Significance
Efficient microbial synthesis in challenging pathways relies on dynamic regulation of multiple metabolic fluxes to balance several competing goals. To address these situations, we developed an autonomous, pathway-independent, and layered regulation tool. By incorporating parts from 2 different quorum-sensing systems, the layers of our system can be tuned independently to ensure generalizability. Application of the regulation system to overcoming 2 different sets of challenges in the naringenin and salicylic acid pathways resulted in significant improvements in titer, demonstrating that the system is an effective tool for improving pathway production.
Keywords: synthetic biology, metabolic engineering, dynamic control
Abstract
Metabolic engineering seeks to reprogram microbial cells to efficiently and sustainably produce value-added compounds. Since chemical production can be at odds with the cell’s natural objectives, strategies have been developed to balance conflicting goals. For example, dynamic regulation modulates gene expression to favor biomass and metabolite accumulation at low cell densities before diverting key metabolic fluxes toward product formation. To trigger changes in gene expression in a pathway-independent manner without the need for exogenous inducers, researchers have coupled gene expression to quorum-sensing (QS) circuits, which regulate transcription based on cell density. While effective, studies thus far have been limited to one control point. More challenging pathways may require layered dynamic regulation strategies, motivating the development of a generalizable tool for regulating multiple sets of genes. We have developed a QS-based regulation tool that combines components of the lux and esa QS systems to simultaneously and dynamically up- and down-regulate expression of 2 sets of genes. Characterization of the circuit revealed that varying the expression level of 2 QS components leads to predictable changes in switching dynamics and that using components from 2 QS systems allows for independent tuning capability. We applied the regulation tool to successfully address challenges in both the naringenin and salicylic acid synthesis pathways. Through these case studies, we confirmed the benefit of having multiple control points, predictable tuning capabilities, and independently tunable regulation modules.
Metabolic engineering seeks to reprogram cells with the goal of producing value-added chemicals that are well-suited to microbial synthesis. Such chemicals vary widely, including ones used in biofuels, plastics, and pharmaceuticals. Enzymes that produce stereochemically pure products simplify downstream separation requirements in fine chemical production. For commodity chemical production, microbial synthesis may allow for utilization of inexpensive or renewable feedstocks. Additionally, microbial synthesis typically occurs under mild conditions, offering safety, energy conservation, and environmental benefits.
Challenges that limit efficient production can arise when heterologous pathways are imported into host organisms. For example, flux through heterologous pathways may inhibit cell growth by generating toxic products or consuming essential endogenous metabolites. Strategies for overcoming these challenges focus on balancing growth and production through controlling reaction fluxes. The static balancing approach aims to maintain fixed reaction fluxes in an intermediate regime such that neither objective dominates, whereas the dynamic approach generally regulates gene expression to favor accumulating biomass or key metabolites early, before diverting metabolic fluxes toward product formation. While more difficult to implement in practice, dynamic control is required for efficient production in some pathways.
Successful dynamic flux regulation strategies have been experimentally demonstrated in a number of production pathways by controlling activity of key pathway enzymes at the transcriptional or posttranslational level. Many recent studies have focused on self-actuating dynamic control methods to minimize required human supervision and to avoid use of exogenous inducers, which can be costly. These control systems couple expression of pathway genes, antisense RNA, CRISPRi components, or proteases to relevant conditions such as external signals (1), internal cell state (metabolites, growth state, and stress state) (2–10), cell density (11–13), glucose concentration (14, 15), or a combination of these (16–18). Control systems which respond to pathway-independent signals such as cell density or O2 level offer the additional advantage of applicability across different synthesis pathways without development of a new sensor for the relevant metabolite in each pathway. However, to our knowledge, there has not yet been a fully pathway-independent dynamic control system for independently regulating multiple metabolic fluxes, which may be required for efficient production of more challenging pathways.
With the goal of developing such a control system, we constructed a circuit containing genetic components from the lux and esaR quorum-sensing (QS) systems. This system contains 2 constitutively expressed genes for the regulator proteins, LuxR and EsaR, which activate and repress the Plux and PesaR promoters, respectively, upon binding. The binding affinity between the regulators and their cognate promoters depends on the level of a common signaling molecule, 3-oxohexanoyl homoserine lactone (AHL), and thus we can dynamically regulate the transcription level of the promoters in a cell density-dependent manner by constitutively expressing the gene encoding the AHL synthase, esaI. To ensure this system can be used to explore a broad metabolic control space, we varied the luxR and esaI expression levels to obtain a range of switching dynamics. The engineered regulatory circuits were applied toward controlling metabolic fluxes in 2 different synthesis pathways with unique trade-offs and metabolic control points. The significant improvement in product titers upon implementation of the control system in both case studies demonstrates the effectiveness of the control circuit for balancing multiple design objectives in synthesis pathways.
Results
Characterization of QS-Based Autonomous Induction.
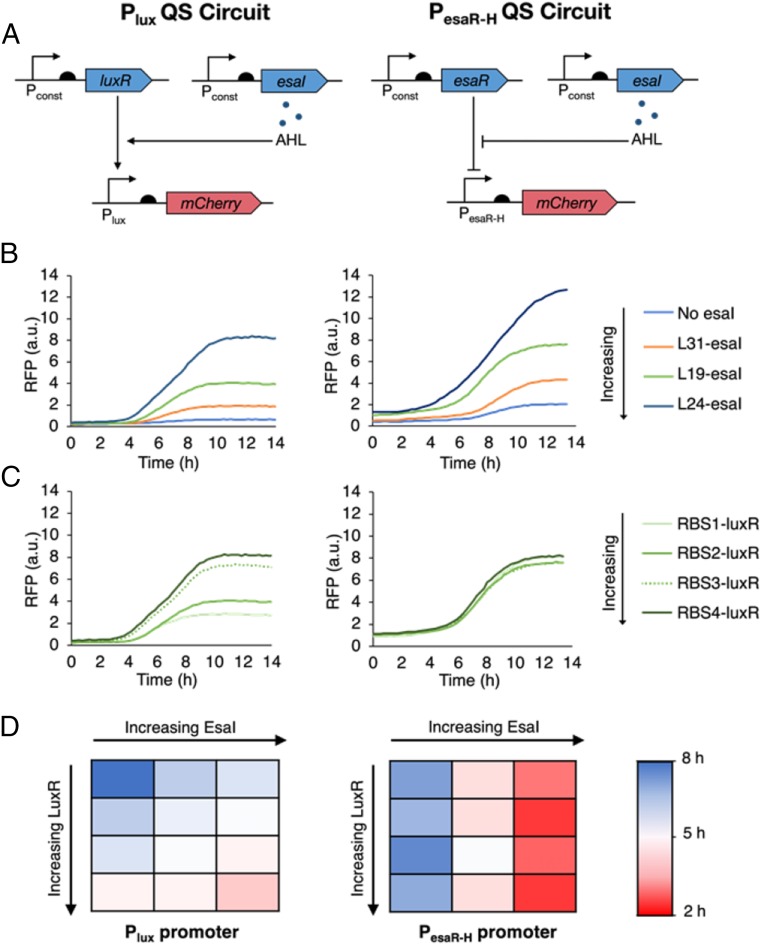
We characterized 2 QS circuits for autonomous and dynamic gene expression control. The first QS circuit uses the transcriptional regulator LuxR, which forms a complex with AHL to activate transcription from the Plux promoter (Fig. 1 A, Left). For the second QS circuit, we constructed a hybrid promoter (PesaR-H) that contains an EsaR binding site (esaO) downstream of the transcription start site of the Ptrc promoter. In the absence of AHL, EsaR binds to the esaO sequence, repressing transcription from PesaR-H. Upon binding to AHL, EsaR can no longer bind to the operator sequence, leading to derepression (Fig. 1 A, Right). Each circuit can be used to dynamically up-regulate the expression of any gene of interest by placing the gene downstream of the Plux or PesaR-H promoters.
Fig. 1.
Overview of lux and esaR QS circuits. (A) Architecture of the Plux (Left) and PesaR-H (Right) QS circuits. Transcription is activated from the Plux promoter when AHL-bound LuxR binds to the promoter. EsaR binds to the PesaR-H promoter to block transcription, and this repression is relieved in the presence of AHL. Here, the arrow and semicircle represent the promoter and RBS, respectively. (B) Representative fluorescence curves showing the response of Plux (Left) and PesaR-H (Right) to varying esaI expression levels. Increasing esaI expression levels results in earlier switching from both promoters. (C) Representative fluorescence curves showing the response of Plux (Left) and PesaR-H (Right) to varying luxR expression level. Increasing luxR expression results in earlier switching from the Plux promoter only. (D) Summary of the trends in switching time from the Plux and PesaR-H promoters varying luxR and esaI expression. Switching time was defined as the time at which fluorescence signal first surpasses a value equal to 90% of the maximum signal from the latest switcher (i.e., the lowest signal).
Gupta et al. (13) showed that the rate of AHL accumulation can be controlled by varying the constitutive expression level of the gene for the AHL synthase, esaI. By changing the AHL accumulation rate, we can tune the switching dynamics of these circuits. When applied to regulating enzyme expression, this tunability corresponds with the ability to vary the schedule of metabolic flux regulation in search for one that suits the desired application. To characterize relative switching dynamics from the Plux and PesaR-H promoters, esaI was integrated into BL21(DE3) under a library of promoter and ribosome binding site (RBS) variants to make the BL21-LXX strain series. A Plux or PesaR-H promoter driving mCherry expression on a medium-copy plasmid (pCOLA-Plux-mCherry or pCOLA-PesaR-H-mCherry) was introduced into the BL21-LXX strain series. Strains testing the Plux promoter required an additional plasmid with the luxR gene constitutively expressed from an Anderson library promoter (BBa_J23114) and varying RBSs on a low-copy plasmid (pACYC-Pcon-RBSX-luxR). The pCOLA-PesaR-H-mCherry vector contains esaR constitutively expressed from a BIOFAB library promoter (apFAB104).
Continuous fluorescence measurements of these strains produced a range of switching times in the up-regulation mode. In general, increasing the esaI expression level leads to earlier switching with both promoters (Fig. 1B) and increasing the luxR expression level results in earlier switching from the Plux promoter only (Fig. 1C). These trends are consistent with expectations based on our understanding of the interactions in the QS circuits (Fig. 1D).
Dynamic Gene Regulation to Control Flux through the Naringenin Pathway.
Naringenin is a natural plant-produced compound that is a common precursor of most flavonoids, natural plant products with a number of desirable therapeutic characteristics including anticancer and antiviral activity (19–21). One naringenin production pathway uses 4 enzymatic reactions to convert l-tyrosine and malonyl-CoA to (2S)-naringenin (Fig. 2A). This pathway has been widely studied in Escherichia coli as a model system with relatively well-characterized challenges that may be addressed through dynamic control (22–26). For this reason, we applied our regulation system toward alleviating the limitations of the naringenin pathway.
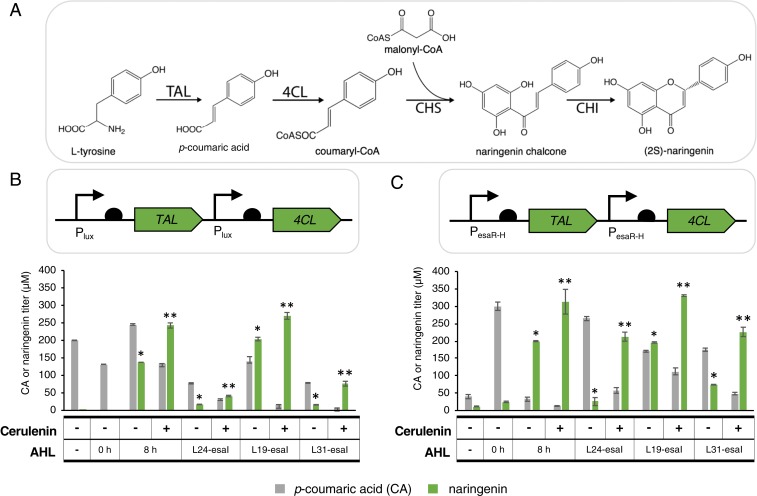
Fig. 2.
Preliminary characterization of the naringenin pathway to confirm the rationale for dual regulation. (A) The naringenin pathway uses 4 heterologous enzymes—TAL, 4CL, CHS, and CHI—to convert l-tyrosine and malonyl-CoA to (2S)-naringenin. Each mole of naringenin requires 1 mol of l-tyrosine and 3 mol of malonyl-CoA. (B) p-coumaric acid and naringenin titers with TAL and 4CL expression controlled under the Plux promoter. Static expression of TAL and 4CL at the leaky expression level (AHL −) or at the fully induced expression level (AHL 0 h) results in low naringenin titers. Exogenous induction of TAL and 4CL expression during midexponential phase (AHL 8 h) improves naringenin titers more than 6-fold; EsaI-mediated induction is able to match that improvement. Addition of cerulenin to increase malonyl-CoA pools results in increased naringenin titers and a decrease in p-coumaric acid titers in all dynamic strains. (C) p-coumaric acid and naringenin titers with TAL and 4CL expression controlled under the PesaR-H promoter. In general, all major trends follow those observed with TAL and 4CL under Plux control. Error bars represent SD of triplicate trials. *P < 0.01 compared to static controls by 2-tailed t test. **P < 0.01 compared to no cerulenin sample at same EsaI level by 2-tailed t test.
Results from previous studies suggest that efficient naringenin production relies on maintaining high levels of chalcone synthase (CHS) and chalcone isomerase (CHI) relative to tyrosine ammonia lyase (TAL) and 4-coumaryl-CoA ligase (4CL), possibly due to an inhibitory interaction of coumaryl-CoA against TAL (22). This balance can be reached by delaying expression of TAL and 4CL, while constitutively expressing CHS and CHI (22). To confirm that QS-based regulation can achieve this balancing effect, TAL and 4CL were expressed from Plux or PesaR-H promoters (pCOLA-Plux-TAL- 4CL or pCOLA-PesaR-H-TAL- 4CL) while CHI and CHS were each expressed from T7 promoters (pET-CHI-CHS), with isopropyl β-d-1-thiogalactopyranoside (IPTG) added at inoculation. These plasmids, along with one that constitutively expresses luxR (pACYC-Pcon-RBSX-luxR), were transformed into the BL21-LXX strain series to produce a set of strains that dynamically up-regulate TAL and 4CL expression at varying cell densities.
Comparison of naringenin titers from dynamically and statically controlled strains confirmed that the naringenin pathway benefits from dynamic regulation of TAL and 4CL expression under both the Plux and PesaR-H promoters (Fig. 2 B and C). With TAL and 4CL expression under the Plux promoter, static expression strains (i.e., with exogenous AHL added at inoculation) produced less than 10 μM naringenin, significantly less than the 204 ± 5 μM naringenin produced from the top strain with autonomous dynamic TAL and 4CL regulation. Similarly, static TAL and 4CL expression controlled from PesaR-H promoters produced less than 30 μM naringenin, also significantly less than the 196 ± 2 μM produced from the top autonomous dynamic strain. In both the Plux- and PesaR-H-controlled systems, an intermediate esaI expression level resulted in the maximum naringenin titer. This trend agrees with our current understanding of the pathway as early-switching strains might be subject to TAL inhibition and late-switching strains might be limited by low pathway fluxes. At the intermediate esaI level, naringenin titers from both QS circuits matched or exceeded those obtained with exogenous AHL addition, suggesting that the autonomous switching strategy can successfully replace exogenous inducer addition in this context.
Previous studies have suggested that naringenin production in E. coli is additionally limited by low endogenous malonyl-CoA levels (22, 25, 26). To confirm that this limitation exists in our system, all strains were cultured with and without cerulenin, an inhibitor of fatty acid synthesis that is known to elevate malonyl-CoA levels (27). Naringenin titers improved in all strains when cultured with cerulenin as expected, confirming that malonyl-CoA pools are limiting (Fig. 2 B and C). In an effort to improve the naringenin titers using the Plux-control system to match the top titer using the PesaR-control system, we explored additional Plux switching dynamics by testing additional esaI and luxR expression levels. However, none resulted in improved naringenin titers (SI Appendix, Figs. S1 and S2) and therefore we decided to further build upon the regulation scheme with the PesaR-H promoter controlling TAL and 4CL expression.
Dynamic Down-Regulation of Endogenous Gene Expression for Malonyl-CoA Accumulation Improves Naringenin Titers.
While effective, cerulenin is not a cost-efficient solution for increasing malonyl-CoA pools, and thus we aimed to dynamically silence gene expression by using the Plux promoter to drive expression of dCas9 and single guide RNA(s) (sgRNA) targeted toward dynamically downregulating the gene(s) of interest. To characterize the down-regulation behavior, 2 Plux promoters driving dCas9 and sgGFP expression on low- and medium-copy plasmids, respectively (pACYC-Plux-dCas9-Pcon10-luxR and pCDF-Plux-sgGFP), were introduced into the BL21-LXX strain series along with a plasmid that expresses degradation-tagged GFP (pTrc-GFP-LVA) (SI Appendix, Fig. S3A). In general, silencing dynamics from the Plux promoter follow the expected trends based on the previous characterization of the lux circuit. That is, increasing esaI or luxR expression level leads to earlier dCas9 and sgRNA expression and earlier down-regulation of the green fluorescent protein (GFP) signal (SI Appendix, Fig. S3 B and C).
dCas9 and sgRNAs targeted toward endogenous genes were expressed from the Plux promoter (Fig. 3C), with combinations of target genes chosen based on results from a previous study that used CRISPRi to elevate malonyl-CoA pools by downregulating competing acetyl-CoA–consuming reactions and fatty acid synthesis cycle reactions (SI Appendix, Fig. S4) (26). The Plux-sgRNA expression cassettes were expressed from the pCDF vector backbone and the Plux-dCas9 cassette and Pcon-luxR variants were expressed from pACYC. To ensure that the down-regulation module was having the desired effects, we first confirmed that transcript levels of the target gene are down-regulated in an EsaI level-dependent manner (SI Appendix, Fig. S5). We additionally confirmed that the decreased transcript levels resulted in elevated malonyl-CoA levels by utilizing a previously developed fluorescence-based sensor (8) (SI Appendix, Fig. S6).
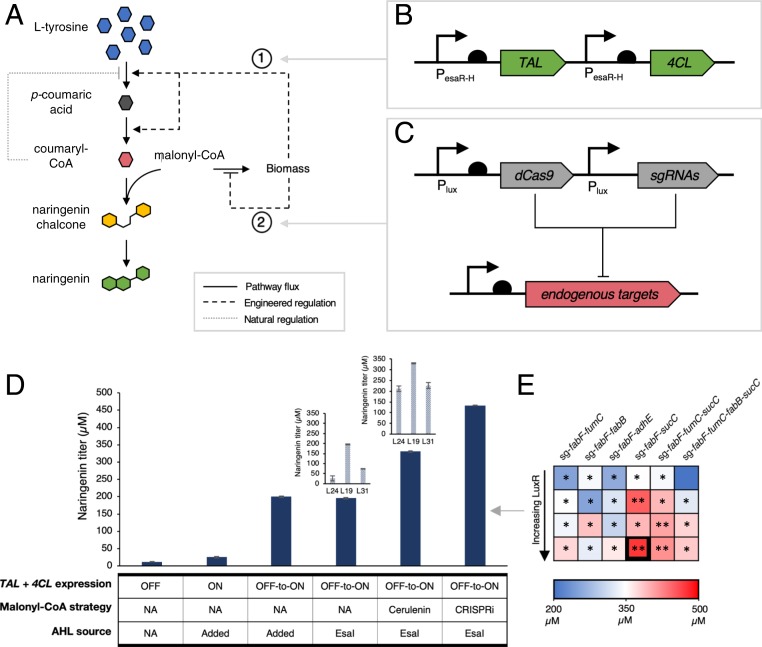
Fig. 3.
Dual regulation in the naringenin pathway. (A) Schematic of the regulatory strategy. Increasing cell density triggers 2 dynamic gene expression switches, one that up-regulates TAL and 4CL expression and a second that down-regulates expression of endogenous genes that are associated with malonyl-CoA accumulation. Dotted lines represent QS circuit responses and solid lines represent metabolic reaction fluxes. (B) Diagram of the TAL and 4CL up-regulation module responsible for actuating the engineered response labeled 1. TAL and 4CL are each under their own PesaR-H promoters. In the presence of constitutive EsaR and EsaI, expression from the PesaR-H promoter turns ON with increasing cell density. (C) Diagram of down-regulation module responsive for actuating the engineered response labeled 2. dCas9 and each sgRNA are expressed from their own Plux promoters. In the presence of constitutive LuxR and EsaI, expression of the target genes turns OFF with increasing cell density. (D) Naringenin titers with different regulatory schemes. Naringenin titers increase 6-fold over static strategies when TAL and 4CL expression are induced autonomously or with the addition of exogenous AHL. Cerulenin treatment improves naringenin titers in all esaI backgrounds and addition of the CRISPRi-mediated down-regulation module results in naringenin titers that are 40% higher than the top cerulenin-treated strain. (E) Heat map indicating naringenin titers with varying LuxR levels and down-regulation target genes. The square corresponding to the top producer is bolded. *P < 0.01 compared to TAL and 4CL controlled strain in L19 background. **P < 0.05 compared to TAL and 4CL controlled strain in L19 background with cerulenin. Error bars indicate SD of triplicate trials.
The plasmids harboring the Plux down-regulation module (Fig. 3C) were then imported into the naringenin-producing strains which control TAL and 4CL expression under the PesaR-H promoter (Fig. 3B). Based on the previous observation that the L19-esaI expression level resulted in the highest naringenin titers under all conditions, we combinatorically tested LuxR levels and sgRNA target genes in the BL21-L19 strain background. Under this dual-regulation scheme (Fig. 3 A–C), increasing cell density leads to dynamic down-regulation of the sgRNA target genes and dynamic up-regulation of coumaryl-CoA–producing reactions. The library of LuxR levels and sgRNA targets resulted in a set of strains that produced varying naringenin titers (Fig. 3E and SI Appendix, Fig. S7). Every strain with dual regulation resulted in higher naringenin titers compared to the nonspecific sgRNA control and half of the library resulted in higher naringenin titers compared to the cerulenin-treated nonspecific sgRNA control, suggesting that the down-regulation module can effectively replace cerulenin addition in this context (Fig. 3D and SI Appendix, Fig. S7). The top producer identified through this screen yields 463 ± 1 μM naringenin, 140% higher than the strain with only the up-regulation module and 40% higher than the cerulenin-treated strain. Fermentation of the top producer at a bench-top bioreactor scale confirms that the improvement achieved through dual-regulation holds across fermentation scales (SI Appendix, Fig. S8).
Dual Regulation for Salicylic Acid Production.
To test the generalizability of the regulatory circuits, we sought to apply the regulation strategy to a different production pathway that might also benefit from 2 points of dynamic control. One such pathway is the salicylic acid production pathway, which converts endogenous chorismate to salicylic acid using the enzymes isochorismate synthase (Ics) and isochorismate pyruvate lyase (Ipl), produced from the genes entC and pchB, respectively (Fig. 4A). Since salicylic acid can burden growth of E. coli, we hypothesized that production could be improved by delaying expression of pathway genes to manage the trade-off between growth and production. Additionally, chorismate is naturally consumed by the cell to produce the aromatic amino acids (AAA) such that knocking out the consumption reactions to divert chorismate toward salicylic acid production results in an auxotrophic strain that requires aromatic amino acid supplementation. To create a salicylic acid producer that does not rely on amino acid supplementation, we used the dual-regulation system to (1) dynamically up-regulate entC and pchB expression to alleviate the growth burden and (2) dynamically down-regulate pheA and tyrA expression to divert chorismate pools toward the salicylic acid production pathway without introducing auxotrophies.
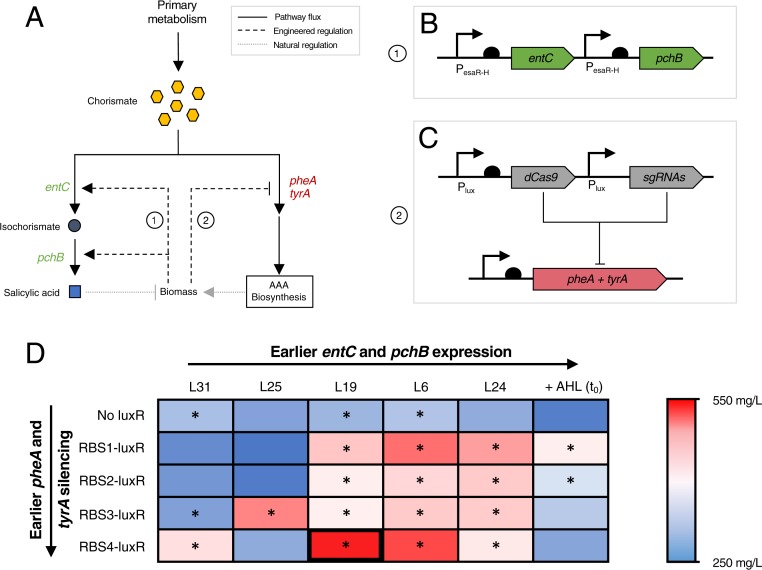
Fig. 4.
Dual regulation in the salicylic acid pathway. (A) Schematic of the regulatory strategy. The genes encoding the salicylic acid pathway enzymes, entC and pchB, are expressed with increasing cell density to balance growth and generation of salicylic acid, a toxic product. pheA and tyrA are silenced with increasing cell density to elevate chorismate levels without introducing an auxotrophy. (B) Diagram of the up-regulation module. entC and pchB are under control of the PesaR-H promoter such that increasing cell density results in increased expression. (C) Diagram of the down-regulation module. dCas9 and sgRNAs targeted toward silencing pheA and tyrA are expressed from the Plux promoter. (D) Heat map of salicylic acid titers at varying EsaI and LuxR expression levels. Values represent the mean of triplicate trials. The rectangle corresponding to the top producer is bolded. *P < 0.01 compared to constitutive entC and pchB control with no AAA pathway down-regulation.
To implement the proposed control strategy, entC and pchB were expressed under the PesaR-H promoter and dCas9 and sgRNAs targeted toward pheA and tyrA were expressed from the Plux promoter. The PesaR-H-entC, PesaR-H-pchB, and Pcon-esaR cassettes were expressed from the pCOLA-duet backbone (pCOLA-PesaR-H-entC-pchB; Fig. 4B). dCas9 was expressed from the same plasmids as in the naringenin experiments (pACYC-Plux-dCas9-Pcon-RBSX-luxR) and the Plux- sgRNA cassette was expressed from the pCDF backbone (pCDF-Plux-sg-pheA-tyrA; Fig. 4C). We tested this set of plasmids in a phenylalanine producer strain background that is commonly used for salicylic acid production (NST74) (28), with genomically integrated esaI (NST74-LXX). Under the proposed regulatory scheme, we can tune the up- and down-regulation modules by varying EsaI level and can tune the down-regulation module only by varying the LuxR level. Rather than holding expression of one of the QS components (EsaI or LuxR) constant in the optimization, we decided to combinatorically vary both since we were only interested in one combination of down-regulation targets in this context.
Dynamically controlling entC and pchB expression only and testing salicylic acid production over a range of esaI expression levels resulted in a maximum observed salicylic acid titer of 291 ± 3 mg/L, a 10% improvement over that static strain with entC and pchB induced at inoculation. Addition of the down-regulation module and exploration of EsaI and LuxR levels resulted in a strain that produced 520 ± 7 mg/L salicylic acid, a 1.8-fold improvement over the static strain (Fig. 4D and SI Appendix, Fig. S9). This application demonstrates the generalizability of our control tool, confirming that independent control of 2 different targets—or sets of targets—can significantly improve production in some pathways.
Conclusions
Dynamic regulation is an important strategy in metabolic engineering for improving production in challenging pathways. We have developed an autonomous, pathway-independent, tunable, and bifunctional gene expression regulation system that can be applied to metabolic flux control. The system was applied to controlling key heterologous and endogenous enzymes in the naringenin and salicylic acid pathways, resulting in production benefits in both case studies. Our work demonstrates the importance of having 2 independently tunable modes of control for dynamic regulation to effectively manage trade-offs and builds on the collection of tools available for developing industrially feasible microbial production strains.
Materials and Methods
All strains and plasmids used in this study are summarized in SI Appendix, Tables S1 and S2, respectively. Sequences for promoters and RBS sequences are provided in SI Appendix, Table S3, the codon-optimized sequences for TAL, 4CL, CHS, and CHI are provided in SI Appendix, Table S4, and guide RNA sequences are provided in SI Appendix, Table S5. For plasmid construction and gene/genome editing, cells were cultured in Luria–Bertani (LB) broth at either 30 °C or 37 °C. Temperature-sensitive plasmids were cured at 42 °C.
Strain Construction.
Synthase expression library integrations.
The esaI expression cassette was integrated into the genome under the control of several different constitutive synthetic promoters (denoted BL2-LXX or NST74-LXX) (13). Integration was performed via “clonetegration” (29). The desired EsaI expression cassette was inserted into the pOSIP-KO backbone using restriction digestion and ligation. The ligation product was used to transform E. coli strain BL21(DE3) or NST74 for integration into the 186 locus. The phage integration genes and antibiotic resistance cassette were cured by transforming with a plasmid containing FLP under control of the Ptet promoter (pTet-FLP), yielding strains BL21-LXX or NST-LXX.
Fluorescence characterization of QS circuits.
The Plux promoter was amplified from pSB1A2-Plux-GFP (30) using primers CD_211 and CD_212, mCherry was amplified from pFM301 (p15A ori, kanamycin resistance, mCherry-BBa_J06504 under constitutive promoter BBa_J23101) using primers CD_213 and CD_214, and the 2 products were joined using splicing by overlap extension PCR to yield Plux-mCherry. This cassette was inserted into a modified pCOLA backbone without the T7 system components using restriction digestion and ligation to yield pCOLA-Plux-mCherry. The Pcon10-luxR cassette was amplified from pSB1A2-Plux-GFP-Pcon10-luxR (30) using primers CD_215 and CD_216 and inserted into the pACYC backbone using restriction digestion and ligation to yield pACYC-Pcon10-luxR. The strength of the RBS driving luxR was decreased using primers lux_R1_a and luxR_R1_b and increased using primers luxR_R3_a, luxR_R3_b, luxR_R4_a, and luxR_R4_b, through Golden Gate cloning.
The Ptrc promoter was amplified from the pTrc99A vector with the esaO operator sequence in place of the lacO sequence to yield the Ptrc-esaO fragment (PesaR-H) using primers esa_RFP_5 and esa_RFP_6. mCherry was amplified from pFM301 using esa_RFP_7 and esa_RFP8, esaR from a constitutive BIOFAB library promoter was amplified from pSB3K3-PesaR-GFP-Pcon-esaR (31) using esa_RFP_3 and esa_RFP_4, and the pCOLA backbone was amplified using esa_RFP_1 and esa_RFP_2. These PCR products were assembled into the vector pCOLA-PesaRH-mCherry using Golden Gate cloning.
A custom synthesized gene fragment which contains BsaI restriction sites followed by the sgRNA scaffold sequence, flanked on both sides with bidirectional terminator B0015 (Genscript), was amplified using primers sg_3 and sg_10. The pCDF backbone was amplified using primer sg_1 and sg_2. These 2 PCR products were combined to make pCDF-BsaI-BsaI-sgRNA using Golden Gate cloning. The 20-bp guide sequence targeting GFPmut3b was appended to Plux using overhang PCR using the template pSB1A2-Plux-GFP and primers A11 and A12 and inserted into pCDF-BsaI-BsaI-sgRNA using Golden Gate cloning to yield pCDF-Plux-sgGFP.
dCas9 was amplified from pdCas9 (32) with SapI sites preceding the gene using primers C9_1 and C9_2 and inserted into the pACYC backbone using restriction digestion and ligation to yield pACYC-SapI-SapI-dCas9. The Plux promoter was PCR-amplified from pSB1A2-Plux-GFP (30) using primers A15 and A16, which add flanking SapI sites, and inserted into pACYC-SapI-SapI-dCas9 using Golden Gate cloning to yield pACYC-Plux-dCas9. The RBS variants of the luxR cassette were amplified using C9_lux_3 and C9_lux_4 and the pACYC-Plux-dCas9 plasmid was amplified using C9_lux_1 and C9_lux_2. The resulting products were assembled using Golden Gate cloning to yield pACYC-Plux-dCas9-Pcon-RBSX-luxR.
Malonyl-CoA biosensor.
The lacI-PT7-fapR cassette was amplified from the pCDM4-fapR plasmid using primers fapR_3 and fapR_4 and the pCOLA backbone was amplified using fapR_1 and fapR_28. The 2 products were assembled using Golden Gate cloning to yield pCOLA-fapR. This modification was to maintain origin of replication compatibility only.
Naringenin pathway.
For QS-based transcriptional control of TAL and 4CL, codon optimized sequences of each gene appended to Plux were inserted into pCOLA using restriction digestion and ligation to yield pCOLA-Plux-TAL-4CL_v1 (Genscript). The final pCOLA-Plux-TAL-4CL plasmid used in this study was obtained using primers T4_R20_ (1–8) using Golden Gate cloning. This modification was carried out to take out the T7 system components from the backbone and to increase the strength of the RBS’s driving TAL and 4CL expression. Codon optimized sequences of CHS and CHI (Genscript) were PCR-amplified using primers CHS_T7F2 and CHS_T7R2 for CHS and CHI_T7F2 and CHI_T7R2 for CHI. Products were digested and ligated into MCS1 and MCS2 of the pET-duet vector to yield pET-CHS-CHI.
Dynamic control of Ics and Ipl in the salicylic acid pathway.
For QS-based transcriptional control of the genes encoding Ics and Ipl, the pCOLA backbone was amplified using EP_1 and EP_2, esaR was amplified from pSB3K3-PesaR-GFP-p104-esaR using EP_3 and EP_4, PesaR-H was amplified from pCOLA-PesaR-H-mCherry using EP_5 and EP_6 along with EP_9 and EP_10, entC (encoding Ics) was amplified from the E. coli genome using EP_7 and EP_8, and pchB (encoding Ipl) was amplified from the Pseudomonas aeruginosa genome using EP_11 and EP_12. The PCR products were assembled into the vector pCOLA-PesaR-H-entC-pchB using Golden Gate cloning.
CRISPRi-mediated control of endogenous enzymes.
For QS-based silencing of endogenous genes, the 20-bp guide sequence was changed from pCDF-Plux-sgGFP by circular polymerase extension cloning using primers [gene name]_luxR_F, [gene name]_luxR_R, sg_CPEC_1 and sg_CPEC_2. The 20-bp guide sequences were obtained from either previous studies (26) or using predictions from ATUM’s gRNA design tool (https://www.atum.bio/). To produce vectors which express multiple guides under the control of individual promoters, the pCDF-Plux-sgRNA vectors which express an sgRNA were used as templates in Golden Gate cloning using primer sg_ (1–10).
Culturing and Fermentations.
Fluorescence characterization.
Switching dynamics over varying expression levels of QS circuit components (esaI and luxR) were quantified using the BioLector microbioreactor system (m2p-labs GmbH). Individual colonies were inoculated in LB medium and grown overnight at 30 °C. One-milliliter cultures were inoculated from these seeds at optical density at 600 nm (OD600) 0.05 into BioLector 48-well flower plates and incubated at 30 °C, 1,200 rpm (3-mm orbit), and 80% relative humidity. The plate was sealed with a gas-permeable sealing foil (m2p-labs GmbH). Cultures were monitored for OD (BioLector units), GFP, and red fluorescent protein (RFP) fluorescence over time.
Fermentations.
Naringenin production trials were performed in glass vials with a 5-mL working volume at 30 °C and 80% humidity with 250 rpm shaking in modified MOPS minimal medium containing 5 g/L d-glucose, 500 mg/L tyrosine, 4 g/L NH4Cl, 1 g/L K2HPO4, 2 mM MgSO4, 0.1 mM CaCl2, 40 mM MOPS, 4 mM tricine, 50 mM NaCl, 100 mM Bis-Tris, 143 μM EDTA, 31 μM FeCl3, 6.2 μM ZnCl3, 0.76 μM CuCl2. 0.42 μM CoCl2, 1.62 μM H3BO3, and 0.081 μM MnCl2. For strains containing plasmids with pET, pCOLA, pACYC, and pCDF vector backbones, the medium was also supplemented with 100 μg/mL carbenicillin, 50 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 100 μg/mL spectinomycin, respectively, for plasmid maintenance. Strains were initially grown in 3 mL of LB medium at 30 °C overnight then diluted 1:100 into 3-mL seed cultures of modified MOPS medium for ∼24 h at 30 °C. These were used to inoculate working cultures at OD600 0.05. Samples were taken periodically for quantification of cell density and extracellular metabolites. Fermentations were carried out for 48 h.
For naringenin bioreactor production trials, colonies were inoculated into 50-mL seed cultures in 250-mL baffled shake flasks and incubated at 30 °C, 250 rpm, 8% humidity for ∼16 h. Seed cultures were then used to inoculate a 3-L Labfors bioreactor (Infors AG; 1-L working volume) at OD600 0.05. The pH was controlled at 7 using 4 M NaOH. Dissolved oxygen (DO) was controlled at 35% of maximum saturation by agitation rate (250 to 1,000 rpm) and constant air sparging at 1 L/min. Batch fermentation was carried out for 48 h, with 5-mL samples removed at 18, 24, and 48 h for OD and titer measurements.
Salicylic acid production trials were performed in BioLector 48-well flower plates (m2p-labs GmbH) with 1-mL working volume and 37 °C and 80% humidity with 900 rpm shaking in M9 minimal medium containing 10 g glycerol, 2.5 g glucose, 6 g Na2HPO4, 0.5 g NaCl, 3 g KH2PO4, 1 g NH4Cl, 245 mg MgSO4⋅7H2O, 14.7 mg CaCl2⋅2H2O, 2 g MOPS, and micronutrients including 2.0 mg vitamin B1, 1.25 mg H3BO3, 0.15 mg NaMoO4⋅2H2O, 0.7 mg CoCl2⋅2H2O, 0.25 mg CuSO4⋅5H2O, 1.6 mg MnCl2⋅4H2O, and 0.3 mg ZnSO4⋅7H2O per liter. For strains containing plasmids with pCOLA, pACYC, and pCDF vector backbones, the medium was supplemented with 50 μg/mL kanamycin, 34 μg/mL chloramphenicol, and 100 μg/mL spectinomycin, respectively, for plasmid maintenance. Strains were initially grown in 1 mL of M9 medium at 37 °C overnight and these cultures were used to inoculate working cultures at OD600 0.05. Samples were taken periodically for quantification of cell density and extracellular metabolites. Fermentations were carried out for 24 h.
Quantification of Metabolites.
Tyrosine, p-coumaric acid, naringenin, and salicylic acid were quantified by high-performance liquid chromatography (HPLC) on an Agilent 1100 series instrument with a ZORBAX Eclipse column (4.6 mm × 150 m × 3.5 μm). The HPLC was run with a mixture of solution A (water + 0.1% trifluoroacetic acid [TFA]) and solution B (acetonitrile + 0.1% TFA) as the eluent at a flow rate of 1 mL/min. The following gradient was used: at 0 min, 90% solution A and 10% solution B; by 10 min, 60% solution A and 40% solution B; by 15 min, 40% solution A and 60% solution B; by 15.5 min, 0% solution A and 100% solution B;, 15.5 to 21 min, 0% solution A and 100% solution B;, by 21.5 min, 90% solution A and 10% solution B; and 21.5 to 28 min, 90% solution A and 10% solution B. Compounds were quantified with 10-μL injections using diode-array detection at 270 nm (tyrosine) or 290 nm (p-coumaric acid, naringenin, and salicylic acid).
Statistics.
All error bars are reported as SDs of replicates. The number of replicates is provided in the corresponding figure legend.
Data Availability.
All data generated and analyzed and analyzed in this study are available from the corresponding author upon reasonable request.
Supplementary Material
Acknowledgments
We thank the C. Voigt laboratory (Massachusetts Institute of Technology, Biological Engineering) for kindly providing the plasmid pFM301 and the M. Koffas laboratory (Rensselaer Polytechnic Institute, Chemical and Biological Engineering) for kindly providing the malonyl-CoA sensor plasmids. This work was supported by the National Science Foundation through the Division of Molecular and Cellular Biosciences (Grants MCB-1517913 and MCB-1817708).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911144116/-/DCSupplemental.
References
- 1.Nevoigt E., et al. , Engineering promoter regulation. Biotechnol. Bioeng. 96, 550–558 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Farmer W. R., Liao J. C., Improving lycopene production in Escherichia coli by engineering metabolic control. Nat. Biotechnol. 18, 533–537 (2000). [DOI] [PubMed] [Google Scholar]
- 3.Kang Z., Wang Q., Zhang H., Qi Q., Construction of a stress-induced system in Escherichia coli for efficient polyhydroxyalkanoates production. Appl. Microbiol. Biotechnol. 79, 203–208 (2008). [DOI] [PubMed] [Google Scholar]
- 4.Liang Q., Zhang H., Li S., Qi Q., Construction of stress-induced metabolic pathway from glucose to 1,3-propanediol in Escherichia coli. Appl. Microbiol. Biotechnol. 89, 57–62 (2011). [DOI] [PubMed] [Google Scholar]
- 5.Zhang F., Carothers J. M., Keasling J. D., Design of a dynamic sensor-regulator system for production of chemicals and fuels derived from fatty acids. Nat. Biotechnol. 30, 354–359 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Dahl R. H., et al. , Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Siedler S., Stahlhut S. G., Malla S., Maury J., Neves A. R., Novel biosensors based on flavonoid-responsive transcriptional regulators introduced into Escherichia coli. Metab. Eng. 21, 2–8 (2014). [DOI] [PubMed] [Google Scholar]
- 8.Xu P., Li L., Zhang F., Stephanopoulos G., Koffas M., Improving fatty acids production by engineering dynamic pathway regulation and metabolic control. Proc. Natl. Acad. Sci. U.S.A. 111, 11299–11304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Doong S. J., Gupta A., Prather K. L. J., Layered dynamic regulation for improving metabolic pathway productivity in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 115, 2964–2969 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., et al. , Sensor-regulator and RNAi based bifunctional dynamic control network for engineered microbial synthesis. Nat. Commun. 9, 3043 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soma Y., Hanai T., Self-induced metabolic state switching by a tunable cell density sensor for microbial isopropanol production. Metab. Eng. 30, 7–15 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Liu H., Lu T., Autonomous production of 1,4-butanediol via a de novo biosynthesis pathway in engineered Escherichia coli. Metab. Eng. 29, 135–141 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Gupta A., Reizman I. M. B., Reisch C. R., Prather K. L. J., Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xie W., Ye L., Lv X., Xu H., Yu H., Sequential control of biosynthetic pathways for balanced utilization of metabolic intermediates in Saccharomyces cerevisiae. Metab. Eng. 28, 8–18 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Bothfeld W., Kapov G., Tyo K. E. J., A glucose-sensing toggle switch for autonomous, high productivity genetic control. ACS Synth. Biol. 6, 1296–1304 (2017). [DOI] [PubMed] [Google Scholar]
- 16.Lo T. M., Chng S. H., Teo W. S., Cho H. S., Chang M. W., A two-layer gene circuit for decoupling cell growth from metabolite production. Cell Syst. 3, 133–143 (2016). [DOI] [PubMed] [Google Scholar]
- 17.He X., Chen Y., Liang Q., Qi Q., Autoinduced and gate controls metabolic pathway dynamically in response to microbial communities and cell physiological state. ACS Synth. Biol. 6, 463–470 (2017). [DOI] [PubMed] [Google Scholar]
- 18.Moser F., et al. , Dynamic control of endogenous metabolism with combinatorial logic circuits. Mol. Syst. Biol. 14, e8605 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fowler Z. L., Koffas M. A. G., Biosynthesis and biotechnological production of flavanones: Current state and perspectives. Appl. Microbiol. Biotechnol. 83, 799–808 (2009). [DOI] [PubMed] [Google Scholar]
- 20.Wang Y., Chen S., Yu O., Metabolic engineering of flavonoids in plants and microorganisms. Appl. Microbiol. Biotechnol. 91, 949–956 (2011). [DOI] [PubMed] [Google Scholar]
- 21.Zhou J., Du G., Chen J., Novel fermentation processes for manufacturing plant natural products. Curr. Opin. Biotechnol. 25, 17–23 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Santos C. N. S., Koffas M., Stephanopoulos G., Optimization of a heterologous pathway for the production of flavonoids from glucose. Metab. Eng. 13, 392–400 (2011). [DOI] [PubMed] [Google Scholar]
- 23.Yang D., et al. , Repurposing type III polyketide synthase as a malonyl-CoA biosensor for metabolic engineering in bacteria. Proc. Natl. Acad. Sci. U.S.A. 115, 9835–9844 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu J., Zhou T., Du G., Zhou J., Chen J., Modular optimization of heterologous pathways for de novo synthesis of (2S)-naringenin in Escherichia coli. PLoS One 9, e101492 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu J., Yu O., Du G., Zhou J., Chen J., Fine-tuning of the fatty acid pathway by synthetic antisense RNA for enhanced (2S)-naringenin production from l-tyrosine in Escherichia coli. Appl. Environ. Microbiol. 80, 7283–7292 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu J., Du G., Chen J., Zhou J., Enhancing flavonoid production by systematically tuning the central metabolic pathways based on a CRISPR interference system in Escherichia coli. Sci. Rep. 5, 13477 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Omura S., The antibiotic cerulenin, a novel tool for biochemistry as an inhibitor of fatty acid synthesis. Bacteriol. Rev. 40, 681–697 (1976). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin Y., Sun X., Yuan Q., Yan Y., Extending shikimate pathway for the production of muconic acid and its precursor salicylic acid in Escherichia coli. Metab. Eng. 23, 62–69 (2014). [DOI] [PubMed] [Google Scholar]
- 29.St-Pierre F., et al. , One-step cloning and chromosomal integration of DNA. ACS Synth. Biol. 2, 537–541 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Shiue E., Improvement of D-glucaric acid production in Escherichia coli (Massachusetts Institute of Technology, 2014). [Google Scholar]
- 31.Gupta A., Dynamic regulation of bacterial metabolic pathways using autonomous, pathway-independent control strategies (Massachusetts Institute of Technology, 2017). [Google Scholar]
- 32.Qi L. S., et al. , Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated and analyzed and analyzed in this study are available from the corresponding author upon reasonable request.