Significance
Although fungi in the air make up a disproportionately large part of particulate matter and affect both local and global biogeochemical cycles, the factors influencing their composition and diversity are less clear, especially over relatively long timescales. Here we leverage the longest time series of aerobiota to date from a remote sampling location on Hawaii Island to examine changes in fungi over time and in response to environmental conditions. Our results provide evidence for a relatively unstable fungal aerobiota, whose composition is likely influenced by the ability to disperse into the system, rather than season or climate. Our finding of core taxa throughout the sampling period indicates resistance among some fungi to natural environmental fluctuations and human-associated global change.
Keywords: fungi, aerobiota, environmental change, time series
Abstract
Fungi are ubiquitous and often abundant components of virtually all ecosystems on Earth, serving a diversity of functions. While there is clear evidence that fungal-mediated processes can influence environmental conditions, and in turn select for specific fungi, it is less clear how fungi respond to environmental fluxes over relatively long time frames. Here we set out to examine changes in airborne fungi collected over the course of 13 y, which is the longest sampling time to date. Air filter samples were collected from the Mauna Loa Observatory (MLO) on Hawaii Island, and analyzed using Illumina amplicon sequencing. As a study site, MLO is unique because of its geographic isolation and high elevation, making it an ideal place to capture global trends in climate and aerobiota. We found that the fungal aerobiota sampled at MLO had high species turnover, but compositional similarity did not decrease as a function of time between samples. We attribute these patterns to neutral processes such as idiosyncratic dispersal timing and trajectories. Furthermore, the composition of fungi at any given point was not significantly influenced by any local or global environmental variables we examined. This, and our additional finding of a core set of persistent fungi during our entire sampling period, indicates some degree of stability among fungi in the face of natural environmental fluctuations and human-associated global change. We conclude that the movement of fungi through the atmosphere is a relatively stochastic process.
Time series data have provided profound insights into phenomena of global importance, such as demonstrating an exponential increase in atmospheric CO2 concentrations during the Anthropocene (1). Dynamics such as atmospheric CO2 concentrations and other global cycles are linked to the activity of microorganisms, whose diversity and function respond to changing environmental conditions. However, in contrast to a detailed record of environmental change, we have a rudimentary understanding of how microbes disperse and how their assemblies change over time. Here we track changes in fungal diversity and identities in air samples collected during a 13-y period, the longest sampled time series for aerobiota to date.
Fungi, like other microorganisms, are globally widespread (2). Among their many functions, fungi play critical roles in symbioses as pathogens and mutualists (3) in hydrologic cycles (4, 5) and in local- and large-scale biogeochemical cycles (6). Thus, the composition of these communities has important implications for global change. Fungi make up a disproportionately large component of the biological portion of particulate matter in the air (aerobiota), emitting upwards of 190 teragrams of spores and hyphal fragments annually on a global scale, which is twice the estimated contribution of pollen (7). However, the identities and distributions of fungal aerobiota are not well understood. Previous time series studies of fungal aerobiota have relied on culture-based sampling methods over the course of a few years (8, 9) or culture-independent (molecular-based) sampling over the course of a week or 2 (10, 11), 6 mo to 1 y (4, 12–14), or 7 y (15). These studies have yielded conflicting results. For example, some show significant changes in fungal diversity over seasons (e.g., refs. 8, 14, and 15), while others show no change in fungal diversity within a year (e.g., refs. 4 and 11). Therefore, generalizing the results from these and other temporal studies of fungal aerobiota is challenging and may be confounded by choice of methods, differences among regional fungal species pools, and relatively short sampling durations that cannot detect long-term patterns. To overcome these limitations, we used a high-throughput amplicon sequencing approach to assess the composition of fungal aerobiota sampled at semiregular intervals for more than a decade, twice the length of prior published studies, from the Mauna Loa Observatory (MLO) on Hawaii Island, one of the most remote locations on earth.
MLO is home to one of the longest running time series of atmospheric measurements. Since 1958, MLO has been collecting data on atmospheric carbon dioxide (CO2) concentrations, which form the backbone for the famous Keeling curve (16). MLO was selected as a study site to examine global trends in atmospheric CO2 because of the absence of local influences; at 3,397 m above sea level, and situated in a volcanic desert, with the nearest continental land mass more than 2,000 kilometers away, it is one of the most isolated climate monitoring stations on earth. Since 1874, Mauna Loa volcano has erupted 33 times, with the most recent eruption in 1984. Today, MLO is surrounded by >650 km2 of lava flows of various ages forming a volcanic desert (SI Appendix, Fig. S1) (17). The climate of MLO is shaped primarily by predictable diurnal patterns of atmosphere mixing from local (within Hawaii Island air during the day) and long-distance (free troposphere air during the night) sources. Seasonal wind patterns such as the dominant northeasterly trade-winds, the less frequent Kona winds that blow from the opposite direction during periods of low pressure, and annual spring dust storms from parts of Asia also shape the climate at MLO (18–20). Shifts among these prevailing patterns should lead to differences in the source pools, and therefore composition of fungal aerobiota.
MLO’s isolation and high elevation make it an ideal site for long-term monitoring of atmospheric constituents in addition to CO2 concentrations, including other climatically important traces gases and aerosol particles. Inadvertently, some of the particulate measurements are collected such that biological specimens including fungi are captured and preserved (SI Appendix, Fig. S2). Average particle density and related aerosol properties have been shown to fluctuate with season, El Niño southern oscillation, and storm events (21–23). However, it remains to be seen whether the aerobiota among these particles change in response to similar variables and over similar timescales. Here we ask whether there are long-term and persistent patterns in fungal aerobiota over time, and if so, whether they relate to local and global meteorological and climatic variables.
Results
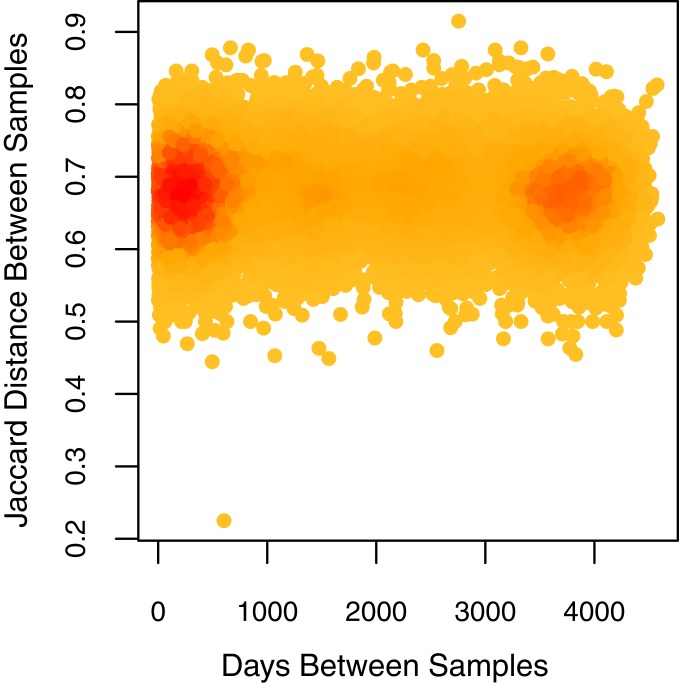
Over the course of our 13-y sampling period, we detected a total of 160 fungal OTUs (operational taxonomic units) from 172 time points. None of the weather or atmospheric variables that we examined predicted composition or OTU abundances (all Wilcox test FDR corrected P values > 0.05). Assemblage dispersion, or the dissimilarity of the fungi present on any 2 filters between any 2 times remained fairly stable over season (quarter) and our 13-y sampling period (Figs. 1 and 2 and SI Appendix, Fig. S3). Pairwise beta diversity (sample dissimilarity) between samples as measured by Jaccard distance, averaged 0.681 (SD = 0.061), with no significant increase with time between samples (Mantel permuted P value = 0.794; Fig. 1). Sample dissimilarity remained stable during our entire sampling period, despite relatively high turnover in fungal taxa among samples (βSIM = 0.986; average distance to the median = 0.480; SD = 0.041; Figs. 1 and 2). Similarly, assemblage dispersion remained stable and beta diversity relatively high within each quarter of the year (SI Appendix, Table S1).
Fig. 1.
Fungal aerobiota at MLO over time. Density plot of pairwise Jaccard distance between fungal aerobiota from each sample by days. The darker the color, the more samples fall in that region. The difference between fungal composition remained constant independent of proximity of sampling periods.
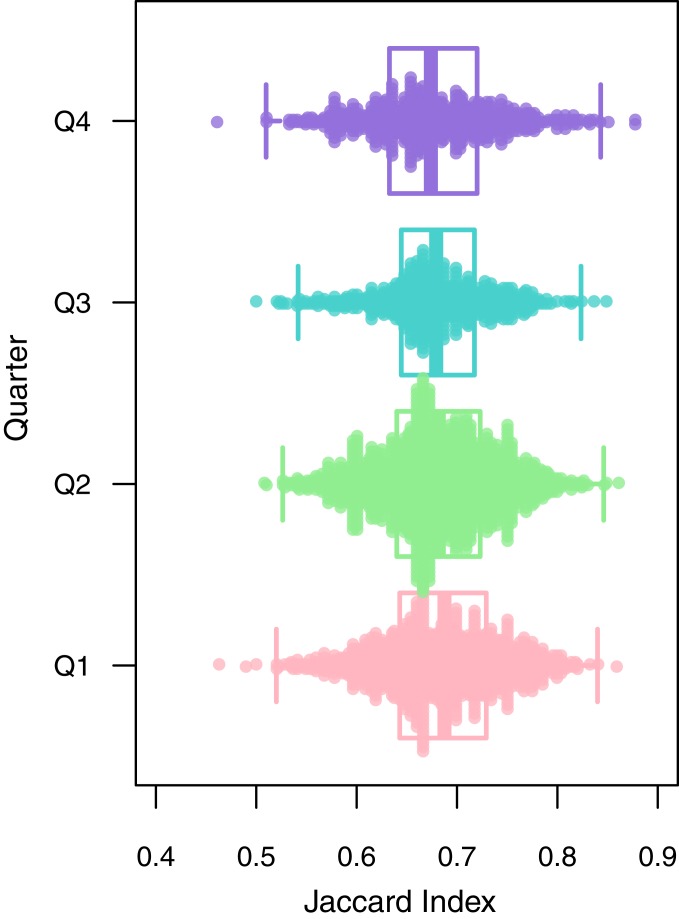
Fig. 2.
Fungal aerobiota at MLO by season box-and-whisker style plots of pairwise Jaccard distance between fungal aerobiota within the same quarter. The boxes cover the interquartile range, and the whiskers cover 1.5 times the interquartile range. The midline indicates the median pairwise distance between samples within the quarter. As the boxes all overlap greatly, the pairwise Jaccard distance between samples within the same quarter did not differ by quarter.
Alpha diversity (the number of taxa per filter) was relatively stable among samples and did not change significantly over season or the entire sampling period (SI Appendix, Fig. S4). Alpha diversity across all samples, as measured by Hill numbers, averaged 0D = 35.23 (SD = 7.49) for richness (Chao1), 1D = 12.39 (SD = 3.25) for entropy (exponential Shannon Index), and 2D = 6.35 (SD = 1.94) for diversity (inverse Simpson’s concentration), with no significant trend over the long-term sampling effort (Mann-Kendall trend test P value = 0.883 for 0D), season (ANOVA P value = 0.763 for 0D), or seasonal trends over time (ANOVA adjusted for year P value = 0.617 for 0D). Similar to beta diversity, alpha diversity did not correlate with sampling time (Spearman P value = 0.192 for 0D; SI Appendix, Fig. S4). Given the high rate of species turnover, the relatively constant number of OTUs observed during the 13 y was unexpected. After evaluating both niche-based and neutral assembly models, we found that the fungi among all samples at MLO follow a neutral assembly model (SI Appendix, Fig. S5), indicating that neutral or random dispersal and stochastic events are influencing the composition of fungi at MLO.
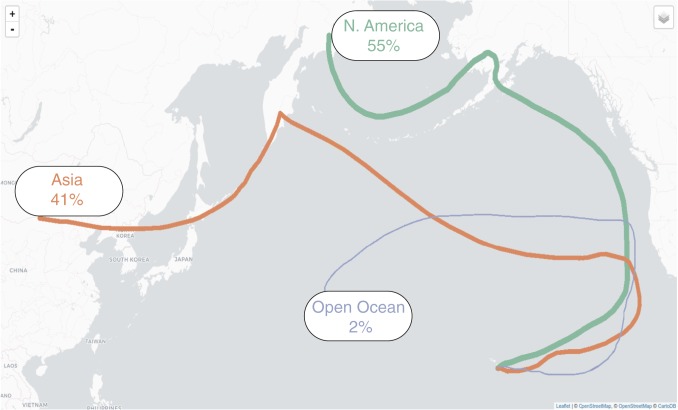
On the basis of prior studies of transport and particle deposition, it takes roughly 10 d for air masses from spring storm events in Asia to reach the west coast of North America (24). Because only nighttime air currents (free-troposphere air) would have contributed long distance air masses to our samples, we examined the back-trajectories of air masses that arrived at MLO at midnight during our sampling period to assess their origin. For all nighttime air currents, the average amount of time since last passing over a landmass was 15.17 d (SD = 7.5 d; Fig. 3). The length of time since passing over a landmass varied by season (ANOVA P value = 0.002), with the shortest times falling in Q1, consistent with the annual fast-moving dust storm events from the Asiatic deserts, and the longest times falling in Q3, consistent with the Pacific hurricane season (SI Appendix, Fig. S6). The continental origins were predominantly North America (55%), including the Aleutian Islands (3,991 km away from MLO), and Asia (41%), including the Kamchatka Peninsula of Russia (5,455 km away from MLO; Fig. 3). The remaining samples originated from a range of locations including Kauai (498 km away from MLO), Peru (9,900 km away from MLO), Indonesia (9,003 km away from MLO), and 4 could not be traced back to a landmass. Only 1 taxon was nominally associated with air masses that were last over Asia: Ruinenia diospyri (adjusted P value = 0.088).
Fig. 3.
Map of the back trajectories of air masses sampled at MLO. Each line shows a representative trajectory of the air masses grouped by the last continental landmass it was over in the 15 d before its arrival at MLO. Air masses having been over North America (55% of samples) are represented by the trajectory arriving at MLO on August 17, 2000, in green. Air masses having been over Asia (41% of samples) are represented by the trajectory arriving at MLO on December 1, 2000, in orange. Air masses that were not over a landmass in the past 60 d, or since formation (2% of samples), are represented by the trajectory arriving at MLO on September 29, 2011, in blue. Samples having last been over South America, Oceania, or Kauai (collectively 2% of samples) are not represented.
Consistent with the dominant tropical trade winds that pass over Hawaii, the majority of winds that occurred during our sampling period (72.3%) came from the northeast or east; only 1 air mass was recorded as having been southeast of MLO, and the remainder (26.6%) came from currents north or northwest of MLO, consistent with annual Kona wind patterns. Of the air masses that came from the north, northeast, or east of MLO, 54% (84 of 155) crossed the California Current, one of the major Pacific Ocean currents that precedes the North Equatorial Current in the North Pacific Gyre. Staninwardia suttonii was the only indicator species nominally associated with any air mass (The California Current, adjusted P value = 0.060).
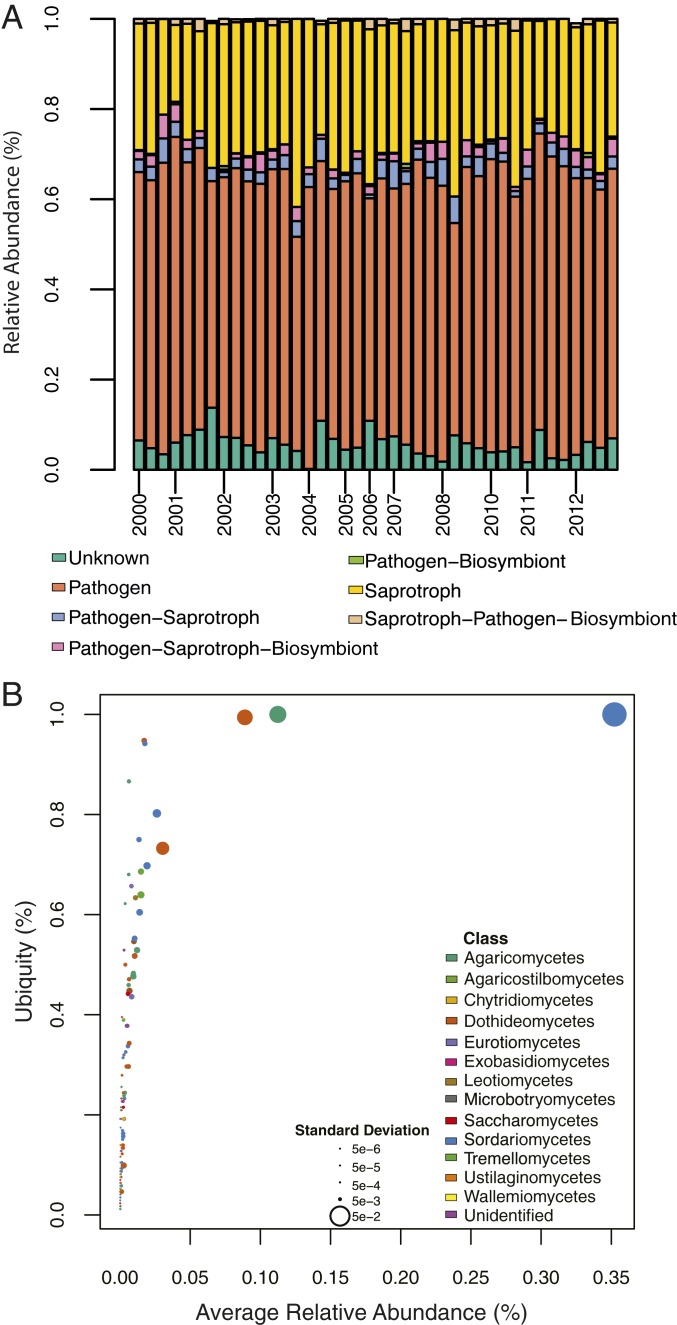
The majority of fungi (76%) belonged to the phyla Ascomycota, followed by Basidiomycota (22%), with the remaining 2% belonging to Chytridiomycota and Zoopagomycota (SI Appendix, Fig. S7). Saprotrophs were the most common trophic mode among all of the fungal guilds, accounting for 86 of the OTUs. However, the most abundant fungi, based on read count (71% relative abundance), belong to pathogens, primarily plant disease-causing fungi, with a few human pathogens as well (Fig. 4A). A large subset of the aerobiota from MLO are mutualists such as mycorrhizal fungi (up to 5.2% of the reads in a given filter). However, these fungi are absent from approximately one-fifth of the filters (19.2%), whereas saprotrophs and plant pathogens are ubiquitous (Fig. 4A). In addition, we found 3 core fungal taxa that are highly ubiquitous (present in >80% of all samples) and highly abundant (>1% of reads in all samples), 2 of which are plant pathogens (Passalora sp. and Ophiostoma breviusculum; Fig. 4B), and the third of which is a saprotroph (Auriscalpium villipes; Fig. 4B).
Fig. 4.
Overview of fungal aerobiota at MLO. (A) Stacked bar graphs of each quarter colored by the average relative abundance of each guild. The majority of reads in each sample were pathogens (in coral), followed by saprotrophs (in yellow). (B) Ubiquity-abundance plot, where each dot represents an OTU, is colored by its taxonomic class, and sized according to the SD of its average relative abundance. The core taxa identified as highly ubiquitous and highly abundant are Ophisoma breviusculum (in blue), Auriscalpium villipes (in green), and Passalora sp. (in orange).
In general, both the sexual and asexual spores of the taxa detected at MLO were small, ellipsoid, smooth, and hyaline (or clear). Compared with all other spores in kingdom Fungi that could be captured at MLO (those with any dimension less than 10 μm), we observed no significant enrichment of shape, texture, or color for asexual or sexual spores from MLO (all P values = 1.000). Spore traits for both asexual and sexual spores also showed no significant differences by quarter, nor trends over time (quarter-adjusted P value between 0.298 and 0.980, and trend-adjusted P values between 0.085 and 1.000).
Discussion
By sampling fungi over such a long time frame and in such a remote high-altitude location that incorporates regional and global inputs, we have gained a more complete picture of the properties of microbial aerobiota (25–28). Unlike more localized aerobiota studies, we examined aggregate changes in both tropospheric fungal aerobiota that represent a portion of the global species pool, as well as regional inputs derived from low-elevation sources below MLO, possibly including agriculture, marine, and industry. By doing so over the course of 13 y, we found that differences among the assemblies of fungi on any given filter did not increase over time, despite relatively high rates of species turnover, even among filters that were sampled a few days apart. In addition, fungal composition did not correlate with season or any local or global climatic, or meteorological conditions that we examined. Even more intriguing is that based on our model of air mass origin, fewer than 5% of the air currents carrying nighttime (troposphere) samples to MLO had passed over land in the last week. Rather, the majority of our samples were carried on air currents that had been over the open ocean for at least 2 wk before being captured at MLO. While most of these samples originated from trade winds (northeasterly), there was no single dominant trade wind current that led to nighttime deposition at MLO. With these results in mind, it is not surprising that our fungal species composition during the entire 13-y sampling period best fit a neutral model for assembly, given that we captured the highly dynamic dispersal of fungi from a broad geographic range including regional and global inputs (Fig. 3).
Despite high species turnover among sampling points, there was no evidence of seasonality or increased compositional divergence among fungi over time. We attribute this result to the influence of primarily neutral processes such as source mixing, long-distance dispersal, and physical similarities among aerobiota (29). For example, physical similarity may be a key characteristic for the fungal aerobiota of MLO, where they must have some combination of the following traits to converge on the same location: the ability to produce large quantities of spores, disperse significant distances, stay uplifted in air currents either moving up from the base of Mauna Loa or across oceans, and tolerate harsh climatic conditions such intense solar radiation (at least to the extent that precludes damage to dsDNA in the ribosomal ITS region). Prior studies that found significant decreases in the similarity of fungal aerobiota during relatively shorter periods (10), or significant associations with specific to environmental conditions or seasons (12, 13, 27), were likely detecting fungi that assemble as a result of more deterministic processes than those at MLO. That we found core fungal aerobiota across our entire sampling period indicates resilience, or the ability to rebound post environmental disturbance of some fungi even in the face of dramatic environmental fluctuations such as El Niño events or the 2008 eruption of Kilauea volcano on Mauna Loa’s flank (30, 31). However, the fact that we detected relatively few taxa overall indicates that our sampling conditions or location limited the total diversity of fungi compared with other studies of aerobiota (32).
Our finding that the alpha diversity of these fungi is relatively low and stable over time could simply be the result of a density-dependent occupancy rate of the filters themselves, or sequencing biases, rather than evidence of an aerobiogeographic pattern (sensu refs. 30 and 31). Additional considerations when interpreting our data are that we did not explicitly test the viability of the fungi from the filters collected at MLO, so their potential contributions to the local Hawaiian species pool remain unknown, and our storage conditions may have favored particularly resilient taxa. Also, because we were unable to parse local versus long-distance origins of the fungi detected at MLO, we hope that future research will address fungal biogeography at this more granular scale.
Our sampling effort provides further evidence for certain groups of fungi such as the Capnodiales being common members of aerobiota. Surprisingly, spores of the fungi from MLO did not conform to prior predictions of traits related to atmospheric dispersal or persistence. For example, prior studies indicate that small, ellipsoid (25), or melanized spores (33) have larger dispersal kernels, but those spore traits were not overly abundant in our long-term sampling regime, indicating that fungal traits or propagules advantageous for dispersal, especially over long distances, are not resolved.
We found that the majority of fungi captured in each sample and across our entire sampling period were plant pathogens, indicating that members of this trophic guild may be particularly well suited to relatively long-distance travel, given that the closest vegetation is >7 km from MLO. Ascomycota dominated our filters, which is a similar finding to other fungal aerobiota studies (4, 7, 9, 15). Among these Ascomycetes was O. breviusculum, a member of our core taxa, which accounted for 35% of our total number of sequences and was present in every sample, including samples from before the species was formally described in 2006 as a putative Japanese endemic associated with bark beetles infecting larch trees (34). Sampling over the course of such a relatively lengthy and continuous time scale also provided rare evidence of long-distance dispersal among other guilds of fungi such as ectomycorrhizal species that form obligate associations with tree roots (35). We detected a Tomentella sp. (present in 0.004% of all reads and observed only twice, 7 y apart) whose only known compatible hosts are the tree Pisonia grandis and Dipterocarp species from the Saraburi province of Thailand (>97.35% identity to GenBank accessions MN077166 and AF020770), and not any of the ectomycorrhizal hosts that exist in Hawaii (36–38). While closer than Thailand, the closest populations of P. grandis are on Palmyra Atoll and a single tree on Lisianski Island (∼1,600 and 2,000 km away from MLO respectively). This finding increases the observed dispersal range (39) by more than an order of magnitude for this genus, highlighting the important aspects of fungal biogeography that can be gleaned from a combination of temporal sampling and high-throughput DNA sequencing studies.
What have we learned about atmospheric fungi by doubling the former collection time frame and sampling from a geographically isolated location situated in a volcanic desert? We have shown that there is a persistent core set of fungi whose origin remains unknown, but likely travel at least several kilometers before arriving at MLO. We have revealed that at least 1 important plant mutualist and possibly some plant pathogens can travel thousands of kilometers before encountering a suitable host or habitat. We have also learned that the fungi that arrive at high elevation do not share any particular spore traits. And we have provided evidence that the composition of aerobiota is determined by stochastic arrival into the system, rather than selective processes, indicating that the movement of microbes through the atmosphere is as variable as the air masses on which they travel.
Materials & Methods
Sample Collection.
Starting in June 2000 and continuing until December 2012, the air filters used in this study were taken from a Particle Soot Absorption Photometer (Radiance Research Inc., Seattle, WA), built to measure light absorption by aerosol particles in the atmosphere (see SI Appendix, Methods for details). In general, the filters were changed every 4 to 7 d, depending on aerosol density. Thus, each filter represents the pooled aerobiota from numerous days of nighttime free-tropospheric air with minimal influence from regional conditions, and daytime air that includes regional sources (19, 20). The estimated volume of air sampled through each Particle Soot Absorption Photometer filter was 6 to 10 m3, depending on particle loads and composition. Filters were stored in individual sealed plastic bags at ambient temperature until DNA extraction.
Downloaded weather and atmospheric conditions that were collected at MLO during the sampling period by the Earth System Research Laboratory, Global Monitoring Division of NOAA and are publicly available (for details, see SI Appendix, Methods). Additional data on the height of the trade-wind inversion layer on the day of sample collection was derived from a long-term data set managed by the laboratory of Thomas Giambelluca at the University of Hawaii at Manoa.
Samples.
Of the ∼1,100 filters collected over the course of 13 y, we chose 383 for sequencing. To make comparisons of fungal composition within and across seasons that correspond with the prevailing winds, we chose to analyze filters from the months that had the greatest number of replicates. These samples were randomly distributed across the 13-y sampling period, with greater density (35%) from the more recent years (2010 to 2012; Dataset S1). In addition, 7 blank filters used as controls for the original Particle Soot Absorption Photometer data were selected from dates spanning from 2006 to 2013 to serve as controls for background contamination. See SI Appendix, Methods for DNA extraction and sequencing protocols.
Taxonomic Assignments.
The seed read for each OTU, also known as the centroid, was considered the representative sequence. These representative sequences were used to assign taxonomy based on consensus from 3 independent approaches: matching to the curated UNITE database using the QIIME assign_taxonomy.py script, matching to the NCBI nuccore database using the BLAST algorithm (40), and identifying the lowest common ancestor using MEGAN5 (41). The final taxonomic assignments were determined using custom python scripts to compare the results of all 3 taxonomic assignment methods (for more details, see SI Appendix, Methods).
All fungi with poor taxonomic resolution were removed incidentally, with OTUs found only in <1% of samples, which were also removed (for more details, see SI Appendix, Methods). Contaminant OTUs were determined from negative controls with the decontam R package, using the prevalence method (42); the 3 OTUs identified as contaminants were either not present in the samples or removed before OTU clustering due to extremely low abundance and/or rarity. This final filtering resulted in 160 fungal OTUs with high-quality taxonomic assignments. Guild and trophic modes were assigned on the basis of final OTU taxonomic assignments using FunGuild v1.0 (43). OTUs that were assigned a final taxonomy but not assigned to a guild were assigned manually using data from Mycobank (44). OTU characteristics including spore traits, taxonomy, trophic mode, guild, and associated references are available in Dataset S2.
Spore Traits.
Sexual and asexual spore traits, including size, shape, texture, and color, were recorded based on our final taxonomic assignments using a spore database representative of kingdom Fungi (see ref. 25). Spore shapes were classified into 4 basic shapes: cylindrical, ellipsoid, fusiform, or spherical. To test for spore traits specific to MLO aerobiota, we compared the spores of taxa from MLO to spores in the database that fit the criteria to be captured at MLO (e.g., <10 μM in diameter), using the Student’s t test for continuous variables and Fisher’s exact test for categorical variables.
Aerobiota Diversity.
Diversity metrics for the fungi from all filters combined were analyzed in R version 2.3.2 (45). Presence-absence based Jaccard distance for beta diversity, dispersion (βSIM), and permutation-based Mantel tests to examine changes in compositional dissimilarity over time were calculated using the vegdist and nestedtemp functions in the vegan package version 2.4–1 (46). Alpha diversities, as measured by the first 3 Hill numbers, were calculated using the iNEXT package version 2.0.12 (ref. 47; for details, see SI Appendix, Methods). A Mann-Kendall trend test was used to examine monotonic trends in alpha diversity over time and calculated using the trend package version 0.2.0 (48). Nonparametric Spearman’s rank correlation between time and alpha diversity was calculated using the cor function as part of base R.
Seasonality was measured by dividing the calendar year into the following quarters: January, February, and March as quarter 1, the time most likely to experience Kona winds; April, May, and June as quarter 2; July, August, and September as quarter 3, which corresponds to hurricane season; and October, November, and December as quarter 4, during which the trade winds generally dominate. Seasonal differences in alpha diversity and among individual OTUs were tested with ANOVA tests, followed by a false discovery rate correction of the P values (49) in base R. Indicator species for seasonality were identified using the indicspecies version 1.7.6 package (50); resulting P values were adjusted using the Benjamini-Hochberg procedure (49). To test the relative importance of stochastic-based (neutral) versus deterministic-based (niche) processes leading to the assembly of MLO fungal aerobiota, both niche-based and neutral assembly models were fit to the fungi’s species (OTU) abundance distributions, using the sads package version 0.3.1 in R (ref. 51, details in SI Appendix, Methods).
Air Mass Trajectories.
The trajectory of the air masses sampled at MLO were calculated using HYSPLIT models (52) in the SplitR package version 0.4 (https://github.com/rich-iannone/SplitR). Starting with the date of sampling and the location of MLO (latitude = 19.5362, longitude = −155.5763), backward trajectories were calculated for 720 h (30 d), using the global NCEP Reanalysis data. The resulting trajectories were mapped to determine over which landmass they last passed, Asia or North America (including the Aleutian Islands). When air masses did not pass over a landmass in the last 30 d (n = 16), trajectories were extended to 1,440 h, 60 d, or until they could not be traced back farther. The air currents were classified as having passed the California Current if they originated east of 130° W longitude in the 7 d before sample collection. Seasonal and monotonic trends in the time since air masses were last over land were tested similarly to diversity metrics. Indicator species for specific air currents were identified using methods similar determining seasonality.
Data Availability.
All metadata (including weather variables, spore traits, taxonomic assignments, and guilds) are available in the SI Appendix, along with custom scripts used for analyses. Raw sequence data have been deposited in the Sequence Read Archive under the accession PRJNA386517.
Supplementary Material
Acknowledgments
The authors would like to acknowledge the A.S.A./N.A.H. laboratory group for valuable feedback on earlier versions of this manuscript. Ryan Longman provided data on the TWIL. N.A.H. is funded by the National Science Foundation award #1556856 and The W. M. Keck Foundation. L.T. is funded by an Alfred P. Sloan Foundation Microbiome of the Built Environment Postdoctoral Fellowship.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: All metadata (including weather variables, spore traits, taxonomic assignments, and guilds) are available in the SI Appendix, along with custom scripts used for analyses. Raw sequence data have been deposited in the Sequence Read Archive under accession no. PRJNA386517.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1907414116/-/DCSupplemental.
References
- 1.Keeling C. D., Whorf T. P., Wahlen M., van der Plichtt J., Interannual extremes in the rate of rise of atmospheric carbon dioxide since 1980. Nature 375, 666–670 (1995). [Google Scholar]
- 2.Brown J. K. M., Hovmøller M. S., Aerial dispersal of pathogens on the global and continental scales and its impact on plant disease. Science 297, 537–541 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Peay K. G., Kennedy P. G., Bruns T. D., Fungal community ecology: A hybrid beast with a molecular master. Bioscience 58, 799–810 (2008). [Google Scholar]
- 4.Fröhlich-Nowoisky J., Pickersgill D. A., Després V. R., Pöschl U., High diversity of fungi in air particulate matter. Proc. Natl. Acad. Sci. U.S.A. 106, 12814–12819 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.DeLeon-Rodriguez N., et al. , Microbiome of the upper troposphere: Species composition and prevalence, effects of tropical storms, and atmospheric implications. Proc. Natl. Acad. Sci. U.S.A. 110, 2575–2580 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ehrlich H. L., “Geomicrobiology: Relative roles of bacteria and fungi as geomicrobial agents” in Fungi in Biogeochemical Cycles , G. M. Gadd, Ed. (Cambridge Core, 2006). 10.1017/CBO9780511550522. (Accessed 11 January 2019). [DOI]
- 7.Fröhlich-Nowoisky J., et al. , Bioaerosols in the Earth system: Climate, health, and ecosystem interactions. Atmos. Res. 182, 346–376 (2016). [Google Scholar]
- 8.Shelton B. G., Kirkland K. H., Flanders W. D., Morris G. K., Profiles of airborne fungi in buildings and outdoor environments in the United States. Appl. Environ. Microbiol. 68, 1743–1753 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jeon E. M., et al. , Impacts of Asian dust events on atmospheric fungal communities. Atmos. Environ. 81, 39–50 (2013). [Google Scholar]
- 10.Fierer N., et al. , Short-term temporal variability in airborne bacterial and fungal populations. Appl. Environ. Microbiol. 74, 200–207 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bowers R. M., et al. , Characterization of airborne microbial communities at a high-elevation site and their potential to act as atmospheric ice nuclei. Appl. Environ. Microbiol. 75, 5121–5130 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barberán A., et al. , Continental-scale distributions of dust-associated bacteria and fungi. Proc. Natl. Acad. Sci. U.S.A. 112, 5756–5761 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woo C., An C., Xu S., Yi S.-M., Yamamoto N., Taxonomic diversity of fungi deposited from the atmosphere. ISME J. 12, 2051–2060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Du P., et al. , Variations of bacteria and fungi in PM2.5 in Beijing, China. Atmos. Environ. 172, 55–64 (2018). [Google Scholar]
- 15.Cáliz J., Triadó-Margarit X., Camarero L., Casamayor E. O., A long-term survey unveils strong seasonal patterns in the airborne microbiome coupled to general and regional atmospheric circulations. Proc. Natl. Acad. Sci. U.S.A. 115, 12229–12234 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keeling C. D., The concentration and isotopic abundances of carbon dioxide in the atmosphere. Tellus 12, 200–203 (1960). [Google Scholar]
- 17.Rhodes J. M., Geochemistry of the 1984 Mauna Loa Eruption: Implications for magma storage and supply. J. Geophys. Res. Solid Earth 93, 4453–4466 (1988). [Google Scholar]
- 18.Ge J. M., Huang J. P., Xu C. P., Qi Y. L., Liu H. Y., Characteristics of Taklimakan dust emission and distribution: A satellite and reanalysis field perspective: Taklimakan dust characteristics. J. Geophys. Res. Atmos. 119, 11,772–11,783 (2014). [Google Scholar]
- 19.Hyslop N. P., Trzepla K., Wallis C. D., Matzoll A. K., White W. H., Technical note: A 23-year record of twice-weekly aerosol composition measurements at Mauna Loa Observatory. Atmos. Environ. 80, 259–263 (2013). [Google Scholar]
- 20.Bodhaine B. A., Aerosol absorption measurements at Barrow, Mauna Loa and the south pole. J. Geophys. Res. 100, 8967–8975 (1995). [Google Scholar]
- 21.Darzi M., Winchester J. W., Aerosol characteristics at Mauna Loa Observatory, Hawaii, after east Asian dust storm episodes. J. Geophys. Res. Oceans 87, 1251–1258 (1982). [Google Scholar]
- 22.Beegum S. N., Lodhi N. K., Singh S., Planetary scale modulations in aerosol properties at Delhi, in the Indo-Gangetic Plain: A quantitative analysis from intra-seasonal to inter-annual timescales. Int. J. Climatol. 36, 3469–3478 (2016). [Google Scholar]
- 23.Li J., Carlson B. E., Lacis A. A., El Niño–Southern Oscillation correlated aerosol Ångström exponent anomaly over the tropical Pacific discovered in satellite measurements. J. Geophys. Res. Atmos. 116, D20204 (2011). [Google Scholar]
- 24.Creamean J. M., et al. , Dust and biological aerosols from the Sahara and Asia influence precipitation in the western U.S. Science 339, 1572–1578 (2013). [DOI] [PubMed] [Google Scholar]
- 25.Kivlin S. N., Winston G. C., Goulden M. L., Treseder K. K., Environmental filtering affects soil fungal community composition more than dispersal limitation at regional scales. Fungal Ecol. 12, 14–25 (2014). [Google Scholar]
- 26.Nguyen N. H., et al. , Ectomycorrhizal fungal diversity and saprotrophic fungal diversity are linked to different tree community attributes in a field-based tree experiment. Mol. Ecol. 25, 4032–4046 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Yeo H.-G., Kim J.-H., SPM and fungal spores in the ambient air of west Korea during the Asian dust (Yellow sand) period. Atmos. Environ. 36, 5437–5442 (2002). [Google Scholar]
- 28.Lymperopoulou D. S., Adams R. I., Lindow S. E., Contribution of vegetation to the microbial composition of nearby outdoor air. Appl. Environ. Microbiol. 82, 3822–3833 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hubbell S. P., The Unified Neutral Theory of Biodiversity and Biogeography (MPB-32) (Princeton University Press, 2001). [Google Scholar]
- 30.Womack A. M., Bohannan B. J. M., Green J. L., Biodiversity and biogeography of the atmosphere. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 3645–3653 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Diehl R. H., The airspace is habitat. Trends Ecol. Evol. 28, 377–379 (2013). [DOI] [PubMed] [Google Scholar]
- 32.Adams R. I., Miletto M., Taylor J. W., Bruns T. D., Dispersal in microbes: Fungi in indoor air are dominated by outdoor air and show dispersal limitation at short distances. ISME J. 7, 1262–1273 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Golan J. J., Pringle A., “Long-distance dispersal of fungi” in The Fungal Kingdom, Heitman J., et al., Eds. (ASM Press, Washington, DC, 2017), pp. 309–333. [Google Scholar]
- 34.Chung W.-H., et al. , Ophiostoma breviusculum sp. nov. (Ophiostomatales, Ascomycota) is a new species in the Ophiostoma piceae complex associated with bark beetles infesting larch in Japan. Mycologia 98, 801–814 (2006). [DOI] [PubMed] [Google Scholar]
- 35.Smith S. E., Read D., “Introduction” in Mycorrhizal Symbiosis, Smith S. E., Read D. J., Eds. (Academic Press, ed. 3, 2008), pp. 1–9. [Google Scholar]
- 36.Chambers S. M., Sharples J. M., Cairney J. W. G., Towards a molecular identification of the Pisonia mycobiont. Mycorrhiza 7, 319–321 (1998). [Google Scholar]
- 37.Hynson N. A., Merckx V. S. F. T., Perry B. A., Treseder K. K., Identities and distributions of the co-invading ectomycorrhizal fungal symbionts of exotic pines in the Hawaiian Islands. Biol. Invasions 15, 2373–2385 (2013). [Google Scholar]
- 38.Hayward J., Hynson N. A., New evidence of ectomycorrhizal fungi in the Hawaiian Islands associated with the endemic host Pisonia sandwicensis (Nyctaginaceae). Fungal Ecol. 12, 62–69 (2014). [Google Scholar]
- 39.Peay K. G., Schubert M. G., Nguyen N. H., Bruns T. D., Measuring ectomycorrhizal fungal dispersal: Macroecological patterns driven by microscopic propagules. Mol. Ecol. 21, 4122–4136 (2012). [DOI] [PubMed] [Google Scholar]
- 40.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 41.Huson D. H., et al. , MEGAN community edition–Interactive exploration and analysis of large-scale microbiome sequencing data. PLoS Comput. Biol. 12, e1004957 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Davis N. M., Proctor D. M., Holmes S. P., Relman D. A., Callahan B. J., Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 226 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nguyen N. H., et al. , FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248 (2016). [Google Scholar]
- 44.Robert V., et al. , MycoBank gearing up for new horizons. IMA Fungus 4, 371–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.R Core Team , R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, 2015). [Google Scholar]
- 46.Oksanen J., et al. , Vegan: Community Ecology Package (R Package Version 2.2-1, 2016).
- 47.Hsieh T. C., Ma K. H., Chao A., iNEXT: an R package for rarefaction and extrapolation of species diversity (Hill numbers). Methods Ecol. Evol. 7, 1451–1456 (2016). [Google Scholar]
- 48.Pohlert T., trend: Non-Parametric Trend Tests and Change-Point Detection (R package Version 0.0.1, 2016).
- 49.Benjamini Y., Hochberg Y., Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57, 289–300 (1995). [Google Scholar]
- 50.Caceres M. D., Legendre P., Associations between species and groups of sites: Indices and statistical inference. Ecology 90, 3566–3574 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Prado P. I., Miranda M. D., Chalom A., sads-package: Maximum Likelihood Models for Species Abundance Distributions (R package Version 0.1, 2014).
- 52.Draxler R. R., Hess G., An Overview of the HYSPLIT _ 4 Modelling System for Trajectories, Dispersion, and Deposition (1998). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All metadata (including weather variables, spore traits, taxonomic assignments, and guilds) are available in the SI Appendix, along with custom scripts used for analyses. Raw sequence data have been deposited in the Sequence Read Archive under the accession PRJNA386517.