Abstract
Myc and p53 proteins are closely associated with many physiological cellular functions, including immune response and lymphocyte survival, and are expressed in the lymphoid organs, which are sites for the development and activation of B-cell malignancies. Genetic alterations and other mechanisms resulting in constitutive activation, rearrangement, or mutation of MYC and TP53 contribute to the development of lymphomas, progression and therapy resistance by gene dysregulation, activation of downstream anti-apoptotic pathways, and unfavorable microenvironment interactions. The cross-talk between the Myc and p53 proteins contributes to the inferior prognosis in many types of B-cell lymphomas. In this review, we present the physiological roles of Myc and p53 proteins, and recent advances in understanding the pathological roles of Myc, p53, and their cross-talk in lymphoid neoplasms. In addition, we highlight clinical trials of novel agents that directly or indirectly inhibit Myc and/or p53 protein functions and their signaling pathways. Although, to date, these trials have failed to overcome drug resistance, the new results have highlighted the clinical efficiency of targeting diverse mechanisms of action with the goal of optimizing novel therapeutic opportunities to eradicate lymphoma cells.
Keywords: B-cell lymphoma, p53, Myc, Molecular mechanisms, Targeted therapy
Introduction
B-cell lymphomas originate from mature B-cells, occurring at many points along with normal B-cell differentiation, including pre-germinal centers (GC), GC, and post GC B cells, which encounter genetic alterations driven by antigens. These alterations include the hierarchy of immunoglobulin (Ig) gene rearrangements and somatic hypermutation of Ig variable region (V) genes, and may cause inactivation of tumor suppressor genes and overexpression of oncogenes that, in turn, result in B-cell transformation. In addition to genetic alterations, microenvironmental factors that deliver positive signals for B-cell survival and growth may contribute to the development and progression of B-cell lymphomas.1 A number of signaling pathways are involved in the initiation and development of B-cell lymphomagenesis. In particular, the functions of several key proteins that represent branching points in the signaling networks are altered as a result of aberrant expression, degradation, and/or accumulation, and these events determine the fate of the affected B-cells. Of these proteins, the most influential transcription factors involved in B-cell lymphomas are Myc and p53, with alterations in these molecules commonly associated with poor prognosis.
Myc aberrations are common in aggressive B-cell lymphoma subtypes. During B-cell lymphomagenesis, oncogenic Myc variants are deregulated via various mechanisms, such as gene translocation, gene amplification, and epigenetic deregulation of expression.2 In most cases, aberrations include translocations or amplifications of the MYC coding region, leading to Myc overexpression and a change in protein function due to aberrations in the amino acid sequence or protein conformation.3 On the other hand, Myc, as a transcription factor, functions as both an activator and a repressor of multiple downstream pathways, promoting proliferation and apoptosis of tumor cells. Myc overexpression adds to the existing oncogenic gene expression profile by enhancing activity of the already active genes in the tumor cells.4 Myc contributes to oncogenic changes and cell transformation; however, its aberration alone is not sufficient to initiate lymphomagenesis. This is consistent with very low or negative Myc protein expression in normal lymphoid cells.
p53 is one of the most important molecules involved in the pathogenesis of cancers, including B-cell lymphomas. Tumor suppression by TP53 occurs via both transcription-dependent and transcription-independent activities. Transcription-dependent activities occur in the nucleus by which p53 regulates transcription of genes involved in the cell cycle, DNA repair, apoptosis, signaling, transcription, and metabolism.5 Transcription-independent activities induce apoptosis and autophagy in the cytoplasm. Mutations in TP53 and dysregulation of the pathway are important in the pathogenesis of many human cancers, including lymphomas. In lymphoid malignancies, the frequency of TP53 deletions and mutations is lower than that in other types of cancers. Nonetheless, the status of TP53 is an independent prognostic factor in most lymphoma types.6
Clinically, each of the Myc or p53 alterations functions as an independent marker of poor prognosis, and alterations in one or the other are detected in a variety of B-cell lymphomas. Notably, lymphomas with co-existent Myc and p53 alterations are synergistic, resulting in more aggressive lymphomas, and patients have a particularly poor prognosis with a short median survival time.7 However, the molecular mechanisms underlying the bidirectional cross-talk between Myc and p53 in B-cell lymphomas have been relatively neglected. Many genes or pathways are involved in the cross-talk between Myc and p53, including Bmi-1, Mel-18, Krueppel-like factor 4 (KLF-4), POXM1, and adenosine diphosphate-ribosylation factor (Arf). Additionally, key microRNAs (miRs) (miR-34a and miR17-92) and the Epstein–Barr virus (EBV) connect the Myc activation to p53, and play a vital role in some B-cell lymphomas, as shown in Table 1. Although identification of the molecular mechanisms between MYC and TP53 is challenging, the results may help to understand how the lymphoma cells escape apoptosis to develop and progress. Understanding these mechanisms will also provide an opportunity to identify new targets and develop novel agents to improve the therapeutic response in patients with various types of lymphomas.
Table 1.
The miRs involved in the cross-talk between p53 and Myc pathways.
| miRs | Functions | Myc | p53 | Types |
|---|---|---|---|---|
| miR-34a | Tumor suppressor | Negative regulation | Positive regulation at transcriptional level; activing TP53 resulting in apoptotic effects mediated by TP53/SIRT1/miR-34a pathway | DLBCL |
| miR-15a/16-1 | Tumor suppressor and diagnostic or prognostic tool | Myc represses miR-15 /16-1 expression through recruitment of HDAC3 | Direct target TP53 in a positive feedback loop | MCL, ALCL |
| miR-17-92 | OncomiRs | Positive regulation at transcriptional level | Repression under hypoxia conditions and at post-transcriptional level | GC-DLBCL, MCL, BL, HCL, FL |
| miR-155 | Tumor suppressor and diagnostic or prognostic tool | Negative regulation at post-transcriptional level | - | DLBCL, MCL, BL, HCL, FL |
| miR-150 | Tumor suppressor | - | Increasing Bim and TP53 | DLBCL, MCL, BL |
| let-7 | Tumor suppressor | Loss of the let-7 (a; c) participates in the genesis and maintenance of the lymphoma phenotype through c-Myc regulation | - | - |
| miR-9 | Tumor suppressor | Positive regulation through direct binding to the miR-9-3 locus | - | DLBCL, BL, HCL, FL |
| miR-26a | - | Negative regulation | - | - |
| miR-29 | - | Negative regulation | - | - |
miR: microRNA; SIRT1: Sirtuin 1; HDAC3: histone deacetylase 3; DLBCL: diffuse large B cell lymphoma; MCL: mantle cell lymphoma; ALCL: anaplastic large cell lymphoma; GC-DLBCL: germinal center - diffuse large B cell lymphoma; BL: Burkitt lymphoma; HCL: hairy cell leukemia; FL: follicular lymphoma; Bim: Bcl-2-interacting mediator of cell death; -: not available.
In this review, we address the functional role and genetic alterations of MYC and TP53 in normal and neoplastic lymphoid cells, the potential clinical impact of these alterations in understanding the clinical and biological heterogeneity of B-cell lymphomas, and the prospects of targeting Myc and p53 as a part of new therapeutic strategies for these lymphomas. Recent advances have greatly enhanced our understanding of MYC and TP53 and have led to new insights into the mechanisms involved in dysregulated gene expression in various subtypes of lymphomas. This has unraveled cellular targets of mechanism-mediated drug resistance and new therapeutic approaches for the treatment of patients with lymphomas.
Myc and P53 function in normal lymphoid tissues
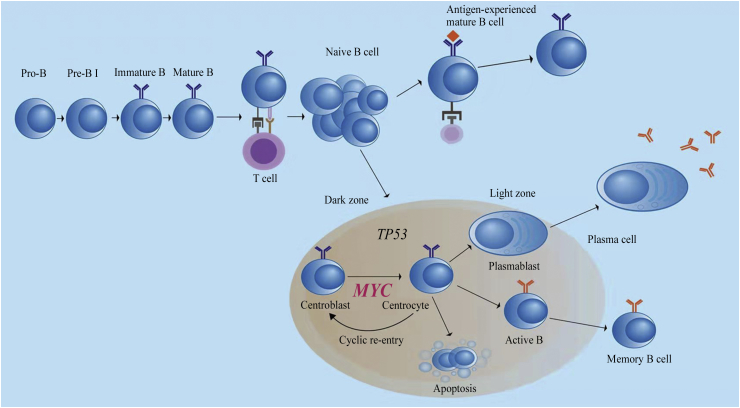
Myc acts as a transcription factor related to numerous physiological functions, and drives the increase in protein synthesis and the cell size of normal pre-transformed B-cells at all stages of B-cell development. Myc is essential and required in the antigen-dependent step of the maturational process of B lymphocytes.8, 9, 10, 11 Antigen naïve B-cells, usually characterized by the expression of surface IgM, encounter specific antigens and are activated with the assistance of T cells, which can trigger the production of the Myc transcription factor and the formation of GC. In the GCs of secondary follicles, B-cells produce the transcription factor B-cell lymphoma 6 (Bcl-6), which suppresses the expression of Myc through binding to the promoter of the MYC gene. In addition, Myc is present in B-cells located in the light zone, acquiring high antigen specificity and ensuring the most appropriate gene antigen affinity, which can suppress Bcl-6 expression and re-express Myc. After interacting with activated T cells, B-cells re-enter the dark zone of the GC to proliferate and undergo a series of divisions. Other B-cells in the light zone of the GC experience a different course of development and differentiation. Normally, these B-cells leave the GCs, do not re-express Myc, downregulate Bcl-6 through the coordinated activity of several signaling pathways, and express B lymphocyte-induced maturation protein-1 (Blimp-1), which further suppresses Myc (Fig. 1). These B-cells become early plasmablasts or eventually memory B cells.2, 9 Notably, Myc is required during the initiation and re-entry into the dark zone from the light zone, whereas it is absent in most GC B-cells.12, 13
Fig. 1.
Physiologic function of MYC and TP53 in normal lymphoid tissues.
p53 mediated critical functions within lymphoid cells including the response to genotoxic stress, differentiation, senescence, and cell death.5, 14 However, unlike Myc, p53 does not act at a specific phase of the B-cell development and differentiation. P53 functions as the guardian of the genome, which is vital for cellular responses to oxidative stresses, and might be a key coordinator in aging and oxidative stress. In spite of a low or high level of oxidative stress, p53 regulates the expression of a panel of genes involved in the cellular response to oxidative stresses. In response to high levels of oxidative stress, p53 shows pro-oxidative activities that further up-regulate the levels of stress, causing cell death. However, at low levels of oxidative stress p53 exhibits antioxidant activities to eliminate oxidative stress and ensures cell survival.15
The pathogenic modes of Myc in B-cell lymphoma
Myc has strong oncogenic potential and participates in lymphomagenesis.2, 8, 16 Translocations of MYC into the IGH or IGK/L loci are present in virtually 100% of Burkitt's lymphoma cases and up to 10%–15% of cases of diffuse large B-cell lymphomas (DLBCL). Myc expression can be detected immunohistochemically in almost 100% of Burkitt's lymphoma and 30%–50% of DLBCL.11, 17 MYC rearrangement, as well as Myc overexpression, affects prognosis, especially in cases of DLBCL with overexpression18 or rearrangement of BCL2. Paradoxically, most lymphomas with MYC alterations originate in cells that usually do not express Myc protein, suggesting that the up-regulation of Myc needs additional oncogenic events to overcome Myc regulatory mechanisms and also its pro-apoptotic functions, such as Bcl-6 in GC cells or Blimp-1 in terminally differentiated B-cells,1 which physiologically repress Myc expression in GC B cells and plasma cells, respectively.
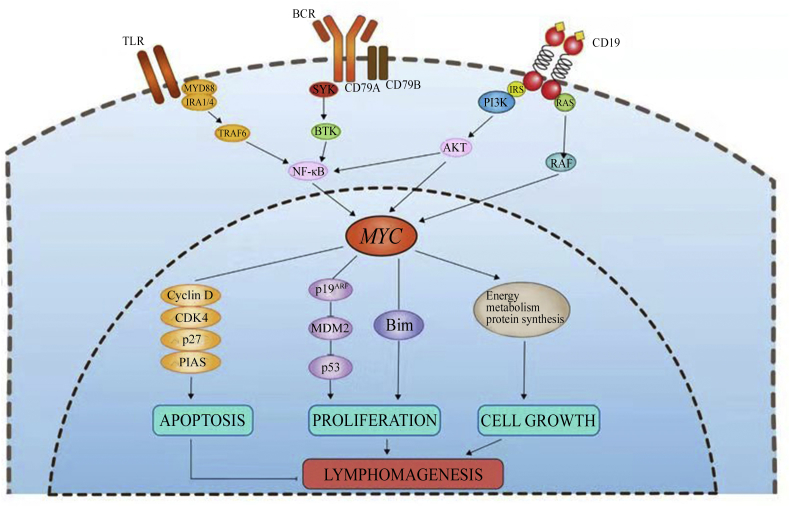
Myc is thought to function as a nonlinear amplifier of expression instead of an on-off specifier of genes, acting universally at active genes.4 B-cell lymphoid malignancies are characterized by an altered signaling of antigen receptors, including, B-cell receptors (BCR), phosphatidylinositol 3′-kinase (PI3K), Toll-like receptors (TLR) and cluster of differentiation 19 (CD19) that are closely associated with Myc18 (Fig. 2). Myc interacting with these targets or environmental antigen receptors provides neoplastic B-cells with growth and/or survival signals that play a vital role in the pathogenesis of lymphomas. In many B-cell lymphomas, Myc is constitutively activated more or less strongly and plays a pathogenic role. To a greater extent, Myc is activated because it functions as the major downstream effector of these pathways, conveying pro-survival and proliferation signals, summarized in the following sections.
Fig. 2.
Role of Myc in B-cell lymphoma pathogenesis. TLR: Toll-like receptors; BCR: B-cell receptors; CD: cluster of differentiation; MYD88: myeloid differentiation factor 88; TRAF: tumor necrosis factor receptor associated factor; SYK: spleen tyrosine kinase; BTK: Bruton's tyrosine kinase; NF-κB: nuclear factor-κB; IRS: insulin receptor substrate; PI3K: phosphatidylinositol 3′-kinase; CDK4: cyclin-dependent kinase 4; PIAS: protein inhibitor of activated signal transducer and activator of transcription; MDM2: mouse double minute 2 homolog; Bim: Bcl-2-interacting mediator of cell death.
The p19ARF–p53 and Myc–Bcl-2-interacting mediator of cell death (Bim) pathways are activated to stimulate proliferation and play a vital role in promoting lymphomagenesis. There are no differences in cell cycle profiles between lymphomas expressing mutant Myc and wild-type, but they both induce lymphoma efficiently. Myc mutants reduce Bim, a BH3-only protein functioning as a member of the Bcl-2 family, to effectively inhibit Bcl-2, leading to abrogation of a parallel apoptotic signal transmitted from Myc to Bim.19 Additionally, myeloid cell leukemia-1 (Mcl-1) has a major role in lymphomas initiating pro-B-cells to oppose Bim, which is upregulated in response to the oncogenic stress.20 Wild-type Myc enforces expression of Bcl-2, or loss of either Bim or p53 function to disrupt the apoptosis of lymphoid cells. On the other hand, wild-type Myc can neutralize Bcl-2 by inducing Bim to trigger apoptosis. In contrast, Myc point mutants fail to induce Bim to evade apoptosis, which promotes lymphomas through escaping both wild-type p53 and p19ARF.21 Another molecular mechanism of MYC involved in the lymphomageneses is through protein inhibitor of activated signal transducer and activator of transcription 1 (STAT 1) (Pias-1) that is often upregulated in B-cell lymphomas. Pias-1 promotes the function of Myc toward the development of mature B-cells, whereas the overexpression of Myc and/or Pias-1 promotes lymphomagenesis, suggesting the possibility of repressing Pias-1 as a therapeutic target22 (Fig. 2).
MiRs interact with Myc acting as a regulatory feedback that can influence the pre- or post-transcriptional expression of multiple genes shown in Table 1. Myc increases expression of both let-7a and miR-34a in DLBCL cells.23 Myc represses several miRs by the recruitment of histone deacetylase (HDAC) inhibitors, including miR15a/16-1, miR26a, miR29, and miR34, which contribute to tumor suppression through deregulation of apoptosis by targeting BCL2 and TP53 (miR15a/16-1 and miR34, respectively), promoting proliferation by targeting CDK6 (miR29a), or cell differentiation by targeting EZH2 (miR26a). On the other hand, Myc itself is also negatively regulated by some miRs, such as miR-34 and miR-494.16 This regulatory loop is complex and still needs elucidation. Further, a positive feedback loop involving miR-26a, miR-29, MYC, and EZH2, exists in both cell lines and primary samples, supporting the presence of a MYC-miR-EZH2 positive feedback loop. Moreover, MYC-miR-26a-EZH2-miR-494 loop and the MYC-EZH2-miR-29 axis have been observed in Myc-expressing lymphoma cell lines and primary lymphoma cells.24
Pathogenic modes and molecular mechanisms of p53 in B-cell lymphoma
TP53 functions as one of the most important genes that regulate the apoptosis. The cellular commitment to apoptosis provides a fundamental benefit to the B-cells, and errors in this process may give rise to lymphoma. Disruption of TP53-dependent apoptosis plays an essential role in the development and progression of lymphoproliferative diseases. Loss of p53 function can result in enhanced rates of cell proliferation, resistance to cell death stimuli, genomic instability, and metastasis.14, 25 TP53 and p53 dysfunction in lymphoid malignancies can occur at the DNA, messenger RNA (mRNA), or protein level. Point mutations and missense mutations are the main mutations in hematologic malignancies, 90% and 79.9% respectively. Many TP53 mutants inhibit wild type (WT)-p53 function acting in a dominant-negative manner. Single allele mutations are frequently followed by loss of heterozygosity, which further induces lymphoma development.26 TP53 mutation acts as a prognosis factor and is linked to worse clinical outcome in patients with DLBCL, chronic lymphocytic leukemia, splenic marginal zone lymphoma, and mantle cell lymphoma.27, 28, 29, 30 Moreover, small TP53 mutated subclones are associated with a worse prognosis and become predominant under conventional treatment, ultimately causing treatment refractoriness,31 suggesting that increasing the frequency of TP53 mutations in lymphomas increases their clinical aggressiveness and the frequency of these neoplasms being refractory to therapy.
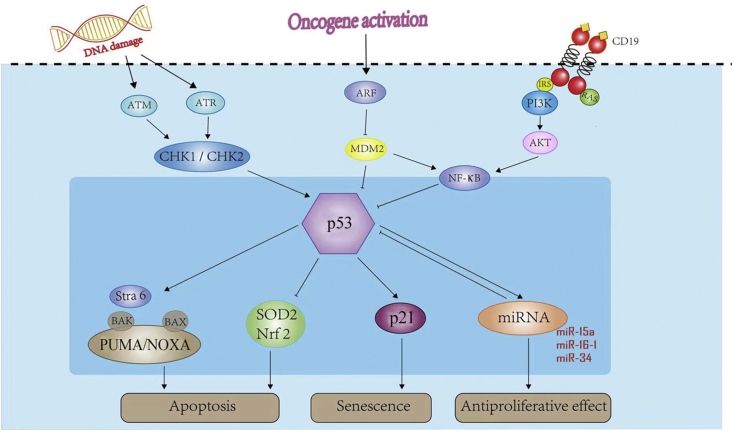
P53 can be activated by oncogenic stress and DNA damage32 by two distinct signaling pathways that involve the kinase mediated phosphorylation of p53 by inhibition of mouse double minute 2 homolog (MDM2) and via p19ARF and the cascade of ATM/CHK2/1, causing p53 protein stabilization, respectively.33, 34, 35 MDM2, an E3 ubiquitin ligase, is the most important negative regulator of p53. By binding directly to p53, MDM2 mediates ubiquitination-dependent degradation of p53, and by ubiquitinating itself, MDM2 targets itself for destruction and promotes the p53 tumor suppressor pathway.25, 34, 36, 37 Heterodimerization with mouse double minute 4 homolog (MDM4) enhances the E3 activity of MDM2 toward p53, whereas MDM4 can also inhibit the E3 ligase activity of MDM2 under some circumstances. Through the direct binding to p53 at its trans-activation domain, MDM4 represses p53 transcriptional activity. Moreover, MDM4 can downregulate the stability of p53 by promoting MDM2-mediated degradation.37 This regulatory mechanism involved in lymphomagenesis has been illustrated (Fig. 3).
Fig. 3.
Role of p53 in B-cell lymphoma pathogenesis. ATM: ataxia-telangiectasia mutated; ATR: ataxia telangiectasia and Rad3-related protein; CHK: checkpoint kinase; ARF: adenosine diphosphate-ribosylation factor; MDM2: mouse double minute 2 homolog; IRS: insulin receptor substrate; PI3K: phosphatidylinositol 3′-kinase; NF-κB: nuclear factor-κB; Stra6: stimulated by retinoic acid 6; BAK: B-cell lymphoma-2 homologous antagonist/killer; BAX: B-cell lymphoma-2-associated X protein; Puma: p53-upregulated modulator of apoptosis; SOD2: superoxide dismutase 2; Nrf2: nuclear factor erythroid 2-related factor 2; miRNA: micro RNA.
TP53, acting as one of the key tumor suppressor genes, induces activation of the intrinsic apoptotic pathway, and the p53 tumor suppressor is one of the first lines of defense against the effects of metabolic changes, hypoxia, genotoxic damage, and oncogene activation.14, 15, 38 Upon transcriptional induction of genes encoding antioxidant and anti-apoptotic proteins, p53, downregulated by nuclear factor-κB (NF-κB), blocks cell apoptosis along the apoptotic pathways.38 Briefly, p53 up-regulates NOXA and p53-upregulated modulator of apoptosis (PUMA; also known as Bcl-2-binding component 3 [BBC3]) which are two proteins of the Bcl-2 family. Next, NOXA and PUMA interact with Bcl-2-associated X protein (Bax) and Bcl-2 homologous antagonist/killer (Bak) at the mitochondrial site, and form the signaling complex or apoptosome to initiate apoptosis, followed by activation of the effector caspases-3, -6 and -7 to complete apoptosis. Both p53-induced apoptosis and an increase of radical oxygen species (ROS) are strongly inhibited by the absence of Bax or PUMA.39 DNA damage upregulates stimulated by retinoic acid 6 (Stra6) in a p53-dependent manner and has a critical role in cell death responses. Stra6 expression promotes significant amounts of apoptosis in normal and cancer cells, and influences p53-mediated cell fate decisions by turning an initial arrest response into cell death,39 shown in Fig. 3.
The pro-oxidative activities of p53 increase cellular oxidative stresses to induce apoptosis through inhibition of the expression of antioxidant genes. P53 can inhibit expression of superoxide dismutase 2 (SOD2)40 and nuclear factor erythroid 2-related factor 2 (Nrf2),41 resulting in the induction of apoptosis or enhanced sensitivity to oxidative stress.15 P53 and manganese-dependent superoxide dismutase (MnSOD) act as sensors of the cellular responses to stress and one of the main anti-oxidant enzymes, and are involved in these processes. P53 could repress SOD2 gene expression at the promoter level whereas overexpression of MnSOD decreases p53-mediated induction of apoptosis.40 Additionally, p53 induces cell cycle arrest via p21, inhibiting proliferation and serving as one of the major transcriptional targets of p53,42, 43 and p21WAF1/Cip1 regulates cell proliferation by inhibiting the activity of cyclin-dependent kinase (CDK) complexes at the G1/S and G2/M cell cycle checkpoints.44
MiRs are also key factors to maintain the normal TP53 pathway, joining forces with MDM2 and MDM4 to guarantee proper p53 activity,45 as shown in Table 1. The dysregulation of these miRs is associated with poor clinical outcomes.46 For example, miRs transactivated by p53 in lymphocytes include miR-15a, miR-16-1 which targets oncogene MYB, antiapoptotic Bcl-2 and Mcl-1,47 and enhances expression of the LEU2 gene,48 a locus deleted in 55% of chronic lymphocytic leukemia (CLL) cases (del13q14). Among the miRs, the miR-34 family (miR-34a, b, c), directly targets p53 transcription and plays a vital role in the TP53 network involved in apoptosis, cell cycle arrest, and senescence, and its loss can cause resistance to p53-mediated apoptosis.46, 49, 50 MiR-34b and miR-34c, targeting Zeta-chain-associated protein kinase 70 (ZAP70), are deleted in 18% of CLL cases (del11q).47 MiR-34a targets FOXP1, BCL2, and can block B-cell development when overexpressed,51 whereas reduced miR-34a expression has been correlated with inferior overall survival (OS) in patients with DLBCL52 and significantly shorter treatment-free survival in CLL patients.53 The regulation of Sirtuin 1 (SIRT1) by miR-34a is part of a positive feedback loop. SIRT1 can deacetylate and inactivate p53 resulting in impaired cell growth arrest and apoptosis in response to oxidative stress, whereas TP53 upregulates miR-34 expression that, in turn, promotes p53 by inhibiting SIRT1,50, 51, 54 suggesting that the precise gene dose regulation by miRs has vital regulatory roles in the pathogenesis of B-cell lymphomas (Fig. 3).
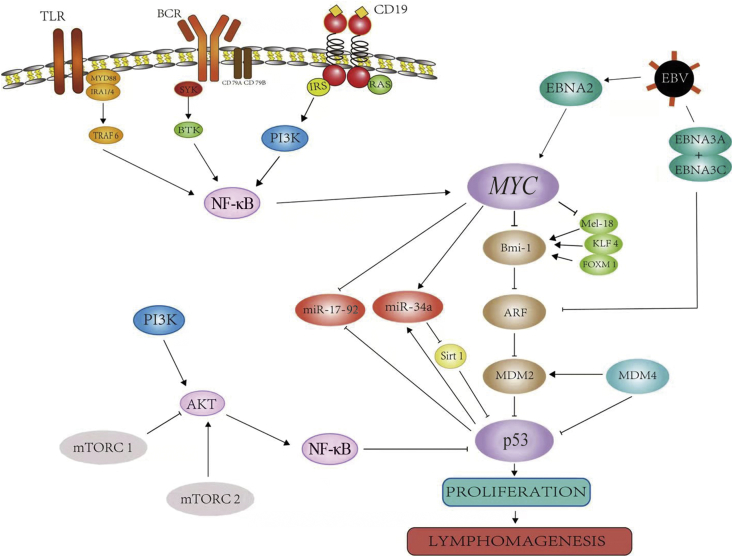
Fig. 4.
Cross-talk between p53 and Myc pathways. TLR; Toll-like receptors; MYD88: myeloid differentiation factor 88; TRAF6: tumor necrosis factor receptor associated factor 6; BCR: B-cell receptors; SYK: spleen tyrosine kinase; BTK: Bruton's tyrosine kinase; CD: cluster of differentiation; IRS: insulin receptor substrate; PI3K: phosphatidylinositol 3′-kinase; NF-κB: nuclear factor-κB; EBNA: Epstein–Barr virus nuclear antigen; EBV: Epstein–Barr virus; KLF4: Krueppel-like factor 4; FOXM1: forkhead box protein M1; miR: micro RNA; ARF: adenosine diphosphate-ribosylation factor; MDM2: mouse double minute 2 homolog; MDM4: mouse double minute 4 homolog; Sirt1: Sirtuin 1; mTORC1: mammalian target of rapamycin complex 1; mTORC2: mammalian target of rapamycin complex 2.
The cross-talk mechanism between Myc and p53
Clinically, MYC-rearrangement (MYC-R), Myc expression, loss of TP53, TP53 mutations, and high expression of p53 are associated with lymphomas that arise from GC B cells. Expression of Myc or p53 is often higher in clinically aggressive non-Hodgkin lymphoma (NHLs).55, 56 Although expression of Myc or P53 individually has been shown to contribute to poor survival in DLBCL patients, the role of MYC-R or Myc expression associated TP53 genetic alterations or p53 expression has been relatively neglected.
Concurrent Myc and p53 expression impacts the prognosis of patients with B-cell lymphoma.35 MYC-R or p53 expression alone are independent prognostic factors for DLBCL patients, whereas those with concurrent p53 expression and MYC-R display a worse prognosis. Ye et al57 and Xie et al58 have reported that Myc expression is similarly distributed between the GC B cell-like (GCB) and activated B cell-like (ABC) subtypes; however, the GCB-DLBCL patients with high Myc expression had a higher frequency of TP53 mutation.59 In other studies, patients with DLBCL without Myc and p53 expression had the best overall survival. In contrast, patients with DLBCL harboring MYC-R or Myc expression and p53 expression had significantly worse OS, regardless of Bcl-2 expression status,7, 60, 61 suggesting that p53 enhances the negative prognostic effect of Myc expression in DLBCL patients.7, 62 Theoretically, dysregulation of Myc and p53 promotes tumor proliferation and apoptosis, and has a synergistic effect on tumor progression and resistance to chemotherapy, similar to the recent reports on double expressor DLBCL with Myc and Bcl-2 co-expression. Data regarding Myc and p53 being involved in bidirectional cross-talk in B-cell lymphomas are summarized in Fig. 4 and Table 2.
Table 2.
Expression and prognoses of p53 and Myc in GCB and non-GCB subtype of diffuse large B-cell lymphoma.
| Items | Authors/published time | p53 and/or Myc expression (cut-off value) | Total, positive/evaluated, n (%) | GCB, positive/evaluated, n (%) | Non-GCB, positive/evaluated, n (%) | OS, months | |
|---|---|---|---|---|---|---|---|
| p53 | Xie et al/201458 | p53 (none) | 8/85 (9.4) | 4/46 (8.7) | 4/39 (10.3) | 88 ± 12a | |
| p53− (<30%) | 42/85 (49.4) | 22/46 (47.8) | 20/39 (51.3) | 57 ± 13a | |||
| p53+ (≥30%) | 27/85 (31.8) | 17/46 (37.0) | 10/39 (25.6) | 74 ± 9a | |||
| p53 (diffuse) | 8/85 (9.4) | 3/46 (6.5) | 5/39 (12.8) | 50 ± 18a | |||
| Wang et al/20177 | p53+ (≥50%) | 67/201 (33.3) | NA | NA | 40b | ||
| p53− (<50%) | 134/201 (66.7) | NA | NA | 110b | |||
| Myc | Ye et al/201657 | Myc+ (>70%) | 249/825 (30.2) | 121/430 (28.1) | 127/390 (32.6) | NA | |
| Xu-Monette et al/201559 | Myc+ (>70%) | 175/535 (32.7) | 76/272 (27.9) | 98/259 (37.8) | NA | ||
| Myc− (<70%) | 360/535 (67.3) | 196/272 (72.1) | 161/259 (62.2) | NA | |||
| Xie et al/201458 | Myc+ (≥40%) | 23/85 (27.1) | 9/46 (19.6) | 14/39 (35.9) | 57 ± 11a | ||
| Myc− (<40%) | 62/85 (72.9) | 37/46 (80.4) | 25/39 (64.1) | 69 ± 9a | |||
| Myc/p53 | Wang et al/20177 | p53+ (≥50%), MYC-R | 23/67 (33.8) | NA | NA | 7.4b | |
| p53+ (≥50%), Myc+ (≥40%), | 47/67 (70.1) | NA | NA | 20b | |||
| p53− (<50%), MYC-R | 33/134 (24.6) | NA | NA | 67b | |||
| p53− (<50%), Myc+ (≥40%), | 59/125 (47.2) | NA | NA | 67b | |||
| Myc+, p53+ | Xu-Monette et al/201559 | Myc+ (>70%), TP53 (MUT) | 39/473 (8.2) | 23/239 (9.6) | 16/231 (6.9) | NA | |
| Xie et al/201458 | Myc+ (>40%), p53+ (>30%) | 16/85 (18.8) | 4/46 (8.7) | 12/39 (30.8) | NA | ||
| Myc+, p53− | Xu-Monette et al/201559 | Myc+ (>70%), TP53 (WT) | 114/473 (24.1) | 43/239 (18.0) | 71/231 (30.7) | NA | |
| Xie et al/201458 | Myc+ (>40%), p53− (<30%) | 7/85 (8.2) | 5/46 (10.9) | 2/39 (5.1) | NA | ||
| Myc−, p53+ | Xu-Monette et al/201559 | Myc− (<70%), TP53 (MUT) | 68/473 (14.4) | 42/239 (17.6) | 26/231 (11.3) | NA | |
| Xie et al/201458 | Myc− (<40%), p53+ (>30%) | 19/85 (22.4) | 16/46 (34.8) | 3/39 (7.7) | NA | ||
| Myc−, p53− | Xu-Monette et al/201559 | Myc− (<70%), TP53 (WT) | 252/473 (53.3) | 131/239 (54.8) | 118/231 (51.1) | NA | |
| Xie et al/201458 | Myc− (<40%), p53− (<30%) | 43/85 (50.6) | 21/46 (45.7) | 22/39 (56.4) | NA | ||
GCB: germinal center diffuse large B cell lymphoma; Non-GCB: non-germinal center diffuse large B cell lymphoma; MYC-R: MYC-rearrangement; MUT: mutation; WT: wild type; OS: overall survival; NA: not available.
Mean ± SE. SE: standard error.
Median.
The first cross-talk mechanism lies in the ability of Myc to inhibit Bmi-1 and Mel-18, and KLF-4 and POXM1 (which regulate Bmi-1); Bmi-1 inhibits Arf. Mounting evidence suggests that Arf has critical p53-independent tumor-suppressor functions, supported by the findings that TP53 and CDKN2A are frequently and simultaneously inactivated in human cancers.63 Moreover, p19ARF downregulates MDM2 and Arf-BP1, which mediates the ubiquitin-dependent degradation of p53, resulting in the accumulation of p53 protein involved in cell cycle arrest, apoptosis, or senescence. Additionally, Bmi-1 serves a key role in this pathway from Myc to p53, and its biological functions including development, cell cycle, DNA damage response, senescence, stem cells, self-renewal, and cancer. In Bmi-1-deficient mice, expression of Ink4a and Arf is greatly increased in the hematopoietic cells, leading to the cell cycle arrest and p53-dependent apoptosis, respectively.64 Bmi-1 collaborates with Myc in inducing proliferation and transformation of primary embryo fibroblasts in an Ink4a–ARF dependent manner, which is associated with prohibiting Myc-mediated induction of p19ARF and apoptosis.65, 66, 67, 68 Bmi-1 can also directly mediate the stability of p53, further negatively regulating cellular proliferation and tumorigenesis through the retinoblastoma protein (pRb)-p53 pathway.67, 69 Meanwhile, Bmi-1 is positively mediated by forkhead box protein M1 (Foxm-1), E2F transcription factor 1 (E2F-1), and Sal-like protein 4 (Sall-4), whereas KLF4, Mel-18, and HDAC inhibitors suppress the expression at the transcriptional level. In addition, Mel-18 reduces Bmi-1 expression by inhibiting Myc, forming a positive regulator of Bmi-1.70
MiR-34a and miR17-92 are the key miRs that connect Myc activation to p53 and play a vital pathogenic role in some B-cell lymphomas. miR-34a is inhibited by Myc and is transcriptionally activated by p53.45, 46 MiR-34a is reported to have tumor-suppressive activity in DLBCL cell lines23 and its expression is mediated by Myc, or through mechanisms that may involve epigenetic silencing of the miR-34a promoter or chromosomal deletion of the miR-34a genomic locus.71, 72, 73 MiR-34a is also closely associated with TP53 through a feedback loop in which TP53 promotes miR-34 expression that, in turn, activates p53 through SIRT1 inhibition. Recently, miR-34a was shown to also target MDM4.45, 74 On the other hand, although most miRs directly regulated by Myc are usually repressed, Myc up-regulates the oncogenic miR17-92 cluster. The miR17-92 polycistron at 13q31 is commonly amplified in several subtypes of aggressive lymphomas, and its oncogenic function is regulated partly by the down-regulation of TP53, PTEN, and E2F1, facilitating the activation of the PI3K/AKT (protein kinase B, PKB) pathway and apoptosis inhibition, respectively.
EBV, a ubiquitous herpes virus, has a tendency to preferentially infect B lymphocytes and plays an important role in some EBV-associated B-cell lymphomas, in which Epstein–Barr viral nuclear antigen 2 (EBNA2) regulates Myc positively whereas EBNA3A and EBNA3C repress Arf, which forms a regulatory loop among EBV, Myc and p53.75, 76 EBV latent antigens (EBNA1, EBNA2 and EBNA3C) are essential for in vitro B-cell immortalization leading to the continuously proliferating lymphoblastoid cell lines (LCLs). DNA damage of LCLs knocked down for EBNA3C experienced a drastic promotion of apoptosis, as a possible outcome of both p53- and E2F1-regulated activities. Mechanistically, EBNA3C inhibits E2F1 transcriptional activity via blocking its DNA binding activity at the responsive promoters of p73 and apoptotic protease activating factor 1 (Apaf-1) apoptosis inducing genes, and also facilitates E2F1 degradation in an ubiquitin-proteasome dependent fashion.77
These results suggest that TP53 and MYC mutations are present in all types of lymphomas, which may help the tumor cells to escape the apoptotic effects of Myc; however, the details of the cross-talk mechanisms between Myc and p53 remain unclear. Through the cross-talk between Myc and p53, a number of genes and pathways are intimately connected in the development and progression of neoplastic B-cells, which participate in the construction of a complex biological network in B-cell lymphomas. Identification of the molecular mechanism between Myc and p53 is difficult as a large number of genes and pathways can be influenced by various factors. However, elucidating these mechanisms will greatly enhance our understanding of Myc and p53, and will lead to new insights into the mechanisms involved in the dysregulated gene expression in various types of B-cell lymphomas, and lead to the development of cellular targeting agents to overcome drug resistance in patients with lymphomas.
Therapeutic approaches and advancements with Myc and p53
Myc and p53 are widely present in normal and tumor cells, and play important roles in lymphomagenesis. However, the use of Myc and p53 inhibitors affects the cycle of normal cells, often clinically, resulting in adverse side effects in the patients. Although it is predicted that long-term treatment of lymphoma patients with Myc and/or p53 inhibitors eventually impairs the immune function, short-term administration of such agents might be possible and manageable. Technically, Myc and p53 can be inhibited by targeting the Myc components directly or indirectly by inhibiting the upstream signaling pathway components, and Myc inhibition also interrupts p53 as a result of being downregulated by Myc. Therefore, combining Myc or p53 inhibitors with cytotoxic chemotherapeutic drugs is a rational and logical therapeutic strategy that is currently being evaluated in trials. Therefore, optimization of the combination and personalized approaches is important and needs to be well defined. Here, we summarize the current research directions for targeted therapy of Myc and p53 pathways, in Fig. 5 and Table 3.
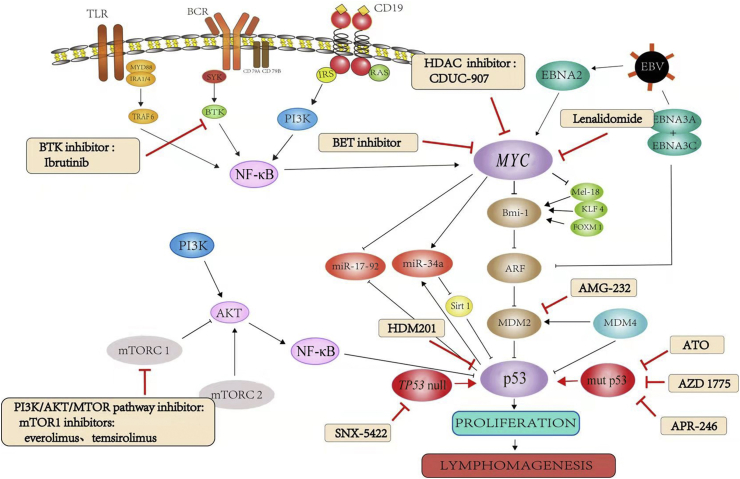
Fig. 5.
Functional mechanisms of novel drugs on Myc and p53 pathways. TLR: Toll-like receptors; BCR: B-cell receptors; CD: cluster of differentiation; MYD88: myeloid differentiation factor 88; TRAF6: tumor necrosis factor receptor associated factor 6; SYK: spleen tyrosine kinase; BTK: Bruton's tyrosine kinase; IRS: insulin receptor substrate; PI3K: phosphatidylinositol 3′-kinase; NF-κB: nuclear factor-ΚB; HDAC: Histone deacetylase; BET: bromodomain and extra-terminal domain; EBNA: Epstein–Barr viral nuclear antigen; Krueppel-like factor 4; FOXM1: forkhead box protein M1; ARF: adenosine diphosphate-ribosylation factor; miR: micro RNA; Sirt1: Sirtuin 1; MDM2: mouse double minute 2 homolog; MDM4: mouse double minute 4 homolog; mTOR: mammalian target of rapamycin; mTORC; mTOR complex.
Table 3.
Ongoing clinical trials of novel agents in patients with different types of cancers with TP53.
| Drug | Targeting gene | Clinical trials | Number | First report time—last report time | Status |
|---|---|---|---|---|---|
| HDM201 | TP53 | To determine and evaluate a safe and tolerated dose of HDM201 in adult patients with selected advanced tumors characterized by wild-type TP53 | NCT02143635 | May 21, 2014–February 23, 2018 | Recruiting |
| SNX-5422 | TP53 | A Single Arm Study of SNX-5422 in Subjects With TP53 Null Cancers | NCT02612285 | November 23, 2015–November 4, 2016 | Terminated |
| AZD-1775 | TP53 | Phase II, Single-arm Study of AZD1775 Monotherapy in Relapsed Small Cell Lung Cancer Patients With MYC Family Amplification or CDKN2A Mutation Combined With TP53 Mutation | NCT02688907 | February 23, 2016–February 7, 2018 | Recruiting |
| AMG-232 | TP53, MDM2 | MDM2 Inhibitor AMG-232 in Treating Patients With Recurrent or Newly Diagnosed Glioblastoma | NCT03107780 | April 11, 2017–March 5, 2018 | Recruiting |
| Anti-p53 protein | TP53 | Phase II Study of Metastatic Cancer That Overexpresses P53 Using Lymphodepleting Conditioning Followed by Infusion of Anti-P53 TCR-Gene Engineered Lymphocytes | NCT00393029 | October 26, 2006–August 15, 2011 | Completed |
MDM2: mouse double minute 2 homolog.
Opportunities for MYC inhibition
Targeting Myc might be an alternative strategy for treating lymphoma patients. HDAC inhibitors are able to reduce the expression of Myc,24, 78 whereas PI3K inhibitors can decrease Myc family protein stability by disrupting their regulation at the post-transcriptional level.79, 80 CUDC-907, an oral small selective molecular inhibitor of both HDAC (class I and II) and PI3K (class Iα, β, and δ) enzymes, has shown a promising outcome with rituximab in the treatment of patients with relapsed and refractory (R/R) DLBCL.81, 82, 83 In a phase I trial, CUDC-907 was used in R/R DLBCL including patients with MYC-alterations, and the results showed an objective response rate in the evaluable patients of 64% (7 of 11; 4 complete responses and 3 partial responses), compared with 29% (2 of 7) in patients with MYC unaltered DLBCL. The median duration of response was 13.6 months in MYC-altered DLBCL versus 6 months in MYC unaltered DLBCL patients. The median progression-free survival (PFS) was 21.8 months (range, 1.0–25.4 months) for MYC-altered DLBCL patients, with a median PFS of 21.8 months (range, 1.0–16.4 months) for patients treated with monotherapy, and was not reached in patients treated with rituximab combined with CUDC-907. In comparison, the median PFS in MYC negative DLBCL patients was 1.3 months (range, 0.4–15.5 months). The most frequently reported grade ≥3 treatment-related events were neutropenia, diarrhea, fatigue, thrombocytopenia, and anemia,81 suggesting that CUDC-907 is well tolerated and long-lasting stabilization of the disease can be obtained in MYC-altered, R/R DLBCL patients, as shown in Fig. 5.
Other potential therapeutic targets of Myc include bromodomain-containing protein 4 (Brd4) and runt-related transcription factor (Runx). Brd4 is essential for MYC transcription to address its regulatory function, and is a member of the bromodomain and extra-terminal domain (BET) subfamily of proteins. Delmore et al68, 84 used JQ1, a small-molecule inhibitor of BET bromodomains, to selectively downregulate MYC transcription in the treatment of human multiple myeloma cells. In myeloma models of MYC-dependent hematologic malignancy, JQ1 shows an active anti-proliferative effect related to cell cycle arrest and cellular senescence. Overall survival was increased in JQ1 treated mice compared to the vehicle control. These studies offer a challenge to treat lymphomas with high Myc expression, and additional clinical trials are needed to better address the role of CUDC-907 in targeting Myc (Fig. 5).
Lenalidomide functions as an immunomodulatory drug, but can also repress Myc and its target genes and proteins in plasma cells harboring MYC rearrangement. In a human tumor xenograft model, interferon regulatory factor 4 (IRF4) was the target gene of the tumoricidal effect of lenalidomide, as the upregulation of IRF4, a hallmark of ABC subtype DLBCL, was associated with high Myc expression. Lenalidomide decreases IRF4 levels in multiple myeloma cell lines and bone marrow samples with high levels of Myc85, 86, 87, 88(Fig. 5). A recent trial showed that lenalidomide combined with rituximab, cyclophosphamide, doxorubicin, vincristine and prednisone (R-CHOP) appears to be particularly beneficial in non-GCB DLBCL, including Myc-positive DLBCL and high-grade B-cell lymphomas (features intermediate between DLBCL and Burkitt's lymphoma with a poor prognosis), showing no difference in the 24-month PFS or OS on the basis of non-GCB and GCB subtype (60% vs. 59% [P = 0.83] and 83% vs. 75% [P = 0.61] at 2 years, respectively). This trial will be one of the first trials to offer up-front adjusted treatment for Myc-positive DLBCL patients, with probably better treatment outcomes in these patients.88 Lenalidomide is synergistic with R-CHOP and overcomes high-risk Myc-positive DLBCL with a poor prognosis, providing valuable insights into the synergistic effect, and this combination is a potential treatment for patients with R/R B-cell lymphomas.
Opportunities for p53 inhibition
Ibrutinib, a first-in-class, oral, irreversible inhibitor of Bruton's tyrosine kinase (BTK), a cytoplasmic signaling molecule situated downstream of the BCR, has shown promising results in the treatment of patients with B-cell malignancies89 (Fig. 5). In a phase II, single-arm study, single-agent ibrutinib was used to treat 51 patients with previously untreated (n = 35) and R/R (n = 16) high-risk CLL with TP53 aberrations.90 This study showed persistent responses in patients with CLL with TP53 aberrations, especially in patients with previously untreated disease. This response to ibrutinib was equivalent to the response in TP53 wild-type CLL cells, suggesting a p53-independent mechanism of action. Furthermore, histological transformation rates of CLL appeared to be lower than in CLL patients treated with conventional chemotherapy when used first line, emphasizing the role of up-front ibrutinib in patients with TP53 aberrations.91 Moreover, in another single-arm phase Ib/II study, the PFS of all patients with TP53 mutation was 78% with an OS of 83.8% at 18 months.91, 92
Other studies support many novel agents that may have potential as therapy for B-cell lymphomas with TP53 mutation.93 Targeting the p53/p21 axis might be an alternative treatment strategy. RO-3306, a cyclin-dependent kinase 1 (CDK1) inhibitor, is able to modulate the balance of p53 functions, which blocks cell-cycle arrest by inhibiting p53-mediated activation of p21, and enhances apoptosis by reducing levels of antiapoptotic Bcl-2, survivin, and MDM2 in acute myeloid leukemia (AML) cell lines with WT-p53 (Fig. 5). X-box binding protein 1 (XBP1) functions as a p53/p21 axis regulator through MDM2 in many cancers regardless of the p53 status (wild-type or mutant). Further, XBP1 suppression induces G0-G1 phase arrest and represses cell proliferation by negatively regulating the p53/p21 axis and enhancing p53 ubiquitination, which in turn down-regulates p21 expression94 as shown in Fig. 5. However, no clinical trials have been performed using inhibitors of the p53/p21 axis in patients with B-cell lymphomas, and additional clinical trials are needed to better address the role of these agents to target p53 in B-cell lymphomas.6, 95
Rapamycin, which targets mammalian target of rapamycin (mTOR), is an important mediator of intracellular signaling in B cells. Rapamycin and other rapalogs, including everolimus and temsirolimus, inhibit the activity of mTOR complex 1 (mTORC1) by binding to mTOR, and it has been verified in vivo that everolimus can downregulate AKT activity in hematopoietic cells96 (Fig. 5). Further, everolimus has been shown to have clinical activity in a variety of B cell lymphomas including CLL with TP53 deletion/mutation.97 R/R CLL patients with TP53 disruption or purine analogue refractory disease had a short survival prior to the recent introduction of BCR and Bcl-2 inhibitor therapy. Clinical trials illustrated that alemtuzumab (anti-CD52 antibody) combined with high dose methylprednisolone therapy used in 22 R/R patients with CLL and TP53 deletion/mutation achieved an objective response rate (ORR) of 77%, median PFS of 6.5 months and median OS of 19.5 months. In this study, everolimus and alemtuzumab reportedly had ORR rates and PFS similar to those of alemtuzumab monotherapy, but with a longer median OS.98 These data highlight that combined later generation mTOR inhibitors with less toxic B cell targeted drugs, such as anti-CD20 monoclonal antibodies or small molecule inhibitors, could provide a direction in designing future clinical trials in CLL or other B cell lymphomas.97
Other agents or small molecular inhibitors have been studied. For example, arsenic trioxide (ATO) has been found to induce expression of Pirh2 E3 ligase at the transcriptional level, degrading mutant p53 for polyubiquitination and subsequent proteasome degradation (Fig. 5). Moreover, ATO can cooperate with heat shock protein 90 (HSP90) or HDAC inhibitors, as well as induce the Pirh2-dependent proteasome pathway to promote mutant p53 degradation and growth suppression in tumor cells.99 Another target, signal transducer and activator of transcription 1 (STAT1), acts as an oncogene in patients with high-risk DLBCL with an active host inflammatory response; targeting STAT1 mediated by BAL1 might be a strategy to increase the sensitivity of DLBCL towards classic therapy.100 In all, these outcomes could pave the way for developing novel therapeutic strategies for clinical trials for the treatment of p53-associated B-cell lymphomas.
Concluding remarks
Both Myc and p53 are closely associated with physiological cellular functions, including immune response and lymphocyte survival, and are expressed in lymphoid organs in which the development and activation of B-cell malignancies occurs. However, Myc and p53 also have an anti-apoptotic role and the activation of these pathways is a common feature in various malignancies, independent of the cell of origin. Aberrant or mutant MYC contributes to lymphomagenesis by interacting with lymphoid cells or environmental antigen receptors, providing neoplastic B-cells with growth and/or survival signals, but MYC aberrations alone are insufficient to induce lymphomagenesis. TP53 dysfunction can result in enhanced rates of cell proliferation, resistance to cell death stimuli, genomic instability, and metastasis. As previously known, many genes and pathways are involved in the process of lymphoma transformation, constituting the regulating molecular web. For these reasons, the B-cell lymphomas function as preferential models for the action of Myc and p53; the cross-talk between Myc and p53 contributes to an inferior prognosis for patients with many B-cell neoplasms. Further knowledge of the cross-talk mechanisms is essential to better define the relationship and effect in each B-cell lymphoma subtype.
MYC and TP53 are an Achilles heel in many B-cell lymphomas, and therapies targeting these pathways have preliminarily shown encouraging therapeutic effects with minimal toxicity in patients with these diseases. Therefore, new agents designed to specifically target the MYC and TP53 pathways are being studied and developed. However, these agents, used as monotherapy or in combination with chemotherapy, have failed to achieve a favorable outcome and overcome tumor resistance. Further studies with the multitude of available agents are needed to create diverse possibilities for targeted therapy, and the outcomes will improve our understanding of the mechanisms of Myc and p53 in each type of B-cell lymphoma, as well as provide an opportunity to improve the therapeutic response for various types of lymphomas. Moreover, the goal in treating patients with B-cell lymphoma should be to combine Myc and p53 targeted inhibitors rationally and synergistically block several pathways, inducing apoptosis, and hopefully achieving efficacious treatment with less toxicity.
Conflicts of interest
All authors declare no conflicts of interest.
Acknowledgments
This work was supported by the National Natural Science Foundation of China 81460030 and 81770221 to LY.
Edited by Pei-Fang Wei
Footnotes
Peer review under responsibility of Chinese Medical Association.
References
- 1.Rui L., Goodnow C.C. Lymphoma and the control of B cell growth and differentiation. Curr Mol Med. 2006;6:291–308. doi: 10.2174/156652406776894563. [DOI] [PubMed] [Google Scholar]
- 2.Korać P., Dotlić S., Matulić M., Zajc Petranović M., Dominis M. Role of MYC in B cell lymphomagenesis. Genes (Basel) 2017;8:E115. doi: 10.3390/genes8040115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Karube K., Campo E. MYC alterations in diffuse large B-cell lymphomas. Semin Hematol. 2015;52:97–106. doi: 10.1053/j.seminhematol.2015.01.009. [DOI] [PubMed] [Google Scholar]
- 4.Nie Z., Hu G., Wei G. c-Myc is a universal amplifier of expressed genes in lymphocytes and embryonic stem cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stiewe T. The p53 family in differentiation and tumorigenesis. Nat Rev Cancer. 2007;7:165–168. doi: 10.1038/nrc2072. [DOI] [PubMed] [Google Scholar]
- 6.Xu-Monette Z.Y., Medeiros L.J., Li Y. Dysfunction of the TP53 tumor suppressor gene in lymphoid malignancies. Blood. 2012;119:3668–3683. doi: 10.1182/blood-2011-11-366062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X.J., Medeiros L.J., Bueso-Ramos C.E. P53 expression correlates with poorer survival and augments the negative prognostic effect of MYC rearrangement, expression or concurrent MYC/BCL2 expression in diffuse large B-cell lymphoma. Mod Pathol. 2017;30:194–203. doi: 10.1038/modpathol.2016.178. [DOI] [PubMed] [Google Scholar]
- 8.Calado D.P., Sasaki Y., Godinho S.A. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nat Immunol. 2012;13:1092–1100. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klein U., Tu Y., Stolovitzky G.A. Transcriptional analysis of the B cell germinal center reaction. Proc NatL Acad Sci USA. 2003;100:2639–2644. doi: 10.1073/pnas.0437996100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Conacci-Sorrell M., McFerrin L., Eisenman R.N. An overview of MYC and its interactome. Cold Spring Harb Perspect Med. 2014;4:a014357. doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taub R., Kirsch I., Morton C. Translocation of the c-myc gene into the immunoglobulin heavy chain locus in human Burkitt lymphoma and murineplasmacytoma cells. Proc NatL Acad Sci USA. 1982;79:7837–7841. doi: 10.1073/pnas.79.24.7837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Basso K., Dalla-Favera R. Germinal centres and B cell lymphomagenesis. Nat Rev Immunol. 2015;15:172–184. doi: 10.1038/nri3814. [DOI] [PubMed] [Google Scholar]
- 13.Dominguez-Sola D., Victora G.D., Ying C.Y. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nat Immunol. 2012;13:1083–1091. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chari N.S., Pinaire N.L., Thorpe L., Medeiros L.J., Routbort M.J., McDonnell T.J. The p53 tumor suppressor network in cancer and the therapeutic modulation of cell death. Apoptosis. 2009;14:336–347. doi: 10.1007/s10495-009-0327-9. [DOI] [PubMed] [Google Scholar]
- 15.Liu D., Xu Y. p53, oxidative stress, and aging. Antioxid Redox Signal. 2011;15:1669–1678. doi: 10.1089/ars.2010.3644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ott G., Rosenwald A., Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122:3884–3891. doi: 10.1182/blood-2013-05-498329. [DOI] [PubMed] [Google Scholar]
- 17.Slack G.W., Gascoyne R.D. MYC and aggressive B-cell lymphomas. Adv Anat Pathol. 2011;18:219–228. doi: 10.1097/PAP.0b013e3182169948. [DOI] [PubMed] [Google Scholar]
- 18.Wang W.G., Liu Z.B., Jiang X.N., Lee J., Zhou X.Y., Li X.Q. MYC protein dysregulation is driven by BCR-PI3K signalling in diffuse large B-cell lymphoma. Histopathology. 2017;71:778–785. doi: 10.1111/his.13287. [DOI] [PubMed] [Google Scholar]
- 19.Hemann M.T., Bric A., Teruya-Feldstein J. Evasion of the p53 tumour surveillance network by tumour-derived MYC mutants. Nature. 2005;436:807–811. doi: 10.1038/nature03845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabow S., Delbridge A.R., Aubrey B.J., Vandenberg C.J., Strasser A. Loss of a single Mcl-1 allele inhibits MYC-driven lymphomagenesis by sensitizing pro-B cells to apoptosis. Cell Rep. 2016;14:2337–2347. doi: 10.1016/j.celrep.2016.02.039. [DOI] [PubMed] [Google Scholar]
- 21.Dang C.V., O'donnell K.A., Juopperi T. The great MYC escape in tumorigenesis. Cancer Cell. 2005;8:177–178. doi: 10.1016/j.ccr.2005.08.005. [DOI] [PubMed] [Google Scholar]
- 22.Rabellino A., Melegari M., Tompkins V.S. PIAS1 promotes lymphomagenesis through MYC upregulation. Cell Rep. 2016;15:2266–2278. doi: 10.1016/j.celrep.2016.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang K., Xu Z., Wang N., Xu T., Zhu M. MicroRNA and gene networks in human diffuse large B-cell lymphoma. Oncol Lett. 2014;8:2225–2232. doi: 10.3892/ol.2014.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X., Zhao X., Fiskus W. Coordinated silencing of MYC-mediated miR-29 by HDAC3 and EZH2 as a therapeutic target of histone modification in aggressive B-Cell lymphomas. Cancer Cell. 2012;22:506–523. doi: 10.1016/j.ccr.2012.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Vousden K.H., Ryan K.M. p53 and metabolism. Nat Rev Cancer. 2009;9:691–700. doi: 10.1038/nrc2715. [DOI] [PubMed] [Google Scholar]
- 26.Peller S., Rotter V. TP53 in hematological cancer: low incidence of mutations with significant clinical relevance. Hum Mutat. 2003;21:277–284. doi: 10.1002/humu.10190. [DOI] [PubMed] [Google Scholar]
- 27.Pospisilova S., Gonzalez D., Malcikova J. ERIC recommendations on TP53 mutation analysis in chronic lymphocytic leukemia. Leukemia. 2012;26:1458–1461. doi: 10.1038/leu.2012.25. [DOI] [PubMed] [Google Scholar]
- 28.Hallek M., Cheson B.D., Catovsky D. Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: a report from the international workshop on chronic lymphocytic leukemia updating the national cancer institute-working group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rivlin N., Brosh R., Oren M., Rotter V. Mutations in the p53 tumor suppressor gene: important Milestones at the various steps of tumorigenesis. Genes Cancer. 2011;2:466–474. doi: 10.1177/1947601911408889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zenz T., Eichhorst B., Busch R. TP53 mutation and survival in chronic lymphocytic leukemia. J Clin Oncol. 2010;28:4473–4479. doi: 10.1200/JCO.2009.27.8762. [DOI] [PubMed] [Google Scholar]
- 31.Rossi D., Khiabanian H., Spina V. Clinical impact of small TP53 mutated subclones in chronic lymphocytic leukemia. Blood. 2014;123:2139–2147. doi: 10.1182/blood-2013-11-539726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Meek D.W. Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer. 2009;9:714–723. doi: 10.1038/nrc2716. [DOI] [PubMed] [Google Scholar]
- 33.Carr M.I., Jones S.N. Regulation of the Mdm2-p53 signaling axis in the DNA damage response and tumorigenesis. Transl Cancer Res. 2016;5:707–724. doi: 10.21037/tcr.2016.11.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nihira N.T., Oqura K., Shimizu K. Acetylation-dependent regulation of MDM2 E3 ligase activity dictates its oncogenic function. Sci Signal. 2017;10 doi: 10.1126/scisignal.aai8026. pii: eaai8026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tessoulin B., Eveillard M., Lok A. p53 dysregulation in B-cell malignancies: more than a single gene in the pathway to hell. Blood Rev. 2017;31:251–259. doi: 10.1016/j.blre.2017.03.001. [DOI] [PubMed] [Google Scholar]
- 36.Linares L.K., Hengstermann A., Ciechanover A., Müller S., Scheffner M. HdmX stimulates Hdm2-mediated ubiquitination and degradation of p53. Proc Natl Acad Sci USA. 2003;100:12009–12014. doi: 10.1073/pnas.2030930100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wade M., Li Y.C., Wahl G.M. MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat Rev Cancer. 2013;13:83–96. doi: 10.1038/nrc3430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yu L., Li L., Medeiros L.J., Young K.H. NF-κB signaling pathway and its potential as a target for therapy in lymphoid neoplasms. Blood Rev. 2017;31:77–92. doi: 10.1016/j.blre.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carrera S., Cuadrado-Castano S., Samuel J. Stra6, a retinoic acid-responsive gene, participates in p53-induced apoptosis after DNA damage. Cell Death Differ. 2013;20:910–919. doi: 10.1038/cdd.2013.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Drane P., Bravard A., Bouvard V., May E. Reciprocal down-regulation of p53 and SOD2 gene expression-implication in p53 mediated apoptosis. Oncogene. 2001;20:430–439. doi: 10.1038/sj.onc.1204101. [DOI] [PubMed] [Google Scholar]
- 41.Faraonio R., Vergara P., Di Marzo D. p53 suppresses the Nrf2-dependent transcription of antioxidant response genes. J Biol Chem. 2006;281:39776–39784. doi: 10.1074/jbc.M605707200. [DOI] [PubMed] [Google Scholar]
- 42.el-Deiry W.S., Tokino T., Velculescu V.E. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- 43.Valente L.J., Grabow S., Vandenberg C.J., Strasser A., Janic A. Combined loss of PUMA and p21 accelerates c-MYC-driven lymphoma development considerably less than loss of one allele of p53. Oncogene. 2016;35:3866–3871. doi: 10.1038/onc.2015.457. [DOI] [PubMed] [Google Scholar]
- 44.Newbold A., Salmon J.M., Martin B.P., Stanley K., Johnstone R.W. The role of p21(waf1/cip1) and p27(Kip1) in HDACi-mediated tumor cell death and cell cycle arrest in the Eμ-myc model of B-cell lymphoma. Oncogene. 2014;33:5415–5423. doi: 10.1038/onc.2013.482. [DOI] [PubMed] [Google Scholar]
- 45.Luo Z., Cui R., Tili E., Croce C. Friend or foe: MicroRNAs in the p53 network. Cancer Lett. 2018;419:96–102. doi: 10.1016/j.canlet.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 46.Hermeking H. MicroRNAs in the p53 network: micromanagement of tumour suppression. Nat Rev Cancer. 2012;12:613–626. doi: 10.1038/nrc3318. [DOI] [PubMed] [Google Scholar]
- 47.Fabbri M., Bottoni A., Shimizu M. Association of a microRNA/TP53 feedback circuitry with pathogenesis and outcome of B-cell chronic lymphocytic leukemia. JAMA. 2011;305:59–67. doi: 10.1001/jama.2010.1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Calin G.A., Dumitru C.D., Shimizu M. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tarasov V., Jung P., Verdoodt B. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 50.Wong M.Y., Yu Y., Walsh W.R., Yang J.L. microRNA-34 family and treatment of cancers with mutant or wild-type p53 (Review) Int J Oncol. 2011;38:1189–1195. doi: 10.3892/ijo.2011.970. [DOI] [PubMed] [Google Scholar]
- 51.Rao D.S., O'Connell R.M., Chaudhuri A.A., Garcia-Flores Y., Geiger T.L., Baltimore D. MicroRNA-34a perturbs B lymphocyte development by repressing the forkhead box transcription factor Foxp1. Immunity. 2010;33:48–59. doi: 10.1016/j.immuni.2010.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roehle A., Hoefig K.P., Repsilber D. MicroRNA signatures characterize diffuse large B-cell lymphomas and follicular lymphomas. Br J Haematol. 2008;142:732–744. doi: 10.1111/j.1365-2141.2008.07237.x. [DOI] [PubMed] [Google Scholar]
- 53.Merkel O., Asslaber D., Piñón J.D., Egle A., Greil R. Interdependent regulation of p53 and miR-34a in chronic lymphocytic leukemia. Cell Cycle. 2010;9:2764–2768. [PubMed] [Google Scholar]
- 54.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 55.Song W., Liu M.G., Zhang J.B., Zhang J.J., Sun M.M., Yu Q.K. Mechanism of action of EBV, Bcl-2, p53, c-Myc and Rb in non-Hodgkin's lymphoma. Eur Rev Med Pharmacol Sci. 2016;20:1093–1097. [PubMed] [Google Scholar]
- 56.Stengel A., Schnittger S., Weissmann S. TP53 mutations occur in 15.7% of ALL and are associated with MYC-rearrangement, low hypodiploidy, and a poor prognosis. Blood. 2014;124:251–258. doi: 10.1182/blood-2014-02-558833. [DOI] [PubMed] [Google Scholar]
- 57.Ye Q., Xu-Monette Z.Y., Tzankov A. Prognostic impact of concurrent MYC and BCL6 rearrangements and expression in de novo diffuse large B-cell lymphoma. Oncotarget. 2016;7:2401–2416. doi: 10.18632/oncotarget.6262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xie Y., Bulbul M.A., Ji L. p53 expression is a strong marker of inferior survival in de novo diffuse large B-cell lymphoma and may have enhanced negative effect with MYC coexpression: a single institutional clinicopathologic study. Am J Clin Pathol. 2014;141:593–604. doi: 10.1309/AJCPPHMZ6VHF0WQV. [DOI] [PubMed] [Google Scholar]
- 59.Xu-Monette Z.Y., Dabaja B.S., Wang X. Clinical features, tumor biology, and prognosis associated with MYC rearrangement and Myc overexpression in diffuse large B-cell lymphoma patients treated with rituximab-CHOP. Mod Pathol. 2015;28:1555–1573. doi: 10.1038/modpathol.2015.118. [DOI] [PubMed] [Google Scholar]
- 60.Swerdlow S.H., Campo E., Pileri S.A. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood. 2016;127:2375–2390. doi: 10.1182/blood-2016-01-643569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li S., Young K.H., Medeiros L.J. Diffuse large B-cell lymphoma. Pathology. 2018;50:74–87. doi: 10.1016/j.pathol.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 62.Fiskvik I., Beiske K., Delabie J. Combining MYC, BCL2 and TP53 gene and protein expression alterations improves risk stratification in diffuse large B-cell lymphoma. Leuk Lymphoma. 2015;56:1742–1749. doi: 10.3109/10428194.2014.970550. [DOI] [PubMed] [Google Scholar]
- 63.Forys J.T., Kuzmicki C.E., Saporita A.J., Winkeler C.L., Maggi L.B., Jr., Weber J.D. ARF and p53 coordinate tumor suppression of an oncogenic IFN-β-STAT1-ISG15 signaling axis. Cell Rep. 2014;7:514–526. doi: 10.1016/j.celrep.2014.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Oguro H., Iwama A., Morita Y., Kamijo T., van Lohuizen M., Nakauchi H. Differential impact of Ink4a and Arf on hematopoietic stem cells and their bone marrow microenvironment in Bmi1-deficient mice. J Exp Med. 2006;203:2247–2253. doi: 10.1084/jem.20052477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haupt Y., Bath M.L., Harris A.W., Adams J.M. bmi-1 transgene induces lymphomas and collaborates with myc in tumorigenesis. Oncogene. 1993;8:3161–3164. [PubMed] [Google Scholar]
- 66.Jacobs J.J., Scheijen B., Voncken J.W., Kieboom K., Berns A., van Lohuizen M. Bmi-1 collaborates with c-Myc in tumorigenesis by inhibiting c-Myc-induced apoptosis via INK4a/ARF. Genes Dev. 1999;13:2678–2690. doi: 10.1101/gad.13.20.2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bhattacharya R., Mustafi S.B., Street M., Dey A., Dwivedi S.K. Bmi-1: at the crossroads of physiological and pathological biology. Genes Dis. 2015;2:225–239. doi: 10.1016/j.gendis.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delmore J.E., Issa G.C., Lemieux M.E. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fan C., He L., Kapoor A. Bmi1 promotes prostate tumorigenesis via inhibiting p16(INK4A) and p14(ARF) expression. Biochim Biophys Acta. 2008;1782:642–648. doi: 10.1016/j.bbadis.2008.08.009. [DOI] [PubMed] [Google Scholar]
- 70.Guo W.J., Datta S., Band V., Dimri G.P. Mel-18, a polycomb group protein, regulates cell proliferation and senescence via transcriptional repression of Bmi-1 and c-Myc oncoproteins. Mol Biol Cell. 2007;18:536–546. doi: 10.1091/mbc.E06-05-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Craig V.J., Cogliatti S.B., Imig J. Myc-mediated repression of microRNA-34a promotes high-grade transformation of B-cell lymphoma by dysregulation of FoxP1. Blood. 2011;117:6227–6236. doi: 10.1182/blood-2010-10-312231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.De Tullio G., De Fazio V., Sgherza N. Challenges and opportunities of microRNAs in lymphomas. Molecules. 2014;19:14723–14781. doi: 10.3390/molecules190914723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tao J., Zhao X., Tao J. c-MYC-miRNA circuitry: a central regulator of aggressive B-cell malignancies. Cell Cycle. 2014;13:191–198. doi: 10.4161/cc.27646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu J., Zhang C., Zhao Y., Feng Z. MicroRNA control of p53. J Cell Biochem. 2017;118:7–14. doi: 10.1002/jcb.25609. [DOI] [PubMed] [Google Scholar]
- 75.Spender L.C., Inman G.J. Developments in Burkitt's lymphoma: novel cooperations in oncogenic MYC signaling. Cancer Manag Res. 2014;6:27–38. doi: 10.2147/CMAR.S37745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Spender L.C., Cornish G.H., Rowland B., Kempkes B., Farrell P.J. Direct and indirect regulation of cytokine and cell cycle proteins by EBNA-2 during Epstein-Barr virus infection. J Virol. 2001;75:3537–3546. doi: 10.1128/JVI.75.8.3537-3546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Saha A., Lu J., Morizur L., Upadhyay S.K., Aj M.P., Robertson E.S. E2F1 mediated apoptosis induced by the DNA damage response is blocked by EBV nuclear antigen 3C in lymphoblastoid cells. PLoS Pathog. 2012;8:e1002573. doi: 10.1371/journal.ppat.1002573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kurland J.F., Tansey W.P. Myc-mediated transcriptional repression by recruitment of histone deacetylase. Cancer Res. 2008;68:3624–3629. doi: 10.1158/0008-5472.CAN-07-6552. [DOI] [PubMed] [Google Scholar]
- 79.Asano T., Yao Y., Zhu J., Li D., Abbruzzese J.L., Reddy S.A. The PI 3-kinase/Akt signaling pathway is activated due to aberrant Pten expression and targets transcription factors NF-kappaB and c-Myc in pancreatic cancer cells. Oncogene. 2004;23:8571–8580. doi: 10.1038/sj.onc.1207902. [DOI] [PubMed] [Google Scholar]
- 80.Kumar A., Marqués M., Carrera A.C. Phosphoinositide 3-kinase activation in late G1 is required for c-Myc stabilization and S phase entry. Mol Cell Biol. 2006;26:9116–9125. doi: 10.1128/MCB.00783-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Oki Y., Kelly K.R., Flinn I. CUDC-907 in relapsed/refractory diffuse large B-cell lymphoma, including patients with MYC-alterations: results from an expanded phase I trial. Haematologica. 2017;102:1923–1930. doi: 10.3324/haematol.2017.172882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Qian C., Lai C.J., Bao R. Cancer network disruption by a single molecule inhibitor targeting both histone deacetylase activity and phosphatidylinositol 3-kinase signaling. Clin Cancer Res. 2012;18:4104–4113. doi: 10.1158/1078-0432.CCR-12-0055. [DOI] [PubMed] [Google Scholar]
- 83.Mondello P., Derenzini E., Asgari Z. Dual inhibition of histone deacetylases and phosphoinositide 3-kinase enhances therapeutic activity against B cell lymphoma. Oncotarget. 2017;8:14017–14028. doi: 10.18632/oncotarget.14876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Aird F., Kandela I., Mantis C., Reproducibility Project: Cancer Biology Replication Study: BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Elife. 2017;6 doi: 10.7554/eLife.21253. pii: e21253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lopez-Girona A., Mendy D., Ito T. Cereblon is a direct protein target for immunomodulatory and antiproliferative activities of lenalidomide and pomalidomide. Leukemia. 2012;26:2326–2335. doi: 10.1038/leu.2012.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lopez-Girona A., Heintel D., Zhang L.H. Lenalidomide downregulates the cell survival factor, interferon regulatory factor-4, providing a potential mechanistic link for predicting response. Br J Haematol. 2011;154:325–336. doi: 10.1111/j.1365-2141.2011.08689.x. [DOI] [PubMed] [Google Scholar]
- 87.Nowakowski G.S., LaPlant B., Macon W.R. Lenalidomide combined with R-CHOP overcomes negative prognostic impact of non-germinal center B-cell phenotype in newly diagnosed diffuse large B-Cell lymphoma: a phase II study. J Clin Oncol. 2015;33:251–257. doi: 10.1200/JCO.2014.55.5714. [DOI] [PubMed] [Google Scholar]
- 88.de Jonge A.V., Roosma T.J., Houtenbos I. Diffuse large B-cell lymphoma with MYC gene rearrangements: current perspective on treatment of diffuse large B-cell lymphoma with MYC gene rearrangements; case series and review of the literature. Eur J Cancer. 2016;55:140–146. doi: 10.1016/j.ejca.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 89.Pan Z., Scheerens H., Li S.J. Discovery of selective irreversible inhibitors for Bruton's tyrosine kinase. ChemMedChem. 2007;2:58–61. doi: 10.1002/cmdc.200600221. [DOI] [PubMed] [Google Scholar]
- 90.Farooqui M.Z., Valdez J., Martyr S. Ibrutinib for previously untreated and relapsed or refractory chronic lymphocytic leukaemia with TP53 aberrations: a phase 2, single-arm trial. Lancet Oncol. 2015;16:169–176. doi: 10.1016/S1470-2045(14)71182-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tucker D.L., Rule S.A. A critical appraisal of ibrutinib in the treatment of mantle cell lymphoma and chronic lymphocytic leukemia. Ther Clin Risk Manag. 2015;11:979–990. doi: 10.2147/TCRM.S73559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Burger J.A., Keating M.J., Wierda W.G. Safety and activity of ibrutinib plus rituximab for patients with high-risk chronic lymphocytic leukaemia: a single-arm, phase 2 study. Lancet Oncol. 2014;15:1090–1099. doi: 10.1016/S1470-2045(14)70335-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bykov V., Eriksson S.E., Bianchi J., Wiman K.G. Targeting mutant p53 for efficient cancer therapy. Nat Rev Cancer. 2018;18:89–102. doi: 10.1038/nrc.2017.109. [DOI] [PubMed] [Google Scholar]
- 94.Huang C., Wu S., Ji H. Identification of XBP1-u as a novel regulator of the MDM2/p53 axis using an shRNA library. Sci Adv. 2017;3:e1701383. doi: 10.1126/sciadv.1701383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kojima K., Shimanuki M., Shikami M., Andreeff M., Nakakuma H. Cyclin-dependent kinase 1 inhibitor RO-3306 enhances p53-mediated Bax activation and mitochondrial apoptosis in AML. Cancer Sci. 2009;100:1128–1136. doi: 10.1111/j.1349-7006.2009.01150.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dienstmann R., Rodon J., Serra V., Tabernero J. Picking the point of inhibition: a comparative review of PI3K/AKT/mTOR pathway inhibitors. Mol Cancer Ther. 2014;13:1021–1031. doi: 10.1158/1535-7163.MCT-13-0639. [DOI] [PubMed] [Google Scholar]
- 97.Zent C.S., Bowen D.A., Conte M.J., LaPlant B.R., Call T.G. Treatment of relapsed/refractory chronic lymphocytic leukemia/small lymphocytic lymphoma with everolimus (RAD001) and alemtuzumab: a Phase I/II study. Leuk Lymphoma. 2016;57:1585–1591. doi: 10.3109/10428194.2015.1113280. [DOI] [PubMed] [Google Scholar]
- 98.Pettitt A.R., Jackson R., Carruthers S. Alemtuzumab in combination with methylprednisolone is a highly effective induction regimen for patients with chronic lymphocytic leukemia and deletion of TP53: final results of the national cancer research institute CLL206 trial. J Clin Oncol. 2012;30:1647–1655. doi: 10.1200/JCO.2011.35.9695. [DOI] [PubMed] [Google Scholar]
- 99.Yan W., Jung Y.S., Zhang Y., Chen X. Arsenic trioxide reactivates proteasome-dependent degradation of mutant p53 protein in cancer cells in part via enhanced expression of Pirh2 E3 ligase. PLoS One. 2014;9:e103497. doi: 10.1371/journal.pone.0103497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Camicia R., Bachmann S.B., Winkler H.C. BAL1/ARTD9 represses the anti-proliferative and pro-apoptotic IFNγ-STAT1-IRF1-p53 axis in diffuse large B-cell lymphoma. J Cell Sci. 2013;126(Pt 9):1969–1980. doi: 10.1242/jcs.118174. [DOI] [PubMed] [Google Scholar]