Significance
Regulatory T (Treg) cells play an essential role in maintaining immune homeostasis. Studying factors that control Treg differentiation and function are critically important to understand immune homeostasis. In this manuscript, we discovered that transcription factor Hhex exerts an inhibitory effect on Treg cell differentiation and function. Hhex-overexpressing Treg cells lose their Foxp3 expression and fail to suppress immune responses. Hhex directly binds to Foxp3 protein and the Foxp3 locus and inhibits expression of Foxp3 and its target genes. Thus, Hhex plays an essential role in inhibiting Treg cell differentiation and function via inhibition of Foxp3. This study will benefit clinical research in developing a therapeutic strategy for Treg cell-related diseases.
Keywords: regulatory T cell, Hhex, Foxp3
Abstract
Regulatory T (Treg) cells play an essential role in maintaining immune homeostasis, but the suppressive function of Treg cells can be an obstacle in the treatment of cancer and chronic infectious diseases. Here, we identified the homeobox protein Hhex as a negative regulator of Treg cells. The expression of Hhex was lower in Treg cells than in conventional T (Tconv) cells. Hhex expression was repressed in Treg cells by TGF-β/Smad3 signaling. Retroviral overexpression of Hhex inhibited the differentiation of induced Treg (iTreg) cells and the stability of thymic Treg (tTreg) cells by significantly reducing Foxp3 expression. Moreover, Hhex-overexpressing Treg cells lost their immunosuppressive activity and failed to prevent colitis in a mouse model of inflammatory bowel disease (IBD). Hhex expression was increased; however, Foxp3 expression was decreased in Treg cells in a delayed-type hypersensitivity (DTH) reaction, a type I immune reaction. Hhex directly bound to the promoters of Foxp3 and other Treg signature genes, including Il2ra and Ctla4, and repressed their transactivation. The homeodomain and N-terminal repression domain of Hhex were critical for inhibiting Foxp3 and other Treg signature genes. Thus, Hhex plays an essential role in inhibiting Treg cell differentiation and function via inhibition of Foxp3.
Regulatory T (Treg) cells are a distinct subset of CD4 T cells that regulate immune responses and maintain immune homeostasis. Treg cells are generated in the thymus (tTreg cells) or differentiate in the periphery (pTreg cells), then circulate through lymphoid organs and reside in tissues to control a variety of immune responses (1–4). However, under inflammatory or pathogenic conditions, Treg cells become unstable, lose their immunosuppressive properties, and become more like effector T cells, resulting in their failure to regulate autoimmune or inflammatory diseases (5). On the other hand, in the tumor microenvironment, accumulation of Treg cells can inhibit antitumor immunity. Tumor-infiltrating Treg cells are a major cause of poor clinical outcomes (6). Thus, with increasing numbers of clinical trials of various immune therapies, studying Treg cell regulators is becoming even more important.
Forkhead box P3 (Foxp3) serves as a lineage specification transcription factor for Treg cells. Foxp3 controls both the identity and function of Treg cells. Ectopic expression of Foxp3 in conventional CD4 T (Tconv) cells induces a regulatory phenotype and function, whereas deletion of the Foxp3 gene in Treg cells results in the loss of their suppressive function (7–10). Foxp3 also induces the expression of Treg signature genes including Il2ra (encodes CD25), Ctla4, Tnfrsf18 (encodes GITR), and Icos (8, 11, 12). CD25 (interleukin-2 [IL-2] receptor alpha-chain) is required for Treg cell survival and IL-2 consumption as part of Treg-mediated suppression (13). CTLA4 mediates Treg-dependent down-regulation of CD80 and CD86 on antigen-presenting cells (14). Along with Foxp3, CD25 and CTLA4 are commonly accepted as markers of Treg cells. Recently, Treg-specific superenhancers in genes such as Foxp3, Il2ra, and Ctla4 have been reported (15). These sites are in a poised state at the early stages of tTreg cell differentiation, which allows other transcription factors to bind and regulate their expression.
Transforming growth factor beta (TGF-β), which is critical for maintaining pTreg cells (16), can also induce Foxp3 in naïve CD4 T cells and promote their differentiation into induced Treg cells (iTreg cells) with suppressive function (17). TGF-β phosphorylates Smad3, resulting in the formation of Smad3/Smad4 heterodimers, which can translocate to the nucleus and bind to the Foxp3 enhancer (conserved noncoding sequence 1 [CNS1]), inducing Foxp3 expression (18, 19).
Many transcription factors have been shown to transactivate the regulatory elements of Foxp3, which include a promoter and 3 intronic enhancers (CNS1, CNS2, and CNS3). Signal transducer and activator of transcription 5 (STAT5; downstream of IL-2), c-Rel, nuclear factor of activated T cells (NFAT), Forkhead box O (Foxo), activator protein-1 (AP-1), and more have been shown to directly bind and activate Foxp3 regulatory elements (4, 20–23).
By contrast, only a few negative regulators of Foxp3 have been reported. GATA-binding protein 3 (GATA3), the master regulator of Th2 cells, and RAR-related orphan receptor γt (RORγt), the master regulator of Th17 cells, bind to the Foxp3 promoter to repress Foxp3 expression during Th2 or Th17 differentiation, respectively (24, 25). In addition, STAT3, which lies downstream of IL-6, competes with STAT5 to down-regulate Foxp3 (23). Our group also identified yin yang 1 (YY1) as an inhibitor of Foxp3 expression and activity (26), but negative regulators of Foxp3 and Treg cell activity and function need to be further studied.
Hematopoietically expressed homeobox (Hhex) is a highly conserved transcription factor belonging to the homeobox protein family. The human and murine Hhex proteins are 94% homologous, with only a single amino acid difference in the homeodomain (27, 28). Hhex was first identified in hematopoietic cells (29, 30). Hhex is expressed in early hematopoietic progenitors and is down-regulated during differentiation (31, 32). Hhex has been reported to play an essential role in B cell lineages, but is not well studied in T cells because of its low expression level (32, 33). Hhex is a homooligomer-forming transcription factor that regulates target genes directly by binding to DNA through homeodomains or indirectly by modulating other transcription factors through protein–protein interactions (27, 34). Hhex can both enhance and repress target genes, but it has been better characterized as a transcriptional repressor (27).
In this study, we examined the role of Hhex in Treg cells. Hhex expression was lower in Treg cells than in Tconv cells, and was down-regulated by TGF-β/Smad3 signaling. Ectopic expression of Hhex impaired the identity and function of Treg cells. Hhex directly bound to the Foxp3 locus and to the promoters of Treg signature genes such as Il2ra and Ctla4. Furthermore, Hhex-overexpressing Treg cells showed lower expression of Foxp3 and Treg signature genes and could not prevent mouse inflammatory bowel disease (IBD). These results strongly suggest that Hhex is an important negative regulator of the Treg lineage.
Results
Expression of Hhex Is Low in Treg Cells.
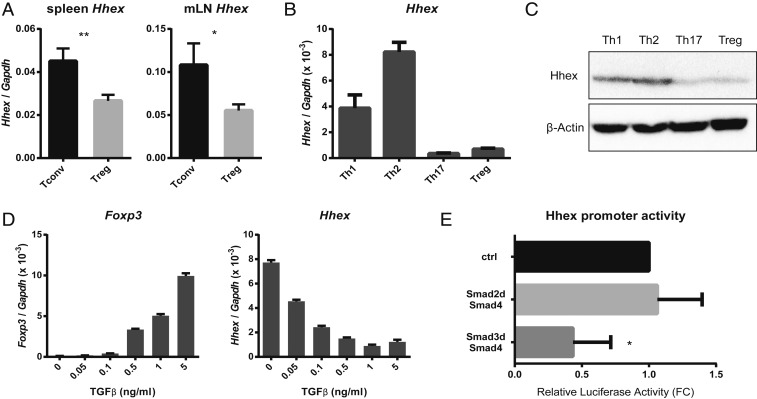
To identify regulators of Treg cells, the transcriptomes of Th2, Th9, and Treg cells were compared by microarray analysis. Naïve CD4 T cells were isolated from mouse spleens and cultured under each differentiation condition. All conditions included anti-CD3/anti-CD28 stimulation and IL-2, with addition of IL-4 for Th2 cells, IL-4 and TGF-β for Th9 cells, and TGF-β for Treg cells. To identify candidates for direct suppressors of Treg differentiation or Foxp3, cell differentiation-related (Gene Ontology Consortium) transcription factors (gene cards) that were expressed at lower levels in Treg cells than in Th2 and Th9 cells were selected (SI Appendix, Fig. S1; Gene Expression Omnibus [GEO] accession no. GSE139297). The homeobox gene Hhex was one of the genes with the largest difference in expression. To confirm the expression of Hhex in CD4 T cells, CD4+ CD25− Tconv cells and CD4+ CD25+ Treg cells were isolated from mouse spleens and mesenteric lymph nodes (mLNs) and Hhex mRNA was evaluated by quantitative reverse transcription PCR (qRT-PCR) (Fig. 1A). Expression of Hhex was significantly lower in Treg cells. Naïve CD4 T cells were also differentiated in vitro, and the expression of Hhex mRNA (Fig. 1B) and protein (Fig. 1C) was measured in each subset. The expression of Hhex was notably low in Th17 and Treg cells.
Fig. 1.
Expression of Hhex is low in Treg cells and is repressed by TGF-β/Smad3 signaling. (A) CD4+ CD25− (Tconv) and CD4+ CD25+ (Treg) cells were sorted from mouse spleens and mLNs. Hhex mRNA expression was measured by qRT-PCR. (B and C) Naïve CD4 T cells were isolated from splenocytes and differentiated into various CD4 T cell subsets (Th1, Th2, Th17, and Treg) for 3 d. Relative Hhex mRNA expression was determined by qRT-PCR (B), and Hhex protein expression was detected by immunoblot analysis (C). (D) Naïve CD4 T cells were cultured for 3 d under Th0 conditions in the presence of the indicated concentrations of TGF-β. Relative expression of Foxp3 and Hhex mRNA was measured by qRT-PCR. (E) Transactivation of the Hhex promoter by Smad proteins was measured by transient reporter assay. EL4 cells were transfected with the Hhex promoter–luciferase (LUC) reporter construct together with an empty vector (ctrl) or expression vectors for Smad2d and Smad4 or Smad3d and Smad4. Promoter activities are shown as the fold change (FC) relative to the ctrl vector. All error bars indicate the SD, and P values were calculated using Student’s t tests. *P < 0.05, **P < 0.01.
Hhex Expression Is Inhibited by the TGF-β/Smad3 Signaling Pathway.
The low expression of Hhex in Th17 and Treg cells, which require TGF-β for differentiation, prompted us to investigate whether TGF-β regulates Hhex expression. Naïve CD4 T cells were cultured under anti-CD3/anti-CD28 stimulation with IL-2 (1 ng/mL) and the indicated concentrations of TGF-β (Fig. 1D), and Hhex and Foxp3 mRNA levels were examined. The expression of Foxp3 was increased by TGF-β, as reported (17), but Hhex expression was significantly decreased by TGF-β in a concentration-dependent manner (Fig. 1D).
In Treg cells TGF-β signaling is transmitted through the phosphorylation of Smad3, but not Smad2, and its binding with a common partner, Smad4 (19, 35). We made a reporter construct containing the Hhex promoter (nucleotides −309/+22, as previously reported) (36) and measured the transactivation of the Hhex promoter after ectopic expression of various Smad proteins (Fig. 1E). Expression of Smad3 along with Smad4 significantly repressed the activity of Hhex promoter, but expression of Smad2 along with Smad4 did not affect the promoter activity. Taken together, these results demonstrate that TGF-β inhibits expression of Hhex in CD4 T cells through Smad3.
Hhex Negatively Regulates the Differentiation and Stability of Treg Cells.
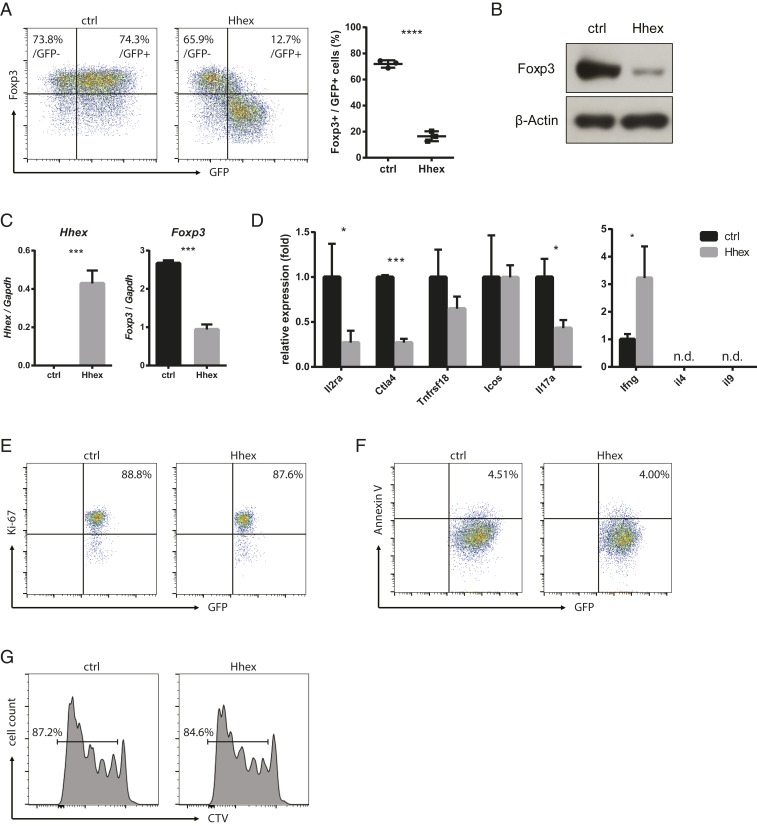
To characterize the role of Hhex in Treg differentiation, naïve CD4 T cells were transduced with a retroviral vector containing GFP and the Hhex gene during their in vitro differentiation into Treg cells. Foxp3 expression was dramatically decreased in Hhex-transduced Treg cells compared with empty vector-transduced (control) Treg cells (Fig. 2 A–C). However, the expression of Foxp3 in GFP (nontransduced) cells was not different between Hhex-transduced and control cells, suggesting that the effect of Hhex on Treg cells is cell intrinsic. In Hhex-transduced cells, the higher the GFP expression, the lower was the expression of Foxp3 (Fig. 2A). Moreover, Treg signature genes, including Il2ra and Ctla4, were markedly reduced in Hhex-overexpressing Treg cells (Fig. 2D). These genes are directly involved in both the development and function of Treg cells (13, 14). The expression of Il17a, a Th17 cytokine, was also decreased by Hhex, but the expression of Ifng, a Th1 cytokine, was increased (Fig. 2D). Cell proliferation and apoptosis of Hhex-transduced Treg cells, examined by Ki-67 and Annexin V staining, were not different from those of controls (Fig. 2 E and F), The proliferation rates examined by CellTrace Violet (CTV) in control and Hhex-overexpressing Treg cells stimulated with anti-CD3/CD28 beads were comparable (Fig. 2G). These results suggest that the decreased Foxp3 expression in Hhex-transduced Treg cells was not due to cell proliferation or death.
Fig. 2.
Hhex overexpression inhibits the expression of Foxp3 and Treg signature genes. (A–F) Naïve CD4 T cells were stimulated for 24 h and transduced with a retroviral vector expressing GFP with or without (ctrl) Hhex. Cells were cultured for an additional 2 d under iTreg differentiation conditions. Foxp3 expression was detected by flow cytometry, and the ratio of Foxp3+ cells among GFP+ (transduced) cells is shown (A). GFP+ cells were sorted, and the relative amount of Foxp3 protein was detected by immunoblot analysis (B). The expression of Hhex, Foxp3, and other Treg signature genes was measured by qRT-PCR (C and D). Proliferation of the GFP+ cells was determined by Ki-67 staining (E). Apoptosis was measured by Annexin V staining (F). (G) Transduced iTreg cells were stained with CTV and cultured for an additional 4 d in the presence of anti-CD3/CD28 beads and IL-2. All error bars represent the SD, and P values were calculated using Student’s t tests. *P < 0.05, ***P < 0.001, ****P < 0.0001. n.d., not detected.
Next, the effect of Hhex overexpression was examined in tTreg cells, which had already developed in vivo. CD25+ CD4+ tTreg cells were isolated from mouse spleens and transduced with the Hhex expression vector. Hhex also reduced Foxp3 expression significantly in tTreg cells, although the reduction was not as great as in iTreg cells (SI Appendix, Fig. S2 A and B). This was an interesting finding, because tTreg cells are known to be very stable (7). Treg signature genes, including Il2ra, Ctla4, and Icos, were also decreased by Hhex (SI Appendix, Fig. S2C). ICOS is involved in Treg cell function and is also used as a marker of highly suppressive T cells (12, 37, 38). By contrast, Ifng expression was increased in response to Hhex expression (SI Appendix, Fig. S2C). Collectively, these data indicate that Hhex interferes with the differentiation of iTreg cells and disrupts the stability of tTreg cells by reducing the expression of Foxp3 and Treg signature genes.
Hhex Promotes a Th1-Like Phenotype in CD4 T Cells.
In both iTreg cells and tTreg cells, ectopic expression of Hhex induced Ifng while reducing Foxp3 (Fig. 2D and SI Appendix, Fig. S2C). Therefore, we examined the effect of Hhex on effector CD4 T cell differentiation by overexpressing Hhex in Th0 cells, obtained by culturing with only anti-CD3/anti-CD28 stimulation and IL-2 (1 ng/mL). Only Ifng was increased, while Il4, Il17a, and Foxp3 were all decreased (SI Appendix, Fig. S3A). Notably, Tbx21 (encoding T-bet) expression was not altered (SI Appendix, Fig. S3A). Hhex was also introduced into Th1 and Th17 cells. Consistent with the above results, expression of IFN-γ was increased in Hhex-transduced Th1 cells, while IL-17a expression was decreased in Hhex-transduced Th17 cells (SI Appendix, Fig. S3 B and C). To examine whether altered IFN-γ expression affects Hhex-mediated suppression of Foxp3, we blocked IFN-γ activity with a neutralizing antibody during iTreg differentiation (SI Appendix, Fig. S4). Anti–IFN-γ antibody only slightly increased Foxp3 expression in both control and Hhex-overexpressing iTreg cells (SI Appendix, Fig. S4A). The inhibition of Foxp3 by Hhex was maintained, regardless of IFN-γ presence (SI Appendix, Fig. S4B), and the extent of the Hhex effect also did not differ much (SI Appendix, Fig. S4C). Together, Hhex increases the expression of IFN-γ in differentiating CD4 T cells, while IFN-γ is not critical for suppression of Foxp3 expression.
Hhex Inhibits the Immunosuppressive Function of Treg Cells.
To examine whether Hhex affects the function of Treg cells, in vitro immunosuppression assays were performed. iTreg and tTreg cells were transduced with either the empty vector or the Hhex expression vector, and GFP+, successfully transduced cells, were sorted. Murine naïve CD4 T cells were labeled with PKH26, a lipophilic red fluorescent dye, and mixed with the transduced Treg cells in various ratios. The cocultures were incubated for 3 d in the presence of anti-CD3/anti-CD28 beads, and the proliferation of the naïve CD4 T cells was measured. Both Hhex-overexpressing iTreg and tTreg cells failed to effectively suppress the proliferation of naïve CD4 T cells, even at a low naïve T cell:iTreg cell ratio (Fig. 3 A and B), suggesting that their suppressive function was impaired.
Fig. 3.
Hhex impairs the immunosuppressive function of Treg cells in vitro and in vivo. (A and B) The in vitro suppressive activity of Treg cells was measured by analyzing the PKH26+ proliferating cells among naïve CD4 T cells cocultured with the indicated ratios of iTreg cells (A) or a 1:1 ratio of tTreg cells (B). Empty vector (ctrl)- or Hhex-transduced (GFP+) Treg cells were sorted for the assay. (C–G) In vivo suppressive activity was assessed in an inflammatory bowel disease model. CD4+ CD25+ tTreg cells were sorted from mouse splenocytes and retrovirally transduced with an empty vector (ctrl) or a Hhex expression vector. Naïve CD4 T (CD4+ CD25− CD62L+ CD45RBhi) cells were administered to RAG1−/− mice alone or together with ctrl-transduced or Hhex-expressing Treg cells. Mice were killed at day 22. (C) Weight loss of mice. (D) Morphology of colons, spleens, and mLNs. (E) Hematoxylin and eosin (H&E) staining of colon sections. (Scale bar, 200 μm.) (F and G) Populations of cytokine-expressing effector T cells in the spleen and mLNs. All error bars represent the SD, and P values were calculated using Student’s t tests. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Next, the function of Hhex-overexpressing Treg cells was examined in vivo using a transfer-induced colitis model, a mouse model of IBD. CD4+ CD62L+ CD45RBhi naïve CD4 T cells were transferred into RAG1-deficient recipient mice alone or in combination with CD4+ CD25+ tTreg cells (control or Hhex overexpressing). Transfer of control tTreg cells along with naïve CD4 T cells efficiently blocked the signs of colitis, including weight loss (Fig. 3C), colon swelling, splenomegaly (Fig. 3D), and infiltration of the lamina propria by inflammatory cells (Fig. 3E). However, Hhex-expressing tTreg cells failed to suppress these inflammatory phenotypes (Fig. 3 B–D). Adoptively transferred naïve CD4 T cells differentiated into effector CD4 T cells, including IFN-γ+ Th1 and IL-17a+ Th17 cells, when they were injected alone (Fig. 3 F and G). Cotransfer of control tTreg cells successfully suppressed the differentiation of naïve CD4 T cells, but transfer of Hhex-overexpressing tTreg cells did not (Fig. 3 F and G), suggesting that Hhex impairs the suppressive function of tTreg cells in vivo. Together, these results suggest that Hhex inhibits the immunosuppressive function of Treg cells both in vitro and in vivo.
Hhex Expression Is Increased under Inflammatory Conditions.
Treg cells can lose Foxp3 expression and suppressive activity under inflammatory conditions (5, 39, 40). Our data above suggest that Hhex may play a role in type I immune responses. To examine the role of Hhex in Treg cells under physiologically relevant conditions, we performed delayed-type hypersensitivity (DTH) reaction, a type I immune response. Sensitization (day 0) and challenge (day 7) with topical application of oxazolone on mice back skin induced epidermal thickening and increased cell infiltration in the skin at day 9 (Fig. 4A). The total number of CD4 T cells and the proportion of IFN-γ+ cells among the CD4 T cells were increased in the draining lymph nodes, indicating that type I inflammatory responses were developed in the mice (Fig. 4 B and C). Treg cells were isolated from the draining lymph nodes of control and DTH-induced mice, and gene expression of Hhex, Foxp3, and Ifng was measured by qRT–PCR. Expression of Hhex was increased in Treg cells under the inflammatory environment (Fig. 4D). In addition, Foxp3 expression was decreased (Fig. 4E) and Ifng expression was increased (Fig. 4F). These results show that Hhex in Treg cells may play a role under physiological inflammatory conditions.
Fig. 4.
Hhex expression is increased under type I inflammatory conditions in vivo. DTH reaction was induced by double topical applications of oxazolone (day 0, sensitization and day 7, challenge) on the mice back skin. The mice were killed at 2 d after challenge and analyzed for disease phenotypes. (A) H&E staining of back skin section. (B) Total CD4 T cell numbers in draining lymph nodes (dLNs) of untreated control and oxazolone-treated DTH groups were counted. (C) Populations of cytokine-expressing CD4 T cells in dLNs. (D–F) CD4+ CD25+ Treg cells were isolated from dLNs of control and DTH mice. Total RNA was extracted and expression of Hhex, Foxp3, and Ifng was measured by qRT-PCR. All error bars represent the SD, and P values were calculated using Student’s t tests. *P < 0.05, **P < 0.01.
Hhex Directly Binds to and Represses Foxp3 and Other Treg Signature Genes.
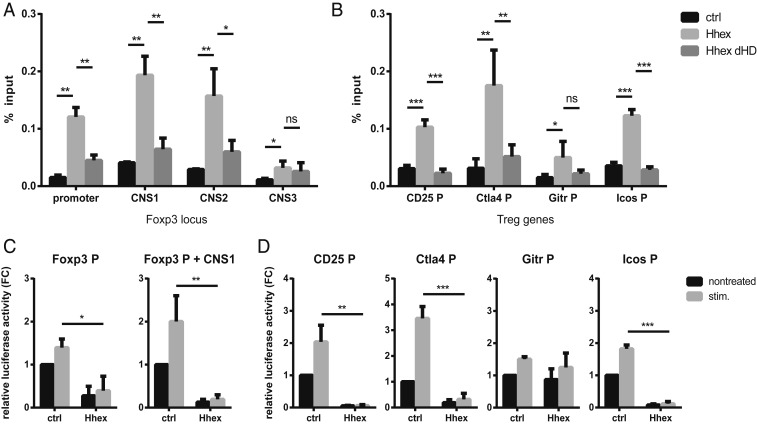
Hhex is a DNA-binding transcription factor that has been characterized mostly as a repressor (27). Hhex binds to DNA with its homeodomain, but the target sequence has yet to be accurately defined. Some studies suggested that repeats of core 5′-ATTA-3′ or 5′-TAAT-3′ sequences are important for the binding specificity of Hhex (31, 34). We found multiple repeats of these sequences at the Foxp3 locus, so we investigated whether Hhex regulates Foxp3 gene expression through direct binding, using chromatin immunoprecipitation (ChIP) assays. A mutant form of Hhex lacking the homeodomain (ΔHD), the DNA-binding domain, was used as a negative control. Both the WT and ΔHD Hhex genes were FLAG tagged at the C terminus. An anti-FLAG antibody was used to precipitate WT or ΔHD Hhex from transduced iTreg cells. WT Hhex, but not ΔHD Hhex, bound directly to the Foxp3 promoter at CNS1 and CNS2, but not at CNS3 (Fig. 5A). Hhex also bound to the promoters of some Treg signature genes, including Il2ra, Ctla4, and Icos (Fig. 5B). These results suggest that Foxp3 and some Treg signature genes are direct targets of the transcription factor Hhex.
Fig. 5.
Hhex binds to and suppresses the Foxp3 locus and promoters of Treg signature genes. (A and B) Binding of Hhex to the Foxp3 locus and promoters of Treg signature genes was measured by ChIP. iTreg cells transduced with empty (ctrl), Hhex-FLAG, or HhexΔHD-FLAG (without the homeodomain) expression vectors were used. Enrichment of the target DNA was quantified by qPCR following immunoprecipitation with a FLAG antibody. (C and D) Transactivation of the promoters of Foxp3 and Treg signature genes by Hhex was measured by transient reporter assay. EL4 cells were transfected with Foxp3P-, Foxp3P+CNS1-, CD25P-, Ctla4P-, GitrP-, or IcosP–LUC reporter constructs together with an empty vector (ctrl) or a Hhex expression vector. After 16 h, cells were left untreated (nontreated) or were stimulated with PMA and ionomycin (stim.). Promoter activity is shown as the fold change (FC) relative to the nontreated ctrl. All error bars indicate the SD, and P values were calculated using Student’s t tests. *P < 0.05, **P < 0.01, ***P < 0.001. ns, not significant.
Next, transient reporter assays were used to examine whether binding of Hhex regulates the activity of target gene promoters. EL4 T cells were transfected with reporter constructs containing the promoter of each gene together with an empty vector or a Hhex expression vector. Stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin was used to mimic activation by TCR signaling and costimulation. With or without CNS1, the promoter activity of Foxp3 was significantly decreased by Hhex overexpression (Fig. 5C). Other binding targets, such as the promoters of IL2ra, Ctla4, and Icos, were also inhibited by Hhex (Fig. 5D). The Tnfrsf18 promoter, where Hhex binding was not detected, was not affected by Hhex transfection (Fig. 5 B and D). Taken together, these results suggest that Hhex regulates the expression of Foxp3, IL2ra, Ctla4, and Icos by directly binding to their gene loci.
Each of the Hhex Domains Has a Specific Role in Inhibiting Treg Differentiation.
Hhex consists of 3 major domains, an N-terminal proline-rich (NT) domain, which represses transcriptional activity, the homeodomain (HD), which binds to DNA, and an acidic C-terminal (CT) domain, which induces transcriptional activity (27, 41, 42). The first 50 amino acids of the NT domain also have a role in the dimerization of Hhex (43). Therefore, Hhex was divided into 4 domains (NT1, NT2, HD, and CT) (Fig. 6A), and 4 mutants were constructed with each domain deleted (ΔNT1, ΔNT2, ΔHD, and ΔCT). To characterize the function of each domain in Treg differentiation, each mutant Hhex construct was transduced into naïve CD4 T cells, and the cells were differentiated into iTreg cells. In contrast to the WT protein, the 4 mutant proteins could not inhibit Foxp3 expression in iTreg cells (Fig. 6 B–E). The NT and HD, but not the CT, domains were essential for the inhibition of the Treg signature genes Il2ra and Ctla4 (Fig. 6E).
Fig. 6.
Domain-deleted forms of Hhex lose their effects in Treg cells. (A) Schematic diagram of Hhex domains showing the NT1 (N-terminal 1), NT2 (N-terminal 2), HD (homeodomain), and CT (C-terminal) domains. (B–E) Retroviral vectors encoding WT Hhex or deletion mutants of Hhex without each domain (ΔNT1, ΔNT2, ΔHD, or ΔCT) were transduced into iTreg cells. Foxp3+ cells were measured by flow cytometry (B), and the ratio of Foxp3+ cells among GFP+ cells is shown (C). Error bars represent the SD, and P values were calculated using Student’s t tests. **P < 0.01, ****P < 0.0001. GFP+ cells were sorted, and the relative amount of Foxp3 protein was measured by immunoblot analysis (D). mRNA levels of Foxp3 and other Treg signature genes were measured by qRT-PCR (E). (F) Binding of Hhex and Foxp3 was determined by co-IP. Hhex-FLAG and Foxp3 were overexpressed in 293T cells. Cell lysates were immunoprecipitated with anti-Foxp3, anti-FLAG (for Hhex), or normal IgG (as a negative control). Then, the proteins were immunoblotted with anti-Hhex or anti-Foxp3 antibodies. (G) WT Hhex-FLAG, ΔNT1-FLAG, ΔNT2-FLAG, ΔHD-FLAG, or ΔCT-FLAG was cotransfected along with Foxp3 into 293T cells. Cell lysates were immunoprecipitated with an anti-FLAG antibody. The immunoprecipitated proteins were detected with an anti-Foxp3 antibody. Input cell lysates were immunoblotted with anti-Foxp3 and anti-FLAG antibodies. (H and I) Transactivation of the promoters of Il2ra and Ctla4 by Foxp3 and Hhex was measured by transient reporter assay. EL4 cells were transfected with CD25P- or Ctla4P–LUC reporter constructs together with a Foxp3 expression vector and a vector expressing full-length or domain-deleted forms of Hhex (dNT1, dNT2, dHD, or dCT). Promoter activities are shown as the fold change (FC) relative to ctrl vector-transduced cells. Error bars represent the SD, and P values were calculated using Student’s t tests. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. ns, not significant.
Hhex can interact with many cellular proteins, mainly through its NT domain (27). We therefore investigated whether Hhex physically interacts with Foxp3 and regulates its activity. Coimmunoprecipitation (co-IP) assays revealed a binding interaction between Hhex and Foxp3 (Fig. 6F). To examine which domain of Hhex is important for binding to Foxp3, co-IPs were performed with full-length Hhex and the 4 mutants. Deletion of the NT2 domain of Hhex caused loss of the interaction with Foxp3 (Fig. 6G), suggesting that Hhex directly binds to Foxp3 through its NT2 domain. To further investigate whether Hhex binding impacts the stability of Foxp3 protein, we coexpressed WT or ΔNT2 Hhex with Foxp3 and measured the stability of Foxp3 by immunoblot assay after treating with a translation inhibitor, cycloheximide (SI Appendix, Fig. S5). The stability of Foxp3 protein was not altered by Hhex binding.
Since Foxp3 has been reported to induce CD25 and CTLA4 expression in Treg cells through direct binding to their genes (11), we examined whether Hhex and its physical interaction with Foxp3 affects Foxp3-dependent transactivation of the Il2ra and Ctla4 genes. The full-length Hhex or 4 domain-deleted mutants of Hhex were transfected into EL4 T cells together with Foxp3 and reporter constructs containing the promoters of Il2ra and Ctla4. Full-length Hhex significantly repressed the activity of the Il2ra and Ctla4 promoters even in the presence of Foxp3 (Fig. 6 H and I). However, the mutant Hhex proteins were only able to partially repress these target genes compared with the full-length Hhex. All 4 domains of Hhex contributed to the regulation of Foxp3 target genes. These results suggest that all 4 domains of Hhex (the HD, as a DNA-binding domain, the NT, as a repression domain, and the CT, limited role in repressing Foxp3 but critical for binding to the promoters of Foxp3 target genes) are critical for inhibiting Foxp3 and Treg signature genes, and NT2 serves as an interface for protein–protein interactions with Foxp3.
Hhex Deficiency Does Not Alter the Development and Function of Treg Cells.
To investigate how Hhex deficiency affects Treg cells, T cell phenotypes were analyzed in Hhexfl/fl Foxp3-Cre conditional knockout (Hhex cKO) mice. Development of CD4, CD8, and Treg cells in the thymus and spleen of Hhex cKO mice was not different from that of Hhexfl/fl littermate control mice (SI Appendix, Fig. S6 A–G). B, NK, and NKT cell populations in the spleen were also not affected (SI Appendix, Fig. S6F). In addition, Hhex deficiency did not affect Foxp3 expression or cell proliferation during in vitro differentiation of Treg cells (SI Appendix, Fig. S6 H and I). The immunosuppressive function of iTreg cells was also not altered by Hhex deficiency (SI Appendix, Fig. S6J). The finding that Hhex deficiency did not affect the differentiation and function of Treg cells may be due to its low expression in Treg cells.
Discussion
Since Foxp3 is the most specific marker for characterizing Treg cells, the terms that indicate the functional status of Treg cells are also based on Foxp3 expression. Treg cell “differentiation” is determined by whether non-Treg cells become Foxp3+ Treg cells; Treg cell “stability” is determined by whether Foxp3+ Treg cells become Foxp3− ex-Treg cells; and Treg cell “plasticity” is determined by whether Treg cells undergo functional changes while maintaining Foxp3 expression (7). In this study, we show that Hhex, as a critical inhibitor of Foxp3, regulates the differentiation, stability, and plasticity of Treg cells. Retroviral overexpression of Hhex inhibited Foxp3 induction during iTreg differentiation. In tTreg cells, Hhex reduced Foxp3+ cell numbers. Finally, Hhex-overexpressing iTreg cells and tTreg cells lost their immunosuppressive functions, gaining a Th1-like phenotype.
Our studies reveal several mechanisms by which Hhex can negatively regulate Treg cells. First, Hhex directly bound to the Foxp3 locus to regulate its expression. Specifically, the activity of the Foxp3 promoter and CNS1 was almost completely inhibited by Hhex binding, which seems to be the most important mechanism for Hhex regulation of Treg cells. Second, Hhex physically interacted with Foxp3. Previous studies suggest that transcription factors including GATA3, RORγt, and YY1 inhibit the function of Foxp3 via protein–protein interactions (26, 44, 45). We add Hhex in the list of Foxp3-interacting proteins. Third, Hhex directly binds to the promoters of Treg signature genes through its HD domain, such as Il2ra and Ctla4. This suggests that Hhex can also act as a Foxp3-independent Treg cell regulator. Foxp3, Il2ra, and Ctla4 have been reported as Treg-specific superenhancers. They may easily be targets of the same transcription factor. It will be interesting to see if Hhex plays a role in the regulation of the superenhancers by directly binding to them.
Our Hhex domain mutant study suggests specific functions of each domain. NT has been shown to exert repressive function by protein–protein interactions (27), and NT2 is shown to be critical for interaction with Foxp3 in our study. HD has been shown to directly bind to DNA (27), and our data support that it is required for Hhex binding to the Foxp3 locus. CT, which has been shown to exert activation function (27), has the lowest effect in Hhex function in our study, since the ΔCT mutant only slightly affected Foxp3 expression. Therefore, it seems that each domain of Hhex plays differential and nonredundant roles in Treg function and it indicates that each mutant has partial effects in Foxp3 expression and its activity. However, summation of these effects may explain the full function of Hhex. The reason that the Hhex mutants did not show dominant-negative effect is not clear. One possible explanation is the low expression of Hhex in Treg cells. Because of low activity of Hhex in Treg cells, additional Hhex mutants may not cause any further effect.
Some of the initial studies on Hhex reported that it is not detected in T cell lineages (30, 32). However, here we detected protein and mRNA expression of Hhex in CD4 T cells. Hhex expression was lower in Treg cells. Consistent with its low expression in Treg cells, Foxp3-dependent Hhex-deficient mice did not show any phenotype. We also determined that TGF-β inhibits Hhex expression in CD4 T cells via Smad3 activation. In CD4 T cells, TGF-β is known to induce Treg differentiation but inhibits Th1 differentiation (17, 46). In this context, it is conceivable that TGF-β suppresses Hhex, which inhibits Treg differentiation, leading to Th1 differentiation. Inhibition of Hhex could be a significant role of TGF-β in the development of Treg cells.
There have been studies in several species on the mechanisms by which TGF-β and Smad proteins regulate Hhex. In human lung fibroblasts, TGF-β1 causes Hhex expression through miR-21–3p induction (47). In a study of bone morphogenetic proteins (BMPs), Smad1, but not Smad2 and Smad3, was found to interact with the BMP-responsive element (BRE) in the mouse Hhex gene to induce Hhex expression (48). In Xenopus tropicalis, ChIP assays using anti-Smad2/3 pulled down the hhex locus (49). Here, we show that Hhex expression is inhibited by TGF-β in mouse CD4 T cells. The promoter activity of Hhex (nucleotides −309/+22, not containing the BRE) was repressed by the active form of Smad3, but not by active Smad2, suggesting a mechanism of TGF-β–dependent regulation of Hhex.
Homeobox proteins are a large family of transcription factors with a highly conserved 60-amino acid long DNA-binding domain known as the homeodomain. However, only a few homeobox proteins have been reported to have a role in CD4 T cells, one of which is Hlx. Hlx is up-regulated during Th1 cell differentiation and induces IFN-γ expression synergistically with T-bet (50). Moreover, retroviral overexpression of Hlx alone also can induce IFN-γ expression in naïve T cells differentiated under Th2-polarizing conditions (51). We obtained similar results when we overexpressed Hhex in CD4 T cells. Hhex induced the expression of IFN-γ in Th0 cells, iTreg cells, and even tTreg cells. The expression of T-bet was not altered. Hlx and Hhex, as homeobox proteins, may play a similar role in CD4 T cells. Further studies on the role of Hhex in Th1 cells and Hlx in Treg cells will be informative.
This study reveals a role of Hhex in mature T cells, while other studies have examined the effect of Hhex overexpression in T cell lineages. Transgene-driven expression of Hhex in thymocytes using the Lck promoter disrupted normal T cell maturation (52). Retrovirally transduced Hhex-overexpressing hematopoietic precursor cells generated immature T lymphocyte-derived lymphomas in bone marrow-recipient mice (17). Thus, we examined the proliferation and apoptosis of Hhex-transduced Treg cells, both of which were found to be similar to those of controls. There were also no side effects in mice adoptively transferred with Hhex-overexpressing tTreg cells.
In conclusion, we identified Hhex as a negative regulator of Treg cells. Hhex directly inhibits Foxp3 and other Treg signature genes. This study suggests a molecular mechanism of Treg cell regulation, which may be relevant for the development of therapeutic strategies for Treg cell-related diseases, including cancer.
Methods
Details of mice, isolation, and culture of mouse CD4 T cells, RNA isolation, and qRT-PCR, immunoblot analysis, transient reporter assay, intracellular flow cytometry, retroviral transduction, in vitro suppression assay, mouse inflammatory bowel disease model, mouse delayed type hypersensitivity model, ChIP assays, and Co-IP assays are described in SI Appendix. All animal experiments were approved by the institutional animal care and use committees of Sogang and Yale universities.
Data Availability.
All data needed to evaluate the conclusions in the paper are present in the paper, SI Appendix, and a dataset in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE139297).
Supplementary Material
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF) grants funded by the Korean government (NRF-2017R1A2B3008621 and NRF-2015M3C9A2054020 to G.R.L., and 2014H1A8A1022457 to S.W.J.). This work was supported by the Howard Hughes Medical Institute (R.A.F.). S.S.H. was supported by a Leslie H. Warner Fellowship from the Yale Cancer Center.
Footnotes
The authors declare no competing interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE139297).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907224116/-/DCSupplemental.
References
- 1.Ohkura N., Kitagawa Y., Sakaguchi S., Development and maintenance of regulatory T cells. Immunity 38, 414–423 (2013). [DOI] [PubMed] [Google Scholar]
- 2.Shevach E. M., Thornton A. M., tTregs, pTregs, and iTregs: Similarities and differences. Immunol. Rev. 259, 88–102 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panduro M., Benoist C., Mathis D., Tissue tregs. Annu. Rev. Immunol. 34, 609–633 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee W., Lee G. R., Transcriptional regulation and development of regulatory T cells. Exp. Mol. Med. 50, e456 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou X., et al. , Instability of the transcription factor Foxp3 leads to the generation of pathogenic memory T cells in vivo. Nat. Immunol. 10, 1000–1007 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Curiel T. J., et al. , Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 10, 942–949 (2004). [DOI] [PubMed] [Google Scholar]
- 7.Sakaguchi S., Vignali D. A., Rudensky A. Y., Niec R. E., Waldmann H., The plasticity and stability of regulatory T cells. Nat. Rev. Immunol. 13, 461–467 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Hori S., Nomura T., Sakaguchi S., Control of regulatory T cell development by the transcription factor Foxp3. Science 299, 1057–1061 (2003). [DOI] [PubMed] [Google Scholar]
- 9.Fontenot J. D., Gavin M. A., Rudensky A. Y., Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 4, 330–336 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Williams L. M., Rudensky A. Y., Maintenance of the Foxp3-dependent developmental program in mature regulatory T cells requires continued expression of Foxp3. Nat. Immunol. 8, 277–284 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Zheng Y., et al. , Genome-wide analysis of Foxp3 target genes in developing and mature regulatory T cells. Nature 445, 936–940 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Herman A. E., Freeman G. J., Mathis D., Benoist C., CD4+CD25+ T regulatory cells dependent on ICOS promote regulation of effector cells in the prediabetic lesion. J. Exp. Med. 199, 1479–1489 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Malek T. R., Castro I., Interleukin-2 receptor signaling: At the interface between tolerance and immunity. Immunity 33, 153–165 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wing K., et al. , CTLA-4 control over Foxp3+ regulatory T cell function. Science 322, 271–275 (2008). [DOI] [PubMed] [Google Scholar]
- 15.Kitagawa Y., et al. , Guidance of regulatory T cell development by Satb1-dependent super-enhancer establishment. Nat. Immunol. 18, 173–183 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Marie J. C., Letterio J. J., Gavin M., Rudensky A. Y., TGF-beta1 maintains suppressor function and Foxp3 expression in CD4+CD25+ regulatory T cells. J. Exp. Med. 201, 1061–1067 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen W., et al. , Conversion of peripheral CD4+CD25- naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J. Exp. Med. 198, 1875–1886 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feng X. H., Derynck R., Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21, 659–693 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Tone Y., et al. , Smad3 and NFAT cooperate to induce Foxp3 expression through its enhancer. Nat. Immunol. 9, 194–202 (2008). [DOI] [PubMed] [Google Scholar]
- 20.Ruan Q., et al. , Development of Foxp3(+) regulatory t cells is driven by the c-Rel enhanceosome. Immunity 31, 932–940 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ouyang W., et al. , Foxo proteins cooperatively control the differentiation of Foxp3+ regulatory T cells. Nat. Immunol. 11, 618–627 (2010). [DOI] [PubMed] [Google Scholar]
- 22.Li X., Liang Y., LeBlanc M., Benner C., Zheng Y., Function of a Foxp3 cis-element in protecting regulatory T cell identity. Cell 158, 734–748 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yao Z., et al. , Nonredundant roles for Stat5a/b in directly regulating Foxp3. Blood 109, 4368–4375 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantel P. Y., et al. , GATA3-driven Th2 responses inhibit TGF-beta1-induced FOXP3 expression and the formation of regulatory T cells. PLoS Biol. 5, e329 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burgler S., et al. , RORC2 is involved in T cell polarization through interaction with the FOXP3 promoter. J. Immunol. 184, 6161–6169 (2010). [DOI] [PubMed] [Google Scholar]
- 26.Hwang S. S., et al. , YY1 inhibits differentiation and function of regulatory T cells by blocking Foxp3 expression and activity. Nat. Commun. 7, 10789 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soufi A., Jayaraman P. S., PRH/Hex: An oligomeric transcription factor and multifunctional regulator of cell fate. Biochem. J. 412, 399–413 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bedford F. K., Ashworth A., Enver T., Wiedemann L. M., HEX: A novel homeobox gene expressed during haematopoiesis and conserved between mouse and human. Nucleic Acids Res. 21, 1245–1249 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Crompton M. R., et al. , Identification of a novel vertebrate homeobox gene expressed in haematopoietic cells. Nucleic Acids Res. 20, 5661–5667 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hromas R., Radich J., Collins S., PCR cloning of an orphan homeobox gene (PRH) preferentially expressed in myeloid and liver cells. Biochem. Biophys. Res. Commun. 195, 976–983 (1993). [DOI] [PubMed] [Google Scholar]
- 31.Pellizzari L., et al. , Expression and function of the homeodomain-containing protein Hex in thyroid cells. Nucleic Acids Res. 28, 2503–2511 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Manfioletti G., et al. , Differential expression of a novel proline-rich homeobox gene (Prh) in human hematolymphopoietic cells. Blood 85, 1237–1245 (1995). [PubMed] [Google Scholar]
- 33.Bogue C. W., Zhang P. X., McGrath J., Jacobs H. C., Fuleihan R. L., Impaired B cell development and function in mice with a targeted disruption of the homeobox gene Hex. Proc. Natl. Acad. Sci. U.S.A. 100, 556–561 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams H., Jayaraman P. S., Gaston K., DNA wrapping and distortion by an oligomeric homeodomain protein. J. Mol. Biol. 383, 10–23 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Yang X., et al. , Targeted disruption of SMAD3 results in impaired mucosal immunity and diminished T cell responsiveness to TGF-beta. EMBO J. 18, 1280–1291 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Denson L. A., McClure M. H., Bogue C. W., Karpen S. J., Jacobs H. C., HNF3beta and GATA-4 transactivate the liver-enriched homeobox gene, Hex. Gene 246, 311–320 (2000). [DOI] [PubMed] [Google Scholar]
- 37.Redpath S. A., et al. , ICOS controls Foxp3(+) regulatory T-cell expansion, maintenance and IL-10 production during helminth infection. Eur. J. Immunol. 43, 705–715 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vocanson M., et al. , Inducible costimulator (ICOS) is a marker for highly suppressive antigen-specific T cells sharing features of TH17/TH1 and regulatory T cells. J. Allergy Clin. Immunol. 126, 280–289, 289.e1-7 (2010). [DOI] [PubMed] [Google Scholar]
- 39.Yang X. O., et al. , Molecular antagonism and plasticity of regulatory and inflammatory T cell programs. Immunity 29, 44–56 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Oldenhove G., et al. , Decrease of Foxp3+ Treg cell number and acquisition of effector cell phenotype during lethal infection. Immunity 31, 772–786 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kasamatsu S., et al. , Identification of the transactivating region of the homeodomain protein, hex. J. Biochem. 135, 217–223 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Tanaka T., et al. , cDNA cloning and expression of rat homeobox gene, Hex, and functional characterization of the protein. Biochem. J. 339, 111–117 (1999). [PMC free article] [PubMed] [Google Scholar]
- 43.Soufi A., Smith C., Clarke A. R., Gaston K., Jayaraman P. S., Oligomerisation of the developmental regulator proline rich homeodomain (PRH/Hex) is mediated by a novel proline-rich dimerisation domain. J. Mol. Biol. 358, 943–962 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Rudra D., et al. , Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat. Immunol. 13, 1010–1019 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang F., Meng G., Strober W., Interactions among the transcription factors Runx1, RORgammat and Foxp3 regulate the differentiation of interleukin 17-producing T cells. Nat. Immunol. 9, 1297–1306 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takimoto T., et al. , Smad2 and Smad3 are redundantly essential for the TGF-beta-mediated regulation of regulatory T plasticity and Th1 development. J. Immunol. 185, 842–855 (2010). [DOI] [PubMed] [Google Scholar]
- 47.Ong J., et al. , Identification of transforming growth factor-beta-regulated microRNAs and the microRNA-targetomes in primary lung fibroblasts. PLoS One 12, e0183815 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang W., Yatskievych T. A., Cao X., Antin P. B., Regulation of Hex gene expression by a Smads-dependent signaling pathway. J. Biol. Chem. 277, 45435–45441 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Chiu W. T., et al. , Genome-wide view of TGFβ/Foxh1 regulation of the early mesendoderm program. Development 141, 4537–4547 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mullen A. C., et al. , Hlx is induced by and genetically interacts with T-bet to promote heritable T(H)1 gene induction. Nat. Immunol. 3, 652–658 (2002). [DOI] [PubMed] [Google Scholar]
- 51.Zheng W. P., et al. , Up-regulation of Hlx in immature Th cells induces IFN-gamma expression. J. Immunol. 172, 114–122 (2004). [DOI] [PubMed] [Google Scholar]
- 52.Mack D. L., et al. , Down-regulation of the myeloid homeobox protein Hex is essential for normal T-cell development. Immunology 107, 444–451 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper, SI Appendix, and a dataset in the Gene Expression Omnibus (GEO) database, http://www.ncbi.nlm.nih.gov/geo (accession no. GSE139297).