Significance
Controlling access to the active sites of kinases using photo-dimerizable domains revolutionizes the types of possible optogenetic perturbations of cell signaling. This technology, however, is useful only when there is strong kinase activity to harness. We report a strategy to systematically enhance and tune functionality of a kinase activating the Extracellular Signal-Regulated Kinase (ERK) pathway, MEK, using gain-of-function point mutations that destabilize MEK. Photoswitching mutant MEK activity by reversibly caging its active site controls a large dynamic range of ERK signaling in vivo. Our approach for optimizing and tuning light-activated protein kinases is not limited to MEK and can be used to systematically extend the capabilities of light-activated regulatory enzymes.
Keywords: optogenetics, ERK signaling, photoswitchable MEK
Abstract
Optogenetic approaches are transforming quantitative studies of cell-signaling systems. A recently developed photoswitchable mitogen-activated protein kinase kinase 1 (MEK1) enzyme (psMEK) short-circuits the highly conserved Extracellular Signal-Regulated Kinase (ERK)-signaling cascade at the most proximal step of effector kinase activation. However, since this optogenetic tool relies on phosphorylation-mimicking substitutions in the activation loop of MEK, its catalytic activity is predicted to be substantially lower than that of wild-type MEK that has been phosphorylated at these residues. Here, we present evidence that psMEK indeed has suboptimal functionality in vivo and propose a strategy to circumvent this limitation by harnessing gain-of-function, destabilizing mutations in MEK. Specifically, we demonstrate that combining phosphomimetic mutations with additional mutations in MEK, chosen for their activating potential, restores maximal kinase activity in vitro. We establish that this modification can be tuned by the choice of the destabilizing mutation and does not interfere with reversible activation of psMEK in vivo in both Drosophila and zebrafish. To illustrate the types of perturbations enabled by optimized psMEK, we use it to deliver pulses of ERK activation during zebrafish embryogenesis, revealing rheostat-like responses of an ERK-dependent morphogenetic event.
Much of our current understanding of signaling networks relies on uniform genetic perturbations using gain- or loss-of-function mutations that broadly disrupt signal transduction. Yet, signaling pathways are reused in many different contexts that require a range of doses, durations, and dynamics (1). With optogenetics, we can generate spatiotemporal signaling patterns that are almost as precise as our measurements of the induced molecular and cellular responses (2, 3). In particular, light-based mechanisms to control signaling through the highly conserved Extracellular Signal-Regulated Kinase (ERK) pathway have begun to reveal important quantitative features of cell-signaling systems such as dynamic encoding of information and temporal control of fate responses in differentiating tissues (4–6).
So far, most opto-ERK–signaling tools activate signal transduction via receptor dimerization or translocation of receptor-proximal pathway components to the plasma membrane (7–10). These optogenetic systems mimic upstream activation steps and are therefore ideal for studies dissecting quantitative features of the downstream signaling cascade (11). With these tools, we can prescribe ligand-like inputs to the pathway with light and use activity of the effector molecule ERK as the output (12). However, to study how the ultimate activation of ERK controls downstream responses, such as target gene induction, it would be ideal to deliver optogenetic inputs as proximally to ERK as possible in order to minimize off-target effects and potentially confounding feedback mechanisms acting within the upstream signal transduction cascade (1, 13, 14). Developing optogenetic tools that work at proximal steps to ERK phosphorylation and activation is therefore an important goal for studying the consequences of pathway activation.
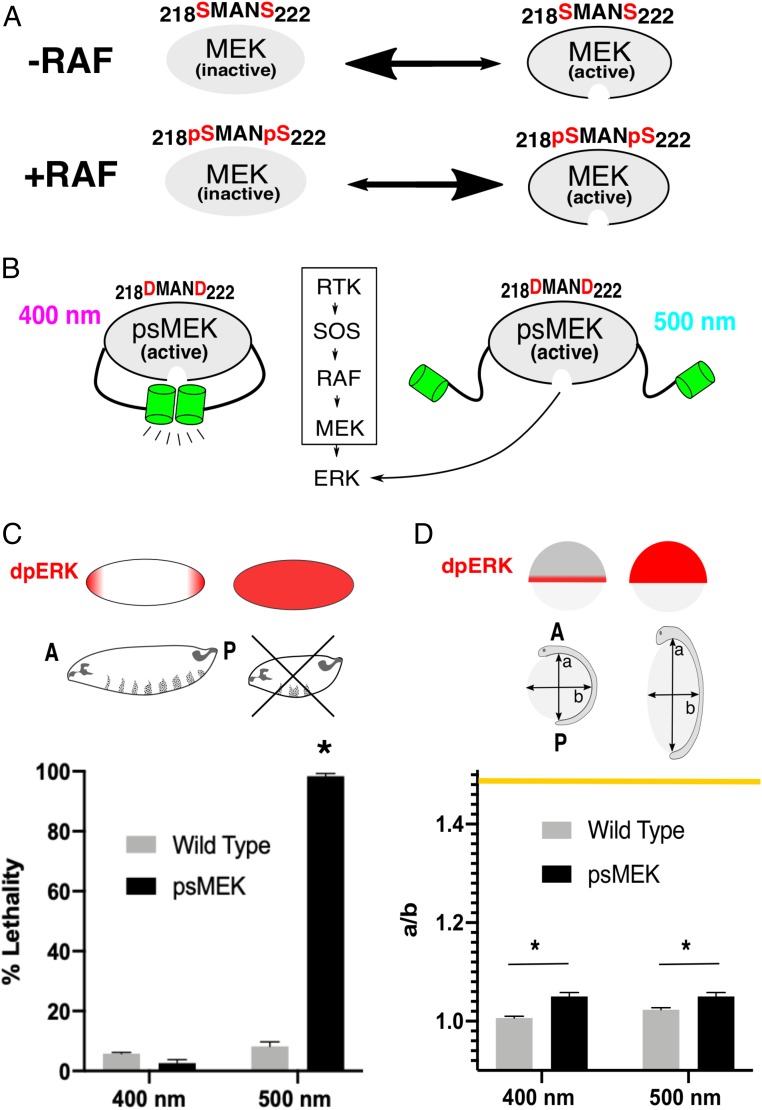
Alternative strategies for more direct activation of ERK have been made possible by the recent development of photoswitchable variants of ERK’s kinase, mitogen-activated protein kinase kinase 1 (MEK1) (15, 16). In a photoswitchable form of MEK1 (reported as psMEK1tight), one domain of engineered photo-dimerizable Dronpa (pdDronpa) has been fused after the N-terminal substrate docking site (amino acids 1 to 60) and another in a flexible region of MEK1 called the FG loop (before amino acid 305) (15, 16). The pdDronpa domains are inserted at these positions so that they can reversibly cage MEK’s active site when the enzyme is in its active conformation. Since this construct is single-chain, intrinsic MEK activity sets the optogenetic input strength. Normally, the kinase RAF phosphorylates 2 serine residues in the activation loop (S218, S222) of MEK to promote a conformational change that allows ERK to access MEK’s active site (17, 18) (Fig. 1A). Serine to aspartic acid (S > D) mutations have been made in the activation loop of psMEK1tight (here referred to as psMEK) to mimic the presence of phosphate groups and constitutively activate the enzyme independently of RAF (15, 19, 20) (Fig. 1B). The constitutive activity of psMEK is silenced when 400 nm light induces pdDronpa dimerization, which forms a physical barrier between MEK and ERK. Five hundred-nanometer light dissociates the pdDronpa domains and restores psMEK function.
Fig. 1.
psMEK effects in vivo. (A) RAF phosphorylates 2 serine residues (red, S218 and S222) in the activation loop of MEK. When MEK is unphosphorylated, its inactive conformation is favored, shown by the larger arrowhead facing to the left. When RAF phosphorylates MEK, the active conformation is favored. (B) psMEK contains S > D mutations in the activation loop that mimic the effects of RAF phosphorylation. The phosphomimetic substitutions maintain psMEK in the active conformation. Constitutively active psMEK is controlled by photoswitchable Dronpa domains (green) that block the active site when dimerized (in 400 nm light) and expose the active site when dissociated (in 500 nm light). (C) ERK is only active (dpERK) at the poles in the 3-h-old Drosophila embryo. Ectopic ERK signaling in the middle of the embryo disrupts formation of the denticle belts in the larvae and is lethal (>36 h p. f.). Embryos expressing psMEK (black bars) exhibited significant lethality when exposed to 500 nm light. Illumination of 400 nm was not lethal. Wavelength of light illumination had no effect on lethality in the WT control embryos (gray bars). Error bars represent means and SDs for 3 replicates. Pooled N numbers are as follows: NOreR, 400 nm = 291, NOreR, 500 nm = 543, NpsMEK, 400 nm = 1166, NpsMEK, 500 nm = 2,179, NpsMEK, dark = 621, and NpsMEK, room light = 607. (D) In zebrafish embryos, ERK is endogenously active in the blastoderm margin at ∼6 h.p.f. ERK signaling in nonmarginal cells causes defective morphogenetic movements that elongate the embryo, measured as the aspect ratio (major axis “a”/minor axis “b”) of the yolk at the tailbud stage (10 h.p.f.). Embryos expressing psMEK via mRNA injection (black bars) do not show a significant wavelength-dependent increase in aspect ratio. Uninjected embryos (gray bars) remained spherical in both 400 and 500 nm light. The yellow line represents the maximum aspect ratio reported in SI Appendix, Fig. S2 and ref. 28. Pooled numbers of measured embryos are as follows: NpsMEK, 400 nm =, 112 NpsMEK, 500 nm = 112, Nuninjected, 400 nm = 114, and Nuninjected, 500 nm = 97. In C and D, *P < 0.05.
Here, we show that psMEK activates ERK only weakly in vivo, based on its light-dependent developmental defects in the Drosophila and zebrafish embryos. While these inconsistent phenotypic effects could be explained by varying expression levels, different sensitivities to active ERK levels, or even differential binding to species-specific homologs of ERK, we find that the MEK-activation strategy using S > D phosphorylation-mimicking mutations is an intrinsically suboptimal design. We characterize the rates of ERK phosphorylation by MEK and demonstrate that phosphomimetic mutations cannot recapitulate phosphorylated MEK activity (15).
We hypothesized that the activity of psMEK can be significantly enhanced by harnessing destabilizing gain-of-function mutations in MEK (20). We established that this strategy works both in vitro, using measurements of MEK-dependent ERK activation kinetics, and in vivo, by perturbing ERK-dependent developmental events in Drosophila and zebrafish. Importantly, our strategy for enhancing the activity of psMEK can be tuned by the choice of destabilizing mutation and does not compromise the reversibility and speed of light-induced activation. Finally, we implement our optimized psMEK to deliver high-amplitude pulses of ERK activity in the early zebrafish embryo, revealing duration-dependent features of ERK sensing that are beyond the dynamic range of the original psMEK.
Results
Phenotypic Effects of psMEK.
We first assayed psMEK function in the early Drosophila embryo, where ERK signaling is normally limited to the anterior and posterior poles to pattern the future head and tail structures (21). Even small signaling increases above basal levels in the middle of the embryo are lethal. This ectopic ERK activity causes aberrant expression of the tightly regulated gap genes that are crucial for formation of 8 complete segments in the larval cuticle (22).
Drosophila embryos expressing psMEK and exposed to 500 nm light exhibited significant lethality compared to 400 nm illumination (Fig. 1C). The larval cuticles of the dead embryos exposed to activating light were mostly destroyed, indicating maximally deregulated ERK signaling (SI Appendix, Fig. S1). The distribution of cuticle phenotypes that we observed is similar to that of strong gain-of-function mutants affecting the ERK pathway and activation with the blue-light–sensitive optoSOS system (7, 23).
We used morphological defects in the early zebrafish embryo as a metric for light-induced ERK activity in a second in vivo context. Constitutive ERK signaling in the zebrafish embryo leads to abnormal convergence and extension movements that cause dose-dependent stretching of the normally spherical yolk along the anterior-to-posterior axis. The result is an elongated embryo (referred to as the “oval embryo” phenotype) that can be quantified by the aspect ratio of the yolk (Fig. 1D) (24–27). This morphological readout reflects the strength of ectopic ERK activation and has been used for ranking the severities of MEK mutations associated with human diseases (28).
Surprisingly, we found that light-activated psMEK did not induce the oval embryo phenotype as expected. We measured a slight increase in aspect ratio for embryos expressing psMEK (∼1.05) versus wild-type (WT) control embryos (∼1) that was not dependent on the wavelength of light (Fig. 1D). Moreover, these effects represent a fraction of the range of possible phenotypes: aspect ratios nearing 1.5 have been measured in cases of strong ectopic ERK activation (28). Overexpression of phosphomimetic MEK (nonphotoswitchable) also displayed less elongation compared to overexpression of stronger MEK variants, such as the one containing the E203K cancer mutation. This suggests that the phosphomimetic substitutions are only weakly activating and are therefore limiting psMEK’s utility to only the most sensitive developmental contexts such as the early Drosophila embryo (SI Appendix, Fig. S2).
Destabilizing Mutations Increase Phosphomimetic MEK Activity.
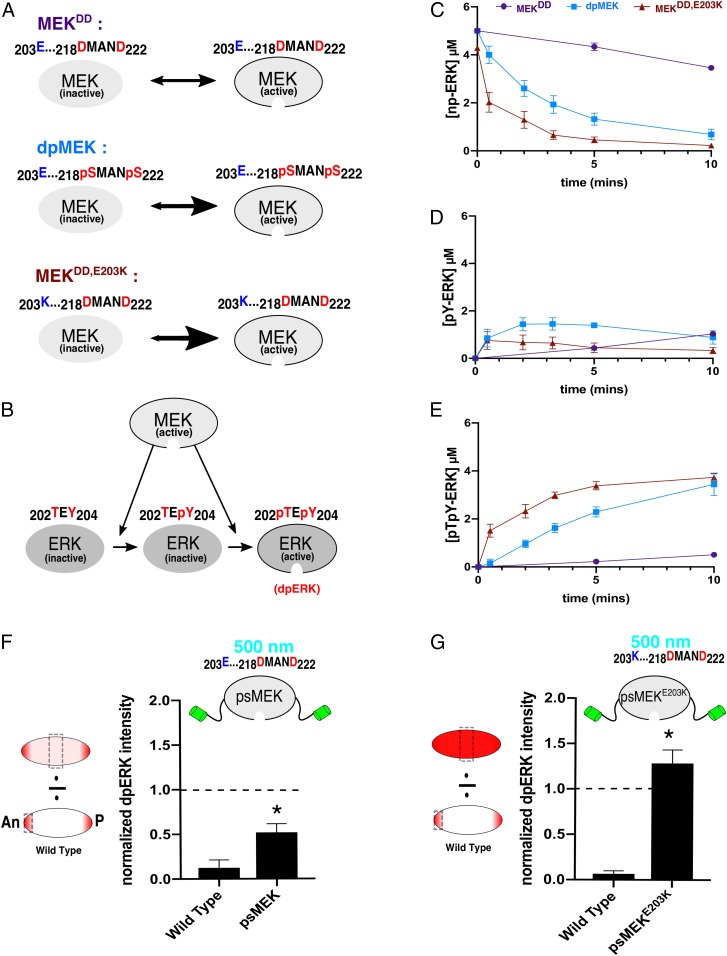
We hypothesized that the activation strategy, using phosphomimetic mutations, may intrinsically limit psMEK function because the aspartic acids (D) do not entirely recapitulate the strongly negatively charged chemical environment of phosphate groups within MEK (29, 30). To improve psMEK, we turned to the idea of combining phosphomimetic substitutions with activating mutations identified by their roles in human diseases caused by abnormally high ERK signaling, including RASopathies and cancer (Fig. 2A). Missense mutations that destabilize MEK and cause constitutive activity have been found in patients with some of these diseases (20, 31). We reasoned that adding these mutations to the phosphomimetic MEK domain of psMEK would increase kinase activity while maintaining independence from upstream activators and ensuring a completely orthogonal optogenetic ERK input. We chose to use the E203K substitution, found in cancer, to enhance kinase activity since it is the top of the list of pathogenic mutations ranked by their effects on zebrafish yolk elongation (28). This substitution disrupts an important interaction known to stabilize a region of the MEK molecule that represses enzymatic activity (31). Consistent with these destabilizing effects, we found that the E203K substitution reduces the thermal stability of MEK (both phosphorylated and unphosphorylated) (SI Appendix, Fig. S3). Furthermore, molecular dynamics simulations show that the E203K variant enters conformations that are very different from those of the WT unphosphorylated MEK, also suggesting destabilization (31). This behavior is similar to a whole class of disease mutations with a range of activation strengths and makes the E203K substitution a good candidate to test our hypothesis for optimizing psMEK (20).
Fig. 2.
Enhancing MEK activity with destabilizing mutations. (A) Phosphomimetic MEK (MEKDD) is predicted to be significantly less active than dpMEK. Destabilizing MEKDD with a distal E203K substitution (blue) creates a different MEK variant (MEKDD,E203K) that we predict will exhibit increased activity. (B) Active MEK catalyzes dual phosphorylation of ERK. (C) Data from in vitro kinase assays showing kinetics of unphosphorylated ERK (np-ERK) levels in the presence of phosphomimetic (purple, MEKDD), phosphorylated (light blue, dpMEK), and phosphomimetic MEK with E203K (dark red, MEKDD,E203K). Data are based on n = 5 for MEKDD and dpMEK and n = 6 for MEKDD,E203K at all time points. (D) Time courses of the monophosphorylated ERK (pY-ERK) levels for 3 different MEK variants. (E) Time courses of the dually phosphorylated ERK (dpERK, pTpY-ERK) levels for 3 different MEK variants. (F) Quantifications of confocal images of immunofluorescence stainings for dpERK in 3-h-old Drosophila embryos. The dpERK intensities in the middle of the embryo (50% embryo length) are presented for WT and psMEK-expressing embryos activated with 500 nm light. All measurements reflect the dpERK levels normalized to the maximum dpERK intensity in the WT control embryos (at the anterior pole). The dashed line drawn at 1.0 represents the maximum dpERK intensity in WT embryos. NWT = 17 and NpsMEK = 25. (G) The same quantifications described for embryos expressing psMEK with an additional E203K mutation, denoted as psMEKE203K. NWT = 18 and NpsMEKE203K = 15. In F and G, *P < 0.05.
Active MEK phosphorylates ERK at a tyrosine and threonine to activate it (Fig. 2B) (32–34). We first measured the kinetics with which the monophosphorylated and dually phosphorylated (dp, active) forms of ERK appear in the presence of MEK with S218D and S222D (referred to as MEKDD), dually phosphorylated MEK (dpMEK), or phosphomimetic MEK with the E203K substitution (MEKDD,E203K) in vitro. MEKDD phosphorylated ERK very slowly compared to dpMEK, confirming that the constitutive activity of the phosphomimetic molecule is indeed suboptimal (Fig. 2 C–E). In contrast, MEKDD,E203K exhibited a dramatic increase in the rate of ERK phosphorylation, even exceeding the rate observed from dpMEK (Fig. 2 C–E). Thus, MEKDD,E203K is more active than dpMEK, but cannot be dephosphorylated and inactivated. Based on these results demonstrating that destabilizing mutations can significantly augment the activity of phosphomimetic MEK in vitro, we proceeded to test if the E203K mutation can enhance psMEK activity in vivo.
Destabilizing Mutations Enhance psMEK Function In Vivo.
We then engineered transgenic flies expressing psMEK, which already contains phosphomimetic mutations, as well as psMEK with an additional E203K mutation (here, denoted as psMEKE203K) in the early embryo. For these experiments, embryos were illuminated with 500 nm light to uncage the Dronpa domains and expose the MEK active site. We performed immunofluorescence stainings for dually phosphorylated ERK (dpERK) and quantified active ERK levels in the middle region of the embryo where all components of the ERK pathway are present but the pathway is not activated by extracellular signals (21). Consistent with the significant increase of enzymatic activity observed in vitro, we discovered that ERK activation, based on dpERK levels caused by activated psMEKE203K, is significantly stronger than dpERK induced by psMEK. Compared to endogenous amounts of dpERK at the poles of the embryo, where ERK is activated by localized receptor tyrosine kinase signaling, psMEK activation induced lower dpERK levels, and psMEKE203K activation induced higher dpERK levels (Fig. 2 F and G). This observation is consistent with our results from the in vitro kinase assays showing a hierarchy of activity from MEKDD, to dpMEK, to MEKDD,E203K. Thus, the E203K substitution hyperactivates psMEK in vivo as well.
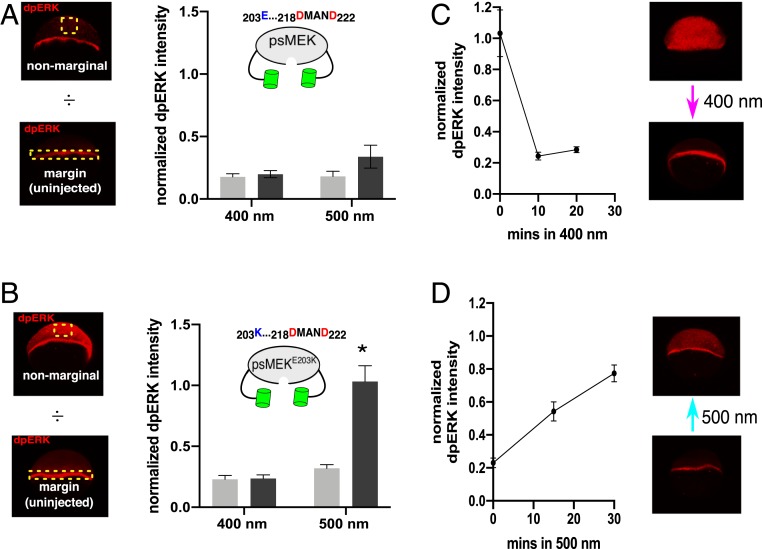
We then asked if the optimized psMEK leaks ERK activation in the caged conformation when illuminated with 400 nm light. We addressed this question in the zebrafish embryo, where leaky activity of the optoSOS signaling tool contributed to a reduction in the dynamic range of ERK-dependent defects (SI Appendix, Fig. S4). We measured dpERK levels in embryos expressing the original psMEK construct at 50% epiboly [∼6 hours post fertilization (h.p.f.)], a stage when endogenous ERK activation is limited to the embryonic margin (23, 35, 36). In 500-nm–illuminated embryos, we detected a small increase in ectopic dpERK for the 500-nm light condition; this increase was still very low when compared to endogenous active ERK at the margin of uninjected embryos (Fig. 3A). These levels may be too low to cause yolk elongation, explaining the weak phenotypic effects reported in the previous section. When the same stimuli were applied to embryos expressing psMEKE203K, 500 nm of light exposure led to much higher levels of ectopic dpERK (Fig. 3B). For both constructs, exposure to 400 nm light still maintained basal dpERK levels in the nonmarginal cells, indicating efficient protection of the active site by the pdDronpa domains in psMEKE203K despite this optimized construct’s hyperactivity (Fig. 3B). Thus, the destabilizing E203K substitution increases the dynamic range of ERK-signaling perturbations that can be made in zebrafish.
Fig. 3.
Effects of optimized psMEK in zebrafish. (A) Quantifications of confocal images of 50% epiboly-stage zebrafish embryos immunofluorescence-stained for dpERK. Embryos expressing psMEK (dark gray) and uninjected WT embryos (light gray) were illuminated with 400 or 500 nm light. For the quantification of psMEK-expressing embryos, dpERK intensities from regions of nonmarginal cells were normalized to the margins of costained uninjected control embryos. Nonmarginal cell regions in the control uninjected embryos were normalized to margins of the same pool of uninjected embryos. For the 400-nm condition, Ninjected = 9 and Nuninjected = 9. For the 500-nm condition, Ninjected = 8 and Nuninjected = 8. (B) The equivalent dpERK quantifications for embryos injected with psMEK containing both the SSDD and E203K mutations (psMEKE203K). For the 400-nm condition, Ninjected = 12 and Nuninjected = 12. For the 500-nm condition, Ninjected = 15 and Nuninjected = 17. *P < 0.05. (C) The high levels of dpERK in psMEKE203K-injected embryos return to normal levels within 10 min when switched to 400 nm light. Quantifications were performed as described in A at each time point. Ninjected = {13,17,15} and Nuninjected = {10,13,17}. N numbers within braces are numbers of embryos measured at each time point in order from 0 to 20 min after the 50% epiboly stage. (D) The low levels of dpERK in 400-nm exposed embryos rise to nearly maximal levels within 30 min. Ninjected = {11,11,12} and Nuninjected = {11,11,12}. N numbers within braces are numbers of embryos measured at each time point in order from 0 to 30 min after the 50% epiboly stage.
Next, we tested whether activation of optimized psMEK is rapidly reversible by switching embryos at 50% epiboly from 500 to 400 nm light and vice versa. The high levels of dpERK induced in zebrafish embryos exposed to 500 nm light were lost within 10 min in response to 400 nm light (Fig. 3C). Conversely, ectopic dpERK in embryos initially exposed to 400 nm light rose to near-maximal levels by 30 min when activated by 500 nm light (Fig. 3D). Thus, destabilizing mutations significantly enhance the potency of psMEK without compromising the speed and reversibility of its activation by light.
For the original psMEK design, 400 nm light was only required to rapidly photo-switch ERK activation and was not required to maintain psMEK inactivity (15). We find that the dark is also sufficient to maintain inactivity of psMEKE203K in vivo. Drosophila embryos expressing either psMEK or psMEKE203K exhibited low lethality when kept in the dark, which strongly indicates that the optimized tool is not leaky in nonilluminated conditions (SI Appendix, Fig. S5A). Zebrafish embryos expressing psMEKE203K and kept in the dark also had only basal levels of ectopic dpERK. Simply removing the 500-nm light source, however, did not rapidly reverse psMEKE203K-induced ectopic ERK activity in the zebrafish embryo, consistent with the properties of the original design (SI Appendix, Fig. S5B) (15).
Tuning psMEK with Mutations.
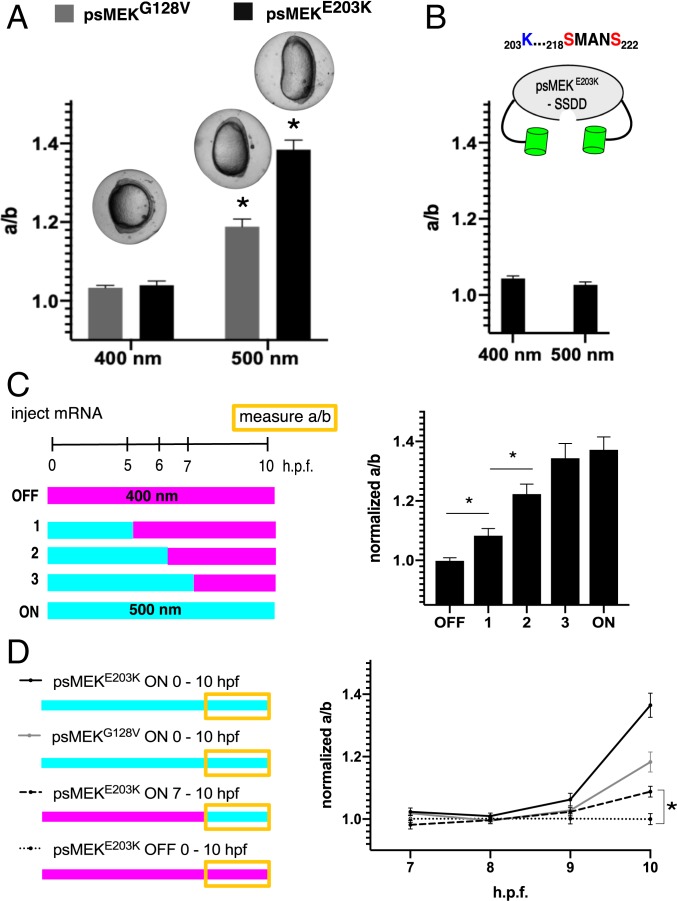
Disease-associated, activating MEK mutations lie on a spectrum of severities that could be used to further tune psMEK activity (28). For example, the E203K cancer-associated mutation is more strongly activating than the G128V mutation found in the group of developmental disorders collectively termed RASopathies (28, 37, 38). To test if the relative strengths of disease mutations are retained when used to optimize psMEK, we used the oval embryo assay to test 2 enhanced psMEK variants containing either an additional G128V (psMEKG128V) or E203K substitution (psMEKE203K). Indeed, psMEKG128V induced a yolk aspect ratio of ∼1.2 upon 500 nm illumination, indicating a lower level of gain-of-function ERK signaling compared to psMEKE203K (an aspect ratio of ∼1.4) under the same conditions (Fig. 4A). Zebrafish embryos injected with a blue-light–dependent optoSOS system displayed only moderate light-induced elongation phenotypes that did not change when the injected messenger RNA (mRNA) dose was increased, further emphasizing that optimized psMEK is a tunable, potent tool for ERK signaling studies in zebrafish embryos (SI Appendix, Fig. S4).
Fig. 4.
Tuning and modulating optimized psMEK. (A) Aspect ratios (anterior-posterior length/dorsal-ventral length) for zebrafish embryos injected with psMEK + G128V (psMEKG128V) or psMEK + E203K (psMEKE203K) at the tailbud stage. Injected embryos were illuminated with 400 or 500 nm light. NpsMEKG128V, 400 nm = 63, NpsMEKG128V, 500 nm = 102, NpsMEKE203K, 400 nm = 99, and NpsMEKE203K, 500 nm = 97. (B) Aspect ratios for psMEKE203K that lacks S > D mutations in the activation loop (denoted as psMEKE203K–SSDD) illuminated with 400 and 500 nm light. NpsMEKE203K-SSDD, 400 nm = 36 and NpsMEKE203K-SSDD, 500 nm = 31. (C) Normalized aspect ratios for embryos exposed to varying durations of 500 nm light. The means and SDs are shown for the bootstrapped aspect ratios of the activated injected embryos to continuously inactivated (400 nm) sibling injected embryos. For perturbations 1, 2, and 3, embryos initially illuminated with 500 nm light were immediately switched to 400 nm light at the indicated time point. All measurements were made at the tailbud stage. Numbers of embryos measured are as follows: NOFF = 47, Npert.1 = 34, Npert. 2 = 39, Npert. 3 = 47, and NON = 30. (D) Dynamics of the aspect ratios of embryos injected with psMEK variants. The traces show emerging elongation for embryos subject to continuous activation of psMEKE203K (black), continuous activation of psMEKG128V (gray), continuous inactivation of psMEKE203K (dotted black), and restricted activation of psMEKE203K from 7 to 10 h.p.f. (dashed black). At each time point, the means and SDs are shown for the bootstrapped aspect ratios of the activated injected embryos to continuously inactivated (400 nm) sibling injected embryos. Numbers of embryos measured for continuous light conditions are as follows: NpsMEKE203K, 500 nm= {11,14,11,14}; NpsMEKE203K, 400 nm = {13,12,9,18}; NpsMEKG128V, 500 nm = {15,19,23,26} ; and NpsMEKG128V, 400 nm = {14,20,25,16}. Numbers of embryos measured for the restricted light condition are the following: NpsMEKE203K, 500 nm 7–10hpf = {33,22,23,23} and NpsMEKE203K, 400 nm = {24,22,25,23}. All numbers within braces are from measurements taken hourly from 7 to 10 h.p.f. In A–D, *P < 0.05.
Since the E203K substitution alone is strong enough to dramatically elongate the embryo when nonphotoswitchable MEK is overexpressed, are the phosphomimetic mutations in the activation loop required within optimized psMEK (SI Appendix, Fig. S2) (28)? To answer this question, we generated a version of psMEK that contained E203K but not S218D and S222D (termed psMEKE203K–SSDD). Surprisingly, this psMEK variant did not induce an oval embryo in activating light (Fig. 4B). Previous studies showed that MEK containing cancer and RASopathy mutations cannot induce an oval embryo when they contain S > A mutations in the activation loop that preclude phosphorylation by RAF, indicating that phosphorylation affects the function of MEK with an E203K substitution (20). The pdDronpa domains could alter psMEK-RAF interactions, which would interfere with the crucial phosphorylation step. This observation suggests that optimized psMEK is truly independent of upstream signals, and both phosphomimetic and destabilizing substitutions like E203K are needed to generate a psMEK variant with a large dynamic range.
Time-Dependent Perturbations Using psMEKE203K.
A prior optogenetic study demonstrated that varying either the level or duration of ERK activity can regulate switch-like cell-fate decisions in the Drosophila embryo (5). With psMEKE203K, we now have the ideal tool to explore a similar correspondence between duration and dose in the zebrafish embryo, where we previously found that an ERK dose provides rheostat-like control of yolk elongation (28). We switched embryos from activating to inactivating light at progressively later time points and found that phenotypic severity increased in proportion to the duration of activating light (Fig. 4C). These perturbations show that the oval embryo phenotype reflects an accumulation of ERK signaling over time. This observation is in contrast to what we found in the Drosophila embryo, where psMEKE203K activation in the first 4 h of embryogenesis was sufficient to cause maximal lethality (SI Appendix, Fig. S6) (7). We observed nearly 100% lethality for 4-h psMEKE203K activation, which is greater than what was observed for 4-h optoSOS stimulation, possibly because of more direct ERK activation at the level of MEK (SI Appendix, Fig. S6). These results are also consistent with those obtained in zebrafish studies implementing pharmacological inhibitors to cut off ectopic ERK signaling at different time points (26).
On the other hand, it was previously shown that exposure to the MEK inhibitor PD-184352 in a 1-h time window from 4.5 to 5.5 h.p.f. was sufficient to suppress the oval embryo phenotype in gain-of-function RAF-mutant embryos (26). This experiment suggests that it is necessary for there to be ectopic ERK signaling in this short time window before 5.5 h.p.f. to generate an elongated embryo. In other words, first exposing ectopic ERK signals after 5.5 h.p.f. did not lead to an oval embryo. By contrast, we found that activating psMEKE203K caused detectable and statistically significant embryo elongation even when light was first delivered as late as 7 h.p.f. (Fig. 4D, dashed line). The effects of ectopic ERK were sensed even as the ranked oval embryo phenotype emerged, implying that the embryo remains sensitive to ectopic ERK activation outside of the time window estimated using pharmacological inhibitors (Fig. 4D). These differences may be due to incomplete washout of the MEK inhibitor, leading to partial inhibition at later time points or to stronger activation attained by direct MEK activation. Modulating psMEKE203K activity with light is also a fundamentally different perturbation from washing in and out small molecules, since our tool controls only the ectopic ERK input and not the endogenous signaling cascade. Thus, optimized psMEK allowed us to discover that ectopic active ERK is in fact sensed throughout the entire 10-h trajectory, not just up until a specific time point.
The oval-embryo phenotype likely reflects altered dynamics of convergence and extension movements that reshape the embryo in the 3-h time window when rapid body-axis elongation occurs (Fig. 4D) (39). Previous studies have shown that these movements are in turn negatively regulated by the bone morphogenetic protein (BMP)-signaling pathway, suggesting that ectopic ERK signaling caused by optimized psMEK could disrupt the endogenous pattern of BMP signaling in the embryo (24, 25, 40, 41). We found that this is indeed the case: light activation disrupted the characteristic ventral-to-dorsal gradient of BMP pathway activation (SI Appendix, Fig. S7). Thus, light-activated MEK can be used to probe how the effects of ERK activation propagate throughout the larger patterning network that controls cell-fate specification and morphogenesis in the embryo.
Discussion
psMEK provides the most direct light-based control of the ERK-signaling cascade (15). In this study, we find that this important technology has a limited strength in vivo. The design of psMEK currently implements engineered Dronpa domains to harness weak constitutive MEK activity provided by phosphomimetic mutations. We discovered this limitation when characterizing light-induced phenotypes that have been extensively implemented as readouts of deregulated ERK signaling. In the fruit fly embryo, where a very low threshold of ectopic ERK signaling causes severe and lethal defects, psMEK enabled light-dependent control of the expected phenotypes (Fig. 1C). The zebrafish embryo seems to be less sensitive to ectopic active ERK levels and does not display light-dependent defects when expressing psMEK (Fig. 1D). Although the zebrafish phenotype could present an anomalous case that requires unusually high levels ERK signaling, we show that the phosphomimetic substitutions are themselves weakly activating and therefore limit the functionality of psMEK (Fig. 2 C–E).
We propose a simple strategy for removing this limitation: take advantage of mutations that destabilize MEK to enhance the potency of psMEK. This approach not only drastically improves phosphorylation-independent MEK activity, but also tunes psMEK for more tailored optogenetic inputs. Optimized psMEK still rapidly controls ERK pathway activity in vivo, enabling us to build synthetic ERK inputs in a wide range of developmental contexts. As an illustration of this approach, we used our optimized psMEK to perturb an ERK-dependent morphogenetic event in zebrafish embryos. Our results indicate that the severity of the resulting morphogenetic defect in this system depends on the duration of ectopic ERK activation.
These timed perturbations were uniquely enabled by optimized psMEK, since optoSOS did not induce strong defects in the zebrafish embryo. SOS-Ras positive feedbacks may be driving pathway adaptation and limiting the system’s overall effects in zebrafish embryos (SI Appendix, Fig. S4) (13, 14). In Drosophila, optoSOS can induce severe defects, but also leaks ERK activity in ambient room light. We found that the psMEKs leak significantly less activity when exposed to room light and may therefore be easier to implement in other systems (SI Appendix, Fig. S8).
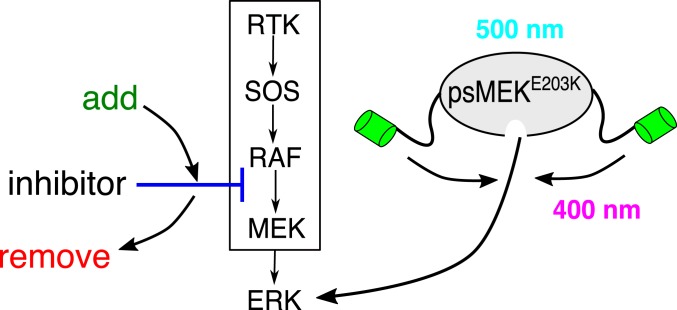
Finally, optimized psMEK enables a broad class of combinatorial perturbations in which light stimulation can be combined with inhibitors that act on signal transduction steps upstream of MEK-ERK interactions (Fig. 5). Such experiments would be an important step toward understanding the interplay between endogenous and ectopic signaling inputs that could underlie complex effects of ERK signaling in developmental systems (23).
Fig. 5.
Optimized psMEK enables combinatorial perturbations. Pharmacological inhibition upstream of MEK (blue) can be combined with optimized psMEK to modulate endogenous (ligand-dependent) and ectopic (optogenetic) ERK inputs independently.
Materials and Methods
Measuring Lethality in Drosophila.
The psMEK sequence was cloned into the transformation vector pTIGER at KpnI and PspXI sites using the TakaraBio in-fusion kit. The constructs were integrated into the second chromosome using Phi-C31 integration at attP sites over a CyO balancer. Virgin females of transgenic flies expressing psMEK were crossed to MTD-GAL4 males. The next generation from this cross was transferred to a breeding cage for embryo collection on an apple juice plate containing baker’s yeast. Cages were kept in a box lined with aluminum foil and exposed to LED plates emitting 405 and 505 nm light. The T1-3/4 through hole LEDs were purchased from https://www.superbrightleds.com/ and were soldered into a custom-built circuit board from SEEED Studio. Approximately 2 W was supplied by the Korad KA3005D power supply, purchased from https://www.amazon.com/. Embryos were collected for 2 to 3 h, after which the apple juice plate was removed from the cage and kept inside the light box for >36 h. Hatch rate at >36 h post collection was measured as a proxy for lethality.
Measuring the Elongated Embryo Phenotype in Zebrafish.
All forms of psMEK were expressed via injection of 100 mRNA transcribed using the Ambion mMessage Machine SP6 kit into the yolk at the one-cell stage. Each injection was made as a 500-pL bolus containing 200 pg/nL mRNA delivered with a PV280 Pneumatic PicoPump (World Precision Instruments). Injected embryos were kept in 5-cm Pyrex glass dishes placed in an aluminum-foil-lined box directly underneath the same LED boards described above to ensure uniform light exposure no more than 20 min after injection and incubated at 30 °C. Yolk aspect ratios were measured when embryos reached the tailbud stage (at 10 h.p.f. under these experiments). Embryos were oriented laterally and imaged on a Leica M205 FA stereomicroscope. All measurements were made using image analysis tools in FIJI. The statistics for the normalized yolk aspect ratios were calculated by generating the distribution of ratios measured for each perturbation to the “off” control (400 nm activation for 10 h) via sampling with replacement. The means and SDs of the generated distributions were calculated. Significance was indicated if the distributions of 2 means lay outside of the 95% confidence interval. All zebrafish protocols adhered to rules and regulations provided by the Princeton University Institutional Animal Care and Use Committee.
Measuring MEK Variant Kinase Activity.
Plasmid construction and protein purification.
The pET28a plasmids containing the N-terminally His6-tagged human MEK (His6-MEK) was obtained as a gift from E. Goldsmith, University of Texas Southwestern Medical Center, Dallas, TX. The phosphomimetic variant MEK-SSDD was constructed as previously described (23). E203K mutation was introduced into the MEK-SSDD plasmid by PCR-amplifying the plasmid with GTCAACTCCCGTGGGaagATCAAGCTCTGTGAC as the forward primer and GTCACAGAGCTTGATcttCCCACGGGAGTTGAC as the reverse primer. Ten nanograms of plasmid, 1 µM of each primer, 250 µM dNTP, 1× PfuUltra II buffer (Agilent Technologies), and 1 µL high-fidelity PfuUltra II Fusion HS DNA polymerase (Agilent Technologies 600674) were incubated together for the PCR (25 cycles, 55 °C denaturing temperature, 2-min extension). The PCR product was digested with DpnI and transformed into a homemade DH5α cell. The presence of the mutations was confirmed via Sanger sequencing. His6-ERK K52R and His6-MEK variants were purified exactly as previously described (23).
In vitro kinase reactions.
All reactions were carried out in the phosphorylation buffer containing 50 mM Hepes, pH 7.2, 20 mM MgCl2, and 5 mM ATP. Raf-activated MEK WT was prepared by reacting 6.5 µM of MEK with 0.05 µM of active c-Raf (c-Raf 14–325-D Millipore) for 1.5 h at 30 °C. For all MEK-ERK reactions, 0.67 µM MEK was reacted with 5 µM ERK K52R. Aliquots for specific time points were collected by adding 4 μL of the reaction to 1.5 μL of 4× sodium dodecyl sulfate (SDS) loading buffer (250 mM Tris⋅HCl, pH 6.8, 10% SDS, 0.01% bromophenol blue, 40% glycerol, 2.86 M β-mercaptoethanol). To break the disulfide bonds, the aliquots were boiled at 60 °C for 10 min, and 5 μL of each aliquot was loaded onto Wako SuperSep Phos-tag 12.5% 17 well gel (Wako 195–17991) and run at 175 V for 2 h and 40 min with Tris/Glycine/SDS running buffer (Bio-Rad 1610732). The gel was stained with Coomassie R staining solution (0.05% wt/vol Coomassie Brilliant Blue R, 25% isopropanol, and 10% acetic acid) for 30 min before destaining with 10% acetic acid until the bands were clearly visible. The phos-tag gels were imaged using the Bio-Rad ChemiDoc MP Imaging System. The intensities of the bands were quantified using ImageJ’s “gels” tool, and the quantified values were normalized so that the sum of all species at each time point was 1. At time 0, the unphosphorylated ERK had a concentration of 5 µM, and the concentration of each species can be calculated by normalizing the quantified gel intensity to that of the unphosphorylated ERK at time 0.
Generating Mutations in psMEK.
The psMEK construct was cloned from the plasmid published in ref. 15 into a PCS2+ vector via In-Fusion cloning (TakaraBio). The G128V mutation was made by Q5 mutagenesis. The E203K and D > S mutations at position 218 and 222 were made using the TakaraBio In-Fusion cloning mutagenesis protocol.
dpERK Immunofluorescence and Quantification in Zebrafish.
dpERK immunofluorescence was performed exactly as described in ref. 23. Dronpa was stained using a mouse antibody (MBL International) added at a dilution of 1:500. Injected and uninjected embryos were stained together in the same tube. Stained embryos were mounted laterally in 1.5% low-melt agarose in glass-bottom dishes. All embryos were imaged in the Nikon A1RSi laser scanning confocal microscope. Z-stacks were rendered in Imaris, and snap-shot images of the renderings were analyzed in FIJI. Embryos positive for Dronpa indicated where to draw the region of interest (ROI) for measuring dpERK staining intensity. Hand-drawn ROIs surrounding the margin in Dronpa-negative images were used to measure dpERK levels in the margins of WT embryos. The quantified dpERK levels in the nonmarginal cells were compared to WT margins by building the distribution of the ratio of levels in the nonmarginal cells to marginal cells after 100 samplings with replacement events (bootstrapping). The means and SDs of the nonmarginal:marginal ratios generated by bootstrapping were calculated. Significance was indicated if the distributions of 2 means were outside of the 95% confidence interval.
dpERK Immunofluorescence and Quantification in Drosophila.
Antibody staining for dpERK was performed as described in ref. 42. Embryos were collected for 1 h in the dark and aged for 2 h and 15 min in the 500-nm light box. Paraformaldehyde fixing was also performed under 500 nm illumination. OreR embryos were stained alongside psMEK and psMEKE203K-expressing embryos as WT controls. dpERK levels at 50% embryo length are reported.
Data Availability Statement.
All data are included in the manuscript and SI Appendix.
Supplementary Material
Acknowledgments
We thank Phillip Johnson and the Laboratory Animal Resources (LAR) staff for zebrafish care; Michael Lin for plasmids; Dr. Gary Laevsky and the Molecular Biology Confocal Microscopy Facility, which is a Nikon Center for Excellence, for microscopy support; and the Alexei Korrennykh laboratory (Molecular Biology, Princeton University) for the use of their qPCR machine and assistance with the thermo shift experiment. We thank Granton Jindal and all members of the S.Y.S. and R.D.B. laboratories for helpful discussions. A.L.P. thanks Rajeshwari Enjeti for her contributions to developing p-Smad5 immunofluorescence and quantification procedures in zebrafish. This work was supported by the National Science Foundation Graduate Research Fellowship Program Grant DGE-1656466 (to A.L.P.). S.Y.S. and R.D.B. were supported by NIH Grant GM086537. J.E.T. was supported by the National Science Foundation CAREER Award 1750663. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912320116/-/DCSupplemental.
References
- 1.Patel A. L., Shvartsman S. Y., Outstanding questions in developmental ERK signaling. Development 145, dev143818 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Johnson H. E., Toettcher J. E., Illuminating developmental biology with cellular optogenetics. Curr. Opin. Biotechnol. 52, 42–48 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang K., Cui B., Optogenetic control of intracellular signaling pathways. Trends Biotechnol. 33, 92–100 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wilson M. Z., Ravindran P. T., Lim W. A., Toettcher J. E., Tracing information flow from Erk to target gene induction reveals mechanisms of dynamic and combinatorial control. Mol. Cell 67, 757–769.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Johnson H. E., Toettcher J. E., Signaling dynamics control cell fate in the early Drosophila embryo. Dev. Cell 48, 361–370.e3 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Toettcher J. E., Gong D., Lim W. A., Weiner O. D., Light-based feedback for controlling intracellular signaling dynamics. Nat. Methods 8, 837–839 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Johnson H. E., et al. , The spatiotemporal limits of developmental Erk signaling. Dev. Cell 40, 185–192 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Toettcher J. E., Gong D., Lim W. A., Weiner O. D., Light control of plasma membrane recruitment using the Phy–Pif System. Methods Enzymol. 497, 409–423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duan L., et al. , Optical activation of TrkA signaling. ACS Synth. Biol. 7, 1685–1693 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnamurthy V. V., et al. , Reversible optogenetic control of kinase activity during differentiation and embryonic development. Development 143, 4085–4094 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toettcher J. E., Weiner O. D., Lim W. A., Using optogenetics to interrogate the dynamic control of signal transmission by the Ras/Erk module. Cell 155, 1422–1434 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bugaj L. J., et al. , Cancer mutations and targeted drugs can disrupt dynamic signal encoding by the Ras-Erk pathway. Science 361, eaao3048 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boykevisch S., et al. , Regulation of ras signaling dynamics by Sos-mediated positive feedback. Curr. Biol. 16, 2173–2179 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Gureasko J., et al. , Membrane-dependent signal integration by the Ras activator Son of sevenless. Nat. Struct. Mol. Biol. 15, 452–461 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou X. X., Fan L. Z., Li P., Shen K., Lin M. Z., Optical control of cell signaling by single-chain photoswitchable kinases. Science 355, 836–842 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou X. X., Chung H. K., Lam A. J., Lin M. Z., Optical control of protein activity by fluorescent protein domains. Science 338, 810–814 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Resing K. A., et al. , Determination of v-Mos-catalyzed phosphorylation sites and autophosphorylation sites on MAP kinase kinase by ESI/MS. Biochemistry 34, 2610–2620 (1995). [DOI] [PubMed] [Google Scholar]
- 18.Mansour S. J., Candia J. M., Matsuura J. E., Manning M. C., Ahn N. G., Interdependent domains controlling the enzymatic activity of mitogen-activated protein kinase kinase 1. Biochemistry 35, 15529–15536 (1996). [DOI] [PubMed] [Google Scholar]
- 19.Mansour S. J., et al. , Transformation of mammalian cells by constitutively active MAP kinase kinase. Science 265, 966–970 (1994). [DOI] [PubMed] [Google Scholar]
- 20.Jindal G. A., et al. , How activating mutations affect MEK1 regulation and function. J. Biol. Chem. 292, 18814–18820 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal Y., Schüpbach T., Shvartsman S. Y., A quantitative model of developmental RTK signaling. Dev. Biol. 442, 80–86 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Furriols M., Casanova J., In and out of Torso RTK signalling. EMBO J. 22, 1947–1952 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goyal Y., et al. , Divergent effects of intrinsically active MEK variants on developmental Ras signaling.Nat. Genet. 49, 465–469 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fürthauer M., Thisse C., Thisse B., A role for FGF-8 in the dorsoventral patterning of the zebrafish gastrula. Development 124, 4253–4264 (1997). [DOI] [PubMed] [Google Scholar]
- 25.Fürthauer M., Van Celst J., Thisse C., Thisse B., Fgf signalling controls the dorsoventral patterning of the zebrafish embryo. Development 131, 2853–2864 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Anastasaki C., Estep A. L., Marais R., Rauen K. A., Patton E. E., Kinase-activating and kinase-impaired cardio-facio-cutaneous syndrome alleles have activity during zebrafish development and are sensitive to small molecule inhibitors. Hum. Mol. Genet. 18, 2543–2554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jopling C., van Geemen D., den Hertog J., Shp2 knockdown and Noonan/LEOPARD mutant Shp2-induced gastrulation defects. PLoS Genet. 3, e225 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jindal G. A., et al. , In vivo severity ranking of Ras pathway mutations associated with developmental disorders. Proc. Natl. Acad. Sci. U.S.A. 114, 510–515 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dephoure N., Gould K. L., Gygi S. P., Kellogg D. R., Mapping and analysis of phosphorylation sites: A quick guide for cell biologists. Mol. Biol. Cell 24, 535–542 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunter T., Why nature chose phosphate to modify proteins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 367, 2513–2516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nikolaev S. I., et al. , Exome sequencing identifies recurrent somatic MAP2K1 and MAP2K2 mutations in melanoma. Nat. Genet. 44, 133–139 (2011). [DOI] [PubMed] [Google Scholar]
- 32.Burack W. R., Sturgill T. W., The activating dual phosphorylation of MAPK by MEK is nonprocessive. Biochemistry 36, 5929–5933 (1997). [DOI] [PubMed] [Google Scholar]
- 33.Ferrell J. E. Jr, Bhatt R. R., Mechanistic studies of the dual phosphorylation of mitogen-activated protein kinase. J. Biol. Chem. 272, 19008–19016 (1997). [DOI] [PubMed] [Google Scholar]
- 34.Piala A. T., Humphreys J. M., Goldsmith E. J., MAP kinase modules: The excursion model and the steps that count. Biophys. J. 107, 2006–2015 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Krens S. F. G., et al. , Distinct functions for ERK1 and ERK2 in cell migration processes during zebrafish gastrulation. Dev. Biol. 319, 370–383 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Wong K.-L., Akiyama R., Bessho Y., Matsui T., ERK activity dynamics during zebrafish embryonic development. Int. J. Mol. Sci. 20, E109 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tidyman W. E., Rauen K. A., The RASopathies: Developmental syndromes of Ras/MAPK pathway dysregulation. Curr. Opin. Genet. Dev. 19, 230–236 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rauen K. A., The RASopathies. Annu. Rev. Genomics Hum. Genet. 14, 355–369 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Glickman N. S., Kimmel C. B., Jones M. A., Adams R. J., Shaping the zebrafish notochord. Development 130, 873–887 (2003). [DOI] [PubMed] [Google Scholar]
- 40.Myers D. C., Sepich D. S., Solnica-Krezel L., Bmp activity gradient regulates convergent extension during zebrafish gastrulation. Dev. Biol. 243, 81–98 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Mullins M. C., et al. , Genes establishing dorsoventral pattern formation in the zebrafish embryo: The ventral specifying genes. Development 123, 81–93 (1996). [DOI] [PubMed] [Google Scholar]
- 42.Lim B., et al. , Dynamics of inductive ERK signaling in the Drosophila embryo. Curr. Biol. 25, 1784–1790 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are included in the manuscript and SI Appendix.