Significance
Neural stem cells (NSCs) generate neurons and glia throughout life. Using global S-acylation profiling in NSCs, we identified BMPR1a, a main driver of embryonic and postnatal development to be S-acylated. We showed that BMPR1a is S-acylated in embryonic stem cells and NSCs, and identified 3 distinct S-acylated sites. We showed that S-acylation of BMPR1a is important for BMPR1a trafficking in vitro and characterized the importance of BMPR1a S-acylation for canonical and noncanonical BMP signaling. We generated a knock-in mouse for BMPR1a, deficient for S-acylation at site 180, and identified BMPR1a S-acylation as a regulatory mechanism controlling oligodendrogenesis in vitro and in vivo. Thus, we here provide insight on how the diverse output of BMPR1a-dependent signaling is regulated throughout development.
Keywords: neural stem cell, neurogenesis, BMP receptor, palmitoylation, oligodendrogenesis
Abstract
Neural stem cells (NSCs) generate neurons and glial cells throughout embryonic and postnatal brain development. The role of S-palmitoylation (also referred to as S-acylation), a reversible posttranslational lipid modification of proteins, in regulating the fate and activity of NSCs remains largely unknown. We used an unbiased screening approach to identify proteins that are S-acylated in mouse NSCs and showed that bone morphogenic protein receptor 1a (BMPR1a), a core mediator of BMP signaling, is palmitoylated. Genetic manipulation of S-acylated sites affects the localization and trafficking of BMPR1a and leads to altered BMP signaling. Strikingly, defective palmitoylation of BMPR1a modulates NSC function within the mouse brain, resulting in enhanced oligodendrogenesis. Thus, we identified a mechanism regulating the behavior of NSCs and provided the framework to characterize dynamic posttranslational lipid modifications of proteins in the context of NSC biology.
The activity and fate of neural stem cells (NSCs) in the developing and adult mammalian brain are tightly regulated by a plethora of cell intrinsic and extrinsic mechanisms ensuring that proper numbers and types of neuronal and glial cells are produced (1). By defining cellular and transcriptional programs that direct the fate of NSCs, substantial progress has been made to understand how NSCs are regulated within their endogenous niches (2–4). However, little is known of how posttranslational modifications (PTMs) of stem cell–expressed proteins affect NSC activity and subsequent fate choices. PTMs, such as phosphorylation, ubiquitination, sumoylation, acetylation, glycosylation, and lipidation among others, have been shown to affect the localization, activity, and stability of a large number of proteins, allowing for fast and partially reversible modification of protein function (5–10). Thus, PTMs represent critical regulators of protein function in a variety of cell types, allowing for fast signal diversification and refined cellular responses to exogenous cues (11). Many proteins are modified by the attachment of lipid moieties such as myristoylation, prenylation, and palmitoylation (also referred to as S-acylation) that modulate protein function (5). Among all known lipid modifications, S-acylation represents the only known reversible form of lipid modification that has been shown to play a pivotal role in protein trafficking, stability, and function (5, 12). 16-Carbon palmitate residues are transferred onto cysteines of target proteins via palmitoyl-acyltransferases (zDHHC1–24) and can be dynamically removed by acyl-protein thioesterases (for example, APT1-2) (13). Indeed, recent data suggest that reversible palmitoylation is involved in a large variety of biological processes such as growth of Arabidopsis thaliana, melanocortin signaling in the skin, synaptic function in the adult central nervous system, and many others (5, 14–16). Thus, over the last years large efforts have been undertaken to define the palmitoyl proteome in a variety of cell types and tissues (17).
Results
Identification of Palmitoylated Proteins in Mammalian NSCs.
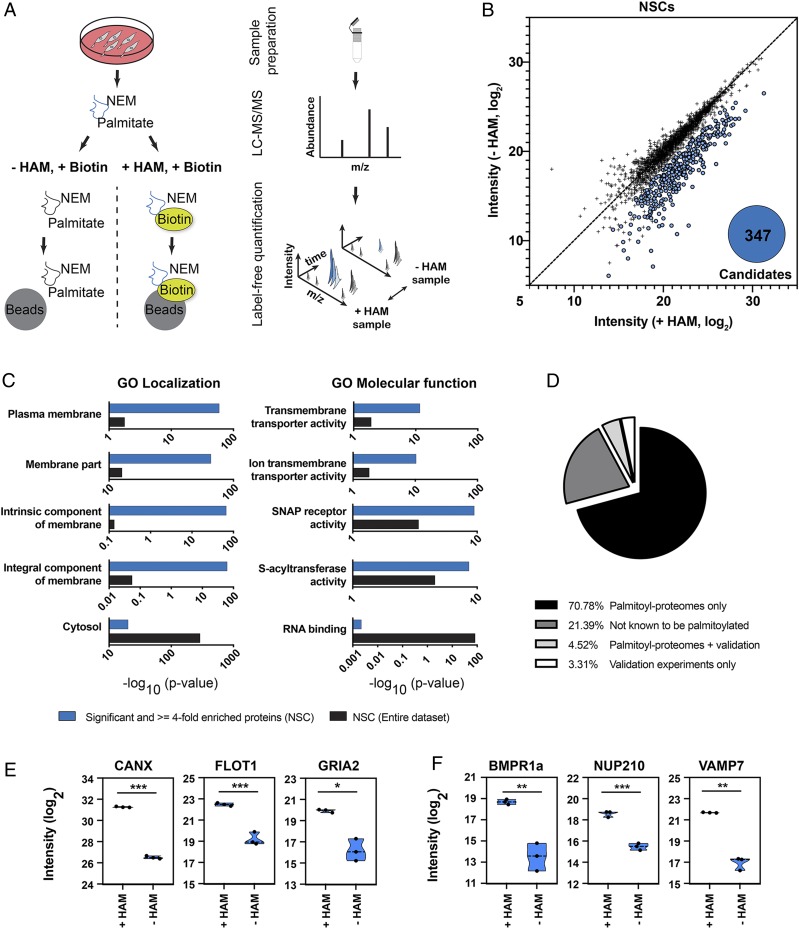
To identify a role for protein palmitoylation in mammalian stem cell function, we used an acyl-biotin exchange (ABE) assay to discover proteins that are S-acylated in NSCs isolated from the hippocampus of 8-wk-old mice (Fig. 1A). NSCs were lysed, and the protein samples were divided into 2 fractions. In the +hydroxylamine (+HAM) sample, the palmitate residue was cleaved off and exchanged with biotin. The −HAM condition served as a negative control. After the ABE reaction was completed, streptavidin beads were used to enrich for biotinylated proteins. Proteins enriched from +HAM and −HAM conditions were identified using mass spectrometry (MS) (Fig. 1B and SI Appendix, Fig. S1A). Proteins with a significant and at least 4-fold higher abundance in the +HAM sample were considered candidate proteins. With this approach, we identified 347 palmitoylated proteins in cultured NSCs (Fig. 1B), with an expected enrichment of identified proteins to be associated with membrane localization and function using Gene Ontology (GO) localization and molecular function analysis (Fig. 1C). Supporting the validity of the assay, we were able to identify previously validated S-acylated proteins (Fig. 1 D and E). Remarkably, 21% of the identified candidate proteins had not previously been described as palmitoylated (Fig. 1D). Given that palmitoyl-proteome screening approaches may yield false-positive results (18), we used protein-specific ABE assays and an alternative method (metabolic labeling of palmitate residues [19]) to verify S-acylation of selected candidate proteins such as NUP210 and VAMP7 (vesicle-associated membrane protein 7) (Fig. 1F and SI Appendix, Figs. S1 B–H and S2A), thereby confirming the validity of the unbiased screening results.
Fig. 1.
Identification of S-acylated proteins in neural stem cells. (A) Schematic describing the enrichment strategy and mass spectrometry-based workflow used for the identification of S-acylated proteins. Mass spectrometry and a label-free analysis were used for identification and quantification of proteins. Protein lysates were divided into 2 fractions. In the +HAM +Biotin condition, the palmitate was cleaved off and exchanged with a biotin. The −HAM +Biotin condition controls for unspecific binding. Streptavidin beads were then used to enrich for biotinylated proteins, followed by mass spectrometry-based identification. NEM, N-ethylmaleimide. (B) ABE enrichment coupled with mass spectrometry–based analysis identified 347 potentially S-acylated proteins (blue). Proteins that were less than 4-fold or not significantly enriched between the −HAM and +HAM conditions are shown in black. (C) Probing identified proteins for GO. “Localization” and “Molecular function” show that a substantial fraction of candidate proteins is associated with membrane compartments (blue, enriched candidates; black, enriched and nonenriched proteins). (D) Comparison of candidates with the SwissPalm database for S-acylated mouse proteins. Of the identified candidate proteins, 21.39% had not been described previously as S-acylated (dark gray). (E) Enrichment and background intensities (blue) of previously confirmed S-acylated proteins identified in the NSC dataset. CANX, calnexin; FLOT1, flotillin-1; GRIA2, AMPA receptor 2. (F) Enrichment and background intensities (blue) of proteins validated in this study. NUP210, nucleoporin 210. Error bars represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
BMPR1a Is Palmitoylated at Multiple Cysteines by zDHHC20.
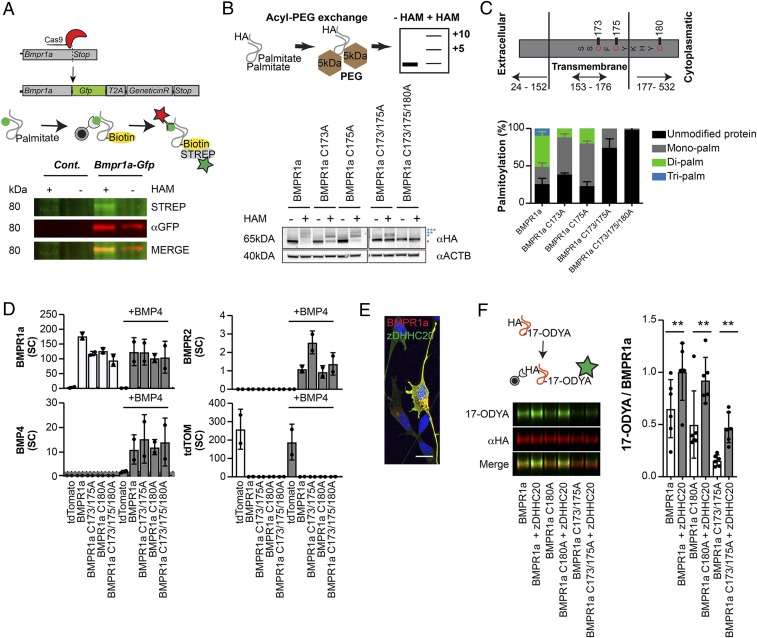
One of the candidate proteins we identified was the bone morphogenic protein receptor 1a (BMPR1a), which together with other components mediates canonical and noncanonical BMP signaling (Fig. 1F) (20, 21). BMP signaling plays a pivotal role in brain development and NSC behavior and has been associated with a large number of related biological processes, among them cell growth, cell differentiation, and cancer (22, 23). We confirmed palmitoylation of BMPR1a by tagging the endogenous Bmpr1a gene with GFP using CRISPR/Cas9 technology, allowing for efficient pull-down of BMPR1a in NSCs and subsequent ABE analysis (Fig. 2A and SI Appendix, Fig. S2B). Analogously, we confirmed palmitoylation of BMPR1a by tagging the endogenous BMPR1a with a hemagglutinin (HA) tag in mouse embryonic stem cells (ESCs) that were subsequently differentiated into NSCs and analyzed using metabolic labeling of palmitoylation sites within BMPR1a (SI Appendix, Fig. S2 C–F). Computational analyses suggested 5 cysteine residues to be palmitoylated within the BMPR1a protein sequence (24). However, using an acyl-PEG (methoxypolyethylene glycol) exchange assay, allowing for determination of the number of S-acylated cysteines, we found that BMPR1a has 3 functional palmitoylation sites (cysteines C173, C175, and C180), which are located close to the transmembrane domain of the receptor (Fig. 2 B and C). Directed mutagenesis showed that S-acylation was absent when cysteine residues were exchanged into alanines from the overexpressed BMPR1a protein (Fig. 2 B and C and SI Appendix, Fig. S2 G and H). Importantly, exchange of palmitoylated cysteines into alanines did not affect binding to the BMP4 ligand, as shown by immunoprecipitation of BMPR1a followed by mass spectrometry of bound proteins (Fig. 2D). We also detected single spectral counts of the interaction partner BMPR2; however, they were not enough to draw any quantitative conclusions. Immunoprecipitated tdTomato samples were used to control for unspecific binding. Coimmunoprecipitation of overexpressed HA-tagged BMPR1a or BMPR1a C173/175A together with FLAG-tagged BMPR1a or BMPR2 in NSCs showed that BMPR1a C173/175A was still able to dimerize with BMPR1a and BMPR2 (SI Appendix, Fig. S3A). We next aimed to identify which of the 24 known palmitoyl-acyltransferases (zDHHC1–24) (25) mediates palmitoylation of BMPR1a. We found zDHHC20 to colocalize with BMPR1a (Fig. 2E). Notably, we found overexpression of zDHHC20 to be sufficient to enhance S-acylation of BMPR1a (Fig. 2F), thus identifying a palmitoyl-acyltransferase that transfers palmitate residues onto BMPR1a.
Fig. 2.
BMPR1a is S-acylated at 3 distinct sites by palmitoyl-transferase zDHHC20. (A, Upper) CRISPR/Cas9-mediated tagging of endogenous Bmpr1a with Gfp, followed by an ABE assay where the palmitate was exchanged for a biotin (yellow). (A, Lower) ABE assay of wild-type and GFP-tagged BMPR1a knock-in NSCs revealed that BMPR1a (red) is S-acylated (green). (B, Upper) During the acyl-PEG exchange, palmitate groups were exchanged for PEG groups, leading to a defined increase in mass for every exchanged group. (B, Lower) Acyl-PEG exchange showed that BMPR1a is S-acylated at 3 distinct sites (blue stars). The red star indicates the unmodified BMPR1a fraction. (C) Schematic showing the localization of S-acylated sites within BMPR1a: 2 of the S-acylated sites (cysteine C173/C175) are based in the transmembrane domain. The site at C180 is located in the cytoplasmatic part of the receptor close to the transmembrane domain. Quantification of acyl-PEG analyses showed that BMPR1a is mainly present in its lipidated form. Unmodified protein (black), monopalmitoylated (gray), dipalmitoylated (green), tripalmitoylated (blue). (D) Coimmunoprecipation and mass spectrometry–based analysis of overexpressed BMPR1a and mutated BMPR1a interaction partners in NSCs showed that impaired S-acylation does not affect the ability of BMPR1a to bind to its ligand, BMP4 (gray). Single spectral counts of interacting BMPR2 were also detected, and tdTomato was used to control for unspecific enrichment. SC, spectral counts. (E) Overexpressed BMPR1a (red) and zDHHC20 (green) colocalized in cultured NSCs. Nuclei were counterstained with DAPI. (F) Overexpression of zDHHC20 (gray; graph) together with wild-type BMPR1a or BMPR1a harboring mutated sites of palmitoylated cysteines led to increased incorporation of 17-ODYA (green; blots) into BMPR1a, BMPR1a C180A, and BMPR1a C173/175A (red; blots), indicating that all 3 S-acylated sites are responsive to zDHHC20 levels. 17-ODYA, alkyne linked palmitate ortholog (17-octadecynoic acid). (Scale bar, 15 µm.) Error bars represent mean ± SD. **P < 0.01.
Palmitoylation of BMPR1a Alters Its Function.
We next probed the functional relevance of BMPR1a palmitoylation by testing if acylation-deficient BMPR1a proteins are sufficient to rescue the complete loss of the function proliferation phenotype of BMPR1a in NSCs (26). Given their positioning within BMPR1a, we analyzed C173/175A and C180A exchanges separately. As expected, we found that CRISPR/Cas9-mediated deletion of BMPR1a reduced proliferation of NSCs in response to BMP4 exposure, as measured using 5-ethynyl-2'-deoxyuridine (EdU) pulse labeling (SI Appendix, Fig. S3 B and C). Whereas wild-type and C180A overexpression normalized proliferation, the proliferation upon C173/175A overexpression was not rescued (SI Appendix, Fig. S3B). Consistent with this, we found that overexpression of C173/175A was not sufficient to transduce canonical BMP signaling, as measured by luciferase assays with BMP response elements (BREs) (SI Appendix, Fig. S3D) (27). Notably, overexpression of zDHHC20 was able to promote canonical BMP signaling in vitro (SI Appendix, Fig. S3E). This finding suggests that palmitoylation of cysteine residues C173/175 is required for canonical BMPR1a function, at least in vitro.
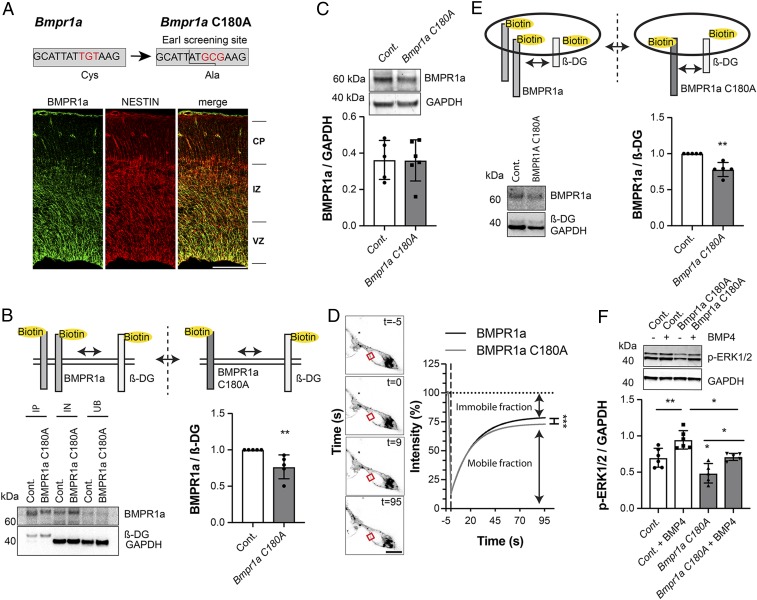
To analyze how palmitoylation of BMPR1a affects NSCs in vivo, we modified acylation sites within BMPR1a and aimed to generate mouse models harboring single (C180A) or double (C173/175A) mutants, again exchanging the palmitoylated cysteines with non-S-acylatable alanines. However, it turned out that C173/175A double mutants could not be obtained, most likely due to early embryonic lethality: out of 767 injected and transferred zygotes, we did not obtain a single viable correct double mutant mouse. This is consistent with the failure of C173/175A BMPR1a to rescue the full BMPR1a knockout in vitro (SI Appendix, Fig. S3B), as full knockouts of BMPR1a die at embryonic day E9.5 (28). However, we were able to generate C180A single mutants (Fig. 3A) and detected expression of BMPR1a using immunohistochemistry of BMPR1a in NSCs in the embryonic and adult brain (Fig. 3A and SI Appendix, Fig. S3F), as described before (26, 29). Thus, we next analyzed how palmitoylation of BMPR1a at position C180 affects its localization and function. Strikingly, we found that embryonic NSCs isolated from embryonic day 17.5 (E17.5) brains of C180A mutant mice showed reduced surface expression of the BMPR1a receptor (Fig. 3B) without affecting overall expression levels (Fig. 3C). In addition, fluorescence recovery after photobleaching (FRAP) analysis revealed an increase in the immobile fraction of overexpressed BMPR1a C180A compared to control BMPR1a (Fig. 3D). In FRAP experiments, the total recovery of fluorescence after photobleaching is influenced by mobile and immobile fractions of the BMP receptor. The mobile fraction undergoes exchange with the photobleached area, whereas the immobile fraction does not. Moreover, we found a robust reduction in endocytosis of BMPR1a in C180A mutant cells (Fig. 3E). Together, these findings indicate that palmitoylation of BMPR1a at position C180 is required for proper localization and mobility of the BMPR1a receptor (30).
Fig. 3.
Palmitoylation at C180 is important for BMPR1a function. (A) Schematic of genomic changes introduced into the Bmpr1a C180A transgenic knock-in mouse. The genetic sequence coding for cysteine 180 of BMPR1a was altered to encode for an alanine. BMPR1a is expressed (green) at E17.5 in the ventricular zone and colocalizes with the stem cell–associated intermediate filament NESTIN (red), as assessed by immunohistochemistry. VZ, ventricular zone; IZ, intermediate zone; CP, cortical plate. (B) BMPR1a cell surface expression is reduced in Bmpr1a C180A–mutated NSCs (gray), as measured by cell surface protein biotinylation. The cell surface protein β-dystroglycan (β-DG) was used for normalization and to compensate for different reaction efficiencies. GAPDH and BMPR1a in the input and unbound fraction revealed a comparable loading and efficient enrichment of BMPR1a. IP, immunoprecipitation; IN, input fraction; UB, unbound fraction. (C) BMPR1a expression is unchanged in NSCs extracted from control (white) and Bmpr1a C180A (gray) embryos (E17.5). (D) The mobile fraction of overexpressed BMPR1a C180A is decreased compared to BMPR1a. Shown are FRAP experiments using BMPR1a-mNeonGreen-3HA (control) and BMPR1a-C180A-mNeonGreen-3HA. Note the reduced mobile fraction in C180A BMPR1a. (E) The rate of BMPR1a endocytosis, as measured by reversible biotinylation, is reduced in NSCs extracted from Bmpr1a C180A (gray) embryos compared to controls (white). The cell surface protein β-DG undergoing regular endocytosis was used as a loading control to normalize between sample-dependent differences in reaction efficiencies. (F) Noncanonical BMP signaling is affected in the Bmpr1a C180A knock-in NSCs (gray) compared to control cells (white), indicated by reduced active ERK 1/2 in knock-in cells. (Scale bars: 100 µm.) Cont, control. Error bars represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
Palmitoylation of C180 Affects Noncanonical BMP Signaling.
To investigate the effects on BMP signaling in C180A mutant cells, we next analyzed signaling activity in proliferating and differentiating BMP4-stimulated cells isolated from C180A mutant mice and controls. We found that stimulation with BMP4 successfully promoted canonical BMP signaling in C180A-derived cells and control cells, as measured by levels of phosphorylated SMAD1/5 (SI Appendix, Fig. S3 G and H). Corroborating these results, we also found BMP4-dependent induction of inhibitor of differentiation 1-4 (Id1-4) gene transcripts at comparable levels in C180A compared to control cells as measured using digital droplet qPCR (SI Appendix, Fig. S3 I and J). This is consistent with the finding that overexpression of C180A does not prohibit canonical BMP signaling as measured using an BRE-dependent luciferase assay (SI Appendix, Fig. S3D). However, the C180A mutation caused a decrease in noncanonical, active extracellular signal-regulated kinase 1/2 (ERK1/2) in nonstimulated and BMP4-stimulated NSCs derived from C180A mice and controls (Fig. 3F) and an increase in active ERK1/2 during in vitro differentiation of C180A-derived cells (SI Appendix, Fig. S3K), which is known to promote oligodendrocyte differentiation (31). These findings indicate that palmitoylation-mediated localization and intracellular trafficking of BMPR1a at site C180 affects noncanonical, ERK-dependent BMP signaling (30).
C180 S-Acylation of BMPR1a Alters Fate of NSCs In Vivo.
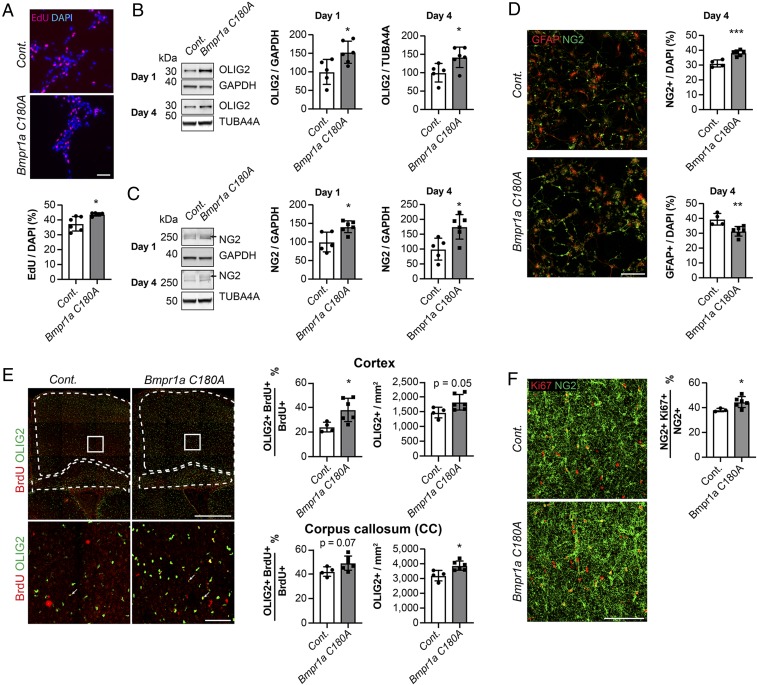
To probe potential effects of altered BMPR1a palmitoylation on NSC activity and fate, we isolated embryonic NSCs at E17.5 and analyzed their functional properties. We found that C180A mutation caused enhanced NSC proliferation in vitro, as measured by EdU pulse labeling (Fig. 4A). Furthermore we found increased levels of key markers of oligodendrocytic differentiation and maturation, OLIG2 (oligodendrocyte transcription factor) and NG2 (neural/glial antigen 2), after 1 and 4 d of in vitro differentiation by Western blotting (Fig. 4 B and C and SI Appendix, Fig. S4A). Analysis by immunocytochemistry against NG2 confirmed that more NG2+ cells were present after 4 d of in vitro differentiation at the expense of a reduced number of GFAP+ (glial fibrillary acidic protein) cells (Fig. 4D), indicating that acylation-deficient C180A cells show an altered NSC behavior with increased oligodendrogenesis. Thus, our findings give direct evidence that BMPR1a S-acylation is an essential modulator of NSC activity and fate choices in vitro.
Fig. 4.
Genetic Bmpr1a C180A exchange promotes oligodendrogenesis in vitro and in vivo. (A) NSCs extracted from Bmpr1a C180A mice (gray) show increased proliferation compared to controls (white), as measured using EdU pulse labeling (red). Nuclei were counterstained with DAPI (blue). (B and C) In vitro differentiation of knock-in NSCs (gray) and control cells (white) revealed enhanced expression of the oligodendrocyte lineage proteins OLIG2 (B) and NG2 (C) after 1 and 4 d. GAPDH was used as a loading control at day 1 and TUBA4A at day 4 due to incompatibility with other antibodies used at day 4. (D) In vitro differentiation of knock-in NSCs (gray) and control cells (white) revealed an elevated number of NG2+ cells (green) and a decreased number of GFAP+ cells (red) after 4 d, as analyzed by immunocytochemistry. (E) Bmpr1a C180A mice (gray) showed a higher density of OLIG2+ cells in the corpus callosum compared to control mice (white) and an increase in the BrdU+/OLIG2+ fraction of BrdU+ cells in the cortex at P7 (white; analyzed cortical and corpus callosum areas are marked). (E, Lower) High-power magnifications of the areas marked by white squares. (F) Bmpr1a C180A mice (gray) show a an increase in the KI67+/NG2+ fraction of NG2+ cells in the cortex at P7. (Scale bars, 50 µm [A and D, Lower], 100 µm [D and F], and 500 µm [D, Upper]). Cont., control. Error bars represent mean ± SD. *P < 0.05, **P < 0.01, ***P < 0.001.
We next analyzed the effects of the C180A BMPR1a mutation within the mouse brain. Given the in vitro phenotype of enhanced oligodendrogenesis, we analyzed the generation of late embryonic/early postnatal oligodendrocytic cells. Therefore, we injected E17.5 C180A and control mice with the thymidine analog BrdU and analyzed the number and fate of BrdU-labeled cells in postnatal brains at postnatal day 7 (P7). Corroborating the in vitro data, we detected an increased density of OLIG2+ cells in the corpus callosum and an increase in the BrdU+/OLIG2+ fraction of BrdU+ cells as well as an increase in the NG2+/KI67+ fraction of NG2+ cells in the cortex of P7 C180A mutant mice compared to controls (Fig. 4 E and F). Thus, the observed increased oligodendrogenesis appears to be based on increased differentiation toward the oligodendrocytic lineage and an increase in proliferation of oligodendrocyte precursor cells.
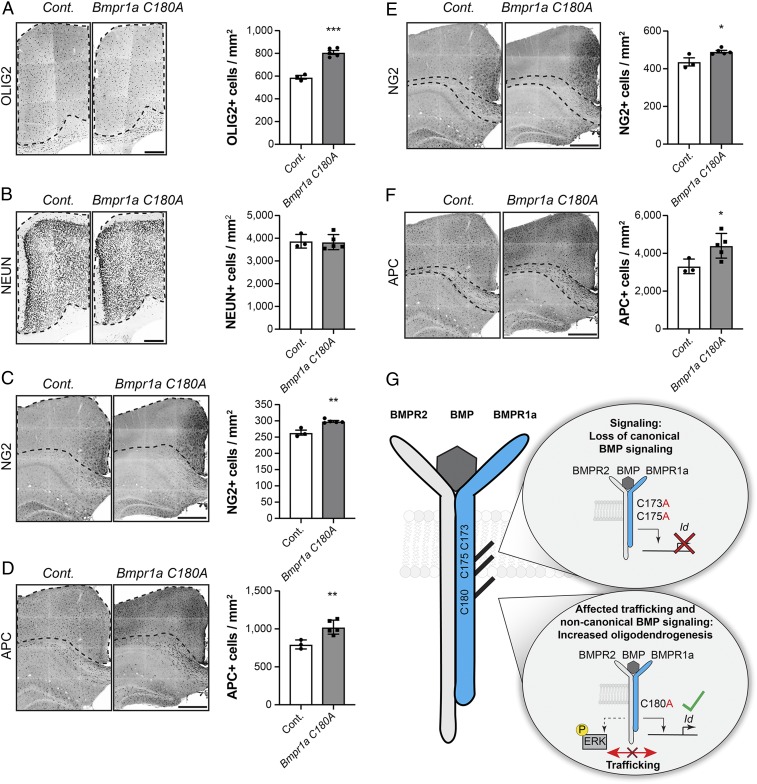
Strikingly, we also found an increased density of OLIG2+, NG2, and APC+ (adenomatous polyposis coli) cells in the cortex (Fig. 5 A, C, and D), corpus callosum (Fig. 5 E and F), and hippocampus of adult mice (SI Appendix, Fig. S4B), confirming and extending the phenotype we had observed in the P7 postnatal brain (Fig. 4E). Interestingly, we found no differences in the number of NeuN-expressing neurons in the cortex (Fig. 5B) or in NSC activity (SI Appendix, Fig. S4C) in the neurogenic niche of the adult hippocampus that were not permissive for oligodendrogenesis (32) in C180A mutant mice compared to controls, suggesting that the palmitoylation-dependent function of BMPR1a requires the physiological oligodendroglial fate potential of endogenous NSCs.
Fig. 5.
S-acylation of BMPR1a at C180 regulates the number of oligodendroglial cells in the adult brain. (A) Adult Bmpr1a C180A mice (gray) show a higher number of OLIG2+ cells per mm2 compared to control mice (white). Black dotted lines indicate the analyzed cortical area. (B) C180A mutation in Bmpr1a C180A mice (gray) does not affect the total number of neurons in the neocortex. Shown is the number of NeuN+ cells (black) per mm2 in mutant mice (gray) and controls (white). (C and D) The number of NG2+ (C, black) and APC+ (D, black) cells increase in the cortex of Bmpr1a C180A mice (gray) compared to control mice (white). (E and F) The number of NG2+ (E, black) and APC+ (F, black) cells in the corpus callosum increases in Bmpr1a C180A mice (gray) compared to controls (white; analyzed area in the corpus callosum is marked). (G) BMPR1a is S-acylated at 3 distinct sites proximal to the transmembrane domain. The substitution of cysteines 173 and 175 with non-S-acylatable alanines affects canonical BMP signaling (Upper Right), while mutating cysteine 180 affects BMPR1a trafficking and noncanonical BMP signaling, leading to increased oligodendrogenesis in vitro and in vivo (Lower Right). (Scale bars: 250 µm [A and B] and 500 µm [C–F]). Cont., control. Error bars represent mean ± SD. *P < 0.05, **P < 0.05, ***P < 0.05.
Discussion
We showed that a large number of proteins are palmitoylated in mammalian NSCs. Thus, we provided a palmitoylation-proteome resource that will be helpful in discovering mechanisms of NSC control in the embryonic and adult mammalian brain. We focused on BMPR1a, newly discovered to be palmitoylated in NSCs, and showed that BMPR1a is modified by zDHHC20–mediated S-acylation of 3 cysteine residues that affect BMP signaling in vitro and in vivo. Thus, we provided direct in vivo evidence of NSC-associated palmitoylation of a target protein that affects the fate of mammalian NSCs. Future studies will aim to probe if enhanced oligodendrogenesis in C180A mice may also cause enhanced remyelination in the context of demyelinating lesions, which may open novel avenues to enhance myelin repair in the adult brain (33).
The identification of palmitoylation of BMPR1a may have broad consequences for our understanding of health and disease given that BMP signaling has been implicated in a large number of biological processes ranging from stem cell–associated differentiation to cancer growth (23, 34). Whether BMPR1a S-acylation is affected in disease states will need to be determined. Notably, palmitoylation of BMPR1a is not specific to NSCs, as we also found acylation of the receptor in pluripotent ESCs (SI Appendix, Fig. S2 C–F). Unfortunately, exchange of cysteines C173/175 into alanines may be nonviable in mice, hindering in vivo analyses at this time. However, the findings that palmitoylation of C173/175 seems to be required for proper embryonic development further support the notion that palmitoylation of BMPR1a is of substantial functional relevance and that C173/175 S-acylation is required for canonical BMP signaling, whereas S-acylation of C180 affects noncanonical BMP signal transduction. Future studies will need to use either conditional or in vitro approaches to exchange S-acylated cysteines on both alleles, which may allow more in-depth functional analyses of the C173/175 sites. As we showed, in vivo manipulation of palmitoylation of BMPR1a seems to selectively affect BMP signaling, resulting in enhanced generation of oligodendrocytes, and our in vitro data suggest that this is based on altered noncanonical BMP signaling.
BMP receptors are present at the cell surface as hetero- or homomeric complexes (35), and while BMPR1a mainly associates with detergent-resistant membrane (DRM) fractions, BMPR2 was reported to localize to DRM and non-DRM compartments (36). However, it remains poorly understood how shifts between canonical and noncanonical BMP signaling are regulated (37–39). A prerequisite for SMAD-independent signaling is the presence of BMPR1 in cholesterol-rich microdomains regulating lateral mobility of type I receptors, which has been described to be crucial for regulating noncanonical BMP signaling without affecting canonical signaling function (36, 40). Thus, future studies are necessary to address the impact of BMPR1a S-acylation on its segregation to specific membrane fractions, its mobility, and the formation of receptor complexes.
We used a screening approach to identify palmitoylated proteins in mammalian NSCs. We showed that BMPR1a is S-acylated and that palmitoylation affects the functionality of the BMP signaling pathway that is critically involved in a variety of cell biological processes (Fig. 5G). Manipulating palmitoylation of BMP signaling components may represent an approach to targeting BMP function in health and disease.
Materials and Methods
Stem Cell Cultures.
Primary adult mouse NSCs were kept in the presence of EGF (20 ng/mL), FGF-2 (20 ng/mL), and heparin (5 mg/mL) in a monolayer culture at 37 °C and 5% CO2. DMEM/F12 GlutaMax supplemented with antibiotics (Anti-Anti) and N2 was used as a standard medium. For some culture conditions, an overnight BMP4 treatment was performed as described earlier (41). For the generation of E17.5 cortical NSC cultures, pregnant Bmpr1a C180A and control dams were killed at E17.5. E17.5 NSCs were kept in DMEM/F12 GlutaMax, N2/B27 supplement, EGF (20 ng/mL), and FGF-2 (20 ng/mL) at 37 °C and 5% CO2. Experiments performed with control or BMPR1a C180A–derived E17.5 NSCs were performed with at least 4 biological replicates, derived from different E17.5 embryos. ESCs (ESC line E14, isolated from mouse strain 129P2/OlaHsd) were cultured in DMEM/F12/GlutaMAX medium supplemented with N2, B27, antibiotics, and LIF (1,000 U/mL), 1 μM PD0325901, and 3 μM CHIR99021 at 37 °C and 5% CO2. Cells were kept in flasks coated with 0.2% (vol/vol) gelatin. Differentiation of ESCs was performed as described earlier (42).
Acyl-Biotin Exchange Assay and Mass Spectrometric Analysis.
The protocol was performed as previously described (18). Please refer to the SI Appendix for a more detailed description. Samples were adjusted to the same volume and concentration (5 mL and 2.2 mg/mL) by the addition of LB. Sample preparation, liquid chromatography (LC) –MS/MS analysis, and data analysis were performed as described previously (41) with some minor changes: raw files were processed with Progenesis QI for proteomics (Nonlinear Dynamics) and Gapdh, Actin, and Tubulin were used for normalization between samples. These proteins bind due to their high abundance to the beads in a palmitoylation-independent manner and can therefore be used to normalize between samples and prevent detection of false-positive proteins. Mascot was used for searching a target-decoy mouse database downloaded from Uniprot (May 31, 2018). Parameters were set as follows: peptide tolerance ±10 ppm, MS/MS tolerance ±0.7 DA, variable modifications: Acetyl (Protein N-term), Carbamidomethyl (C), Gln → pyro Glu (N-term), N-Ethylmaleimide, Oxidation (M). Resulting Mascot data files were imported into Scaffold 4 (Proteome Software). The threshold was set for peptides at 95% and for proteins at 99%. Decoy hits were excluded, and protein quantification and statistics were exported as CSV files. To visualize the data, raw data with the value 0 were replaced with the value 10 and maximal fold chance was limited to 250, followed by a log2 transformation of all values. Log2 intensities between 5 and 35 were included in the graph. Gene names, P values, and normalized protein abundance were exported to Metacore 6.37 build 69500 (Clarivate Analytics), and an enrichment analysis was performed. GO localization and GO molecular function results were then exported as an XLS file. Data were visualized using GraphPad Prism 8. Identified candidate proteins (that were significantly and at least 4-fold enriched over −HAM) were compared to the SwissPalm database (April 24, 2019). Experiments were performed in triplicates. For mass spectrometric analysis of coimmunoprecipitations, the same setup and protocol were used as described earlier, with some modifications: When performing the mascot search, Carbamidomethyl (C) was set as fixed and acetyl (Protein N-term), Gln → pyro Glu (N-term), and Oxidation (M) as variable modifications. Block randomization was performed, and samples were measured in duplicates. Search results were then analyzed by spectral counting using Scaffold 4 (Proteome Software). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014355.
Image Analysis.
Image analysis was performed using Fiji imaging analysis software (43), ZEN 2 (Carl Zeiss), or Imaris (Bitplane). All quantifications were performed in a blind manner. The detailed procedures are provided in the SI Appendix.
Statistical Analysis.
Statistical analysis was performed using Prism 8.1 (GraphPad). For the comparison of 2 samples, we used unpaired t tests. If the variance of the 2 compared samples was significantly different, unpaired t tests with Welch’s correction were used instead. Experiments where the controls were set to the value 1 were analyzed by nonparametric testing using a Kolmogorov-Smirnov test. For comparisons involving more than 2 samples, we used 1-way ANOVA analysis followed by Sidak’s multiple comparison test.
Other materials and methods are provided in the SI Appendix, Materials and Methods. All data are presented in the main file and the SI Appendix, Figs. S1–S4 and Table S1. The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD014355.
Supplementary Material
Acknowledgments
We thank Peter Gehrig, Bernd Roschitzki, Jonas Grossmann, Claudia Fortes, and Paolo Nanni from the Functional Genomics Center Zurich and Heide Oller from the Basel Center for Transgenic Models for experimental help. We thank Annina Denoth-Lippuner and Ana Stankovic for comments on the manuscript. This work was supported by the European Research Council (STEMBAR), the Swiss National Science Foundation (BSCGI0 157859), and the Zurich Neuroscience Center. T.W. was supported by a fellowship of the Boehringer Ingelheim Fonds and a Candoc Forschungskredit of University of Zurich.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The mass spectrometry proteomics data have been deposited in the publicly accessible database ProteomeXchange Consortium via the Proteomics Identifications (PRIDE) partner repository with the dataset identifier PXD014355.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912671116/-/DCSupplemental.
References
- 1.Urbán N., Guillemot F., Neurogenesis in the embryonic and adult brain: Same regulators, different roles. Front. Cell. Neurosci. 8, 396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bjornsson C. S., Apostolopoulou M., Tian Y., Temple S., It takes a village: Constructing the neurogenic niche. Dev. Cell 32, 435–446 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andreotti J. P., et al. , Neural stem cell niche heterogeneity. Semin. Cell Dev. Biol., S1084-9521(18)30057-0 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shin J., et al. , Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen B., Sun Y., Niu J., Jarugumilli G. K., Wu X., Protein lipidation in cell signaling and diseases: Function, regulation, and therapeutic opportunities. Cell Chem. Biol. 25, 817–831 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendriks I. A., Vertegaal A. C., A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Moremen K. W., Tiemeyer M., Nairn A. V., Vertebrate protein glycosylation: Diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Narita T., Weinert B. T., Choudhary C., Functions and mechanisms of non-histone protein acetylation. Nat. Rev. Mol. Cell Biol. 20, 156–174 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Rape M., Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Ubersax J. A., Ferrell J. E. Jr, Mechanisms of specificity in protein phosphorylation. Nat. Rev. Mol. Cell Biol. 8, 530–541 (2007). [DOI] [PubMed] [Google Scholar]
- 11.Linder M. E., Deschenes R. J., Palmitoylation: Policing protein stability and traffic. Nat. Rev. Mol. Cell Biol. 8, 74–84 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Chamberlain L. H., Shipston M. J., The physiology of protein S-acylation. Physiol. Rev. 95, 341–376 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Won S. J., Cheung See Kit M., Martin B. R., Protein depalmitoylases. Crit. Rev. Biochem. Mol. Biol. 53, 83–98 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kang R., et al. , Neural palmitoyl-proteomics reveals dynamic synaptic palmitoylation. Nature 456, 904–909 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar M., et al. , S-acylation of the cellulose synthase complex is essential for its plasma membrane localization. Science 353, 166–169 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Chen S., et al. , Palmitoylation-dependent activation of MC1R prevents melanomagenesis. Nature 549, 399–403 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blanc M., et al. , SwissPalm: Protein palmitoylation database. F1000 Res. 4, 261 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wan J., Roth A. F., Bailey A. O., Davis N. G., Palmitoylated proteins: Purification and identification. Nat. Protoc. 2, 1573–1584 (2007). [DOI] [PubMed] [Google Scholar]
- 19.Martin B. R., Nonradioactive analysis of dynamic protein palmitoylation. Curr. Protoc. Protein Sci. 73, Unit 14.15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazono K., Maeda S., Imamura T., BMP receptor signaling: Transcriptional targets, regulation of signals, and signaling cross-talk. Cytokine Growth Factor Rev. 16, 251–263 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Sieber C., Kopf J., Hiepen C., Knaus P., Recent advances in BMP receptor signaling. Cytokine Growth Factor Rev. 20, 343–355 (2009). [DOI] [PubMed] [Google Scholar]
- 22.Wang R. N., et al. , Bone morphogenetic protein (BMP) signaling in development and human diseases. Genes Dis. 1, 87–105 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bond A. M., Bhalala O. G., Kessler J. A., The dynamic role of bone morphogenetic proteins in neural stem cell fate and maturation. Dev. Neurobiol. 72, 1068–1084 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie Y., et al. , GPS-lipid: A robust tool for the prediction of multiple lipid modification sites. Sci. Rep. 6, 28249 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Globa A. K., Bamji S. X., Protein palmitoylation in the development and plasticity of neuronal connections. Curr. Opin. Neurobiol. 45, 210–220 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Panchision D. M., et al. , Sequential actions of BMP receptors control neural precursor cell production and fate. Genes Dev. 15, 2094–2110 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Korchynskyi O., ten Dijke P., Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the Id1 promoter. J. Biol. Chem. 277, 4883–4891 (2002). [DOI] [PubMed] [Google Scholar]
- 28.Mishina Y., Suzuki A., Ueno N., Behringer R. R., Bmpr encodes a type I bone morphogenetic protein receptor that is essential for gastrulation during mouse embryogenesis. Genes Dev. 9, 3027–3037 (1995). [DOI] [PubMed] [Google Scholar]
- 29.Mira H., et al. , Signaling through BMPR-IA regulates quiescence and long-term activity of neural stem cells in the adult hippocampus. Cell Stem Cell 7, 78–89 (2010). [DOI] [PubMed] [Google Scholar]
- 30.Ehrlich M., Endocytosis and trafficking of BMP receptors: Regulatory mechanisms for fine-tuning the signaling response in different cellular contexts. Cytokine Growth Factor Rev. 27, 35–42 (2016). [DOI] [PubMed] [Google Scholar]
- 31.Galabova-Kovacs G., et al. , Essential role of B-Raf in oligodendrocyte maturation and myelination during postnatal central nervous system development. J. Cell Biol. 180, 947–955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jessberger S., Toni N., Clemenson G. D. Jr, Ray J., Gage F. H., Directed differentiation of hippocampal stem/progenitor cells in the adult brain. Nat. Neurosci. 11, 888–893 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plemel J. R., Liu W. Q., Yong V. W., Remyelination therapies: A new direction and challenge in multiple sclerosis. Nat. Rev. Drug Discov. 16, 617–634 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Bach D. H., Park H. J., Lee S. K., The dual role of bone morphogenetic proteins in cancer. Mol. Ther. Oncolytics 8, 1–13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gilboa L., et al. , Bone morphogenetic protein receptor complexes on the surface of live cells: A new oligomerization mode for serine/threonine kinase receptors. Mol. Biol. Cell 11, 1023–1035 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hartung A., et al. , Different routes of bone morphogenic protein (BMP) receptor endocytosis influence BMP signaling. Mol. Cell. Biol. 26, 7791–7805 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ali I. H., Brazil D. P., Bone morphogenetic proteins and their antagonists: Current and emerging clinical uses. Br. J. Pharmacol. 171, 3620–3632 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li W., et al. , Membrane targeting of inhibitory Smads through palmitoylation controls TGF-β/BMP signaling. Proc. Natl. Acad. Sci. U.S.A. 114, 13206–13211 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nohe A., et al. , The mode of bone morphogenetic protein (BMP) receptor oligomerization determines different BMP-2 signaling pathways. J. Biol. Chem. 277, 5330–5338 (2002). [DOI] [PubMed] [Google Scholar]
- 40.Guzman A., et al. , SMAD versus non-SMAD signaling is determined by lateral mobility of bone morphogenetic protein (BMP) receptors. J. Biol. Chem. 287, 39492–39504 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Knobloch M., et al. , A fatty acid oxidation-dependent metabolic shift regulates adult neural stem cell activity. Cell Rep. 20, 2144–2155 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Conti L., et al. , Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 3, e283 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindelin J., et al. , Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.