Significance
B and T lymphocytes collaborate during immune responses to antigens. B cells use membrane-bound antibody as part of their antigen receptor while T cells use a different receptor that recognizes antigen fragments bound to MHC molecules. We show here that T cells can recognize the variable parts of the B cell receptor when these are presented on MHC molecules. A prerequisite for such receptor cross-talk is that the B cell receptor binds antigen. The cross-talk results in collaboration between B and T cells and production of antibodies directed against the antigen. The findings have implications for basic immune regulation. The results may also help us understand the mechanism behind the development of SLE-like autoimmune diseases and B cell lymphomas.
Keywords: idiotype-driven T–B collaboration, M315-like BCR, idiotypic peptide: MHCII, V-gene modified mouse model, BCR ligation by antigen
Abstract
The B cell receptors (BCRs) for antigen express variable (V) regions that are enormously diverse, thus serving as markers on individual B cells. V region-derived idiotypic (Id) peptides can be displayed as pId:MHCII complexes on B cells for recognition by CD4+ T cells. It is not known if naive B cells spontaneously display pId:MHCII in vivo or if BCR ligation is required for expression, thereby enabling collaboration between Id+ B cells and Id-specific T cells. Here, using a mouse model, we show that naive B cells do not express readily detectable levels of pId:MHCII. However, BCR ligation by Ag dramatically increases physical display of pId:MHCII, leading to activation of Id-specific CD4+ T cells, extrafollicular T–B cell collaboration and some germinal center formation, and production of Id+ IgG. Besides having implications for immune regulation, the results may explain how persistent activation of self-reactive B cells induces the development of autoimmune diseases and B cell lymphomas.
Each B cell expresses unique BCR variable (V) regions due to V(D)J recombination and somatic hypermutation (1). The highly diversified V regions express idiotypic (Id) determinants that can be recognized by antibodies (2) and by CD4+ T cells (3).
B lymphoma cells constitutively antigen-process their BCR and present Id peptides on their MHC class II molecules (pId:MHCII) to Id-specific CD4+ T cells (4, 5). Consistent with this, Id peptides were eluted from MHC class II molecules of tumor B cells (6). On the basis of these results, it was proposed in 1993 that Id-specific T cells help B cells that display pId:MHCII complexes on their surface (7). Such Id-driven T–B collaboration appears to be limited to rare Id peptides that express somatic mutations or unique N-region sequences (3, 8–11), since T cells are tolerized to germline-encoded V region sequences (8, 12). The existence of Id-driven T–B collaboration has been supported by studies using paired Ig/TCR-transgenic mice in 2 independent models (7, 13, 14). Chronic Id-driven T–B collaboration in these models has been associated with development of SLE-like autoimmune disease (13–16) and even B cell lymphomas (17).
The relevance of Id-driven T–B collaboration to disease development has been supported by recent observations in humans. First, bioinformatic analysis has indicated that human Ig V-regions are enriched for sequences that bind MHC molecules (18). Second, signs of Id-driven T–B collaboration have been observed in multiple sclerosis patients (19–21). Third, evidence of Id-driven T–B collaboration was obtained in chronic lymphatic leukemia (CLL) patients (22). Fourth, Id peptides were readily isolated from MHC class II molecules of mantle cell B lymphomas (23), as well as follicular B cell lymphomas, diffuse large B cell lymphoma, and CLL (24).
As an explanation for the pathogenicity of Id-driven T–B collaboration, it has been hypothesized that autoreactive B cells, in lieu of help from self antigen-specific T cells (that are tolerized), could instead receive help from Id-specific CD4+ T cells (13, 25). It was further hypothesized that BCR ligation by self antigen could contribute to such pathogenic Id-driven T–B collaboration (13, 25). In support of the hypothesis, BCR ligation caused a GC reaction and isotype switch; however, the experiments employed memory B cells and Th2 cells, not naive cells (13).
To investigate the unresolved issue of whether BCR ligation is required for Id-dependent collaboration between naive B and T cells in vivo, we have here generated a strain of mice that have a low frequency of B cells with a BCR that (i) can be deliberately ligated by antigen and (ii) contains a particular Id sequence in its V region. The model employs a type of V gene segment-modified mice that yields physiological expression of the Id sequence only subsequent to a Vλ2 → Jλ2 rearrangement in developing B cells. The surface display of pId:MHCII was physically detected by a staining reagent. The results show that naive B cells do not express detectable levels of pId:MHCII. However, BCR ligation induces pId:MHCII display, thereby enabling Id-driven T–B collaboration. The findings support a mechanism where BCR ligation by self antigen is required for display of pId:MHCII on autoreactive B cells and solicitation of help from Id-specific T cells.
Results
A Model System for Studying the Influence of BCR Ligation on Id-Driven T–B Collaboration.
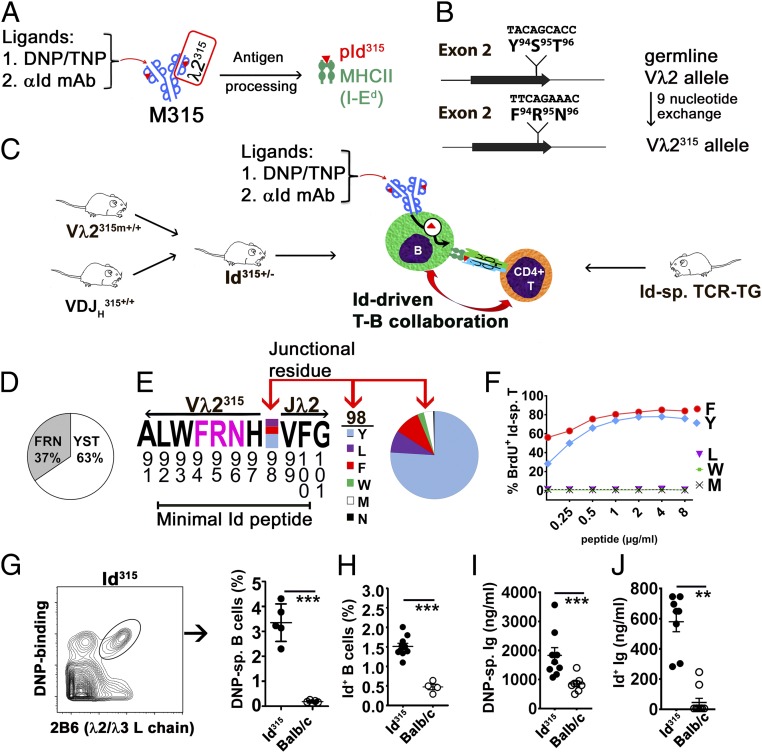
The M315 myeloma protein produced by the MOPC315 myeloma cell line binds defined ligands such as anti-Id mAbs (26) and DNP/TNP haptens (27) (Fig. 1A). Its L chain (λ2315) contains a mutated Id sequence that spans the V–J junction in the CDR3 loop. Upon antigen processing by APCs, the Id peptide is released and binds to the MHC class II molecule I-Ed for presentation to Id-specific CD4+ T cells (3, 4) (Fig. 1A).
Fig. 1.
A model system for studying the influence of BCR ligation on idiotype-driven T–B collaboration. (A) The M315 monoclonal Ig binds the indicated ligands. The λ2315 L chain expresses a mutated CDR3 Id peptide (residues 92 to 100; red triangle) that, after antigen processing, is presented on the MHC class II molecule I-Ed. (B) In Vλ2315m mice, the germline Vλ2 gene segment was modified by the exchange of 9 nucleotides so that the modified Vλ2 allele encodes the mutated residues F94R95N96 of the central part of the Id sequence. (C) Offspring from a Vλ2315m × VDJH315 cross, called Id315 mice (Left), should have a low frequency of B cells with an Id+ M315-like BCR that can be deliberately ligated. Id-driven T–B collaboration (Center) can be tested using Id-specific CD4+ T cells from previously established TCR-transgenic mice (Right). (D) The prevalence of germ line (YST) and mutated (FRN) sequences in λ2 encoding mRNAs in splenic CD19+ B cells of heterozygous Vλ2315m mice. (E) Map of the Vλ2315m → Jλ2 junction containing the mutated FRN sequence (pink letters) within the minimal Id peptide. The prevalence of various amino acids at the junctional position 98 is indicated. (F) In vitro response (BrdU incorporation) of Id-specific T cells to synthetic peptides with varying junctional residues presented by splenic APCs. (G) Frequency of Id315 BCR+ B cells in the circulation of Id315 and BALB/c mice detected using biotinylated DNP-albumin and λ2/3 L chain-specific (2B6 mAb) staining (n = 5 per group). (H) Frequency of Id315 BCR+ B cells detected using anti-Id mAb (Ab2-1.4) that requires association of VDJH315 and Vλ2315m chains for its binding (n = 10 Id315; n = 4 BALB/c). (I and J) DNP-specific (I) or Ab2-1.4-specific (J) antibodies in the sera of Id315 and BALB/c mice (n = 6 to 9 per group). Statistical comparisons: unpaired t tests (G–J). **P ≤ 0.01, ***P ≤ 0.005.
To study the role of BCR ligation in Id-driven T–B collaboration, we developed two strains of mice that express the VH and the VL of M315, respectively. Upon cross-breeding, the offspring should express an M315-like BCR on a low proportion of their B cells. For the VH, we made a conventional BCR knock-in mouse where a rearranged and moderately mutated VDJH315 was placed in the JH locus (SI Appendix, Figs. S1 and S2 A and B). For the VL, we generated a type of V gene segment-modified mouse where nine nucleotides encoding the amino terminal part of the mutated Id peptide replaced the corresponding germline nucleotides in the Vλ2 gene segment (Fig. 1B and SI Appendix, Fig. S3 A and B). In rare B cells, where the modified Vλ2315m rearranges to the Jλ2, a recombined VJ is generated that should encode the entire mutated Id315 peptide. The F1 progeny of these two strains, called Id315 mice, should express an M315-like BCR on a small subset of their B cells. Such rare Id+ B cells were tested for collaboration with Id-specific CD4+ T cells from previously described TCR-transgenic mice (28), either in the presence or the absence of BCR ligation by defined ligands (Fig. 1C).
Characterization of Gene-Modified Strains of Mice.
In the VDJH315 mouse, allelic exclusion was pronounced, and VDJH315 was expressed by most B cells (SI Appendix, Fig. S2 C–H). B cell development in the bone marrow was slightly accelerated (SI Appendix, Fig. S3G), as is commonly found in VDJ knock-in mice. In the Vλ2315m mouse, codons 94, 95, and 96 of the germline Vλ2 were exchanged with those of Vλ2315expressed by MOPC315. Since the FRT recombination left a short residual sequence within the intron immediately downstream of Vλ2315m (SI Appendix, Fig. S3A), the recombination frequency to Jλ2 could have been influenced. To test this, we analyzed the frequencies of Vλ2 → Jλ2 rearrangements on the WT and Vλ2315m chromosomes in heterozygous Vλ2315m mice by cDNA amplicon sequencing (29). Both transcripts were expressed, although Vλ2315m–Jλ2 joints were found at a slightly lower frequency than Vλ2–Jλ2 joints (Fig. 1D). At the Vλ2–Jλ2 junction, residue 98 varies due to junctional diversity (30). We found that the rearranged WT and Vλ2315m alleles had an identical frequency of different amino acids in position 98 with the ranking order Tyr>>Phe∼Leu (Fig. 1E). Since the minimal Id peptide (residues 92 to 100) spans the VJ junction, we tested synthetic peptides with naturally occurring amino acids at position 98 for their ability to stimulate Id-specific CD4+ T cells. Peptides with both Tyr98 and Phe98 residues were fully compatible with stimulation of Id-specific CD4+ T cells, while Leu98 was not (Fig. 1F). This result indicates that most B cells in which a Vλ2315m → Jλ2 rearrangement occurs should potentially be able to stimulate Id-specific T cells. The frequencies and specific lineages of λ2/3+ B cells (detected by a Cλ2/Cλ3 cross-reactive mAb, 2B6) and the levels of total λ2/3+ serum Ig did not differ between Vλ2315m and BALB/c mice (SI Appendix, Fig. S3 D–G). These results indicate that the V gene segment modification in Vλ2315m did not influence B cell development, so the mouse strain displayed a physiological B cell compartment.
Offspring of homozygous VDJH315 and Vλ2315m mice, called Id315 mice, had a slightly accelerated development of B cells in the bone marrow and a small increase in λ2/λ3+ T2 cells in the spleen, but were otherwise normal (SI Appendix, Fig. S3 F and G). These changes are most likely due to the prearranged VDJH315 component of Id315 mice (SI Appendix, Fig. S2A). In Id315 mice, ∼3 to 4% of circulating B cells expressed an M315-like BCR detected by binding of DNP (Fig. 1G), while ∼1.5% were detected by the anti-Id IgG1 Ab2-1.4 (26) (Fig. 1H), consistent with the fine specificity differences between these two BCR ligands (SI Appendix, Table S1) (26) (also detailed later). The result with the highly specific Ab2-1.4 mAb (Fig. 1H) is as expected, since Vλ2315m → Jλ2 rearrangements should only occur in ∼1 to 2% of peripheral B cells (30). Id315 mice had about ∼1 to 2 μg/mL of serum IgG that bound DNP (Fig. 1I) and ∼0.3 to 0.8 μg/mL that bound anti-Id Ab2-1.4 mAb (Fig. 1J), again consistent with the fine specificity of these reagents (SI Appendix, Table S1) (26). Collectively, these results indicate that VDJH315, Vλ2315m, and Id315 mice displayed the expected features.
Id315 mice were used in most of the in vivo experiments described herein. However, in some experiments (e.g., adoptive transfers to CD45.1+ congenic BALB/c mice), large numbers of Id315 B cells (hereafter called Id+ B cells) were needed. For this purpose, VDJH315 mice were crossed with previously described λ2315 transgenic mice (31). In the progeny, as much as 70 to 80% of peripheral B cells expressed an M315 BCR (SI Appendix, Fig. S4 A and B). In λ2315TG × VDJH315 mice, splenic B cells had slightly reduced IgM expression levels (SI Appendix, Fig. S4C), and marginal zone (MZ) B cells were increased relative to follicular (FO) B cells (SI Appendix, Fig. S4D). Nevertheless, Id+ B cells from the progeny were fully responsive to stimulation. Pre-B and pro-B cells in the bone marrow were reduced, but mature B cells were found to be at normal levels (SI Appendix, Fig. S4E). Since activated B and T cells may modulate their surface antigenic receptors, the use of CD45.1+ congenic mice facilitated the detection of the transferred lymphocyte populations upon recovery.
Characterization of Specificity of BCR Ligands.
The amino acid exchanges introduced in positions 94, 95, and 96 of Vλ2315m, as well as the junctional variation in position 98 in Vλ2315m-Jλ2 joints (Fig. 1E), could influence the binding of anti-Id mAb Ab2-1.4 and DNP/TNP used as ligands for the Id+ BCR (Fig. 1A). To investigate this, we established a panel of recombinant IgGs that express VH315 together with various Vλ2-regions and tested these for binding to the anti-idiotypic mAb Ab2-1.4 as well as DNP and TNP haptens (SI Appendix, Table S1). In brief, the Ab2-1.4 bound VH315 associated with Vλ2315m but not with germline Vλ2. Moreover, Ab2-1.4 binding was compatible with the three most frequent amino acids found in position 98: Tyr, Phe, and Leu (Fig. 1E). The germline equivalent of VH315, VH3-6*02, was also compatible with binding, while a quite different VH, VHA20, was not. However, VH3-6*02 is probably poorly expressed in Id+ mice due to the prerearranged VDJH315. In conclusion, Ab2.1.4 is a highly specific ligand for the BCR of Id+ B cells and should bind all BCRs independent of junctional variation. Its affinity for Id+ M315 is KA = 7.7 × 106 M−1 (32).
TNP and DNP BCR ligands have a broader specificity since either of the haptens bound to VH315 and VH3-6*02 (but not VHA20) associated with either Vλ2315m or Vλ2 (SI Appendix, Table S1). Junctional variation in position 98 did not influence binding. These results extend a previous study demonstrating that TNP and DNP bind VH315 associated with either λ1, λ2, and λ3 light chains, but not κ light chains (26). Thus, TNP/DNP should bind BCRs that express endogenous λ-chains together with VH315 in Id315 mice. The affinity of DNP for M315 has been measured to be in the range of KA= 1.6 × 105 to 3.9 × 106 M−1 (33), and the affinities of DNP and TNP are in a similar range (34).
In the course of the experiments described later, TNP/DNP and Ab2-1.4 were used not only as BCR ligands but also to measure B cell and antibody responses. Somatic hypermutation could have influenced binding and detection, which is difficult to control for. However, this does not seem to have been a major issue, since potent responses were detected by the BCR ligands over prolonged periods of time.
BCR Ligation Is Required for Id-Driven T–B Collaboration in Vitro.
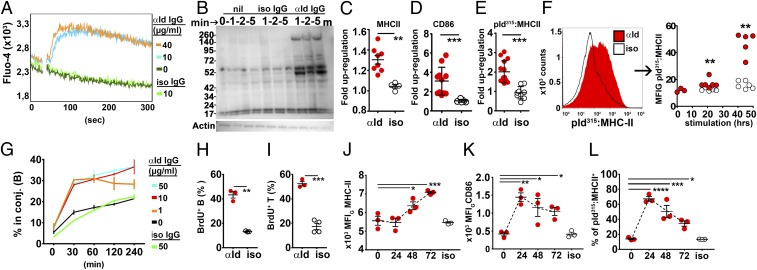
We performed a series of in vitro experiments on naïve splenic Id+ B cells enriched through immunomagnetic depletion. Exposure of Id+ B cells to Ab2-1.4 (hereafter referred to as anti-Id IgG) as BCR ligand resulted in intracellular Ca2+ mobilization (Fig. 2A), protein phosphorylation (Fig. 2B), and up-regulation of MHC class II (Fig. 2C) and CD86 (Fig. 2D), but not CD80, cell surface expression. Similar, although slightly lower, responses were obtained with DNP-OVA and TNP-OVA as BCR ligands (SI Appendix, Fig. S5). Importantly, BCR ligation resulted in an increased display of Id peptide–MHC class II complexes on the B cell surface, detected by an scFv reagent specific for the pId315:I-Ed complex [hereafter called pId:I-Ed or pId:MHCII; the reagent detecting this complex is hereafter called a TCR mimetic (TCRm); Fig. 2E; see also SI Appendix, Fig. S6]. Expression of pId:I-Ed was first detected after 18 to 20 h and remained elevated for at least 45 h after ligation (Fig. 2F and SI Appendix, Fig. S6G). When naïve Id+ B cells were mixed with naïve Id-specific T cells, BCR ligation induced T cell–B cell conjugate formation within 30 min (Fig. 2G). In cocultures of Id+ B cells and Id-specific T cells, ligation of the Id+ BCR by anti-Id IgG resulted in the proliferation of B cells and T cells (Fig. 2 H and I). These in vitro results show that BCR ligation enhances Id-driven T–B collaboration and that up-regulation of pId:MHCII is a likely contributor.
Fig. 2.
Ligation of the Id+ BCR by antigen induces downstream signaling, Id peptide presentation on MHC-II, and stimulation of Id-specific T cells. (A–I) In vitro experiments. Id+ B cells (enriched from λ2315TG × VDJH315 mice by negative selection) were stimulated with anti-Id IgG or added isotype-matched control IgG (10 µg/mL unless otherwise indicated). (A) Ca2+ flux assay. (B) Phosphotyrosine blots of cell lysates. (C–E) MHC-II (I-Ad/I-Ed), CD86, and pId315:I-Ed expression after 18 to 21 h in culture. The data represent fold increases in the geometric mean of FI signal of Id+ B cells added either anti-Id IgG or isotype control IgG. MFIG values from nonstimulated Id+ B cells in each experiment were taken as a baseline. Fold up-regulation values were pooled from 2 to 3 individual in vitro assays. (F, Left) Histogram plot of pId315:I-Ed expression on Id+ B cells measured after 48 h using the TCR mimetic scFv reagent. (F, Right) pId315:I-Ed presentation on Id+ B cells after 21 and 45 h. (G) Conjugate formation between Id+ B cells and Id-sp. T cells labeled with CFSE and CTV. (H and I) BrdU incorporation into Id+ B cells and Id-specific T cells, measured during the last 14 h of a 4-d coculture. (J–L) In vivo experiments. Id315 mice were injected i.v. with 60 µg BCR ligand anti-Id IgG or isotype control IgG, and splenic Id+ B cells were analyzed for expression of MHC-II (J), CD86 (K), and pId315:I-Ed (L; FACS plots shown in SI Appendix, Fig. S6F). Statistical comparisons: unpaired t tests (C–E, H, and I), Mann–Whitney U tests for BCR ligation versus control at each time point (F), and Dunnett’s multiple comparisons test for baseline levels versus the different time points (J–L). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001.
It was of interest to assess whether pId:MHCII expression increased only as a consequence of a general up-regulation of MHC class II molecules, and whether BCR ligation was required. To investigate this possibility, Id+ B cells were BCR-ligated with TNP-OVA and compared with stimulation by the TLR4 ligand LPS (SI Appendix, Fig. S7). LPS stimulation increased the MFI signal of MHCII expression more than 5-fold, whereas pId:MHCII expression was essentially unaltered. The inverse was observed with TNP-OVA as BCR ligand. These results argue that BCR ligation resulted in a specific up-regulation of pId presentation on MHC class II molecules.
BCR Ligation Is Required for Id-Driven T–B Collaboration In Vivo.
We first tested the effect of BCR ligation on Id+ B cells in vivo in the absence of Id-specific T cells. A cohort of Id315 mice were injected i.v. with anti-Id IgG or isotype-matched specificity control mAb, and splenic Id+ B cells were analyzed by FACS at the indicated time points. Within 24 to 48 h, BCR ligation enhanced the surface expression of MHC class II molecules, CD86, and pId:MHCII complexes on Id+ B cells (Fig. 2 J–L and SI Appendix, Fig. S6F). The increased expression was still detectable after 72 h. These results suggest that in vivo provision of BCR ligand prepares Id+ B cells for efficient interaction with Id-specific T cells through surface display of both pId:MHCII and costimulatory molecules. Up-regulation of pId:MHCII was observed already at 24 h (Fig. 2L), while a general up-regulation of MHCII was seen first after 48 h (Fig. 2J), again arguing that BCR ligation has a specific contribution to increased display of pId:MHCII.
To probe the sensitivity of the model for Id-driven T–B collaboration, we titrated the amounts of Id-specific T cells, anti-Id IgG, and Id+ B cells needed to obtain Id+ IgG responses in Id315 and in CD45.1 congenic BALB/c recipients. Transfer of 15,000 naive T cells and 4 μg anti-Id IgG sufficed to elicit full Id+ Ig responses in Id315 recipients (SI Appendix, Fig. S8 A and B). Even as little as 1,500 Id-specific T cells induced Id+ IgG levels above background levels. Assuming a 10 to 15% parking efficiency, this cell dose is estimated to result in a physiological frequency of the Id-specific T cells in the recipient (35). Naive Id+ B cells were titrated by transfer into CD45.1+ congenic BALB/c mice together with saturating dose of naive Id-specific T cells and anti-Id IgG. About 105 Id+ B cells were required to elicit Id+ IgG responses (SI Appendix, Fig. S8C).
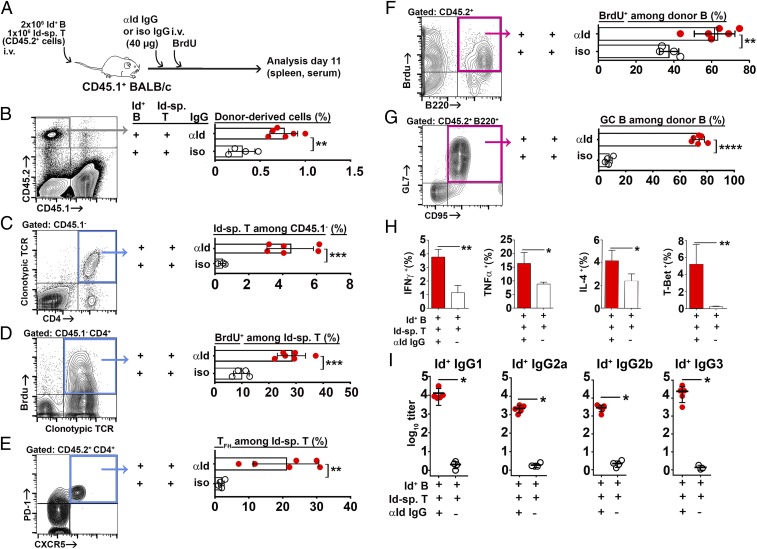
Based on these results, we transferred naïve Id-specific T cells and Id+ B cells, enriched through immunomagnetic depletion of nondesired populations, into CD45.1+ congenic BALB/c mice, followed by anti-Id IgG or isotype-matched control IgG and a continuous BrdU administration (Fig. 3A). A number of conclusions could be made based upon analysis of recipient spleens (Fig. 3) and lymph nodes (SI Appendix, Fig. S9). In the spleen, BCR ligation increased the numbers of (i) donor-derived CD45.2+ cells (Fig. 3B), (ii) Id-specific CD4+ T cells that had incorporated BrdU (Fig. 3 C and D), (iii) follicular T helper cells (TFH; Fig. 3E), (iv) Id+ B cells that had incorporated BrdU (Fig. 3F), and (v) germinal center (GC) B cells (Fig. 3G). Intracellular staining of recovered Id-specific T cells revealed that BCR ligation increased expression of IFN-γ, TNF-α, and IL-4 cytokines and the transcription factor T-Bet (Fig. 3H), whereas GATA-3 and Foxp3 were not detected. Responses were stronger in the spleen than in the lymph nodes (SI Appendix, Fig. S9). BCR ligation induced a 3- to 4-log10 increase in serum levels of Id+ IgG of all subclasses (Fig. 3I).
Fig. 3.
BCR ligation is required for idiotype-driven T–B collaboration in the spleen. (A) Experimental setup (Id-sp. T/anti-Id IgG, n = 6; Id-sp. T/isotype control IgG, n = 4). (B–G) FACS analysis. The gated populations are shown: B cells, pink gates; T cells, blue gates. (B) Frequencies of donor lymphocytes (CD45.2+ CD45.1−). (C) Frequencies of Id-specific CD4+ T cells detected by the TCR clonotype-specific mAb GB113. (D) BrdU incorporation into Id-specific T cells. (E) Follicular T helper cell differentiation of Id-specific T cells. (F) BrdU incorporation into Id+ B cells (CD45.2+ B220+). (G). Germinal center differentiation of Id+ B cells. (H) Intracellular cytokine and intranuclear transcription factor expression by Id-specific CD4+ T cells. (I) Serum titers of Id+ (anti-Id Ig-reactive) IgG antibody subclasses on day 11. Statistical comparisons: unpaired t tests (B–G), Mann–Whitney U tests (H and I). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.005.
Some of the anti-Id mAb Ab2-1.4 (IgG1) used in the aforementioned experiments could have been internalized via FcγRIIb on B cells rather than through receptor-mediated uptake, which could have contributed to pId:MHCII presentation. To exclude this possibility, we generated F(ab)2 fragments of the anti-Id IgG (SI Appendix, Fig. S10). F(ab)2 fragments were at least as stimulatory as intact anti-Id mAb to initiate Id-driven T–B cell collaboration, demonstrating that the Fc region and uptake via FcγRIIb is dispensable for responsiveness to BCR ligation (SI Appendix, Fig. S11).
In these experiments, the necessity of BCR ligation for initiation of Id-driven T–B collaboration was demonstrated with naive B and T cells. However, it has been previously demonstrated that naive B cells, although unable to stimulate naive T cells, can stimulate memory T cells (36). Consistent with this, it was shown that naive B cells from λ2315-transgenic mice could stimulate Id-specific Th2 cells in the absence of BCR ligation (37). We therefore tested if naive Id+ B cells in Id315 mice could collaborate with in vitro-polarized Id-specific Th2 cells after adoptive transfer. The results show that naive Id+ B cells can collaborate with Th2 cells in vivo; however, addition of BCR ligation clearly enhanced responses (SI Appendix, Fig. S12).
Localization of Id-Driven T–B Collaboration.
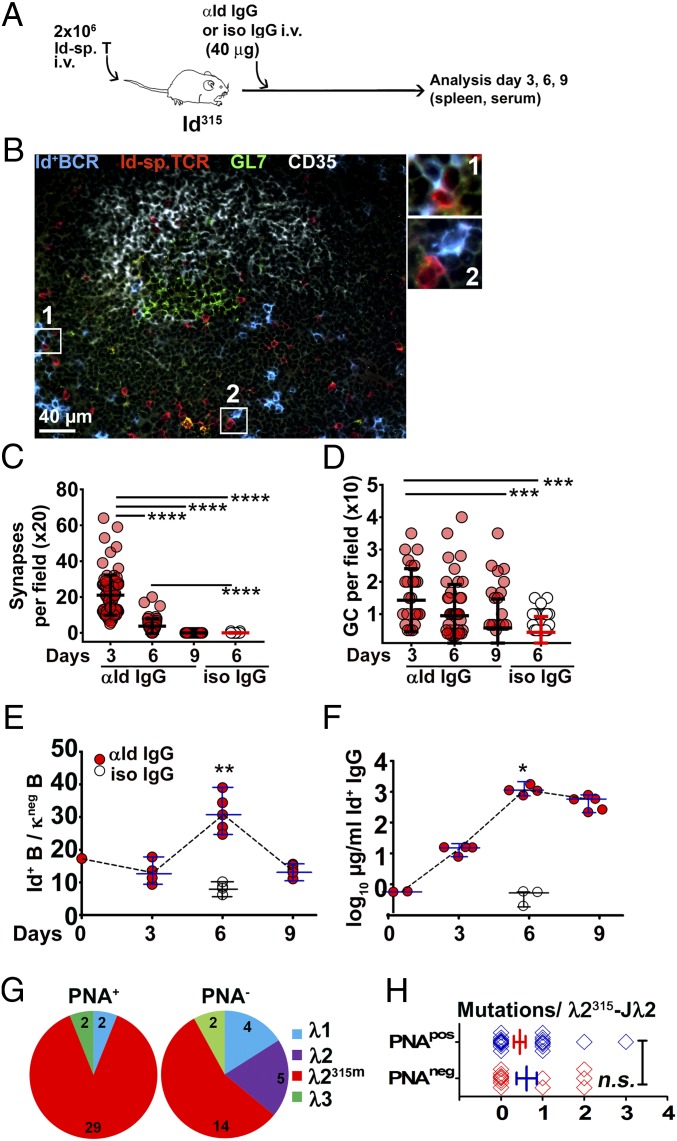
To study the histological correlates of BCR ligation, we transferred naïve Id-specific T cells into Id315 mice, injected anti-Id IgG or isotype-matched control IgG the next day, and analyzed spleens at different time points after ligation (Fig. 4 and SI Appendix, Figs. S13 and S14). Staining of sections for the GL7 lymphocyte activation marker showed that BCR ligation induced a robust extrafollicular activation of B and T cells on day 3 and day 6; however, activation had considerably subsided by day 9. Germinal centers (GCs) also peaked early (day 3), but these still remained on day 9, although they were fewer (SI Appendix, Fig. S13A and Fig. 4D; further detail provided later). By immunofluorescence analysis using mAbs specific for the Id+ BCR, the Id-specific TCR, the follicular dendritic cell light zone marker CD35, and GL7, we found considerable numbers of Id+ B cells and Id-specific T cells localized in regions surrounding GL7+/CD35+ GCs. Moreover, direct contacts (synapses) were observed between Id+ B cells and Id-specific T cells outside GCs (Fig. 4B and SI Appendix, Fig. S14A). GL7+ GC B cells stained only weakly with Ab2-1.4 anti-Id mAb (SI Appendix, Fig. S14B, Top), most likely due to BCR down-regulation. In contrast, stronger staining for the Id+ Ig, sometimes suggestive of cytosolic accumulation, was detected in B cells outside the GC (Fig. 4B and SI Appendix, Fig. S14A). In addition to their predominantly extrafollicular location, Id-specific T cells were also observed, albeit more scarcely, in the GC (Fig. 4B and SI Appendix, Fig. S14B). Staining for Ki-67 corroborated that proliferation occurred both in GCs and in extrafollicular B cell clusters on day 3 postligation (SI Appendix, Fig. S14C).
Fig. 4.
Germinal centers form rapidly in Id315 mice upon ligation of the Id+ BCR. (A) Experimental setup (Id-sp. T/anti-Id IgG, n = 14; Id-sp. T/isotype control IgG, n = 4). (B) Representative germinal center in the spleen of a recipient mouse on day 3. Id-specific TCR (clonotype-specific mAb GB113, red) and Id+ BCR (anti-Id BCR Ab2-1.4 mAb, blue) staining shows an area of T cell–B cell interaction. Germinal center B cells were identified as GL7+ clusters (green) with the light zone indicated by the presence of follicular dendritic cells (CD35, white). Examples of T–B synapses are magnified in 1 and 2. SI Appendix, Fig. S14B shows single stains contributing to the overlay. (C) Quantification of specific T–B synapses based on transgenic receptor expression in sections. Each data point represents the number of Id+ B/Id-sp. T synapses counted in one ×20 field. (D) Quantification of germinal centers from spleen cryosections (means per ×10 field). (E) Id+ B cells (Ab2-1.4-reactive) among κ light chain negative splenic B cells by FACS analysis. (F) Id+ IgG serum levels. (G) Frequencies of the different λ transcripts recovered among PNA+ and PNA− fractions of sorted B cells. The B cells were sorted on day 7 from spleens of Id315 mice primed with Id-specific T cells and anti-Id F(ab)2 fragments (SI Appendix, Fig. S11). (H) Comparisons of mutational load among Vλ2315m-Jλ2 L chain transcripts recovered from the PNA-positive and PNA-negative fractions. Statistical comparisons: Dunnett’s multiple comparisons test (C and D), unpaired t tests (E, F, and H). *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.005, and ****P ≤ 0.001; n.s., not significant.
Quantification revealed that BCR ligation increased specific T–B synapses as well as GCs in spleen sections (Fig. 4 C and D). Synapses as well as GCs were most frequent already on day 3, and declined thereafter (Fig. 4 C and D). Synapses and GC formation preceded the peak expansion of Id+ B cells (day 6; Fig. 4E) and the peak of Id+ serum IgG (Fig. 4F). In summary, Id-driven T–B collaboration appeared to occur predominantly in extrafollicular sites and to a smaller extent in GCs. The latter appeared small and of short duration, perhaps indicative of abortive GC reactions. Analysis of stained sections (Fig. 4B and SI Appendix, Figs. S13 and S14) revealed less robust GC responses than did the FACS analysis (Fig. 3), where a preponderance of GC B cells and TFH was found. Of note, for the FACS analyses, we used the CD45.2 congenic marker, which is expressed irrespective of down-modulation of the BCR or the TCR, thereby facilitating recovery and enumeration of a larger proportion of the transferred cells. Furthermore, the anatomical location of Id+ B cells could differ in Id315 mice (natural positioning) compared to their location after injection i.v. into CD45.1 congenic recipients, thereby influencing the results.
Analysis of Id-Driven T–B Collaboration by Ig Gene Sequencing.
On day 7 of Id-driven T–B collaboration, using anti-Id F(ab)2 fragments as BCR ligand (SI Appendix, Fig. S11), we purified PNA+ and PNA− B cells from Id315 mouse spleens and PCR-amplified and sequenced light chain transcripts with primers specific for λ1, λ2, and λ3 chains. The PNA+ fraction contained 20% GL7+ CD95+ cells by FACS, indicating ∼15 fold enrichment of GC B cells. Sequence analysis showed that the PNA+ population was almost completely dominated by Vλ2315m sequences having the idiotypic FRN sequence in positions 94 to 96 (Fig. 4G). A similar but less pronounced result was obtained for the PNA− population. These observations demonstrate a strong expansion of Vλ2315m+ B cells, not only in the GC (PNA+) population, but also in the non-GC (PNA−) population. The results support the functional data (Figs. 3 and 4). Interestingly, the junctional amino acid in Vλ2315m-Jλ2 sequences was exclusively F98 even though Y98 was by far the most frequent junctional residue in Vλ2315m-Jλ2 sequences of unstimulated B cells (Fig. 1E). The level of nucleotide substitutions was surprisingly low, with most sequences being unmutated or having only one mutation (Fig. 4H and SI Appendix, Fig. S15). The level of nucleotide substitutions in Vλ2315m-Jλ2 transcripts was not significantly different between the PNA+ population and the PNA− population (Fig. 4H). The paucity of mutations in the PNA+ cell population is surprising and appears consistent with a limited GC response.
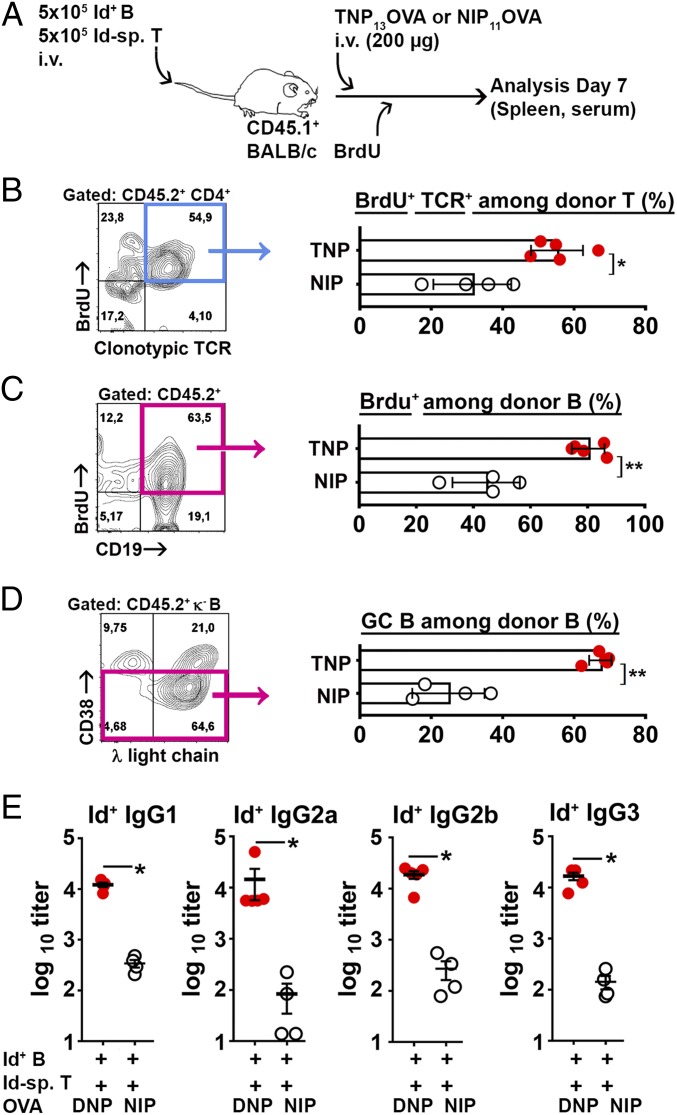
Ligation of the Id+ BCR Using a Hapten–Protein Conjugate Promotes Id-Driven T–B Collaboration.
M315 has a specificity for the structurally similar haptens DNP and TNP (27), but does not bind the NIP hapten. We therefore tested if BCR ligation by DNP- and TNP-conjugated proteins could promote Id-driven T–B collaboration. CD45.1+ BALB/c mice were transferred with Id+ B cells and Id-specific T cells, followed by TNP-OVA or NIP-OVA (specificity control). Thereafter, mice continuously received BrdU (Fig. 5A). High doses of hapten–OVA conjugates were used (200 μg), since preliminary experiments suggested that DNP-OVA is relatively inefficient at promoting Id-driven T–B collaboration in vivo compared to anti-Id IgG (SI Appendix, Fig. S16). Several factors could contribute to this difference. First, OVA is filtrated in the kidneys (38), while the higher-MW anti-Id IgG is not. Second, B cells expressing VDJH315 together with other λ chains than λ2315m could bind DNP/TNP conjugates (SI Appendix, Table S1) and thus serve as a sink for DNP/TNP-OVA. Finally, the Ig structure of the anti-Id IgG ligand could be particularly efficient at cross-linking BCR due to matching distances between antigen binding sites of Id+ and anti-Id Igs.
Fig. 5.
Ligation of the Id+ BCR by TNP-OVA enables Id-driven T–B collaboration. (A) Experimental setup (n = 5 Id+ B/Id-sp. T/TNP-OVA; n = 4 Id+ B/Id-sp. T/NIP-OVA). (B–D) FACS analysis. The gated populations are shown; B cells, pink gates; T cells, blue gates. (B) BrdU+ Id-specific T cells among transferred T cells. (C) BrdU+ donor B cells. (D) Id+ GC (CD38lo/-) B cells. (E) Serum titers of Id+ IgG antibody subclasses on day 7. Statistical comparisons: unpaired t tests (B–D), Mann–Whitney U test (E). *P ≤ 0.05, **P ≤ 0.01.
Despite the decreased sensitivity, BCR ligation by TNP-OVA in vivo increased BrdU incorporation into both Id-specific T cells and Id+ B cells compared to that seen with NIP-OVA (Fig. 5 B and C). Further, BCR ligation increased the frequency of GC-like Id+ B cells (Fig. 5D). Finally, BCR ligation enhanced the serum levels of Id+ IgG of all subclasses (Fig. 5E). These results show that BCR ligation with TNP-OVA enhances Id-driven T–B collaboration.
BCR Ligation with a T Cell-Independent Type 2 Antigen Enhances Id-Driven T–B Collaboration.
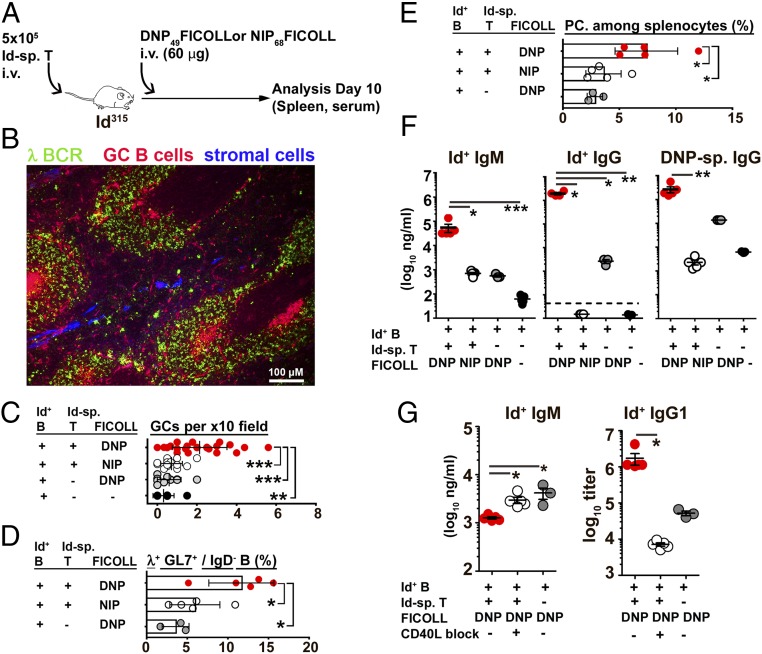
In the experiments described here earlier, endogenous T cell responses directed against the BCR ligand, either anti-Id IgG or OVA, could have influenced the results. To exclude this possibility, we tested DNP-FICOLL as a BCR ligand, the rationale being that FICOLL is a polysaccharide that should not be presented on MHC class II molecules to endogenous T cells. Moreover, since DNP-FICOLL is a T cell-independent type 2 (TI-2) antigen, the experiment addresses whether Id-specific CD4+ T cells influence B cell responses to a TI-2 antigen.
Id-specific T cells were transferred to Id315 mice followed by DNP-FICOLL or NIP-FICOLL the next day (Fig. 6A). Responses in the spleens were analyzed on day 10. Immunohistochemical staining revealed an enhanced generation of GCs on day 10 when using DNP-FICOLL compared to NIP-FICOLL (Fig. 6B), which was verified by quantitative assessment (Fig. 6C). In the absence of T cell help, BCR ligation by DNP-FICOLL elicited a slight expansion of Id+ (λ+) B cells, including GC B cells and plasma cells, as determined by flow cytometry (Fig. 6 D and E and SI Appendix, Fig. S17). Addition of Id-specific T cells enhanced the expansion of Id+ B cells and promoted their differentiation into GC B cells and plasma cells (Fig. 6 D and E and SI Appendix, Fig. S17). Finally, the concentrations of serum IgM and IgG that bound anti-Id mAb or DNP were enhanced in the presence of DNP-FICOLL and Id-specific T cell help compared to control groups (Fig. 6F). Taken together, the results show that a TI-2 BCR ligand alone stimulates Id+ B cell responses weakly, but that provision of Id-specific T cell help significantly augments responses. Further, recognition of the BCR ligand by endogenous polyclonal T cells is not required for Id-driven T–B collaboration.
Fig. 6.
B cell responses to a BCR-specific T-independent type-2 antigen is enhanced by Id-specific T cells. (A) Experimental setup (n = 5 Id-sp. T/DNP-FICOLL; n = 5 Id-sp. T/NIP-FICOLL; n = 3 DNP-FICOLL). Spleens were analyzed by IHC (B and C) or by FACS (D and E). (B) Representative cryosection from a recipient mouse spleen immunostained using anti-λ BCR L chain Alexa Fluor 488 (green), peanut agglutinin Alexa Fluor 594 (red), and rat anti-mouse reticular fiber Ab (ER-TR7, blue). (C) GCs quantified from immunostained spleen sections. (D) Germinal center B cells. GL7+ cells with λ L chains were gated among IgD− B cells (SI Appendix, Fig. S17). (E) Plasma cells (CD138+) among splenocytes. (F) Serum concentrations of anti-Id Ig-reactive IgM, IgG, and DNP-reactive IgG on day 10. (G) CD40L blockade by repetitive injection of anti-CD40L mAb or isotype-matched control. Effects on serum levels of Id+ IgM (day 5) and IgG1 (day 7). A detailed description and analysis is given in SI Appendix, Fig. S18. Statistical comparisons: Tukey’s multiple comparisons tests (C–E), Dunn's multiple comparisons test (F and G). *P ≤ 0.05, **P ≤ 0.01, and ***P ≤ 0.005.
To test if the CD40L–CD40 axis was involved in Id-driven T–B collaboration, we next tried to block responses to DNP-FICOLL by repetitious injections of anti-CD40L mAb (Fig. 6G and SI Appendix, Fig. S18). The results show that CD40L blockade increased the levels of Id+ IgM while Id+ IgG of all subclasses were decreased (Fig. 6G and SI Appendix, Fig. S18). Thus, CD40L is involved in the IgM → IgG switch in the presence of Id+ B and Id-specific T cells. Anti-CD40L mAb did not inhibit the IgM response by Id+ B cells induced by DNP-FICOLL in the absence of Id-specific T cells, consistent with DNP-FICOLL being a TI-2 antigen (SI Appendix, Fig. S19).
The experiment was extended to adoptive transfer of Id+ LPS blasts and naïve Id-specific T cells to CD45-congenic recipients, followed by injection of DNP-FICOLL (SI Appendix, Fig. S20A). BCR ligation enhanced responses of Id+ LPS blasts, both in terms of plasma cell differentiation and IgG production (SI Appendix, Fig. S20 C–E). A slight expansion of Id-specific T cells was observed in response to LPS blasts in the absence of ligation (SI Appendix, Fig. S20B), but this did not result in IgG serum levels above that observed in mice transferred LPS blasts only (SI Appendix, Fig. S20 D and E). These results indicate that nonspecific activation of Id+ B cells through TLR4 is not sufficient to induce collaboration with naïve Id-specific T cells, but that BCR ligation is required. This may well relate to the importance of recognition of pId:MHCII by Id-specific T cells, since the level of pId:MHCII remained unchanged after LPS stimulation (SI Appendix, Fig. S7).
Ligation of the Id+ BCR Promotes the Development of Memory B Cells, Bone Marrow Plasma Cells and Id-Specific Regulatory T Cells.
After having evaluated early responses to BCR ligation, we performed experiments where responses were analyzed at later time points, e.g., day 28 (SI Appendix, Fig. S21). Id+ B cells and Id-specific T cells were transferred to CD45.1+ BALB/c mice followed by specific BCR ligation (DNP-FICOLL) or nonspecific control antigen (NIP-FICOLL). Four weeks later, the spleens and bone marrow cells from femurs were analyzed (SI Appendix, Fig. S21A). Id-specific regulatory T cells were found on day 28, and had increased to ∼2% of recovered Id-specific T cells in the spleen (SI Appendix, Fig. S21B). The development of Tregs required BCR ligation. Formation of Id+ memory B cells (CD73+ CD273+) in the presence of BCR ligation was also increased (SI Appendix, Fig. S21C). Finally, bone marrow plasma cells were significantly expanded in recipients that had received the BCR ligand (SI Appendix, Fig. S21D), and serum antibodies specific for anti-Id IgG and DNP were significantly elevated (SI Appendix, Fig. S21 E–H). Id+ plasma cells secreting Id+ IgM and Id+ IgG were also found on day 40 in another experiment, where anti-Id IgG was used as BCR ligand (SI Appendix, Fig. S22). These results show that BCR ligation in Id-driven T–B collaboration has a long-lasting influence.
Discussion
The present results demonstrate that BCR V region-derived Id peptides are undetectable on MHC class II molecules of naïve B cells, but that BCR ligation induces display. Further, in the presence of naive Id-specific CD4+ T cells, BCR ligation initiates the full panoply of events of T cell–B cell collaboration, including T–B synapse formation, mutual activation and proliferation, GC formation with generation of GC B cells and TFH, extrafollicular T–B cell responses, and an increase in plasma cells and antibody production. Despite the expansion of Id+ B cells, the level of mutations in their BCR L chain V region was surprisingly small. Whether BCR-ligated Id-driven T–B cell collaboration requires dendritic cells has not been addressed in the current work, although DCs were dispensable during conventional T–B collaboration using anti-Id BCR knock-in mice and Id+ M315 as antigen (39).
These results seemingly contradict previous functional studies in mice (4, 5, 37) and MHCII elution studies in mice (6) and humans (23, 24, 40), which have demonstrated spontaneous pId:MHCII presentation without deliberate BCR ligation. However, these previous studies employed malignant B cells of unknown specificity cultured in vitro, and a number of uncontrolled factors could have contributed to constitutive pId:MHCII display. In more physiologically relevant experiments, naive B cells from λ2315 Ig L-transgenic mice stimulated Id-specific Th2 cells, but not naive Id-specific CD4+ T cells (37). In a second Ig L-chain transgenic model, B cells had to be activated in order to stimulate Id-specific T cell hybridomas in vitro (41). In these studies, the Ig L chain transgenic B cells had a polyclonal H chain repertoire, thus making it difficult to exclude a contribution of BCR ligation by unknown antigens to pId:MHCII display. The present study removes many of the ambiguities of these previous studies. First, a V gene segment-modified mouse was created that had a low frequency of B cells, which, subsequent to a Vλ2 → Jλ2 rearrangement, had a physiological expression of a defined Id sequence in the BCR L chain. Second, these Id+ B cells expressed a defined H chain (from VDJH knock-in mice), and thus had a BCR of known specificity, equivalent to that of myeloma protein M315. Third, naive Id+ B cells and naive Id-specific T cells were employed. Fourth, relatively low (but still unphysiological [35]) numbers of B and T cells were used in the cell transfer experiments. Fifth, three different BCR ligands, including a TI-2 ligand, were employed, with similar results. Sixth, display of pId:MHCII was physically measured by a staining reagent, in addition to functional readouts.
The mechanism by which BCR ligation by antigen enhances pId:MHCII expression is unknown, but is most likely due to enhanced endocytosis and processing of ligated BCR. Thus, we propose that ligation not only induces endocytosis, processing, and presentation of Ag (42) but also of the BCR itself. This makes sense, since the BCR and Ag components of the BCR–Ag complex should follow the same track in the endocytic pathway and should therefore be exposed to the same proteolytic enzymes in endosomes. The idea is consistent with antigen receptor-directed antigen “handover” to class II MHC molecules with the addition that BCR fragments are also generated and transferred (43, 44). Cathepsins L and S have been described to be involved in proteolytic fragmentation of Ag (45) and may also be involved in processing of the BCR. Interesting in this respect, in silico analysis indicated that Cathepsin B, L, and S sites are enriched within and immediately upstream of CDR3 regions in intrathecal IgG from multiple sclerosis patients (21).This observation suggests that CDR3 peptides could be preferentially released by antigen processing of the BCR in endosomes.
However, other possibilities for the cellular origin of the pId component of the pId:MHCII complex cannot be excluded. It has previously been described that nascent Ig L chains experimentally retained in the ER/Golgi can be the source of pId displayed on MHCII of transfected B lymphoma cells (5). Since BCR ligation resulted in an up-regulated expression of MHCII, this mechanism could possibly increase the display of pId derived from ER/Golgi. Moreover, since BCR ligation enhanced levels of costimulatory molecules, pId:MHCII complexes on B cells could more potently elicit T cell responses.
Ligation-induced display of pId:MHCII on B cells could be of importance in immune regulation. Thus, whenever Ag ligates the BCR of a B cell, that B cell is anticipated to display both pAg:MHCII and pId:MHCII complexes and thus be regulated by both Ag-specific and Id-specific T cells. Such a mechanism would effectively merge conventional (42, 46) and Id-driven (7, 13) T–B collaboration.
The observation that DNP- and TNP-conjugated OVA were somewhat less potent at inducing B cell responses than was anti-Id IgG may be related to the slightly lower affinity of the haptens for the Id+ BCR. Further, filtration of TNP/DNP-OVA in the kidney, and a binding of TNP/DNP to BCRs having irrelevant (non-Id) λ chains, could also contribute. DNP-FICOLL was highly efficient, but this may relate to the TI-2 nature of FICOLL. It should be noted that DNP-FICOLL elicited much more potent responses in the presence of Id-specific T cells, even when employing LPS-stimulated B cells.
It has been hypothesized that Id-driven T–B collaboration could cause development of autoimmune diseases, since autoreactive B cells that have their BCR ligated by autoantigen could receive help from Id-specific T helper cells (13, 25). Such Id-driven T–B collaboration could easily become chronic, since self-antigens are not readily eliminated, in contrast to exogenous antigens. The persistent nature of Id-driven T–B collaboration could cause development of autoimmune diseases and, after acquisition of oncogenetic events, B cell lymphomas. Supporting the hypothesis, chronic Id-driven T–B collaboration has been shown to elicit SLE-like disease in mice (16, 25), with hallmarks of human SLE (15). Moreover, Id-driven T–B collaboration has been associated with multiple sclerosis in humans (19–21). Finally, chronic Id-driven T–B collaboration initiated development of B cell lymphomas in mice (17) and possibly CLL in humans (22). The influence of BCR ligation could not be addressed in these studies. Here, in support of the hypothesis, we demonstrate that BCR ligation is required for efficient Id-driven T–B cell collaboration. This suggests that (i) chronic ligation of the BCR by autoantigen and (ii) persistent help from Id-specific T cells may synergize in causing chronic proliferation of B cells and the development of disease. This scenario is consistent with the observations that not only autoimmune diseases (47) but also B cell cancers (48–51) are associated with self-reactive BCRs. It should be stressed that the hypothesis relates to the induction of B cell disorders. In later phases of the disease, the requirement for BCR ligation and for Id-specific T cell help could diminish as the pathologic B cells attain an increasingly autonomous state.
Autoimmune diseases and B cell malignancies are relatively rare disorders. Therefore, in most individuals, Id-driven T–B collaboration does not escalate beyond control but is down-regulated over time. Such down-modulation might occur through a number of suppressive mechanisms, such as peripheral exhaustion of Id-specific T cells (52), deletion of Id-specific thymocytes (12), and perhaps through the action of Id-specific T suppressor cells (53). We here demonstrate the induction of Foxp3+ Id-specific regulatory T cells in the wake of Id-driven T–B collaboration; such cells could contribute to down-regulation of the response. Presumably, only when one or more of these suppressive mechanisms fail may unchecked Id-driven T–B collaboration result in development of autoimmunity and B cell cancers.
Experimental Outline.
Extended materials and methods have been added at the end of the SI Appendix after SI Appendix, Table S1 (p. 37–41). SI Appendix describes the generation of VDJH315 mice (SI Appendix, Fig. S2, p. 5) and Vλ2315m mice (SI Appendix, Fig. S3, p.7). Pages 7 and 8 describe the Id315 and λ2315 TG × VDJH315 mice. Page 7 describes FACS characterization of splenic and bone marrow B cell lineages. SI Appendix, Fig. S6 (p. 13) shows generation of the anti-pId315:I-Ed scFv. SI Appendix, Fig. S10 (p. 17) shows preparation and validation of the anti-Id F(ab)2. SI Appendix, Figs.S13 and S14 show GL7 immunostaining (p. 21) and immunofluorescence (p. 23) procedures. SI Appendix, Fig. S15 (p. 26) shows germinal center B cell sorting and sequencing procedures. Cell enrichment, in vitro stimulation cultures, and in vivo transfer experiments are described in SI Appendix, Extended Materials and Methods (p. 37–38). ELISAs for Id+ IgM and IgG are described in SI Appendix, Extended Materials and Methods (p. 38–39). All antibodies for flow cytometry are described in SI Appendix, Extended Materials and Methods (p. 39). λ Amplicon sequencing in naïve Vλ2315 mice is described in SI Appendix, Extended Materials and Methods (p. 39). Analyses of in vitro B cell responses, including of Ca2+ flux measurements, and phosphotyrosine Western blotting are described in SI Appendix, Extended Materials and Methods (p. 40).
Data and Materials Availability.
The V-gene modified mice and the TCRm reagent may be obtained on a collaborative basis. Sequencing raw data from λ amplicon sequencing from the Vλ2315m+/− mouse are available at the Sequence Read Archive. Identifiers are BioSample SAMN10220898; sample name, VL2-315 B+/−; SRA, SRS3891429; BioProject, PRJNA495162.
Supplementary Material
Acknowledgments
Hilde Omholt, Peter Hofgaard, Keith M. Thompson, Marte Fauskanger, Kristina Randjelovic, Elisabeth Vikse, Nicolay Rustad Nilssen, and Olaf F. Schreurs are thanked for technical help; Vegard Nygaard and Eivind Hovig at the Oslo University Hospital Bioinformatics Core Facility for help with analyzing sequence data; Omri Snir for help with mRNA QC; and the staff at the Department of Comparative Medicine, Rikshospitalet, for assistance with mouse experiments. We are indebted to Drs. Robert Bremel and Jane Homan for critically reviewing the manuscript. Funding: The Norwegian Research Council (NFR, project 221709, to B.B.) and South-East Health Authority (Helse Sør-Øst, project 2017082, to B.B.) are acknowledged for funding.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Sequence Read Archive accession ID PRJNA495162.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1902836116/-/DCSupplemental.
References
- 1.Tonegawa S., Somatic generation of antibody diversity. Nature 302, 575–581 (1983). [DOI] [PubMed] [Google Scholar]
- 2.Sirisinha S., Eisen H. N., Autoimmune-like antibodies to the ligand-binding sites of myeloma proteins. Proc. Natl. Acad. Sci. U.S.A. 68, 3130–3135 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bogen B., Malissen B., Haas W., Idiotope-specific T cell clones that recognize syngeneic immunoglobulin fragments in the context of class II molecules. Eur. J. Immunol. 16, 1373–1378 (1986). [DOI] [PubMed] [Google Scholar]
- 4.Weiss S., Bogen B., B-lymphoma cells process and present their endogenous immunoglobulin to major histocompatibility complex-restricted T cells. Proc. Natl. Acad. Sci. U.S.A. 86, 282–286 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiss S., Bogen B., MHC class II-restricted presentation of intracellular antigen. Cell 64, 767–776 (1991). [DOI] [PubMed] [Google Scholar]
- 6.Rudensky A. Yu., Preston-Hurlburt P., al-Ramadi B. K., Rothbard J., Janeway C. A. Jr, Truncation variants of peptides isolated from MHC class II molecules suggest sequence motifs. Nature 359, 429–431 (1992). [DOI] [PubMed] [Google Scholar]
- 7.Bogen B., Weiss S., Processing and presentation of idiotypes to MHC-restricted T cells. Int. Rev. Immunol. 10, 337–355 (1993). [DOI] [PubMed] [Google Scholar]
- 8.Bogen B., Jørgensen T., Hannestad K., T helper cell recognition of idiotopes on lambda 2 light chains of M315 and T952: Evidence for dependence on somatic mutations in the third hypervariable region. Eur. J. Immunol. 15, 278–281 (1985). [DOI] [PubMed] [Google Scholar]
- 9.Bogen B., Lambris J. D., Minimum length of an idiotypic peptide and a model for its binding to a major histocompatibility complex class II molecule. EMBO J. 8, 1947–1952 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eyerman M. C., Wysocki L., T cell recognition of somatically-generated Ab diversity. J. Immunol. 152, 1569–1577 (1994). [PubMed] [Google Scholar]
- 11.Eyerman M. C., Zhang X., Wysocki L. J., T cell recognition and tolerance of antibody diversity. J. Immunol. 157, 1037–1046 (1996). [PubMed] [Google Scholar]
- 12.Bogen B., Dembic Z., Weiss S., Clonal deletion of specific thymocytes by an immunoglobulin idiotype. EMBO J. 12, 357–363 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Munthe L. A., Os A., Zangani M., Bogen B., MHC-restricted Ig V region-driven T-B lymphocyte collaboration: B cell receptor ligation facilitates switch to IgG production. J. Immunol. 172, 7476–7484 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Snyder C. M., et al. , Activation and tolerance in CD4(+) T cells reactive to an immunoglobulin variable region. J. Exp. Med. 200, 1–11 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aas-Hanssen K., Funderud A., Thompson K. M., Bogen B., Munthe L. A., Idiotype-specific Th cells support oligoclonal expansion of anti-dsDNA B cells in mice with lupus. J. Immunol. 193, 2691–2698 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Zangani M., et al. , Tracking early autoimmune disease by bioluminescent imaging of NF-kappaB activation reveals pathology in multiple organ systems. Am. J. Pathol. 174, 1358–1367 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zangani M. M., et al. , Lymphomas can develop from B cells chronically helped by idiotype-specific T cells. J. Exp. Med. 204, 1181–1191 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bremel R. D., Homan E. J., Frequency patterns of T-cell exposed amino acid motifs in immunoglobulin heavy chain peptides presented by MHCs. Front. Immunol. 5, 541 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hestvik A. L., et al. , T cells from multiple sclerosis patients recognize multiple epitopes on Self-IgG. Scand. J. Immunol. 66, 393–401 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Holmøy T., Vartdal F., Hestvik A. L., Munthe L., Bogen B., The idiotype connection: Linking infection and multiple sclerosis. Trends Immunol. 31, 56–62 (2010). [DOI] [PubMed] [Google Scholar]
- 21.Høglund R. A., et al. , In silico prediction analysis of idiotope-driven T-B cell collaboration in multiple sclerosis. Front. Immunol. 8, 1255 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Os A., et al. , Chronic lymphocytic leukemia cells are activated and proliferate in response to specific T helper cells. Cell Rep. 4, 566–577 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Khodadoust M. S., et al. , Antigen presentation profiling reveals recognition of lymphoma immunoglobulin neoantigens. Nature 543, 723–727 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khodadoust M. S., et al. , B cell lymphomas present immunoglobulin neoantigens. Blood 133, 878–881 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Munthe L. A., Corthay A., Os A., Zangani M., Bogen B., Systemic autoimmune disease caused by autoreactive B cells that receive chronic help from Ig V region-specific T cells. J. Immunol. 175, 2391–2400 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Kristoffersen G., Hannestad K., Hansen T., Two M315 idiotopes defined by isologous monoclonal antibodies: One depends on germline and the other on mutated murine lambda 2 light chain sequences. Scand. J. Immunol. 26, 535–546 (1987). [DOI] [PubMed] [Google Scholar]
- 27.Eisen H. N., Simms E. S., Potter M., Mouse myeloma proteins with antihapten antibody acitivity. The protein produced by plasma cell tumor MOPC-315. Biochemistry 7, 4126–4134 (1968). [DOI] [PubMed] [Google Scholar]
- 28.Bogen B., Gleditsch L., Weiss S., Dembic Z., Weak positive selection of transgenic T cell receptor-bearing thymocytes: Importance of major histocompatibility complex class II, T cell receptor and CD4 surface molecule densities. Eur. J. Immunol. 22, 703–709 (1992). [DOI] [PubMed] [Google Scholar]
- 29.Huszthy P. C., Gopalakrishnan R. P., Bogen B., Lambda amplicon sequencing in V-gene modified mice that express the MOPC315 Idiotope. NCBI Sequence Read Archive. https://www.ncbi.nlm.nih.gov/sra/?term=PRJNA495162. Deposited 9 October 2018.
- 30.Eisen H. N., Reilly E. B., Lambda chains and genes in inbred mice. Annu. Rev. Immunol. 3, 337–365 (1985). [DOI] [PubMed] [Google Scholar]
- 31.Bogen B., Weiss S., A rearranged lambda 2 light gene chain retards but does not exclude kappa and lambda 1 expression. Eur. J. Immunol. 21, 2391–2395 (1991). [DOI] [PubMed] [Google Scholar]
- 32.Jacobsen J. T., et al. , B-cell tolerance to the B-cell receptor variable regions. Eur. J. Immunol. 43, 2577–2587 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Azuma T., Sakato N., Fujio H., Characterization of the combining site of mouse myeloma protein M315. Biochemistry 27, 6116–6120 (1988). [DOI] [PubMed] [Google Scholar]
- 34.Dower S. K., Gettins P., Jackson R., Dwek R. A., Givol D., The binding of 2,4,6-trinitrophenyl derivatives to the mouse myeloma immunoglobulin A protein MOPC 315. Biochem. J. 169, 179–188 (1978). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moon J. J., et al. , Tracking epitope-specific T cells. Nat. Protoc. 4, 565–581 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ronchese F., Hausmann B., B lymphocytes in vivo fail to prime naive T cells but can stimulate antigen-experienced T lymphocytes. J. Exp. Med. 177, 679–690 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Munthe L. A., Kyte J. A., Bogen B., Resting small B cells present endogenous immunoglobulin variable-region determinants to idiotope-specific CD4(+) T cells in vivo. Eur. J. Immunol. 29, 4043–4052 (1999). [DOI] [PubMed] [Google Scholar]
- 38.Lawrence M. G., et al. , Permeation of macromolecules into the renal glomerular basement membrane and capture by the tubules. Proc. Natl. Acad. Sci. U.S.A. 114, 2958–2963 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jacobsen J., et al. , Naive idiotope-specific B and T cells collaborate efficiently in the absence of dendritic cells. J. Immunol. 192, 4174–4183 (2014). [DOI] [PubMed] [Google Scholar]
- 40.Chicz R. M., et al. , Specificity and promiscuity among naturally processed peptides bound to HLA-DR alleles. J. Exp. Med. 178, 27–47 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Snyder C. M., Zhang X., Wysocki L. J., Negligible class II MHC presentation of B cell receptor-derived peptides by high density resting B cells. J. Immunol. 168, 3865–3873 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Lanzavecchia A., Antigen-specific interaction between T and B cells. Nature 314, 537–539 (1985). [DOI] [PubMed] [Google Scholar]
- 43.Davidson H. W., Watts C., Epitope-directed processing of specific antigen by B lymphocytes. J. Cell Biol. 109, 85–92 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watts C., The endosome-lysosome pathway and information generation in the immune system. Biochim. Biophys. Acta 1824, 14–21 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hsieh C. S., deRoos P., Honey K., Beers C., Rudensky A. Y., A role for cathepsin L and cathepsin S in peptide generation for MHC class II presentation. J. Immunol. 168, 2618–2625 (2002). [DOI] [PubMed] [Google Scholar]
- 46.Mitchison N. A., The carrier effect in the secondary response to hapten-protein conjugates. II. Cellular cooperation. Eur. J. Immunol. 1, 18–27 (1971). [DOI] [PubMed] [Google Scholar]
- 47.Vinuesa C. G., Sanz I., Cook M. C., Dysregulation of germinal centres in autoimmune disease. Nat. Rev. Immunol. 9, 845–857 (2009). [DOI] [PubMed] [Google Scholar]
- 48.Young R. M., et al. , Survival of human lymphoma cells requires B-cell receptor engagement by self-antigens. Proc. Natl. Acad. Sci. U.S.A. 112, 13447–13454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hervé M., et al. , Unmutated and mutated chronic lymphocytic leukemias derive from self-reactive B cell precursors despite expressing different antibody reactivity. J. Clin. Invest. 115, 1636–1643 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu C. C., et al. , Many chronic lymphocytic leukemia antibodies recognize apoptotic cells with exposed nonmuscle myosin heavy chain IIA: Implications for patient outcome and cell of origin. Blood 115, 3907–3915 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Catera R., et al. , Chronic lymphocytic leukemia cells recognize conserved epitopes associated with apoptosis and oxidation. Mol. Med. 14, 665–674 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bogen B., Peripheral T cell tolerance as a tumor escape mechanism: Deletion of CD4+ T cells specific for a monoclonal immunoglobulin idiotype secreted by a plasmacytoma. Eur. J. Immunol. 26, 2671–2679 (1996). [DOI] [PubMed] [Google Scholar]
- 53.Heiser R. A., Snyder C. M., St Clair J., Wysocki L. J., Aborted germinal center reactions and B cell memory by follicular T cells specific for a B cell receptor V region peptide. J. Immunol. 187, 212–221 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.