8 T-cell responses. Immunology 129, 496–505 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hein Z., et al. , Peptide-independent stabilization of MHC class I molecules breaches cellular quality control. J. Cell Sci. 127, 2885–2897 (2014). [DOI] [PubMed] [Google Scholar]
- 51.Hein Z., Borchert B., Abualrous E. T., Springer S., Distinct mechanisms survey the structural integrity of HLA-B*27:05 intracellularly and at the surface. PLoS One 13, e0200811 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toor J. S., McShan A. C., Tripathi S. M., Sgourakis N. G., Crystal structure of H-2Dd with C84-C139 disulfide in complex with gp120 derived peptide P18-I10. RCSB Protein Data Bank. https://www.rcsb.org/structure/6NPR. Deposited 18 January 2019.
- 53.Schoenhals G. J., et al. , Retention of empty MHC class I molecules by tapasin is essential to reconstitute antigen presentation in invertebrate cells. EMBO J. 18, 743–753 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Garstka M. A., et al. , Tapasin dependence of major histocompatibility complex class I molecules correlates with their conformational flexibility. FASEB J. 25, 3989–3998 (2011). [DOI] [PubMed] [Google Scholar]
- 55.Colbert R. A., Tran T. M., Layh-Schmitt G., HLA-B27 misfolding and ankylosing spondylitis. Mol. Immunol. 57, 44–51 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Genest O., Wickner S., Doyle S. M., Hsp90 and Hsp70 chaperones: Collaborators in protein remodeling. J. Biol. Chem. 294, 2109–2120 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ilca F. T., et al. , TAPBPR mediates peptide dissociation from MHC class I using a leucine lever. eLife 7, e40126 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Purcell A. W., et al. , Quantitative and qualitative influences of tapasin on the class I peptide repertoire. J. Immunol. 166, 1016–1027 (2001). [DOI] [PubMed] [Google Scholar]
- 59.Badrinath S., Kunze-Schumacher H., Blasczyk R., Huyton T., Bade-Doeding C., A micropolymorphism altering the residue triad 97/114/156 determines the relative levels of tapasin independence and distinct peptide profiles for HLA-A(*)24 allotypes. J. Immunol. Res. 2014, 298145 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bailey A., et al. , Selector function of MHC I molecules is determined by protein plasticity. Sci. Rep. 5, 14928 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo J., Zhou H.-X., Protein allostery and conformational dynamics. Chem. Rev. 116, 6503–6515 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Robinson J., et al. , The IPD and IMGT/HLA database: Allele variant databases. Nucleic Acids Res. 43, D423–D431 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ilca F. T., Drexhage L. Z., Brewin G., Peacock S., Boyle L. H., Distinct polymorphisms in HLA class I molecules govern their susceptibility to peptide editing by TAPBPR. Cell Reports 29, 1621–1632 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ayres C. M., Corcelli S. A., Baker B. M., Peptide and peptide-dependent motions in MHC proteins: Immunological implications and biophysical underpinnings. Front. Immunol. 8, 935 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hawse W. F., et al. , Peptide modulation of class I major histocompatibility complex protein molecular flexibility and the implications for immune recognition. J. Biol. Chem. 288, 24372–24381 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizvi S. M., Raghavan M., Direct peptide-regulatable interactions between MHC class I molecules and tapasin. Proc. Natl. Acad. Sci. U.S.A. 103, 18220–18225 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pedersen L. O., et al. , The interaction of beta 2-microglobulin (beta 2m) with mouse class I major histocompatibility antigens and its ability to support peptide binding. A comparison of human and mouse beta 2m. Eur. J. Immunol. 25, 1609–1616 (1995). [DOI] [PubMed] [Google Scholar]
- 68.Neerincx A., Boyle L. H., Preferential interaction of MHC class I with TAPBPR in the absence of glycosylation. Mol. Immunol. 113, 58–66 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mandal T. K., Mukhopadhyay C., Effect of glycosylation on structure and dynamics of MHC class I glycoprotein: A molecular dynamics study. Biopolymers 59, 11–23 (2001). [DOI] [PubMed] [Google Scholar]
- 70.Song Y., et al. , High-resolution comparative modeling with RosettaCM. Structure 21, 1735–1742 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.