Significance
The TLS pathway plays a vital role in maintaining genome integrity during DNA replication. Here we describe a previously unknown TLS regulatory mechanism mediated by HSCARG, a cellular redox sensor. HSCARG enhances the interaction between PCNA and USP1 and thus inhibits PCNA ubiquitination, which further impairs the recruitment of POLH and promotes the generation of DSBs. Consequently, the up-regulation of HSCARG exacerbates mammary tumorigenesis. Importantly, the cellular redox state regulates HSCARG dimerization and subcellular localization and influences the efficiency of HSCARG in regulating the TLS pathway. Our studies discover a regulator of the TLS pathway and identify a crosstalk between the cellular redox status and the DNA damage response.
Keywords: HSCARG, PCNA, translesion synthesis, redox status, breast carcinoma
Abstract
The translesion synthesis (TLS) pathway is a double-edged sword in terms of genome integrity. Deficiency in TLS leads to generation of DNA double strand break (DSB) during replication stress, while excessive activation of the TLS pathway increases the risk of point mutation. Here we demonstrate that HSCARG, a cellular redox sensor, directly interacts with the key protein PCNA in the TLS pathway. HSCARG enhances the interaction between PCNA and the deubiquitinase complex USP1/UAF1 and inhibits the monoubiquitination of PCNA, thereby impairing the recruitment of Y-family polymerases and increasing cell sensitivity to stimuli that trigger replication fork blockades. In response to oxidative stress, disaggregation of HSCARG dimers into monomers and the nuclear transport of HSCARG activate the regulatory function of HSCARG in the TLS pathway. Moreover, HSCARG, which is highly expressed in breast carcinoma, promotes the accumulation of DSBs and mutations. HSCARG knockout PyMT transgenic mice exhibit delayed mammary tumorigenesis compared with that in HSCARG wild-type or heterozygous PyMT mice. Taken together, these findings expand our understanding of TLS regulatory mechanisms and establish a link between the cellular redox status and the DNA damage response (DDR).
Proliferating cell nuclear antigen (PCNA) is a key protein involved in DNA replication. PCNA homotrimer forms a ring-shaped structure that acts as a clamp to assist replicative polymerases in sliding along DNA strands, which evidently enhances the processivity of DNA replication (1). Unfortunately, replication forks are easily stalled by pyrimidine dimers induced by UV radiation or base adducts that result from genotoxic chemical agents during S phase, as high-fidelity polymerases delta and epsilon are not able to accommodate these types of damages. To avoid replication fork collapse and increasingly severe DSBs, cells activate the TLS pathway, a type of error-prone postreplication repair (2, 3). During TLS, PCNA is monoubiquitinated at Lys164 by the Rad6B/Rad18 complex (4, 5). Monoubiquitinated PCNA promotes the recruitment of Y-family polymerases, including DNA polymerase eta (POLH) that contains ubiquitin-binding zinc-finger (UBZ) domain (6, 7). In contrast to compact polymerases delta and epsilon, Y-family polymerases are capable of synthesizing DNA past damaged sites. In such instances, damaged sites are temporarily retained and are subsequently repaired by the BER or NER pathways. However, unscheduled activation of TLS during normal replication is likely to introduce point mutations as Y-family polymerases exhibit low fidelity due to the lack of proofreading activity (7, 8). Therefore, once replication forks have bypassed damaged sites, USP10 is recruited to deubiquitinate PCNA, thus releasing POLH and terminating TLS activity (9). Under normal conditions, deubiquitinase USP1 abrogates the ubiquitination of PCNA to prevent unnecessary TLS. USP1 is up-regulated during S phase and forms a heterodimer with its cofactor UAF1, which greatly increases its catalytic activity (10, 11). In response to UV radiation, USP1 is autocleaved and degraded, enabling TLS initiation (12, 13). In addition to USP1 and USP10, USP7 has also been reported to suppress the ubiquitination of PCNA induced by hydrogen peroxide throughout interphase (14).
HSCARG, also known as NMRAL1, is a sensor protein of intracellular redox status (15). HSCARG consists of an N-terminal NmrA domain containing a Rossmann fold and a C-terminal domain that mediates its dimerization (16, 17). Under normal physiological conditions, HSCARG exists as an asymmetric dimer with 1 subunit that bonds to 1 molecule of the reduced coenzyme NADPH via its Rossmann fold. Decreases in the NADPH/NADP+ ratio lead to disaggregation of HSCARG dimers into monomers and promote the nuclear localization of HSCARG (15, 18), while a nuclear export signal (NES) in the C terminus of HSCARG mediates its transport from the nucleus to the cytoplasm (19). As a cellular redox sensor protein, HSCARG prevents overproduction of nitric oxide (NO) and reactive oxygen species (ROS) through inhibiting the catalytic activity of argininosuccinate synthetase (AS) and down-regulating the expression of NADPH oxidase subunit NCF1 (p47phox) (20, 21). Recently, it has been revealed that HSCARG also functions in innate immunity and DDR. The regulatory functions of HSCARG in these pathways are strongly associated with ubiquitination of important scaffold and functional proteins (22–24). Here we show that HSCARG is a PCNA binding partner that negatively regulates the TLS pathway and contributes to the development of breast carcinoma.
Results
HSCARG Interacts Directly with PCNA.
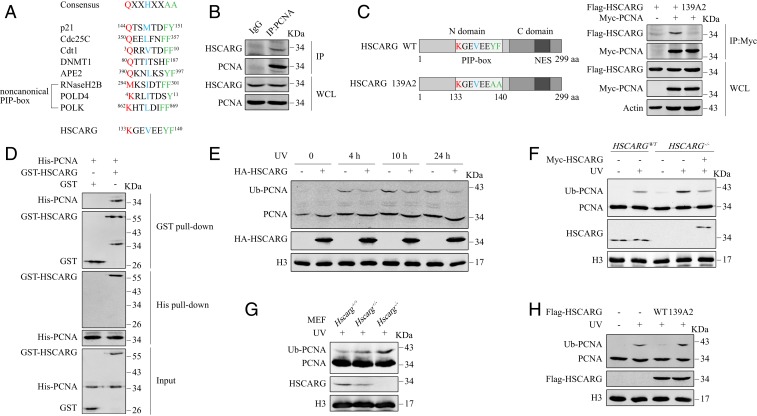
To elucidate the regulatory function of the cellular redox sensor protein HSCARG, we performed immunoprecipitation assays with Flag M2-Agarose beads in HEK 293T cells transfected with Flag-HSCARG and treated with or without NO donor S-nitroso-N-acetylpenicillamine (SNAP). Mass spectrometry (MS) analysis showed that PCNA was immunoprecipitated by Flag-HSCARG (SI Appendix, Fig. S1A and Datasets S1 and S2). Interestingly, we also discovered that HSCARG contains a potential PCNA-Interacting Protein-box (PIP-box), 133KGEVEEYF140 (Fig. 1A). Coimmunoprecipitation (co-IP) assays confirmed that HSCARG interacted with PCNA in vivo (Fig. 1B and SI Appendix, Fig. S1B). We then explored whether the interaction of HSCARG with PCNA depends on the potential PIP-box identified. Considering that the last 2 aromatic amino acid residues within the PIP-box are crucial for binding PCNA, we mutated Y139 and F140 to alanines to generate the HSCARG 139A2 mutant (Fig. 1C). In contrast to wild-type (WT) HSCARG, the 139A2 mutant failed to interact with PCNA (Fig. 1C). In addition, an in vitro pull-down assay demonstrated that HSCARG interacted with PCNA in vitro (Fig. 1D). Based on these results, we conclude that HSCARG interacts with PCNA directly via its PIP-box.
Fig. 1.
HSCARG interacts directly with PCNA and down-regulates the ubiquitination of PCNA. (A) Sequence alignment of the PIP-box in HSCARG and other previously reported PCNA binding partners. (B) Endogenous interaction between HSCARG and PCNA was assessed using an anti-PCNA antibody. (C) A schematic diagram of HSCARG PIP-box mutant 139A2. Interactions between PCNA and WT HSCARG or PIP-box mutant were assessed by co-IP. (D) His-PCNA and GST-HSCARG proteins were purified from Escherichia coli. An in vitro pull-down assay was performed with GST protein as a negative control. (E) HEK 293T cells transfected with the indicated plasmids were exposed to 20 J/m2 UV and subjected to chromatin extraction assays at different time points after irradiation. (F) HSCARGWT and HSCARG−/− HEK 293T cells transfected as indicated were exposed to 20 J/m2 UV radiation, and a chromatin extraction assay was performed after 10 h. (G) MEF cell lines were exposed to 5 J/m2 UV radiation and subjected to a chromatin extraction assay after 10 h. (H) HEK 293T cells were transfected with equal amounts of WT Flag-HSCARG or mutant 139A2 and exposed to 20 J/m2 UV. Chromatin fractions were extracted after 10 h of recovery.
HSCARG Down-Regulates the Monoubiquitination of PCNA.
Previous studies have revealed that the regulatory function of HSCARG is associated with ubiquitination of substrate proteins that function in important cellular processes. Therefore, we speculated that since HSCARG is a direct binding partner of PCNA, it may influence the monoubiquitination of PCNA, which is a key step in the TLS pathway. To test this hypothesis, we exposed HEK 293T cells to UV radiation to activate PCNA ubiquitination, and a His-ubiquitin pull-down analysis showed that overexpression of HSCARG significantly inhibited the ubiquitination of PCNA (SI Appendix, Fig. S1D). Meanwhile, chromatin extraction assays revealed that overexpression of HSCARG reduced the level of monoubiquitinated PCNA at different time points after UV exposure (Fig. 1E and SI Appendix, Fig. S1C). We then repeated the assays in HSCARG−/− HEK 293T cells and observed that HSCARG−/− cells displayed enhanced PCNA ubiquitination (Fig. 1F and SI Appendix, Fig. S1 E and F). This phenomenon was rescued by transfection of cells with exogenous HSCARG (Fig. 1F and SI Appendix, Fig. S1F). Consistent with the results in HEK 293T cells, the intensity of PCNA ubiquitination was negatively related to the expression level of HSCARG in MEF cells generated from Hscarg+/+, Hscarg+/−, and Hscarg−/− mice (Fig. 1G). Knowing that the interaction between HSCARG and PCNA is mediated by the PIP-box, we then explored the influence of HSCARG 139A2, the PIP-box mutant, on PCNA ubiquitination. Both His-ubiquitin pull-down and chromatin extraction assays showed that in contrast to WT HSCARG, the 139A2 mutant had no inhibitory effect on the monoubiquitination of PCNA (Fig. 1H and SI Appendix, Fig. S1G). This suggests that the regulatory effects of HSCARG on PCNA are interaction-dependent.
HSCARG Impairs the TLS Pathway.
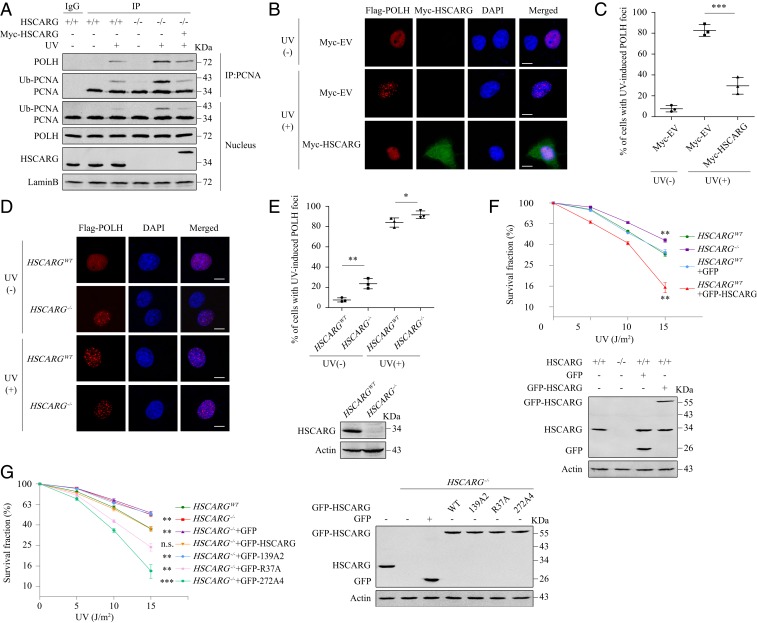
Based on the results that HSCARG negatively regulates PCNA ubiquitination, we speculated that HSCARG may consequently impair the TLS pathway. To test this hypothesis, we performed co-IP assays to detect the influence of HSCARG on the interaction between PCNA and POLH, which is the best characterized Y-family polymerase in mammalian cells. Similar to the results of previous studies (25), overexpression of POLH activated the monoubiquitination of PCNA in the absence of UV radiation. Meanwhile, we could also detect the interaction between POLH and PCNA. Both phenomena were promoted by exposure to UV radiation but inhibited by overexpression of HSCARG (SI Appendix, Fig. S2A). Consistently, the interaction between endogenous PCNA and POLH, which could only be detected after UV radiation, was enhanced in HSCARG−/− cells (Fig. 2A). To further confirm that HSCARG inhibited the recruitment of POLH to replication stalling sites, we carried out immunofluorescence assays and discovered that overexpression of HSCARG significantly impaired the formation of UV-induced POLH foci in HeLa cells (Fig. 2 B and C and SI Appendix, Fig. S2 B and C). In addition, deletion of HSCARG increased the percentages of POLH foci positive cells under both normal conditions and UV radiation (Fig. 2 D and E).
Fig. 2.
HSCARG impairs the TLS pathway. (A) HSCARGWT and HSCARG−/− HEK 293T cells transfected as indicated were exposed to UV (20 J/m2). After 10 h, the interaction between PCNA and POLH was assessed by co-IP. (B–E) HeLa cells transfected as indicated were treated with or without 20 J/m2 UV radiation. Ten hours later, immunofluorescence assays were performed using antibodies against Flag (red) and Myc (green). Nuclei were stained with DAPI (blue). Immunofluorescence images (B and D) and percentage of cells with POLH foci ≥10 (C and E) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 3). (Scale bar, 10 μm.) (F and G) HSCARGWT and HSCARG−/− HeLa cells transfected with equal amounts of the indicated plasmids were exposed to the indicated intensities of UV radiation. After 12 d, cell clones were stained with crystal violet and counted. Statistical analysis was performed to compare each group with HSCARGWT cells using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001, n.s., nonsignificant.
During replication stress, defects in polymerase switch result in stalled and collapsed replication forks, which lead to S phase arrest and cell apoptosis. The impairment of HSCARG on the formation of POLH foci led us to detect the influence of HSCARG on cell cycle and cell viability. HSCARGWT and HSCARG−/− HEK 293T cells, as well as HEK 293T cells stably transfected with Flag/HA double-tagged-HSCARG (pOZN-HSCARG), were treated with or without UV radiation, after which the percentages of cells in different phases were determined. In comparison to HSCARGWT cells, pOZN-HSCARG and HSCARG−/− cells showed no difference in cell cycle distribution under normal physiological conditions. However, after exposure to UV radiation, the percentage of S phase cells was positively related to the level of HSCARG (SI Appendix, Fig. S2 D and E), suggesting that up-regulation of HSCARG inhibits cells from entering the G2/M phase from the S phase under replication stress.
Next, we tested the influence of HSCARG on cell sensitivity to stimuli that trigger replication fork blockade. HSCARGWT and HSCARG−/− HeLa cells were exposed to different intensities of UV radiation or treated with different dosages of cisplatin or hydroxyurea (HU) and subjected to clonogenic survival assays. As expected, HSCARG−/− cells exhibited greater resistance to all 3 stimuli, whereas overexpression of HSCARG increased cell death (Fig. 2F and SI Appendix, Fig. S2G). The viability of HSCARG−/− cells was reduced to a level similar to that of WT cells after reintroduction of a moderate amount of WT GFP-HSCARG but not after reintroduction of the PIP-box mutant 139A2 (Fig. 2G and SI Appendix, Fig. S2H), suggesting that the influence of HSCARG on cell sensitivity to these stimuli is dependent on its interaction with PCNA. Similar to the observations in HeLa cells, depletion of HSCARG in MEF cells also resulted in a significantly increased survival rate after exposure to UV radiation (SI Appendix, Fig. S2F). Together, these results demonstrate that HSCARG reduces cell viability by interrupting the normal functioning of the TLS pathway.
HSCARG Inhibits PCNA Ubiquitination in a USP1-Dependent Manner.
Next, we wanted to elucidate the molecular mechanism underlying HSCARG inhibition of PCNA monoubiquitination. First, we considered whether HSCARG functions as a deubiquitinase of PCNA. In most cases, the enzyme activity of a deubiquitinase depends on a catalytic triad consisting of a histidine, a cysteine, and an aspartic acid or asparagine, among which the cysteine is the most crucial residue (26). We therefore mutated each of the 3 cysteine residues in HSCARG to alanine. A His-ubiquitin pull-down analysis revealed that the 3 point mutants inhibited PCNA ubiquitination as efficiently as WT HSCARG (SI Appendix, Fig. S3A), proving that HSCARG is not a deubiquitinase of PCNA.
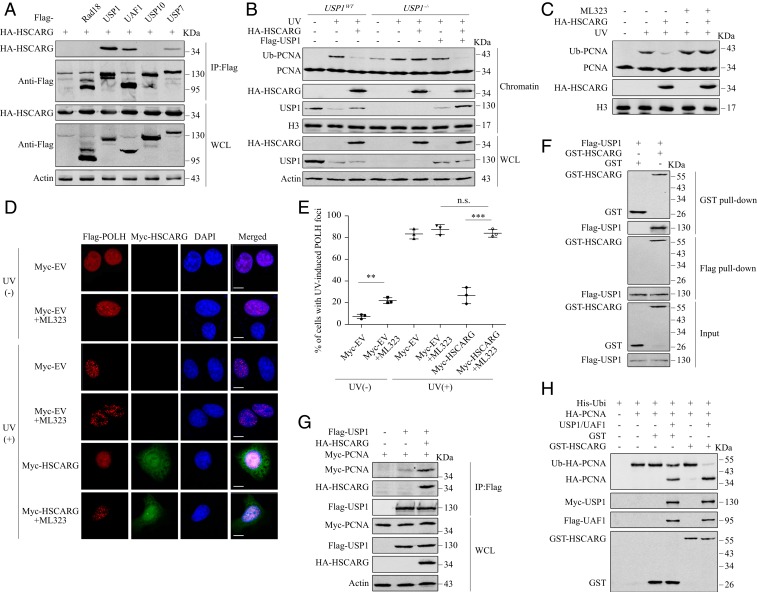
We next focused on whether the regulatory function of HSCARG depends on enzymes that target PCNA. We examined the interactions between HSCARG and E3 ligase Rad18, deubiquitinases USP1, USP7, USP10, and USP1 cofactor UAF1. We detected a band corresponding to HSCARG in precipitates of USP1, UAF1, and USP7 (Fig. 3A). We first examined the influence of HSCARG on PCNA ubiquitination in USP7−/− cells. As expected, knockout of USP7 led to a slight increase in the level of ubiquitinated PCNA after UV exposure. However, overexpression of HSCARG impaired PCNA ubiquitination both in the presence and absence of USP7 (SI Appendix, Fig. S4A). Besides PCNA, USP7 has been reported to target other TLS effectors. Specifically, USP7 inhibits the ubiquitination of Rad18 and POLH, thus suppressing their proteasome-mediated degradation (25, 27). Interestingly, knockout of USP7 also increases POLH stability through a p53-mediated mechanism (25). Consistent with previous studies, we found that deletion of USP7 led to decreased level of Rad18 and increased levels of p53 and POLH, while both phenomena were rescued by MG132 treatment. However, we did not observe similar effects in HSCARG−/− cells (SI Appendix, Fig. S4C). In addition, unlike that of USP7, HSCARG overexpression did not enhance the stability of POLH in p53−/− cells (SI Appendix, Fig. S4D). Indeed, a co-IP assay revealed that HSCARG had no influence on the interactions between USP7 and these 2 substrates (SI Appendix, Fig. S4B). Taken together, these results suggest that HSCARG is not involved in USP7-mediated TLS regulation.
Fig. 3.
HSCARG inhibits PCNA ubiquitination dependently on the USP1/UAF complex. (A) Interactions between HSCARG and the indicated enzymes were detected by co-IP. (B) USP1WT and USP1−/− HEK 293T cells were exposed to 20 J/m2 UV radiation followed by a chromatin extraction assay 10 h later. (C) HEK 293T cells treated with or without 30 μM ML323 were transfected as indicated and exposed to UV (20 J/m2). Ten hours later, the level of PCNA ubiquitination was detected by a chromatin extraction assay. (D and E) HeLa cells cotransfected with Flag-POLH and Myc-HSCARG or empty vector were treated with or without 30 μM ML323 and 20 J/m2 UV radiation. An immunofluorescence assay was performed using antibodies against Flag (red) and Myc (green). Nuclei were stained with DAPI (blue). Immunofluorescence images (D) and the percentage of cells with POLH foci ≥10 (E) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 3). (Scale bar, 10 μm.) **P < 0.01, ***P < 0.001, n.s., nonsignificant. (F) An in vitro pull-down assay was performed with Flag-USP1 protein extracted from HEK 293T cells and GST-HSCARG protein purified from E. coli. (G) HEK 293T cells were transfected as indicated and subjected to co-IP. (H) Ubiquitinated PCNA was precipitated from HEK 293T cells using Ni2+ beads. The indicated proteins were added, and the mixtures were incubated at 30 °C for 1 h. The amount of ubiquitinated PCNA was detected by Western blotting.
In contrast to the results in USP7−/− cells, the inhibitory effects of HSCARG on PCNA ubiquitination induced by either UV radiation or cisplatin treatment were completely abolished in USP1−/− cells, and the phenomena could be rescued by USP1 reintroduction (Fig. 3B and SI Appendix, Fig. S3B). We further confirmed the results using a recently discovered inhibitor of USP1 named ML323 (28). Under ML323 treatment, HSCARG overexpression no longer inhibited PCNA ubiquitination (Fig. 3C and SI Appendix, Fig. S3D), nor did it block the accumulation of POLH at replication stalling sites (Fig. 3 D and E). Collectively, these results indicate that HSCARG impairs the TLS pathway in a USP1-dependent manner.
HSCARG Stimulates the Deubiquitination of PCNA by the USP1/UAF1 Complex.
As HSCARG interacted with both USP1 and its cofactor UAF1 in vivo (Fig. 3A), we conjectured that HSCARG is able to interact with the USP1/UAF1 complex rather than binding to monomers of each subunit independently. To confirm this speculation, we incubated purified Myc-USP1/Flag-UAF1 dimers with GST-HSCARG in vitro and performed gel filtration analysis. In the gel filtration profile of the mixture, an elution peak before the peaks corresponding to the USP1/UAF1 dimer and excess GST-HSCARG appeared. Western blot analysis of the eluted fractions showed that this peak contained all 3 proteins, demonstrating the presence of the USP1/UAF1/HSCARG triple complex (SI Appendix, Fig. S5). Endogenous co-IP and in vitro pull-down assays showed that HSCARG interacted directly with USP1 (Fig. 3F and SI Appendix, Fig. S6A), whereas the interaction between UAF1 and HSCARG could only be detected in the presence of USP1 (SI Appendix, Fig. S6 B and C). These results indicate that HSCARG binds to the USP1 subunit of the USP1/UAF1 heterodimer; hence, USP1 acts as an intermediate in the interaction between HSCARG and UAF1.
In response to UV radiation, USP1 is autocleaved and degraded, which abolishes its inhibition of PCNA ubiquitination. We observed that HSCARG overexpression partly rescued the level of USP1 in the chromatin fraction but not in whole-cell lysate after UV exposure (Fig. 3B). Knowing that functional PCNA is located in chromatin, we speculated that HSCARG, as a direct binding partner of both USP1 and PCNA, might enhance the interaction between them. We therefore carried out co-IP and in vitro pull-down assays and observed that both overexpression of HSCARG in vivo and addition of HSCARG in vitro enhanced the interaction between USP1 and PCNA (Fig. 3G and SI Appendix, Fig. S6F). Moreover, the HSCARG PIP-box mutant 139A2, which was incapable of binding PCNA, still interacted with USP1 (SI Appendix, Fig. S6G), suggesting that HSCARG binds PCNA and USP1 via different regions. Furthermore, we carried out an in vitro deubiquitination assay to detect the influence of HSCARG on the catalytic efficiency of the USP1/UAF1 complex. Although HSCARG alone was unable to deubiquitinate PCNA, the deubiquitination efficiency of the USP1/UAF1 complex was significantly increased in the presence of HSCARG (Fig. 3H). Taken together, these results demonstrate that HSCARG enhances the interaction between deubiquitinase USP1 and its substrate PCNA and therefore stimulates the deubiquitination of PCNA by USP1/UAF1.
HSCARG NADPH-Unbound and NES Mutants Inhibit PCNA Ubiquitination More Efficiently than WT HSCARG.
Next, we studied the relationship between the regulatory function of HSCARG in TLS and its role as a cellular redox sensor. HSCARG contains an N-terminal Rossmann fold that binds to the reduced coenzyme NADPH and a C-terminal domain that mediates its homodimerization. Therefore, we generated HSCARG truncation mutants 1–153 and 154–299 and examined their interactions with PCNA and USP1. Consistent with the fact that the PIP-box of HSCARG is located at its N terminus (Fig. 1C), full-length HSCARG and truncation mutant 1–153 interacted with PCNA, whereas truncation mutant 154–299 did not (SI Appendix, Fig. S7A). Interestingly, we discovered that the region of HSCARG that binds USP1 is also located in its N terminus (SI Appendix, Fig. S7B). Accordingly, the N-terminal truncation 1–153 alone was capable of inhibiting the monoubiquitination of PCNA in vivo, whereas the C-terminal truncation 154–299 showed no influence (SI Appendix, Fig. S7C). However, under normal physiological conditions, the N-terminal Rossmann fold of HSCARG is occupied by NADPH. Previous studies have revealed that binding of HSCARG to NADPH suppresses its nuclear translocation (15), which leads to spatial separation of HSCARG from functional PCNA. Moreover, in our previous MS analysis, we detected more peptides corresponding to PCNA in the precipitate of HSCARG under treatment with SNAP (SI Appendix, Fig. S1A). Based on these observations and analyses, we hypothesized that NADPH binding impaired the ability of HSCARG to down-regulate PCNA ubiquitination. We then tested the influence of the 2 previously discovered NADPH-unbound HSCARG point mutants, R37A and Y81A, on PCNA ubiquitination (18). As expected, dimerization of these 2 NADPH-unbound mutants was weakened, while the nuclear localization of these 2 mutants was mildly enhanced compared with that of the WT protein (SI Appendix, Fig. S8 A and B). Consistent with our hypothesis, the interactions of these 2 mutants with both PCNA and USP1 were stronger than that of WT HSCARG (SI Appendix, Fig. S8 C and D). Accordingly, these 2 mutants were more capable of enhancing the interaction between PCNA and USP1 and down-regulating UV-induced PCNA ubiquitination (SI Appendix, Fig. S8 D–F).
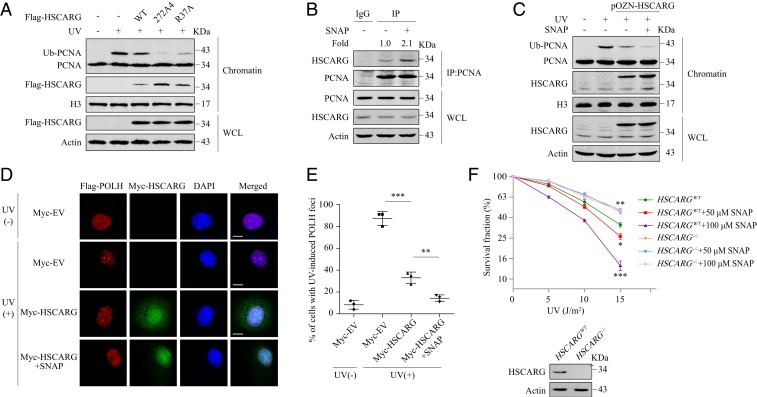
Apart from NADPH binding, the subcellular distribution of HSCARG is also regulated by the NES located in its C terminus (26). Substitution of all 4 leucines or isoleucines within the NES with alanines resulted in clear increases in nuclear and chromatin localization of HSCARG (Fig. 4A and SI Appendix, Fig. S9A). Correspondingly, the NES mutant 272A4 exhibited greater inhibitory effect on the level of ubiquitinated PCNA than WT HSCARG and NADPH-unbound mutant R37A (Fig. 4A and SI Appendix, Fig. S9B). A clonogenic survival assay showed that in comparison with HSCARG−/− cells transfected with WT GFP-HSCARG, HSCARG−/− cells transfected with an equal amount of mutant R37A or 272A4 were less resistant to stimuli that trigger replication fork blockade (Fig. 2G and SI Appendix, Fig. S2H).
Fig. 4.
Cellular redox status influences the efficiency of the regulation of the TLS pathway by HSCARG. (A) HEK 293T cells transfected with the indicated plasmids were exposed to UV (20 J/m2) and recovered for 10 h, then the level of PCNA ubiquitination was detected by a chromatin extraction assay. (B) HEK 293T cells were treated with 100 μM SNAP or DMSO. After 24 h, cells were lysed and immunoprecipitated with IgG or anti-PCNA antibodies. (C) HSCARGWT and pOZN-HSCARG HEK 293T cells transfected with the indicated plasmids were treated with 100 μM SNAP or DMSO. After 14 h, cells were exposed to UV (20 J/m2). After 10 h, a chromatin extraction assay was performed. (D and E) HeLa cells cotransfected with Flag-POLH and Myc-HSCARG or empty vector were treated with or without 100 μM SNAP and 20 J/m2 UV. An immunofluorescence assay was performed using antibodies against Flag (red) and Myc (green). Nuclei were stained with DAPI (blue). Immunofluorescence images (D) and percentage of cells with POLH foci ≥10 (E) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 3). (Scale bar, 10 μm.) (F) HSCARGWT and HSCARG−/− HeLa cells exposed to different intensities of UV radiation were treated with the indicated dosages of SNAP for 24 h. After 12 d, cell clones were stained with crystal violet and counted. Statistical analysis was performed to compare each group with the HSCARGWT cells using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 3). *P < 0.05, **P < 0.01, ***P < 0.001.
Cellular Redox Status Influences the Efficiency of the Regulation of the TLS Pathway by HSCARG.
The results with the NADPH-unbound and NES mutant indicate that dissociation from NADPH and nuclear transport enhances the ability of HSCARG to inhibit the TLS pathway. To further examine the regulatory efficiency of HSCARG under different cellular redox status, we treated cells with dehydroepiandrosterone (DHEA) that decreased the cellular NADPH/NADP+ ratio or SNAP that is an NO donor, which promoted the translocation of HSCARG from the cytoplasm to the nucleus (SI Appendix, Fig. S10 A and B). This phenomenon was intensified when the concentration of SNAP reached 100 μM (SI Appendix, Fig. S10A). Similar to the results shown for the NADPH-unbound and NES mutants, SNAP or DHEA treatment led to increased interaction between HSCARG and PCNA and enhanced inhibitory effect of HSCARG on PCNA ubiquitination (Fig. 4 B and C and SI Appendix, Fig. S10 C–F). An IF assay revealed that treatment of HeLa cells with SNAP resulted in the nuclear translocation of HSCARG (Fig. 4D), which is consistent with the effect observed in HEK 293T cells. Meanwhile, we detected a further reduction in the number of UV-induced POLH foci (Fig. 4 D and E). In addition, we exposed HSCARGWT and HSCARG−/− HeLa cells to different intensities of UV radiation and cultured them in medium with different dosages of SNAP for 24 h. A clonogenic survival assay performed after an additional 12 d of culture showed that treatment of WT cells with 50 μM SNAP led to impaired cell survival after UV exposure; the phenomenon was more pronounced in cells treated with 100 μM SNAP (Fig. 4F). Given that SNAP treatment alone did not decrease the viability of HSCARGWT cells (SI Appendix, Fig. S10G), a reasonable explanation for the reduced survival rate resulting from UV exposure is that SNAP treatment enhanced cell sensitivity to UV radiation. This effect was HSCARG-dependent as we did not observe a significant influence of SNAP on the viability of UV-radiated HSCARG−/− cells (Fig. 4F). Taken together, these results suggest that the cellular redox status fine-tunes HSCARG dimerization and subcellular localization and influences its efficiency in down-regulating the TLS pathway.
HSCARG Promotes the Generation of DSBs and the Accumulation of Mutations.
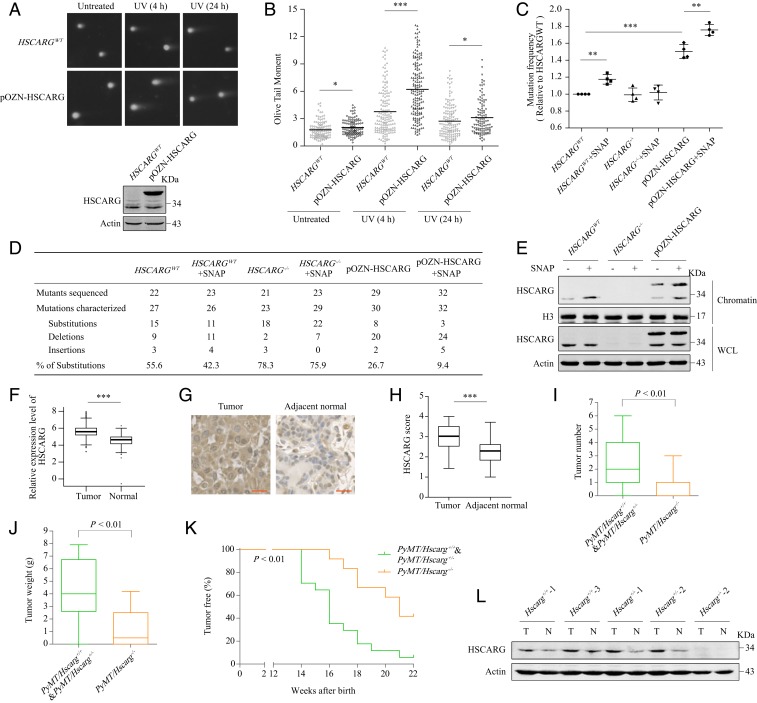
The above results suggest that HSCARG impairs the TLS pathway, so we proposed that HSCARG might further promote the generation of DSBs and the accumulation of mutations during DNA replication stress. A neutral comet assay showed that HEK 293T cells stably transfected with pOZN-HSCARG possessed more obvious comet tails than HSCARGWT cells, especially at 4 h after UV radiation (Fig. 5 A and B). Consistently, deletion of HSCARG in MEF cells led to a significant decrease in comet tail length after UV exposure (SI Appendix, Fig. S11 A and B). Moreover, a mutation frequency assay indicated that overexpression of HSCARG, as well as chromatin enrichment of HSCARG caused by SNAP treatment, resulted in elevated mutation frequencies and increased percentages of base pair insertions or deletions (Fig. 5 C–E). In contrast, HSCARG deletion promoted introduction of base pair substitutions, which is the consequence of irregular activation of TLS (Fig. 5 D and E). Previous research has revealed that HSCARG also influences the ubiquitination of histone H2A (24), which plays a critical role in recruitment of downstream DSB repair proteins. We therefore examined the influence of HSCARG on DSB repair efficiency and found that HSCARG decreased the repair efficiency of HR and increased the repair efficiency of NHEJ (SI Appendix, Fig. S11 C and D). Unfortunately, overuse of the error-prone NHEJ pathway leads to production of frameshift mutations and chromosome translocations. Taken together, these results indicate that excessive expression of HSCARG is detrimental to genomic stability.
Fig. 5.
HSCARG promotes the accumulation of mutations and is involved in the genesis of breast carcinoma. (A and B) HSCARGWT and pOZN-HSCARG HEK 293Tcells were exposed to UV (20 J/m2) and subjected to neutral comet assays. Approximately 120 cells were counted in each group. Representative images (A) and quantified data presented in scatterplots (B) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. (C–E) UV-damaged pZ189 plasmids were transfected into HSCARGWT, HSCARG−/−, and pOZN-HSCARG HEK 293T cells treated with DMSO or SNAP (100 μM). Mutation frequencies (C) and the distribution of the mutations in different classes (D) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. Data are presented as the mean ± SD (n = 4). Cell lines and effectiveness of SNAP were confirmed by a chromatin extraction assay (E). (F) Bioinformatics analysis of the TCGA dataset for the expression of HSCARG in breast carcinoma and adjacent normal tissue samples. Statistical analysis was performed using a 2-tailed Student’s t test. Data are presented in box plots. (G and H) IHC analysis of breast carcinoma and adjacent normal tissue samples was performed using anti-HSCARG antibodies. Representative images of IHC staining (G) and quantified data presented in box plots (H) are shown. Statistical analysis was performed using a 2-tailed Student’s t test. (Scale bar, 20 μm.) (I and J) Tumor numbers (I) and tumor weights (J) in PyMT mice at 150 d of age. Multiple tumors in 1 individual were added together. Statistical analysis was performed using the Mann–Whitney test. (K) Mammary adenocarcinoma incidence in PyMT/Hscarg+/+ or PyMT/Hscarg+/− (n = 18) and PyMT/Hscarg−/− mice (n = 12) is depicted as the percentage of tumor-free mice. Statistical analysis was performed using the Gehan–Breslow–Wilcoxon test. (L) Whole-cell lysates of breast tumors and adjacent normal tissues from PyMT mice were extracted and analyzed by Western blotting. *P < 0.05, **P < 0.01, ***P < 0.001.
HSCARG Is Related to the Genesis of Breast Carcinoma.
Mutation accumulation is the basic cause of cancer development. We therefore explored the biological relationship between dysregulation of HSCARG and tumorigenesis. First, we searched the TCGA dataset and found that HSCARG was more highly expressed in tumors of several types of cancer than in adjacent normal tissue, including invasive breast carcinoma (Fig. 5F and SI Appendix, Fig. S11E). Next, we performed tissue microarrays and immunohistochemistry (IHC) staining to analyze HSCARG expression in 45 paired samples of breast carcinoma tissue and adjacent normal tissue. The breast carcinoma specimens exhibited increased staining of HSCARG by IHC compared to that in samples from adjacent normal tissue (Fig. 5 G and H).
To further clarify the relevance of HSCARG to mammary tumorigenesis, we crossed Hscarg+/+, Hscarg+/−, and Hscarg−/− mice with MMTV-polyomavirus middle T antigen (PyMT) transgenic model mice. As expected, we observed a significant delay in the development of mammary tumors in PyMT/Hscarg−/− mice (Fig. 5K). Specifically, all but 1 (17/18) of the PyMT/Hscarg+/+ and PyMT/Hscarg+/− mice spontaneously developed mammary tumors by 150 d of age. In contrast, only approximately half (7/12) of the PyMT/Hscarg−/− mice had started to develop tumors by 150 d of age. In addition, both the tumor number and the total tumor weight in PyMT/Hscarg−/− mice were significantly lower than those in PyMT/Hscarg+/+ and PyMT/Hscarg+/− mice (Fig. 5 I and J). We further detected the levels of HSCARG in tissues from PyMT mice. Overall, the levels of HSCARG in tumors from PyMT/Hscarg+/+ and PyMT/Hscarg+/− mice were similar (Fig. 5L and SI Appendix, Fig. S11F), which is consistent with the observation that there was no significant difference in the incidence and development of mammary carcinoma in PyMT/Hscarg+/+ and PyMT/Hscarg+/− mice. The level of HSCARG in tumor tissue from Hscarg+/− mouse 3 was lower than that in the other heterozygous mice and WT mice (SI Appendix, Fig. S11F). This mouse started to develop mammary carcinoma at 19 wk of age, whereas the other mice detected in SI Appendix, Fig. S8D, developed tumors by 16 wk of age. Similar to the results of the bioinformatics and IHC analyses of human tissues, HSCARG protein levels in mouse mammary carcinoma tissues were higher than those in adjacent normal tissues (Fig. 5L).
Previous studies revealed that HSCARG inhibits cellular ROS generation, suggesting a potential tumor suppressor role of HSCARG. Mechanistically, HSCARG impairs the expression of NCF1, a subunit of NADPH oxidase, through repressing NF-kappaB activity in the cytoplasm (21). Here we found that in contrast to WT HSCARG, the NES mutant 272A4, which accumulated in the nucleus and down-regulated the TLS pathway more efficiently, had no significant suppressive effects on ROS and NCF1 levels (SI Appendix, Fig. S12 A and B). These observations suggest that the subcellular localization of HSCARG counterbalances its oncogenic and tumor suppressive functions. We then detected the distribution of HSCARG in tissues from 3 PyMT mice with multiple tumors that developed at different time; one of the tumors detected was developed at ∼15 wk of age (old tumor), and another tumor detected was developed at ∼21 wk of age (new tumor). Although both the old tumors and the new tumors exhibited increased levels of HSCARG in comparison to those in normal tissues, the highly expressed HSCARG protein was enriched in the nucleus only in cells in new tumors. As a result, the levels of HSCARG and NCF1 in the cytoplasm of cells in new tumors were similar (SI Appendix, Fig. S12C). The nuclear enrichment of HSCARG in the new tumors might have resulted from oxidative stress in the microenvironment, while the old tumors suffered hypoxia due to their enlarged size and distance from blood vessels, thus attenuating the nuclear transport of HSCARG. As the microenvironment and redox status of new tumors should be similar to that of tissues undergoing cancer development, the above results indicated that only the oncogenic function of HSCARG was activated during tumorigenesis. Collectively, these results demonstrate that the up-regulation and nuclear transport of HSCARG promote the genesis of breast cancer.
Discussion
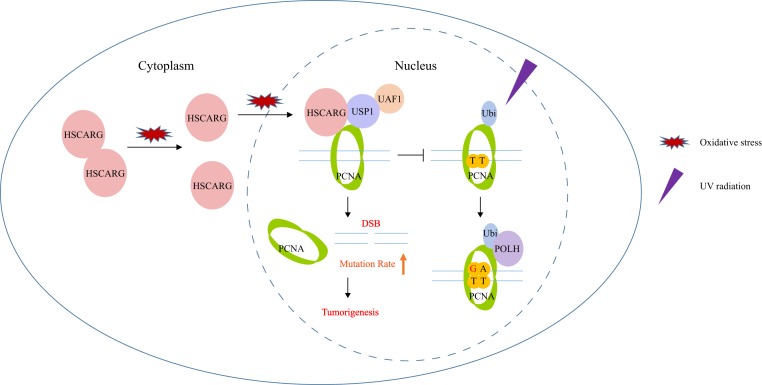
Processivity in DNA replication is a basic requirement for cell proliferation and development. However, replication forks are easily blocked by DNA lesion sites, including UV-induced cyclobutane pyrimidine dimers. Under such circumstances, monoubiquitination of PCNA rapidly initiates the TLS pathway, which recruits lesion-tolerant Y-family polymerases to replication stalling sites. In this study, we discovered that the cellular redox sensor protein HSCARG plays an important regulatory role in the TLS pathway. Specifically, HSCARG binds to both PCNA and its deubiquitinase USP1 directly and thereby stimulates the deubiquitination of PCNA by the USP1/UAF1 complex. Once PCNA ubiquitination is impaired, Y-family POLH cannot be recruited to synthesize DNA past DNA lesion sites, leading to collapse of replication forks and generation of DSBs. Oxidative stress results in disaggregation of HSCARG dimers into monomers and translocation of HSCARG from the cytoplasm to the nucleus, where it becomes active in blocking the TLS pathway and exacerbating tumorigenesis (Fig. 6).
Fig. 6.
A proposed model for HSCARG regulation of the TLS pathway. Oxidative stress leads to disaggregation of HSCARG dimers into monomers and the translocation of HSCARG from the cytoplasm to the nucleus. In the nucleus, HSCARG stimulates the deubiquitination of PCNA via the USP1/UAF1 complex and thereby abrogates the recruitment of POLH and promotes the generation of DSBs, thus elevating mutation rate and exacerbating mammary tumorigenesis.
Recently, the deubiquitinase USP1 was reported to be a potential therapeutic target for osteosarcoma and nonsmall cell lung cancer (28, 29). USP1 binds tightly with its cofactor UAF1 to form a functional heterodimer (11). The USP1/UAF1 complex also targets other proteins that participate in the DDR, such as FANCD2, which participates in the Fanconi pathway, and RAD51AP1, which participates in the HR pathway (10, 30). However, little is known about what determines the substrate specificity of USP1. In the current study, we found that HSCARG functions as a direct binding partner of USP1 and stimulates the deubiquitination of substrate PCNA by USP1. Moreover, the 2 regulatory factors UAF1 and HSCARG utilize different mechanisms to enhance USP1 function. Co-IP assays revealed that overexpression of HSCARG or UAF1 had no influence on the interaction between USP1 and the other protein (SI Appendix, Fig. S6 D and E), indicating that UAF1 and HSCARG interact with USP1 independently of each other. Overexpression of HSCARG alleviated UV-induced decreases in chromatin-bound USP1 but not global USP1 levels (Fig. 3B), suggesting that HSCARG does not increase USP1 stability as UAF1 does. Instead, HSCARG interacts with PCNA through its PIP-box (Fig. 1 A and C) and enhances the interaction between PCNA and USP1 (Fig. 3G). We also investigated the function of USP1 in HSCARG−/− cells. USP1 could inhibit the ubiquitination of PCNA in the absence of HSCARG, and its catalytic efficiency was improved via simultaneous coexpression of UAF1. However, the level of UV-induced monoubiquitination of PCNA in HSCARG−/− cells remained higher than that in WT cells even when both USP1 and UAF1 were overexpressed (SI Appendix, Fig. S3E). These results indicate that in addition to being regulated by enzymatic activity, the function of USP1 in the TLS pathway is also strictly regulated by substrate binding affinity. UAF1 increases the global enzyme activity of USP1, while HSCARG specifically enhances the interaction between USP1 and its substrate PCNA. This work adds to the understanding of the regulatory mechanisms of deubiquitinase USP1.
Importantly, in this study, we show that coordination exists between the 2 roles of HSCARG as a sensor of the intracellular redox status and a regulator of the TLS pathway. HSCARG binds to coenzyme NADPH via its Rossmann fold, and this binding stabilizes HSCARG as an asymmetric dimer in the cytoplasm (15). Oxidative stress leads to disaggregation of HSCARG dimers into monomers and the nuclear translocation of HSCARG, bringing it spatially close to PCNA (SI Appendix, Figs. S10 A and B and S12E). Apart from regulating the TLS pathway, HSCARG has also been reported to prevent overproduction of NO and ROS, which promote cell resistance against oxidative stress (20, 21). As these functions of HSCARG depend on its interaction with AS and inhibition of NF-kappaB activity in the cytoplasm, the nuclear accumulated HSCARG NES mutant 272A4 had no significant influence on ROS generation (SI Appendix, Fig. S12A). Meanwhile, HSCARG 272A4 mutant impaired the TLS pathway with an increased efficiency (Figs. 2G and 4A). Interestingly, the NADPH-unbound mutant R37A inhibited the levels of NCF1 and ROS more effectively than WT HSCARG despite its mildly increased nuclear localization (SI Appendix, Figs. S8B and S12 A and B), and previous studies also revealed that HSCARG NADPH-unbound mutants exhibited stronger binding affinity to AS, a rate-limiting enzyme in NO synthesis which is located in the cytoplasm (18). These results demonstrate that abolition of NADPH binding activates the functions of HSCARG both in the cytoplasm and in the nucleus. The paradoxical findings for the NADPH-unbound mutants and the NES mutant could be explained by their differential subcellular localization. Although both mutants showed increased nuclear and chromatin localization, the NADPH-unbound mutants showed this localization to a lesser degree; thus, there was still a sufficient amount of protein in the cytoplasm (SI Appendix, Fig. S12F). Cell fractionation assays of endogenous HSCARG revealed that SNAP treatment finely regulated the enrichment of HSCARG in the nucleus over the cytoplasm in a dose-dependent manner (SI Appendix, Fig. S10A). Further studies confirmed that 50 μM SNAP, which was sufficient to abolish the dimerization of HSCARG, resulted in mildly increased nuclear localization of WT HSCARG to a level similar to that of the R37A mutant (SI Appendix, Fig. S12 E and F). Accordingly, HSCARG impaired both ROS generation and TLS more efficiently in cells treated with 50 μM SNAP (Fig. 4F and SI Appendix, Fig. S12D). In contrast, when the concentration of SNAP reached 200 μM, HSCARG was overenriched in the nucleus as was the NES mutant 272A4, which attenuated its function in terms of inhibiting ROS generation (SI Appendix, Figs. S10A and S12 D and F). Consistent with our observations, previous studies on the function of HSCARG in NO synthesis showed that decreased NADPH/NADP+ ratio caused by DHEA treatment enhanced the interaction between HSCARG and AS, while an excess of SNAP (500 μM) weakened this interaction (20). These findings collectively demonstrate that HSCARG plays a dual role in regulating cell response to increased oxidative stress. When cells are exposed to mild oxidative stress, disaggregation of HSCARG dimers into monomers activates the interaction of HSCARG with other binding partners, and cytoplasmically localized HSCARG prevents the generation of ROS and NO, thereby facilitating cell recovery to normal redox status. However, when the oxidative stress becomes too intense, excessive enrichment of HSCARG in the nucleus attenuates its functions in the cytoplasm and further elevates the impairment of the TLS pathway, resulting in reduced cell viability and an increased mutation rate.
In summary, we identify the cellular redox sensor protein HSCARG as a binding partner of PCNA. HSCARG impairs ubiquitination of PCNA in a USP1-dependent manner. Oxidative stress enhances the nuclear localization of HSCARG and promotes its negative regulation of the TLS pathway. Overall, this work enriches our understanding of TLS regulatory mechanisms and provides insights into the connection between the cellular redox status and the DDR.
Materials and Methods
All mouse experiments were approved by the Peking University Laboratory Animal Center and performed in accordance with the “Principles for the Utilization and Care of Vertebrate Animals” (31) and ref. 32. Bioinformatics analysis of the expression of HSCARG in breast carcinoma and adjacent normal tissues was performed using the TCGA database. The data were downloaded using the FireBrowse RESTful API with R package FirebrowseR. Details of materials and methods including plasmids, reagents, antibodies, cell lines, sources of HSCARG knockout and MMTV-PyMT mice, in vitro pull-down and deubiquitination, and all cell-based assays are described in SI Appendix, SI Methods.
Data Availability.
All data discussed in the paper have been made available to readers.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (grants 81730080 and 31670786) and the National Key Research and Development Program of China (grant 2016YFC1302401). We sincerely thank Prof. Lingqiang Zhang for providing the MMTV-PyMT transgenic mice; Prof. Jun Huang for providing the pZ189 plasmid and E. coli strain MBM7070; Prof. Huadong Pei for providing the HR and NHEJ reporter systems; and Prof. Wensheng Wei for providing the CRISPR/Cas9 related plasmids. We are grateful to the Core Facilities at School of Life Sciences, Peking University, for assistance with the protein MS analysis work. We also appreciate the assistance of Dong Liu, Guilan Li, Hongxia Lv, and Xiaochen Li from the Core Facilities of the School of Life Sciences at Peking University.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. J.W. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1910250116/-/DCSupplemental.
References
- 1.Moldovan G. L., Pfander B., Jentsch S., PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007). [DOI] [PubMed] [Google Scholar]
- 2.Leung W., Baxley R. M., Moldovan G. L., Bielinsky A. K., Mechanisms of DNA damage tolerance: Post-translational regulation of PCNA. Genes (Basel) 10, E10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee K. Y., Myung K., PCNA modifications for regulation of post-replication repair pathways. Mol. Cells 26, 5–11 (2008). [PMC free article] [PubMed] [Google Scholar]
- 4.Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S., RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002). [DOI] [PubMed] [Google Scholar]
- 5.Bailly V., Lamb J., Sung P., Prakash S., Prakash L., Specific complex formation between yeast RAD6 and RAD18 proteins: A potential mechanism for targeting RAD6 ubiquitin-conjugating activity to DNA damage sites. Genes Dev. 8, 811–820 (1994). [DOI] [PubMed] [Google Scholar]
- 6.Kannouche P., et al. , Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 15, 158–172 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sale J. E., Lehmann A. R., Woodgate R., Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat. Rev. Mol. Cell Biol. 13, 141–152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fuchs R. P., Fujii S., Translesion DNA synthesis and mutagenesis in prokaryotes. Cold Spring Harb. Perspect. Biol. 5, a012682 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Park J. M., et al. , Modification of PCNA by ISG15 plays a crucial role in termination of error-prone translesion DNA synthesis. Mol. Cell 54, 626–638 (2014). [DOI] [PubMed] [Google Scholar]
- 10.Nijman S. M., et al. , The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell 17, 331–339 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Cohn M. A., et al. , A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell 28, 786–797 (2007). [DOI] [PubMed] [Google Scholar]
- 12.García-Santisteban I., Peters G. J., Giovannetti E., Rodríguez J. A., USP1 deubiquitinase: Cellular functions, regulatory mechanisms and emerging potential as target in cancer therapy. Mol. Cancer 12, 91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang T. T., et al. , Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8, 339–347 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kashiwaba S., et al. , USP7 is a suppressor of PCNA ubiquitination and oxidative-stress-induced mutagenesis in human cells. Cell Rep. 13, 2072–2080 (2015). [DOI] [PubMed] [Google Scholar]
- 15.Zheng X., et al. , Restructuring of the dinucleotide-binding fold in an NADP(H) sensor protein. Proc. Natl. Acad. Sci. U.S.A. 104, 8809–8814 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stammers D. K., et al. , The structure of the negative transcriptional regulator NmrA reveals a structural superfamily which includes the short-chain dehydrogenase/reductases. EMBO J. 20, 6619–6626 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dai X., Gu X., Luo M., Zheng X., Protein expression, crystallization and preliminary X-ray crystallographic studies on HSCARG from Homo sapiens. Protein Pept. Lett. 13, 955–957 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Dai X., et al. , NADPH is an allosteric regulator of HSCARG. J. Mol. Biol. 387, 1277–1285 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Zhang M., et al. , A CRM1-dependent nuclear export signal controls nucleocytoplasmic translocation of HSCARG, which regulates NF-κB activity. Traffic 13, 790–799 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Zhao Y., et al. , An NADPH sensor protein (HSCARG) down-regulates nitric oxide synthesis by association with argininosuccinate synthetase and is essential for epithelial cell viability. J. Biol. Chem. 283, 11004–11013 (2008). [DOI] [PubMed] [Google Scholar]
- 21.Xiao W., et al. , HSCARG inhibits NADPH oxidase activity through regulation of the expression of p47phox. PLoS One 8, e59301 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Peng Y., Xu R., Zheng X., HSCARG negatively regulates the cellular antiviral RIG-I like receptor signaling pathway by inhibiting TRAF3 ubiquitination via recruiting OTUB1. PLoS Pathog. 10, e1004041 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li T., Guan J., Li S., Zhang X., Zheng X., HSCARG downregulates NF-κB signaling by interacting with USP7 and inhibiting NEMO ubiquitination. Cell Death Dis. 5, e1229 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu B., Li S., Zhang X., Zheng X., HSCARG, a novel regulator of H2A ubiquitination by downregulating PRC1 ubiquitin E3 ligase activity, is essential for cell proliferation. Nucleic Acids Res. 42, 5582–5593 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qian J., et al. , USP7 modulates UV-induced PCNA monoubiquitination by regulating DNA polymerase eta stability. Oncogene 34, 4791–4796 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nijman S. M., et al. , A genomic and functional inventory of deubiquitinating enzymes. Cell 123, 773–786 (2005). [DOI] [PubMed] [Google Scholar]
- 27.Zlatanou A., et al. , USP7 is essential for maintaining Rad18 stability and DNA damage tolerance. Oncogene 35, 965–976 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Liang Q., et al. , A selective USP1-UAF1 inhibitor links deubiquitination to DNA damage responses. Nat. Chem. Biol. 10, 298–304 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J., et al. , Gene silencing of USP1 by lentivirus effectively inhibits proliferation and invasion of human osteosarcoma cells. Int. J. Oncol. 49, 2549–2557 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Cukras S., et al. , The USP1-UAF1 complex interacts with RAD51AP1 to promote homologous recombination repair. Cell Cycle 15, 2636–2646 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.US Office of Science and Technology Policy , Principles for the utilization and care of vertebrate animals. Fed Regist 50, 20864–20865 (1985). [PubMed] [Google Scholar]
- 32.National Research Council , Guide for the Care and Use of Laboratory Animals (National Academies Press, Washington, DC, ed. 8, 2011). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in the paper have been made available to readers.