Significance
Protein structures fluctuate owing to thermal motion and in response to functional changes such as ligand binding. As a consequence, it is challenging to determine which protein motions are functionally most important at equilibrium. Enzymes that are transiently covalently modified during catalysis offer a way to identify functional motions, as the modification can trigger catalytically important conformational changes. The covalent modification of the active-site cysteine in isocyanide hydratase weakens a critical hydrogen bond required for reactivity. Hydrogen bond disruption triggers a cascade of conformational changes whose modulation by mutation is detrimental to enzyme turnover. Most enzymes that form catalytic intermediates will experience similar transient changes in active-site electrostatics, suggesting that modification-gated conformational dynamics is common in enzymes.
Keywords: X-ray crystallography, cysteine modification, enzyme conformational dynamics, XFEL, radiation damage
Abstract
How changes in enzyme structure and dynamics facilitate passage along the reaction coordinate is a fundamental unanswered question. Here, we use time-resolved mix-and-inject serial crystallography (MISC) at an X-ray free electron laser (XFEL), ambient-temperature X-ray crystallography, computer simulations, and enzyme kinetics to characterize how covalent catalysis modulates isocyanide hydratase (ICH) conformational dynamics throughout its catalytic cycle. We visualize this previously hypothetical reaction mechanism, directly observing formation of a thioimidate covalent intermediate in ICH microcrystals during catalysis. ICH exhibits a concerted helical displacement upon active-site cysteine modification that is gated by changes in hydrogen bond strength between the cysteine thiolate and the backbone amide of the highly strained Ile152 residue. These catalysis-activated motions permit water entry into the ICH active site for intermediate hydrolysis. Mutations at a Gly residue (Gly150) that modulate helical mobility reduce ICH catalytic turnover and alter its pre-steady-state kinetic behavior, establishing that helical mobility is important for ICH catalytic efficiency. These results demonstrate that MISC can capture otherwise elusive aspects of enzyme mechanism and dynamics in microcrystalline samples, resolving long-standing questions about the connection between nonequilibrium protein motions and enzyme catalysis.
Protein dynamics plays an important role in enzyme catalysis (1–4). Many enzymes form covalent catalytic intermediates that can alter enzyme structure and conformational dynamics (5–8). Because catalytic intermediates transiently modify enzymes, they may activate a new protein motion regime that facilitates later steps of catalysis. Determining how these transient changes in enzyme structure and dynamics facilitate passage along the reaction coordinate is a topic of intense interest in structural enzymology (4, 8). Of all amino acids in proteins, reactive cysteine residues are subject to the most diverse set of covalent modifications (9, 10). Moreover, the targeted modification of cysteine residues proximal to active sites has become a frontline strategy for the development of covalent enzyme inactivators as tools for chemical biology and drugs (11). Therefore, understanding how the H-bonding environment of active-site cysteine residues modulates their reactivity and how the covalent modification of cysteine thiolate anions controls protein conformational dynamics can provide general insights into the functions of many proteins whose activities require or are regulated by covalent modifications.
X-ray crystallography provides an atomically detailed ensemble- and time-averaged view of protein conformational dynamics in the lattice environment (12–15). Technological advances in X-ray sources, detectors, and sample delivery have enabled a new class of crystallography experiments that report on nonequilibrium protein motions in response to external perturbations (16). Because these perturbations can be selected for functional relevance (e.g., by the infusion of substrate or induction of a particular modification), the resulting nonequilibrium changes often correspond to functionally important protein motions. In parallel, advances in computer modeling of multiple conformational substates now enable visualizing minor populations of crystalline protein structural ensembles. These approaches have permitted identification of networks of side-chain disorder (17), allowed refinement of whole-protein ensemble models (18), aided identification of minor binding modes of therapeutic ligands (19), and characterized spatial correlations in protein mobility (20).
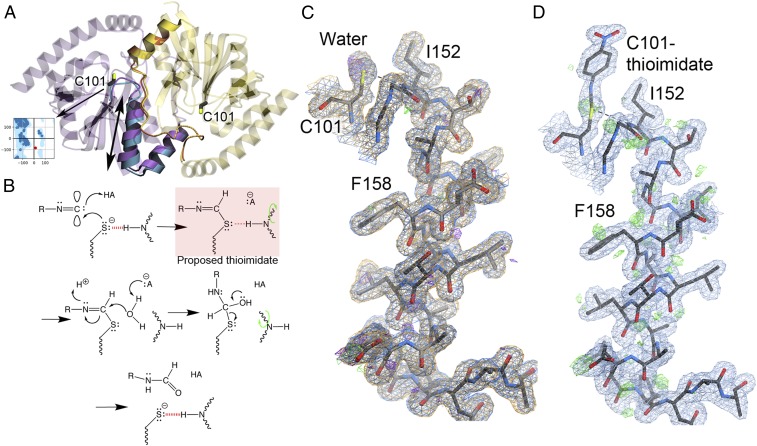
The enzyme studied here is isocyanide hydratase (ICH; EC 4.2.1.103), a homodimeric cysteine-dependent enzyme in the DJ-1 superfamily that irreversibly hydrates many isocyanides to yield N-formamides (21) (Fig. 1A). Prior crystallographic studies of ICH showed evidence of helical mobility near the active-site Cys101 residue (Fig. 1A), which coincided with photooxidation of C101 to Cys101-SOH (22). Cys101 accepts an S−-HN hydrogen bond from the severely strained (φ = 14°, ψ = −83°) backbone amide of Ile152. When this hydrogen bond is disrupted in an engineered C101A mutant (Fig. 1A), Ile152 relaxes to an unstrained backbone conformation concomitant with displacement of the helix (22, 23). The relevance of these motions for ICH function was not established in these prior studies.
Fig. 1.
Catalytic intermediate captured with MISC. (A) The ICH dimer is shown as an overlay of WT ICH (purple) and C101A ICH (blue). The “B” protomer is shown in yellow. The mobile helix H in C101A and areas exhibiting correlated backbone–side-chain disorder are rendered opaque. Ile152 is a Ramachandran outlier (Inset) whose backbone torsion angles move with helical displacement. (B) Postulated reaction mechanism for ICH, beginning with Cys101 thiolate attack at the electrophilic carbenic carbon atom of isocyanide substrates and proceeding in the direction of the arrows. A previously postulated thioimidate intermediate (red box) eliminates the charge on Cys101 and weakens the H-bond to Ile152 (dashed red line). This relieves backbone torsional strain at Ile152 and permits sampling of shifted helix H conformations (green curved arrow), allowing water access to the intermediate for hydrolysis. (C) ICH completes a full catalytic cycle in the crystal during MISC. Helix H is shown with 2mFo-DFc electron density (0.8 rmsd) prior to the introduction of substrate (blue) and after substrate has been exhausted (orange). These maps overlap almost perfectly, indicating that ICH is fully restored to its resting conformation after catalysis. The hydrogen bond between the peptide backbone of Ile152 and Cys101 is shown in a dotted line. The helix is not mobile in these resting structures, indicated by the absence of features in the mFo-DFc difference electron density (2.5 rmsd prior to substrate [green] and after catalysis is complete [purple]) (D) MISC confirms that ICH forms a covalent Cys101-thioimidate intermediate 15 s after substrate mixing. Difference mFo-DFc electron density contoured at 2.5 rmsd (green) supports sampling of shifted helix H conformations upon intermediate formation.
Here, we use X-ray crystallography, molecular dynamics simulation, rigidity theory, mutagenesis, and enzyme kinetics to characterize a set of correlated conformational changes that remodel the ICH active site and propagate across the dimer interface in response to cysteine modification. Using time-resolved mix-and-inject serial crystallography (MISC) at an X-ray free electron laser (XFEL), we directly observed formation of a covalent intermediate and transient, functionally important motions during ICH catalysis. Pre-steady-state kinetics of wild-type and mutant ICH suggests that cysteine-gated conformational changes are important for water to access and hydrolyze the catalytic intermediate. The prominence of conformational dynamics in ICH and the tractability of this protein using multiple biophysical techniques make ICH a valuable model system for understanding how cysteine modification can modulate functional protein dynamics.
Results
Structural Characterization of a Thioimidate Intermediate and Helix Motion during ICH Catalysis.
We examined the role of active-site structural dynamics in ICH turnover by directly monitoring catalysis in ICH microcrystals using MISC at the Linac Coherent Light Source (LCLS) XFEL at 298 K. We infused the substrate p-nitrophenyl isocyanide (p-NPIC; SI Appendix) into a stream of unmodified ICH microcrystals (Materials and Methods) and collected diffraction data at carefully selected time delays (SI Appendix, Fig. S1). The concentration of p-NPIC in the stream allowed only a single turnover of crystalline ICH before substrate depletion. The proposed reaction mechanism for ICH postulates that the catalytic Cys101 nucleophile attacks organic isocyanides at the electron-deficient carbene-like center, followed by proton abstraction from a nearby general acid to generate a covalent thioimidate intermediate (Fig. 1B). An enzyme-linked thioimidate, which has not been previously observed, eliminates the negative charge of the Cys101 thiolate (S−) and is proposed to reduce the strength of the Cys101-Ile152 hydrogen bond. Because the “diffraction before destruction” principle ensures that serial XFEL diffraction data are minimally affected by X-ray radiation damage (24), the enzyme suffered no radiation-induced oxidation of Cys101 (Fig. 1C). After 15 s of mixing, 2mFo-DFc (Fig. 1D) and omit mFo-DFc electron density maps (SI Appendix, Fig. S2A) unambiguously revealed the formation of a thioimidate covalent intermediate in both ICH protomers. Once the intermediate is formed, positive mFo-DFc difference electron density appears around helix H (Fig. 1D) and its B-factors increase (SI Appendix, Fig. S2 B–D), indicating that it samples a new conformational ensemble. This difference density and the elevated B-factors are absent both prior to catalysis and after ICH has exhausted substrate 5 min after mixing (Fig. 1C and SI Appendix, Fig. S2 B–D). Therefore, helix H becomes transiently mobile upon intermediate formation during ICH catalysis, dynamically changing the active-site microenvironment.
Cys101 Modification Triggers Conformational Changes that Remodel the ICH Active Site.
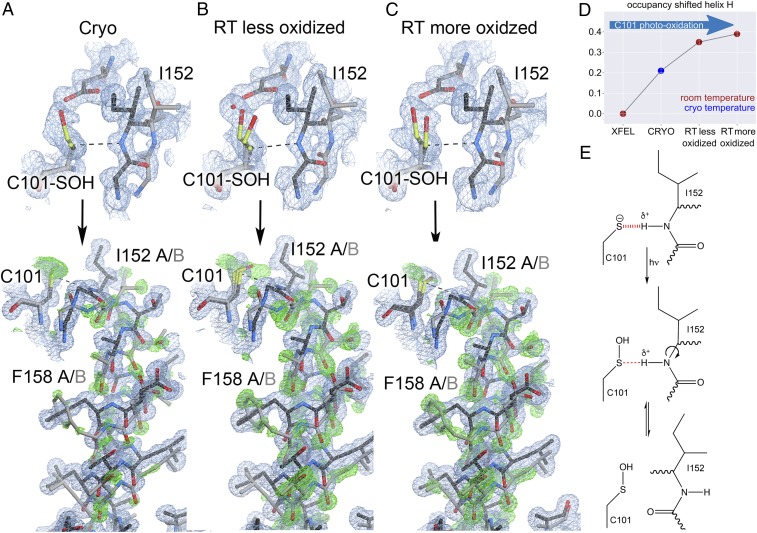
We reasoned that the on-pathway thioimidate intermediate weakens the Cys101-Ile152 hydrogen bond that holds helix H in a strained conformation similarly to the X-ray–induced Cys101-SOH modification in the resting enzyme (22). Consistent with this idea, the cryogenic dataset collected at 100 K (Cryo) showed evidence of radiation-driven Cys101-SOH formation and a corresponding increase in helical mobility (Fig. 2A) (22). To enrich populations of the shifted conformation, we therefore collected a series of X-ray diffraction datasets of wild-type (WT) ICH at increasing absorbed doses of X-ray radiation (Fig. 2 A–C). Two ambient-temperature (274–277 K) synchrotron radiation datasets at 1.20–1.15 Å revealed radiation-dose–dependent Cys101 oxidation and helical displacement with increasing occupancy (Fig. 2D). Refined occupancies of alternate conformations of helix H confirmed a population shift toward the displaced helix position along this series of datasets that correlates with the extent of absorbed X-ray dose and concomitant Cys101 oxidation. The shifted helix conformations across datasets are highly similar, suggesting that the observed electron density features resulted from coupling between this helix and specific, radiation-induced photochemistry at Cys101.
Fig. 2.
X-ray–induced cysteine oxidation drives helical motion in ICH. (A–C, Upper) The environment of Cys101 with varying degrees of oxidation to Cys101-sulfenic acid. 2mFo-DFc electron density is contoured at 0.7 rmsd (blue), and the hydrogen bond between the peptide backbone of Ile152 and Cys101 is shown in a dotted line. “Cryo” is synchrotron data collected at 100 K (PDB 3NON); “RT less oxidized” is synchrotron data collected at 274 K with an absorbed dose of 2.4 × 104 Gy; and “RT more oxidized” is synchrotron data collected at 277 K with an absorbed dose of 3.7 × 105 Gy. (Lower) helix H in its strained (black) and relaxed, shifted conformations (gray). 2mFo-DFc electron density is contoured at 0.8 rmsd (blue) and omit mFo-DFc electron density for the shifted helical conformation is contoured at 3.0 rmsd (green). At 274–277 K, increased Cys101 oxidation disrupts the hydrogen bond to Ile152 and results in stronger difference electron density for the shifted helix conformation. (D) The refined occupancy of helix H in each X-ray dataset indicates that increases in temperature and Cys101 oxidation result in higher occupancy for the shifted (relaxed) helix conformation. (E) Mechanism of X-ray–induced covalent modification of C101 and weakening of the S−-HN hydrogen bond.
Notably, these stronger density peaks correspond to the difference electron density observed in the MISC experiment during ICH catalysis (Figs. 1D and 2 A–C). In the ambient-temperature synchrotron radiation datasets, the helical shift is caused by radiation-induced oxidation of Cys101, which eliminates the negative charge on the Sγ thiolate (Cys101-S−) and thus weakens the hydrogen bond between the amide H of Ile152 and Cys101-S−. This hydrogen bond goes from −2.2 kcal/mol with a Cys101-S− acceptor to −0.91 kcal/mol with a Cys101-SOH acceptor (Fig. 2E and SI Appendix, Fig. S3). Similar loss of Cys101 thiolate negative charge occurs during formation of the on-pathway thioimidate intermediate, which also weakens the Cys101-Ile152 hydrogen bond (Fig. 1B).
Molecular Dynamics Simulations Support a Thiolate-Hydrogen Bond Restraint Mechanism.
To investigate the local dynamical response of ICH to cysteine modification, we used molecular dynamics (MD) simulations of the reduced (Cys101-S−) and Cys101-SOH crystals of ICH. We simulated the Cys101-SOH adduct rather than the thioimidate catalytic intermediate because it reports on the effects of charge neutralization (Fig. 2E and SI Appendix, Fig. S3) more directly without the structural effects of the larger bound thioimidate intermediate or biases from thioimidate parameterization. Simulations of Cys101-S− were started from the XFEL crystal structure (“XFEL” simulation, Fig. 3 A and B, red) while the Cys101-SOH–oxidized structure was similar to the synchrotron radiation (SR-)–oxidized structure (“SR” simulation, Fig. 3 A and B, blue). For the XFEL simulation, the Ile152-Cys101-S− hydrogen bond was predominantly maintained in the simulated crystal for the full 1-μs length of the MD trajectory (davg(C101SG,I152H) = 2.8 Å; Fig. 3A). By contrast, in simulations of the SR Cys101-SOH state the hydrogen bond typically dissociated very early in the trajectory (davg(C101SG,I152H) = 4.3 Å; Fig. 3A). Moreover, several protomers in the simulated Cys101-SOH crystal experienced a shift of helix H similar to that observed in the ambient-temperature synchrotron radiation datasets (mean shift 0.69 Å; Fig. 3B and Movie S1). These helical shifts were observed less frequently in simulations with the Cys101-S− (mean shift 0.60 Å; Fig. 3B), consistent with the hypothesis that local redistribution of cysteine electrostatic charge modulates these long-range motions.
Fig. 3.
Cysteine modification results in a protein-wide conformational response. (A) Fluctuations of the C101SG-I152H distance in simulations of ICH crystals in the Cys101-S− (red) or Cys101-SOH (blue) state. Each ICH dimer is represented by a semitransparent line. Opaque red and blue lines denote the average C101SG-I152H distance across the dimers. (B) Conformational shift of helix H in the Cys101-S− (red) or Cys101-SOH (blue) state. The dark gray lines in A and B represent the trajectory selected for Movie S1. (C) rmsfs calculated from MD simulations indicate highest fluctuations in linker I′-J′ of the B protomer. Helix H of the A protomer just underneath the linker also shows elevated rmsfs. (D) An isomorphous Fo(SR) – Fo(XFELFREE) difference map reveals features (green, positive; red, negative) distributed throughout the dimer, suggesting broadly altered structure and dynamics upon formation of the Cys101-SOH in the 274-K synchrotron radiation (FoRT) dataset. The “A” conformer is shown in slate blue, and the “B” conformer is shown as a gray semitransparent cartoon. Helices H and I are opaque in both conformers. The catalytic nucleophile is shown in spheres. Difference electron density features are nonuniformly distributed, with stronger features near helix H in the A protomer, and along region B169–B189, which contacts the N-terminal end of helix H. Maps are contoured at ±3.0 rmsd. (E) CONTACT analysis identifies allosteric coupling across the dimer interface, in striking agreement with isomorphous difference maps and rmsfs from D. The A protomer is color-coded in blue; the B protomer, in red. Residues identified in the CONTACT analysis are projected onto the cartoon.
Widespread Dynamic Conformational Responses to Cysteine Modification in ICH.
Root-mean-square fluctuations (rmsfs) calculated along a simulation trajectory show widespread movement across the entire dimer (Fig. 3C). Consistent with this simulation result, an Fo(SR) – Fo(XFELFREE) isomorphous difference electron density map of the Cys101-SOH SR dataset (Fo (SR)) and the Cys101-S− XFEL dataset of the free enzyme before substrate was introduced (Fo(XFELFREE)) reveal widely distributed difference features (Fig. 3D). A difference map calculated between the 15-s thioimidate intermediate XFEL dataset and the free-enzyme XFEL dataset (Fo(XFEL15s) – Fo(XFELFREE)) shows overlapping but weaker features, confirming that formation of the thioimidate intermediate during ICH catalysis shifts the conformational ensemble similarly to cysteine oxidation (SI Appendix, Fig. S4). The Fo(SR) – Fo(XFELFREE) difference map features are most prominent in protomer A near the active site and the mobile helix H (Fig. 3D). Difference features at the active site correspond to the dramatic conformational change of a conserved diglycyl motif, G150-G151 and Ile152, while those near the interface of helix H and the β-sheet in protomer A reflect residues adjusting their position in response to helix H motion. There are far fewer peaks in protomer B, consistent with the absence of mobility of helix H′ in that protomer. However, the Fo(SR) – Fo(XFELFREE) map reveals significant features along the C terminus of helix I′ at the dimer interface in protomer B, which directly contacts ThrA153 through TyrB181 (Fig. 3D). These peaks likely report on conformational changes allosterically propagating from protomer A through the I′J′ linker (residues 181–185, where primes indicate that the linker is contributed by protomer B) into protomer B. By contrast, the difference map around the C terminus of helix I and linker IJ in protomer A (IJA, which packs against the stationary helix H of protomer B) is comparatively featureless.
Notably, the simulations indicate that the conformational shift in linker I′J′ toward the unshifted helix H′ is larger than observed in the crystal structure (Movie S1). Our simulations suggest that the I′J′ conformational shift can precede relaxation of the strained Ile152 conformation and subsequent shift of helix H by several nanoseconds, allosterically communicating dynamical changes in protomer A across the dimer interface into protomer B, reminiscent of interprotomer communication observed in other systems (4). We further examined dynamical communication across the dimer interface in ICH using CONTACT (contact networks through alternate conformation transitions) network analysis of the electron density maps. CONTACT elucidates pathways of collective amino acid main- and side-chain displacements by mapping van der Waals conflicts in multiconformer qFit models (25) that would result from side-chain conformational disorder if correlated motions were not considered (17). CONTACT identified a large network of correlated residues in protomer A (with the mobile helix) that connects with a smaller network in protomer B (Fig. 3E), corroborating the isomorphous difference maps. We then used rigidity analysis (SI Appendix, Fig. S5) and kinematic flexibility analysis (KFA; ref. 26) to analyze how the Cys101-Ile152 hydrogen bond modulates motion modes accessible to the enzyme (SI Appendix, Fig. S6). Using KFA, we compared motion modes corresponding to the lowest mode-specific free energies in the XFEL structure when the H-bond is intact in both A and B protomers (C101-I152A&B) to those present when this H-bond is disrupted in protomer A (C101-I152B) (SI Appendix, Fig. S6). These altered motion modes are those most sensitive to disruption of the hydrogen bond. Perturbations in the hydrogen-bonding network are propagated primarily to the IJ linkers, helices H and I and helices J and J′, consistent with the MD simulations. Strikingly, KFA predicted large rmsfs within the IJA linker near the active site in protomer B, suggesting that the two sites are in allosteric communication. Considered together, the isomorphous difference electron density map, CONTACT analysis, KFA, and long-time MD simulations indicate that catalytically important local changes in hydrogen bonding at Cys101 initiate a cascade of dynamical changes that propagate across the entire ICH dimer in an asymmetric manner (4).
Cysteine Modification–Gated Motions Increase ICH Catalytic Efficiency.
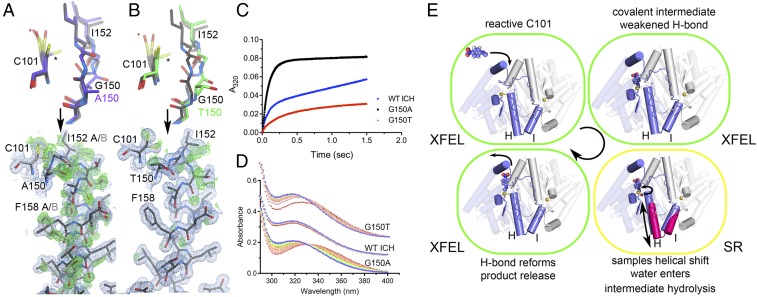
To modulate helical mobility and measure the impact on ICH catalysis, we designed two mutations, G150A and G150T. G150 is part of a highly conserved diglycyl motif that moves ∼3 Å to accommodate helical motion in ICH. Ambient-temperature (274–277 K) crystal structures of G150A and G150T ICH show that the added steric bulk at this position biases the helix toward its relaxed conformation (Fig. 4 A and B). In G150A ICH, helix H samples both conformations in both protomers whereas only one protomer of WT ICH shows helical motion (Fig. 4A and SI Appendix, Fig. S7). By contrast, the G150T structure shows helix H constitutively shifted to its relaxed position (Fig. 4B). Both G150A and G150T have an alternate Cys101 side-chain conformation which conflicts with the unshifted conformation of Ile152 and helix H (Fig. 4 A and B, asterisk), indicating that the helix must move partially independently of Cys101 modification in these mutants. As in wild-type ICH, G150A shows evidence of Cys101-SOH oxidation in the electron density (Fig. 4A). By contrast, Cys101 in G150T ICH is not modified by comparable exposure to X-rays (Fig. 4B), indicating that the Ile152-NH-Cys101-S− hydrogen bond is important for enhancing Cys101 reactivity.
Fig. 4.
Mutations at Glu150 alter helical mobility and reduce ICH catalytic turnover. (A and B) The environment of Cys101 in Gly150A and G150T ICH (Top). 2mFo-DFc electron density is contoured at 0.7 rmsd (blue) and the hydrogen bond appears as a dotted line. Both G150 mutations permit unmodified Cys101 to sample conformations (asterisk) that sterically conflict with Ile152 in the strained helical conformation (black), requiring the helix to sample shifted conformations (gray) in the absence of Cys101 modification. (A and B, Lower) The helix in its strained (black) and relaxed, shifted conformations (gray). 2mFo-DFc electron density is contoured at 0.8 rmsd (blue) and omit mFo-DFc electron density is contoured at +3.0 rmsd (green). (C) Pre-steady-state enzyme kinetics of WT (blue circles), G105A (black squares), and G150T (red triangles) ICH at 160 μM p-NPIC shows a pronounced burst phase for each protein with differing burst and steady-state rate constants. The divergent pre-steady-state profiles indicate that G150A impacts steps after the first chemical step, while G150T affects both early and later steps. (D) Single-turnover spectra of ICH enzymes with p-NPIC substrate shown from early (red) to later (blue) timepoints in 5-s increments. At early times, G105A and G150T accumulate a species with λmax = 335 nm, likely the thioimidate intermediate that resolves to product in the blue spectra with λmax = 320 nm. (E) The catalytic cycle of ICH. Substrate binds with Cys101-S− poised for nucleophilic attack and with the Cys101-Ile152 H-bond intact. Formation of the thioimidate intermediate weakens the Cys101-Ile152 H-bond and causes helical motion (magenta helices H and I) that promotes intermediate hydrolysis (red sphere; curved arrow). Hydrolysis of the thioimidate intermediate restores the reactive C101-S− and strengthens the Cys101-Ile152 H-bond, favoring the strained helical conformation.
Unlike the C101A mutant, which shows similar helical displacement but is catalytically inactive, G150A and G150T ICH are catalytically active. This allowed us to investigate the role of helical displacement in the ICH catalytic cycle. Steady-state enzyme kinetics of the G150A and G150T mutants measured using p-NPIC as the substrate showed a ∼6-fold reduction in kcat for both mutants compared to the wild-type enzyme but largely unchanged KM values (SI Appendix Fig. S8 and Table S1). In contrast to their similar steady-state kinetic behavior, the G150 mutants have divergent pre-steady-state kinetic profiles in stopped-flow mixing. ICH exhibits “burst” kinetics (Fig. 4C), indicating that the rate-limiting step for ICH catalysis comes after formation of the thioimidate intermediate. G150A ICH has a burst exponential rate constant k of ∼11 s−1 that is comparable to the wild-type enzyme, but G150T has a reduced burst rate constant of ∼4 s−1, indicating a slower chemical step (Fig. 4C and SI Appendix, Fig. S8 and Table S1). WT and G150A ICH have comparable second-order rate constants for the burst phase, while G150T is markedly slower (SI Appendix, Fig. S8 and Table S1). In addition, the burst amplitude of G150A is approximately twice as large as that of the wild-type ICH despite equal concentrations of enzyme (Fig. 4C), which correlates with both protomers of G150A having a mobile helix H compared to only one protomer of wild-type ICH (SI Appendix, Fig. S7). This suggests communication between the two active sites in the ICH dimer that is dynamically gated by helix H.
Although both the G150A and G150T mutations impair ICH catalysis, the kinetic effect of the G150A mutation is predominantly in steps after formation of the intermediate, while G150T impairs both the rate of intermediate formation and, later, the rate-limiting steps. Furthermore, we observed spectral evidence for the thioimidate intermediate in single-turnover UV-visible spectra (Fig. 4D). G150A ICH accumulates a species whose absorbance maximum is 335 nm, while WT ICH completes a single turnover and accumulates the 320-nm formamide product in the ∼30-s deadtime of manual mixing (Fig. 4D). G150T ICH accumulates less of the 335-nm intermediate than G150A ICH due to a closer match between the rates of formation and consumption of the intermediate in G150T ICH. In G150A ICH, the 335-nm species slowly converts to the 320-nm product over ∼40 s (Fig. 4D), consistent with the slow rate of product formation observed after the burst in G150A pre-steady-state kinetics (Fig. 4C). These data support the conclusion that the 335-nm species is the ICH-thioimidate intermediate observed in the MISC experiment and that G150A is impaired in hydrolyzing this covalent intermediate from the active-site Cys101 due to the perturbed dynamics of helix H.
Discussion
Covalent modification is a common and physiologically important perturbation to proteins. Reactive residues such as cysteine are prone to diverse covalent modifications with catalytic or regulatory consequences. Moreover, cysteine residues are attractive targets for covalent inhibitors, which hold great promise for the generation of highly potent and selective drugs (11). We found that cysteine modification in ICH remodels active-site H-bonding networks and gates catalytically important changes in protein dynamics with a clearly defined mechanism. Modification of the active-site cysteine thiolate during catalysis neutralizes its negative charge and weakens a key H-bond, activating correlated motions that span the entire ICH dimer. Transiently increased flexibility of the active site facilitates water entry and attack at the thioimidate to form a proposed tetrahedral intermediate. The H-bond between Cys101 and Ile152 then reforms, stabilizing the nascent Cys101 thiolate and making it a better leaving group, thereby coupling dynamical changes in the enzyme to progress along the reaction coordinate (Fig. 4E). Notably, all cysteine thiolates experience a similar loss of negative charge upon covalent bond formation. Because cysteine residues can have multiple roles in a protein, including catalytic nucleophile, metal ligand, acylation target, redox target, covalent inhibitor target, and others, many different modification-mediated signals may be transduced through altered cysteine electrostatics to impact protein dynamics and function, expanding the ways in which cysteine can couple protein biophysical properties to cellular needs. Integrating recent developments in serial and conventional room-temperature (RT) crystallography with advanced computational methods (27) can offer exciting opportunities to reveal enzyme mechanisms and interrogate the consequences of cysteine modification for protein dynamics.
Materials and Methods
Pseudomonas fluorescens ICH was expressed in E. coli as a thrombin-cleavable, N-terminally 6xHis-tagged protein and purified as previously described (22). Crystals in space group P21 were grown using hanging drop vapor equilibration for synchrotron datasets and by batch microseeding for the XFEL data collections. Synchrotron data were collected at the Stanford Synchrotron Radiation Laboratory (SSRL) from capillary-mounted crystals at 274–277 K. XFEL data were collected at the Macromolecular Femtosecond Crystallography (MFX) end station at the LCLS at 298 K. Models were fitted into electron density maps in COOT (28) and refined in PHENIX (29). Crystallographic data and model statistics are provided in the SI Appendix, Tables S2 and S3. Molecular dynamics simulations were performed using the Schrödinger 2018–3 software suite. Steady-state and pre-steady-state enzyme kinetics was measured for recombinant ICH proteins using the model substrate p-NPIC, and formamide product formation was monitored by absorption at 320 nm. Detailed methods are provided in the SI Appendix.
Data Availability.
All coordinates and structure factors are freely available for download from the RCSB Protein Data Bank (http://www.rcsb.org).
Supplementary Material
Acknowledgments
We thank Dr. Donald Becker (University of Nebraska) and Dr. Joseph Barycki (North Carolina State University) for helpful discussions and Lauren Barbee (University of Nebraska) for assistance with ICH kinetic data and analysis. R.G.S. acknowledges the support of the Office of Basic Energy Sciences through the Atomic, Molecular, and Optical Sciences Program program within the Chemical Sciences, Geosciences, and Biosciences Division and of the US Department of Energy (DOE) through the SLAC Laboratory Directed Research and Development Program. A.S.B. and N.K.S. were supported by National Institutes of Health (NIH) grant GM117126 to N.K.S. for data-processing methods. M.C.T. and J.S.F. were supported by the National Science Foundation (NSF) (STC-1231306), by a Ruth L. Kirschstein National Research Service Award (F32 HL129989) to M.C.T., and by awards to J.S.F. from the NIH (GM123159, GM124149), the David and Lucile Packard Foundation (Packard Fellowship), and the University of California (UC) Office of the President Laboratory Fees Research Program (LFR-17-476732). M.E.W. was supported by the Exascale Computing Project (17-SC-20-SC), a collaborative effort of the DOE, Office of Science, and the National Nuclear Security Administration, and the UC Office of the President Laboratory Fees Research Program (LFR-17-476732). S.H.P.d.O. and H.v.d.B. are supported by award NIH GM123159 to H.v.d.B. H.v.d.B. is supported by a Mercator Fellowship from the Deutsche Forschungsgemeinschaft (DFG). M.A.W. was supported by the Nebraska Tobacco Settlement Biomedical Research Development Fund. Use of the SSRL, SLAC National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE, Office of Biological and Environmental Research, and by the NIH, National Institute of General Medical Sciences (NIGMS) (including P41GM103393). Use of the LCLS, SLAC National Accelerator Laboratory, is supported by the DOE, Office of Science, Office of Basic Energy Sciences, under Contract No. DE-AC02-76SF00515. The helium-rich ambient system for in-helium experiments at MFX was developed by Bruce Doak and funded by the Max Planck Institute for Medical Research. This research used resources of the Advanced Photon Source, a DOE, Office of Science, User Facility operated for the DOE, Office of Science, by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of BioCARS was also supported by NIH NIGMS under grant number R24GM111072. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. N.S.S. is a guest editor invited by the Editorial Board.
Data deposition: Refined atomic coordinates and X-ray crystallographic data have been deposited in the RCSB Protein Data Bank, http://www.rcsb.org (accession nos. 6NI4–6NI9, 6NIA, 6NPQ, 6UND, and 6UNF).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1901864116/-/DCSupplemental.
References
- 1.Danyal K., Mayweather D., Dean D. R., Seefeldt L. C., Hoffman B. M., Conformational gating of electron transfer from the nitrogenase Fe protein to MoFe protein. J. Am. Chem. Soc. 132, 6894–6895 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Henzler-Wildman K. A., et al. , Intrinsic motions along an enzymatic reaction trajectory. Nature 450, 838–844 (2007). [DOI] [PubMed] [Google Scholar]
- 3.Aviram H. Y., et al. , Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions. Proc. Natl. Acad. Sci. U.S.A. 115, 3243–3248 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim T. H., et al. , The role of dimer asymmetry and protomer dynamics in enzyme catalysis. Science 355, eaag2355 (2017). [DOI] [PubMed] [Google Scholar]
- 5.Hammes G. G., Multiple conformational changes in enzyme catalysis. Biochemistry 41, 8221–8228 (2002). [DOI] [PubMed] [Google Scholar]
- 6.Kale S., et al. , Efficient coupling of catalysis and dynamics in the E1 component of Escherichia coli pyruvate dehydrogenase multienzyme complex. Proc. Natl. Acad. Sci. U.S.A. 105, 1158–1163 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mehrabi P., et al. , Time-resolved crystallography reveals allosteric communication aligned with molecular breathing. Science 365, 1167–1170 (2019). [DOI] [PubMed] [Google Scholar]
- 8.Stiers K. M., et al. , Structural and dynamical description of the enzymatic reaction of a phosphohexomutase. Struct. Dyn. 6, 024703 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pace N. J., Weerapana E., Diverse functional roles of reactive cysteines. ACS Chem. Biol. 8, 283–296 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Reddie K. G., Carroll K. S., Expanding the functional diversity of proteins through cysteine oxidation. Curr. Opin. Chem. Biol. 12, 746–754 (2008). [DOI] [PubMed] [Google Scholar]
- 11.Lonsdale R., Ward R. A., Structure-based design of targeted covalent inhibitors. Chem. Soc. Rev. 47, 3816–3830 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Furnham N., Blundell T. L., DePristo M. A., Terwilliger T. C., Is one solution good enough? Nat. Struct. Mol. Biol. 13, 184–185 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Fraser J. S., et al. , Accessing protein conformational ensembles using room-temperature X-ray crystallography. Proc. Natl. Acad. Sci. U.S.A. 108, 16247–16252 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Smith J. L., Hendrickson W. A., Honzatko R. B., Sheriff S., Structural heterogeneity in protein crystals. Biochemistry 25, 5018–5027 (1986). [DOI] [PubMed] [Google Scholar]
- 15.Wilson M. A., Brunger A. T., The 1.0 A crystal structure of Ca(2+)-bound calmodulin: An analysis of disorder and implications for functionally relevant plasticity. J. Mol. Biol. 301, 1237–1256 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Martin-Garcia J. M., Conrad C. E., Coe J., Roy-Chowdhury S., Fromme P., Serial femtosecond crystallography: A revolution in structural biology. Arch. Biochem. Biophys. 602, 32–47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van den Bedem H., Bhabha G., Yang K., Wright P. E., Fraser J. S., Automated identification of functional dynamic contact networks from X-ray crystallography. Nat. Methods 10, 896–902 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burnley B. T., Afonine P. V., Adams P. D., Gros P., Modelling dynamics in protein crystal structures by ensemble refinement. eLife 1, e00311 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Zundert G. C. P., et al.. , qFit-ligand reveals widespread conformational heterogeneity of drug-like molecules in X-ray electron density maps. J. Med. Chem. 61, 11183–11198 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wall M. E., Wolff A. M., Fraser J. S., Bringing diffuse X-ray scattering into focus. Curr. Opin. Struct. Biol. 50, 109–116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goda M., Hashimoto Y., Shimizu S., Kobayashi M., Discovery of a novel enzyme, isonitrile hydratase, involved in nitrogen-carbon triple bond cleavage. J. Biol. Chem. 276, 23480–23485 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Lakshminarasimhan M., Madzelan P., Nan R., Milkovic N. M., Wilson M. A., Evolution of new enzymatic function by structural modulation of cysteine reactivity in Pseudomonas fluorescens isocyanide hydratase. J. Biol. Chem. 285, 29651–29661 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brereton A. E., Karplus P. A., Native proteins trap high-energy transit conformations. Sci. Adv. 1, e1501188 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chapman H. N., Caleman C., Timneanu N., Diffraction before destruction. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Bedem H., Dhanik A., Latombe J. C., Deacon A. M., Modeling discrete heterogeneity in X-ray diffraction data by fitting multi-conformers. Acta Crystallogr. D Biol. Crystallogr. 65, 1107–1117 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Budday D., Leyendecker S., van den Bedem H., Kinematic flexibility analysis: Hydrogen bonding patterns impart a spatial hierarchy of protein motion. J. Chem. Inf. Model. 58, 2108–2122 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van den Bedem H., Fraser J. S., Integrative, dynamic structural biology at atomic resolution—It’s about time. Nat. Methods 12, 307–318 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 29.Adams P. D., et al. , PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All coordinates and structure factors are freely available for download from the RCSB Protein Data Bank (http://www.rcsb.org).