Abstract
Dissociation of a protein from DNA is often assumed to be described by an off rate that is independent of other molecules in solution. Recent experiments and computational analyses have challenged this view by showing that unbinding rates (residence times) of DNA-bound proteins can depend on concentrations of nearby molecules that are competing for binding. This “facilitated dissociation” (FD) process can occur at the single-binding site level via formation of a ternary complex, and can dominate over “spontaneous dissociation” at low (sub-micromolar) concentrations. In the crowded intracellular environment FD introduces new regulatory possibilities at the level of individual biomolecule interactions.
Keywords: DNA-protein interactions, transcription factor, facilitated dissociation, binding kinetics
Introduction
All activities of a living cell are ultimately controlled by patterns of transcription which are in turn controlled by the binding of proteins to DNA. Binding and unbinding of transcription factors (TFs) and other DNA-interacting proteins (e.g., DNA-bending and looping “architectural” or “genome folding/packaging”) proteins take place in a molecularly crowded environment where there is exposure of a DNA binding site to an array of competing potential binding partners. A TF bound to a regulatory site must have a long enough residence time for it to affect transcription; at the same time, the TF may need to be removed when its regulatory effect is to be ended. TF-DNA interactions therefore ought to be stable enough to survive chemical and mechanical “molecular noise”, while remaining malleable enough for disassembly to occur when an appropriate physiochemical signal arrives.
While protein-DNA binding kinetics are established on diffusive mechanisms where binding rate goes up with concentration [1, 2], mechanisms of dissociation of a protein from a protein-DNA complex have been less heavily studied. Lifetimes of protein-DNA complexes are usually assumed to be independent of concentrations of proteins in solution; this assumption is often implicit in the identification Kd = koff/γ where γ is the association rate constant. This lack of concentration dependence implictly assumes dissociation to involve crossing of one free energy barrier, which is likely inappropriate for a protein bound to DNA by an array of weak, non-covalent electrostatic and hydrophobic interactions.
Recent experiments in vivo and in vitro show that a variety of nucleic-acid-binding proteins more quickly leave DNA binding sites with increasing concentrations of solution-phase competitor proteins [3•, 4, 5•, 6•, 7•, 8, 9•, 10•, 11] (also reviewed in Ref. ([12]), for concentrations in the tens to hundreds of nanomolar rage (at or below physiological values estimated for many DNA-binding proteins in vivo). This acceleration of dissociation by a competing molecule is an instance of “facilitated dissociation” (FD) [13, 14, 15, 3•]. The idea is general, and posits that instead of having only one “bound” state, a protein-DNA complex can stochastically and intermittently visit a partially-bound state, in which a competitor molecule may be able to grab part of the exposed binding site, facilitating removal of the originally bound molecule (Figure 1A,B). The basic idea of FD has been long understood in the chemical kinetics literature [16], and FD effects have been long reported to occur for biomolecules, e.g., in transfer of ethidum bromide between DNA molecules [17]. The notion of transient protein-DNA-protein complexes being involved in facilitation of dissociation of proteins from double-stranded DNA has attracted attention only relatively recently [18, 19].
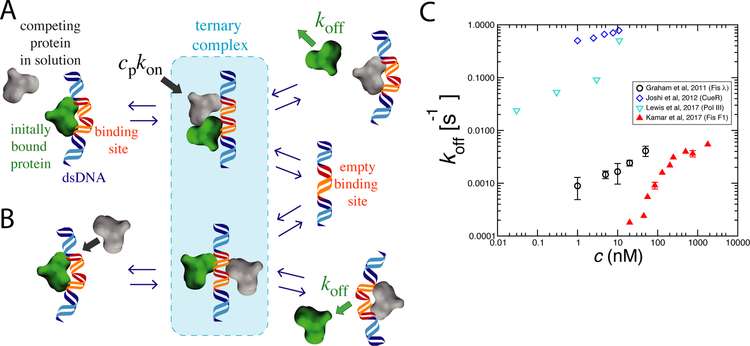
Figure 1: Kinetic pathways for facilitated dissociation (FD) and experimental data.
A) Competing protein from solution (gray) binds to partially exposed DNA site arising from thermal fluctuation of previously bound protein (green). In the ternary complex, each protein is weakly bound, which leads to dissociation of one or both proteins.
B) Within the ternary complex, proteins can compete may interfere with one another’s binding via allostery through the substrate as well as via sterically blocking one anothers’ interactions with the DNA.
C) Experimental data showing protein off-rate increasing with bulk protein concentration for a few DNA-binding proteins (see text for details). In each case the off-rate increases with solution concentration of that protein, the hallmark of facilitated dissociation.
Here, we briefly review experiments which have shown FD for DNA-protein interactions. We also discuss potential consequences of FD in vivo, as well as the potential for FD to play a role in other kinds of biomolecular interactions (e.g., protein-protein interactions) and its potential role in design of pharmaceutical and functional materials.
Evidence for FD of proteins from DNA in vitro
Recent experiments have reported competing-protein-concentration-dependent off rates koff (i.e., inverse of residence time) for proteins bound to DNA. Surface plasmon resonance (SPR) experiments showed that the heterodimeric TF NF−κB had dissocation from DNA binding sites accelerated by increasing concentration of IκBαs [20] (which competes with the host DNA to bind NF−κB). Subsequent experiments showed that the disassembly rate of the DNA−NF-κB−IκBα complex showed a linear increase with increasing IκBα concentrations up to 10 µM [20, 10•].
Strong FD effects were observed for the homodimeric E. coli TF Fis, which interacts nonspecifically and specifically with DNA and is found at high concentrations in rapidly growing E. coli cells. A set of non-sequence-specific experiments used fluorescence imaging of GFP-Fis on long (48 kb) λ-phage DNAs [18] extended using magnetic tweezers. It was found that pre-bound GFP-labeled Fis could be efficiently stripped off DNA by Fis (Figure 1C, black circles), as well as by other DNA-binding proteins introduced into solution (E. coli HU and human HMGB1 proteins), the latter experiments indicating that FD can be heterotypic. Further experiments on bacterial chromosomes isolated in vitro [4] showed remarkably similar effects, with both experiments measuring a sequence-averaged FD exchange rate constant (kexch) of 104 M−1s−1.
FD effects have been reported for the homodimeric bacterial TF CueR (Copper Efflux Regulator), which regulates transcriptional response to copper ions. Single molecule FRET measurements for CueR revealed that increased free CueR concentration promotes dissociation of CueR specifically bound to its 25bp-long DNA binding site [6•] (Figure 1C, blue diamonds). Nonspecifically-bound CueR did not exhibit FD, suggesting a role of the sequence-specific complex structure in FD. The FRET data of that study also indicated that dissociation of CueR can be facilitated through displacement or direct exchange processes (corresponding roughly to the two pathways shown in Figure 1 A and B) contribute to CueR-DNA interactions.
Single-molecule fluorescence studies of Fis interacting with specific, very tightly binding 21 bp “F1” DNA sites over a wide concentration range revealed both the initial linear concentration dependence, and saturation of off-rate at concentrations above about 200 nM, roughly the physiolgical value expected in vivo [3•] ((Figure 1C, filled red triangles). The observation of saturation of off rate is in accord with a mechanism based on transient formation of a less stable Fis-DNA complex, consistent with the suggestion of off-rate saturation in earlier Fis experiments on isolated bacterial chromosomes [4]. Molecular dynamics (MD) simulations of a coarse-grained bead-spring model recapitulated the experimental observations, and suggested that FD can occur for a very wide range of binding affinities, and without dependence on details of chemical structure of the molecules involved [3•, 21].
Multiprotein complexes have also been reported to exhibit a concentration-dependent dissociation mechanism [9•, 8] (Figure 1C, cyan triangles, data for DNA polymerase from Ref. [?]. Single-molecule fluorescence experiments showed that T7 bacteriophage DNA polymerase components at a replication fork can dynamically exchange [9•]. Furthermore, exposing the wild-type pre-bound polymerase with a mutated, less efficient polymerase (Y526F) led to a decrease in replicated DNA, in a Y526F-concentration-dependent manner, again suggesting a dynamic exchange mechanism [9•]. Experimental data suggest that FD is readily observable, with large changes in off-rate, over a wide range of affinities, types of protein, binding lifetimes and protein concentrations (Fig. 1C).
FD of proteins from DNA in living cells
Experiments have begun to address the challenging goal of quantifying FD effects in vivo. Using in vivo fluorescence imaging, faster unbinding rates for CueR and its Zn+2 sensitive version ZntR have been observed with increasing concentration of each protein type in vivo [5•]. A relation between dissociation rate and chromosome condensation levels was also reported: residence times for passive (i.e., apo) forms of the metalloregulators were reported to be longer than those of active (i.e., metal-bound) forms in condensed chromosomes, with the opposite trend in cells with more loosely compacted chromosomes [5•]. Similar effects for the zinc-responsive uptake regulator Zur in E. coli cells were also reported [22].
Dynamics of fluorescently labeled DNA polymerases have been studied in live E. coli cells, leading to the observation that the Polymerase III* complex (holoenzyme lacking the β2 sliding clamp) has a residence time of 4 ± 2 s [7•], in accord with other in vivo studies [8]. These measurements are consistent with in vitro exchange times measured at physiologically-relevant concentrations of polymerase, which are on the order of 10 nM for E. coli cells under growth conditions. Recently, it has been reported that the binding-site-residency time of CTCF decreased when its expression was increased (Fig. 2 Supplement 2B of [23]), possibly extending in vivo observations of FD to human nuclei.
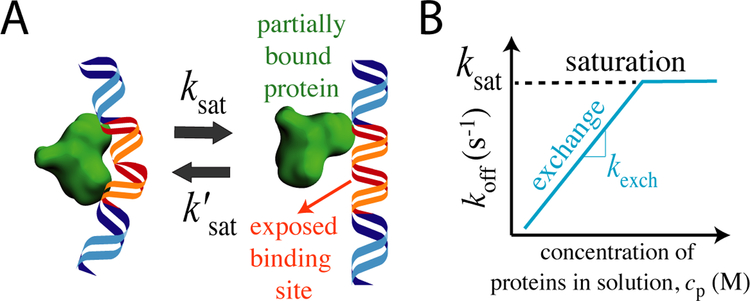
Figure 2:
A) Partially bound protein on DNA binding site. The time window during which the binding site is exposed to invading proteins from solution sets the limiting FD off rate approached at high competitor concentration. B) Schematic behavior of protein concentration-dependence of off rate: off rate increases linearly at low concentration with slope of kexch, while at higher concentrations the off rate saturates at the limiting off rate.
FD of proteins from DNA by competing nucleic acids
Dissociation of a DNA bound protein does not necessarily require an invading protein from solution. Competing DNA segments are well known to be able to facilitate dissociation of proteins [24, 25, 26, 27, 28, 29, 30, 31, 32•, 33, 34, 35, 36, 37, 11, 38], a process often called “intersegmental transfer [1]. Recent magnetic-tweezer assays probing dissociation of DNA-bound Fis through changes in effective DNA persistence length observe a strong dependence of off rate on solution concentration of free nucleic acid segments [39]. As for competing protein, the off rate for Fis from DNA shows an initial linear increase for competitor DNA concentration, which then saturates at ≈ 1 × 10−2 s−1 above dsDNA concentrations ≈20 ng/µl, remarkably close to the limiting rates obtained for Fis dissociating from bacterial chromosomes [4].
Single strand binding (SSB) proteins also exhibit FD characteristics
Despite the very different flexibility of single-stranded DNA (ssDNA, ≈ 1 nm persistence length) relative to dsDNA (50 nm persistence length), FD has been observed for sequence-nonspecific binding of ssDNA-binding proteins. Homotetrameric fluorescently-labeled E. coli single-strand-binding protein (SSB) from E. coli exhibited faster dissociation from ssDNA binding site with increased SSB in solution [40]. Following a linear dependence of dissociation rate for SSB concentration of up to 4 µM, a hyperbolic behavior was observed, which was interpreted as rate-limiting step due to conformational arrangement of SSB on the ssDNA [40]. Similarly, single-molecule imaging experiments studying unbinding of heterodimeric Replication Protein A (RPA) showed that 10000 nM solution-phase RPA can reduce the residence of pre-RPA from roughly two hours to less than a minute [41]. RPA-ssDNA complexes were also observed to disassemble in the presence of other single-stranded-binding proteins such as yeast Rad51 and E. coli SSB proteins indicating heterotypic FD/exchange [41]. Weak FD effects have also been observed during dissociation of ssRNAs [42], attributed to diffusive return kinetics rather than to formation of a stable ternary complex [42, 39].
Molecular picture of FD
Ternary intermediate model of FD
Single-molecule observations of strong FD effects [3•, 6•] suggest an explanation involving dynamics of a single protein-DNA complex, rather than via many-protein interactions (e.g., cooperative binding). A ternary complex of two proteins sharing one DNA binding site is likely to be less stable than a well-bound protein-DNA complex [43] (in the DNA-protein-DNA “direct transfer” case, one protein is shared by two nucleic acid binding sites). Assuming that partially-bound states can occur (even rarely visited states of this type are sufficient) may allow weak binding of a second protein. The thus-partially bound “invader” protein may block the ability of the originally bound protein to return to its fully-bound state, shortening its average residence time and accelerating its dissociation. If the original protein dissociates, the invader may stay behind, and the process is one of “exchange”, but it is possible for both proteins to leave the binding site as well (in that latter case, one could anticipate rapid filling of the empty binding site by some other molecule). The key point is that instead of there being a single barrier between binding and unbinding of the original protein, there is at least one partially bound state, on which the competitor can act to reduce the residence time of the original protein.
This ternary complex model leads to Michaelis–Menten–like kinetics whereby dissociation of the originally bound protein is sped up by the invader [14, 44, 15, 3•]. This model describes the initial increase and saturation of off rate with competitor concentration observed experimentally and computationally [3•, 21]. This type of model does not depend on (or specify) the molecular details of the competition, which may result from steric or other interactions between the two proteins (e.g., direct binding site occlusion, Figure 1A), or from allostery through the DNA without direct protein-protein interaction [45](Figure 1B).
Given that a number of FD observations involve dimeric proteins (Fis [3•, 18, 4], HU [18], CueR [6•, 5•]) one might wonder if this is a requirement for the effect; certainly a dimeric protein involves two DNA-binding surfaces. However, observation of FD for single proteins in multiprotein complexes [9•, 7•] and for the monomeric NHP6A [3•, 18], as well as similar effects for transcription activation domains indicate that dimericity is not a requirement [46].
Salt concentration only weakly affects FD
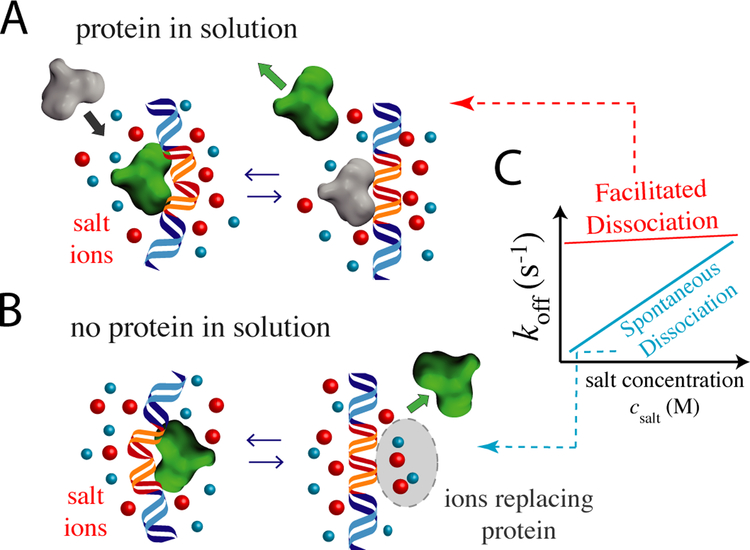
FD can be probed in vitro in experiments performed at varied salt concentrations (Figure 3). For spontaneous dissociation, any counterions associated with the unbound DNA or protein must bind from solution, driving the strong salt concentration effects typically seen for nucleic acid-protein interactions [47, 48, 35]. Conversely, for homotypic FD/exchange, there should be no net change in bound ions and minimal salt effects. In accord with this, the zero-protein-concentration off rate for Fis [3•] shows a strong salt effect with a Hill slope of about 2 [3•]. For an “ion release” picture, this suggests a net binding of about 2 ions to dissociating DNA and protein. At elevated protein concentration, FD with almost no salt effect is observed [3•] (Figure 3). MD simulations show the same effects as a function of salt concentration [21]. Detailed molecular modeling of Fis-DNA interactions indicates the reduction of electrostatic interaction strength alone leads to partially unbound state, suggesting that electrostatic interaction strength can be used to adjust the nature of the partially unbound protein states and ternary complex [43].
Figure 3:
Effects of salt ions on unbinding. A) Salt weakly affects off rates via FD since there is no net ion adsorption or release. B) For spontaneous dissociation, many ions may bind the dissociating protein and DNA, resulting in a strong effect of salt concentration. C) Schematics of salt concentration versus the off rate data for the two cases [3•, 21].
Perspective
Given the dynamic variations in the concentration ranges of proteins in the nucleus or bacterial cell volume it is likely that FD may regulate residence times of DNA-bound proteins, and it is plausible that FD for TFs has been selected to improve reliability of gene regulation. It may be advantageous to the cell for a TF to stay stably bound for a long time in order to allow a given process to be completed [49, 50], or to provide “memory” of a previous transcriptional state, robustly against thermal noise. At the same time, it may be advantageous for that protein to promptly unbind in response to a regulatory molecular signal. By using FD to trigger unbinding of the initially bound TF, one can enjoy the advantages of stable binding and rapid switching, relative to the non-FD case where one must wait for spontaneous dissociation of the TF to occur. Similarly FD could accelerate the response of TF-sensors that respond to metabolites or toxins [5•, 6•, 22].
FD may also play a role in accelerating the action of enzymes acting processively along DNA. Turnover of DNA replisome components [7•] may help overcome DNA lesions and other obstacles to replication [8, 51]. Chromatin structure may well be modified by FD, for example via turnover of proteins that bend or crossbridge chromatin segments. Notably the presence of nucleosomes has been observed to affect TF off rates [11]. Chromatin compaction levels are known to affect residence times of DNA-bound proteins [5•]; the extended residence times of eukaryotic polycomb [52] and trithorax [53] group proteins during mitosis relative to interphase may be due to suppression of FD via chromatin compaction.
FD needs to be considered in experiments seeking to quantify DNA-protein binding kinetics. One cannot take for granted that an off rate at zero protein concentration is the same as the off rate at elevated concentrations of competitor molecules, and indeed FD has been observed for a wide variety of types of protein-DNA interactions (Figure 1C), suggesting that it may well be generic. One might expect FD to lead to systematically larger estimates of faster off rates and therefore higher “equilibrium” Kd’s in titration experiments (e.g., EMSA “gel shifts”), versus kinetically-based dilution-dissociation (e.g., SPR “Biacore”) experiments [32•].
FD may well be characteristic of a wide range of ligand-receptor interactions. In situations where effectiveness of a receptor-ligand interaction depends on its residence time [49], it may be favorable to have a long intrisic residence time, but to rapidly turn over binding when a competing ligand is available. A therapeutically-relevant example is enhanced IgE-receptor dissociation by solution-phase variants of IgE [54]. IgE antibodies bind to high-affinity cell-surface receptors as part of immune response activation: drugs engineered to use FD to rapidly displace tightly bound IgEs promise to be useful in treatment of a wide range of allergic responses.
Finally, being a rather basic chemical-kinetic phenomenon, FD has broad application to fields outside of biology. FD may be useful in the design of responsive soft materials, e.g., polymer gels whose crosslinkers can undergo FD so as to rapidly adapt their crosslinking density, correlation length and stiffness in response to molecules in solution [55]. Such materials include associating polymers and vitrimers where crosslinking topology is fluctuating: FD could accelerate and make more precise crosslink reorganization, by allowing the combination of stable crosslinks with rapid dissocation kinetics in the presence of suitable competitor molecules.
Acknowledgement
This work was supported by NIH grants GM105847, CA193419 (PS-ON), and by subcontract to DK107980 (4DN). AE acknowledges H. Basak Senergin help with the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].von Hippel PH Berg OG: Facilitated target location in biological systems. J Biol Chem 1989, 264: 675–678. [PubMed] [Google Scholar]
- [2].Halford SE Marko JF: How do site-specific DNA-binding proteins find their targets? Nucl Acids Res 2004, 32: 3040–3052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3•].Kamar RI, Banigan EJ, Erbaş A, Giuntoli RD, Olvera de la Cruz M, Johnson RC, Marko JF: Facilitated dissociation of transcription factors from single DNA binding sites. Proc Nat Acad Sci USA 2017, 114: E3251–E3257.•Single-molecule study of FD for Fis and other proteins interacting with a short 27 bp DNA, showing large increase of off rate with increasing free protein concentration, with saturation at large concentration.
- [4].Hadizadeh N, Johnson RC, Marko JF: Facilitated dissociation of a nucleoid protein from the bacterial chromosome. J Bact 2016, 198: 1735–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5•].Chen TY, Santiago AG, Jung W, Krzemiński Å, Yang F, Martell DJ, Helmann JD, Chen P: Concentration- and chromosome-organization-dependent regulator unbinding from DNA for transcription regulation in living cells. Nat Comm 2015, 6: 7445.•In vivo observation of FD of a bacterial transcription factor.
- [6•].Joshi CP, Panda D, Martell DJ, Andoy NM, Chen TY, Gaballa A, Helmann JD, Chen P: Direct substitution and assisted dissociation pathways for turning off transcription by a MerR-family metalloregulator. Proc Nat Acad Sci USA 2012, 109: 15121–15126.•Single-molecule observation of facilitated dissociation for a bacterial transcription factor, showing clearly that the FD effects occur at the level of a single binding site.
- [7•].Beattie TR, Kapadia N, Nicolas E, Uphoff S, Wollman AJM, Leake MC, Reyes-Lamothe R: Frequent exchange of the DNA polymerase during bacterial chromosome replication. eLife 2017, 6: e21763.•Observation in vivo of DNA polymerase replacement at rates similar to those observe in vitro.
- [8].Lewis JS, Spenkelink LM, Jergic S, Wood EA, Monachino E, Horan NP, Duderstadt KE, Cox MM, Robinson A, Dixon NE, et al. : Single-molecule visualization of fast polymerase turnover in the bacterial replisome. eLife 2017, 6: e23932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9•].Loparo JJ, Kulczyk AW, Richardson CC, van Oijen AM: Simultaneous single-molecule measurements of phage T7 replisome composition and function reveal the mechanism of polymerase exchange. Proc Nat Acad Sci USA 2011, 108: 3584–3589.•Observation of rapid exchange kinetics for polymerase subunits.
- [10•].Alverdi V, Hetrick B, Joseph S, Komives EA: Direct observation of a transient ternary complex during IκBα-mediated dissociation of NF-κB from DNA. Proc Nat Acad Sci USA 2014, 111: 225–230.•Observation of a ternary complex during a “protein-stripping” reaction involving key eukaryoate transcription factors.
- [11].Luo Y, North JA, Rose SD, Poirier MG: Nucleosomes accelerate transcription factor dissociation. Nucl Acids Res 2014, 42: 3017–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Chen T, Cheng Y, Huang P, Chen P: Facilitated unbinding via multivalency-enabled ternary complexes: new paradigm for protein-DNA interactions. Acc. Chem. Res 2018, 51: 860–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Ha T: Single-Molecule Approaches Embrace Molecular Cohorts. Cell 2013, 154: 723–726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Åberg C, Duderstadt KE, van Oijen AM: Stability versus exchange: a paradox in DNA replication. Nucl Acids Res 2016, 44: 4846–4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Sing CE, Olvera de la Cruz M, Marko JF: Multiple-binding-site mechanism explains concentration-dependent unbinding rates of DNA-binding proteins. Nucl Acids Res 2014, 42: 3783–3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Bernasconi CF: 1976, . Relaxation Kinetics (Academic Press, New York: ). [Google Scholar]
- [17].Bresloff JL Crothers DM: DNA-ethidium reaction kinetics: demonstration of direct ligand transfer between DNA binding sites. . J Mol Biol 1975, 95: 103–23. [DOI] [PubMed] [Google Scholar]
- [18].Graham JS, Johnson RC, Marko JF: Concentration-dependent exchange accelerates turnover of proteins bound to double-stranded DNA. Nucl Acids Res 2011, 39: 2249–2259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Chen P, Keller A, Joshi C, Martell D, Andoy N, Benitez J, Chen T, Santiago A, Yang F: Single-molecule dynamics and mechanisms of metalloregulators and metallochaperones. Biochem 2013, 52: 7170–7183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Bergqvist S, Alverdi V, Mengel B, Hoffmann A, Ghosh G, Komives EA: Kinetic enhancement of NF-κB dissociation by IκBα. Proc Nat Acad Sci USA 2009, 106: 19328–19333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Erbaş A, de la Cruz MO, Marko JF: Effects of electrostatic interactions on ligand dissociation kinetics. Phys Rev E 2018, 97: 022405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Jung W Chen P: Biphasic unbinding of Zur from DNA for transcription (de)repression in Live Bacteria. Bioaxiv.org 2018, : 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Hansen AS, Pustova I, Cattoglio C, Tjian R, Darzacq X: CTCF and cohesin regulate chromatin loop stability with distinct dynamics. . eLife 2017, 6: e25776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Schneider RJ Wetmur JG: Kinetics of transfer of Escherichia coli single strand DNA binding protein between single-stranded DNA molecules. Biochemistry 1982, 21: 608–615. [DOI] [PubMed] [Google Scholar]
- [25].Fried MG Crothers DM: Kinetics and mechanism in the reaction of gene regulatory proteins with DNA. Journal of molecular biology 1984, 172: 263–282. [DOI] [PubMed] [Google Scholar]
- [26].Menetski JP Kowalczykowski SC: Transfer of recA protein from one polynucleotide to another. Effect of ATP and determination of the processivity of ATP hydrolysis during transfer. J Biol Chem 1987, 262: 2093–2100. [PubMed] [Google Scholar]
- [27].Aragay A, Diaz P, Daban J: Association of nucleosome core particle DNA with different histone oligomers. Transfer of histones between DNA-(H2A,H2B) and DNA-(H3,H4) complexes. J Mol Biol 1988, 204: 141–54. [DOI] [PubMed] [Google Scholar]
- [28].Ruusala T Crothers DM: Sliding and intermolecular transfer of the lac repressor: kinetic perturbation of a reaction intermediate by a distant DNA sequence. Proc Nat Acad Sci USA 1992, 89: 4903–4907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lieberman BA Nordeen SK: DNA intersegment transfer, how steroid receptors search for A target site. J Bio Chem 1997, 272: 1061–1068. [DOI] [PubMed] [Google Scholar]
- [30].Lorch Y, Zhang M, Kornberg RD: Histone octamer transfer by a chromatin-remodeling complex. Cell 1999, 96: 389–92. [DOI] [PubMed] [Google Scholar]
- [31].Kozlov AG Lohman TM: Kinetic mechanism of direct transfer of Escherichia coli SSB tetramers between single-stranded DNA molecules. Biochemistry 2002, 41: 11611–11627. [DOI] [PubMed] [Google Scholar]
- [32•].Skoko D, Wong B, Johnson RC, Marko JF: Micromechanical analysis of the binding of DNA–bending proteins HMGB1, NHP6A, and HU reveals their ability to form highly stable DNA–protein complexes. Biochemistry 2004, 43: 13867–13874.•Single-DNA experiment observing very slow off-rates for HU, HMGB1 and NHP6A from DNA at zero concentration, but fast off-rates in the presence of competitor DNA, using a fluorescence-label-free DNA mechanics assay.
- [33].Zimmerman J Maher LJ III: Transient HMGB protein interactions with B-DNA duplexes and complexes. Biochem Biophys Res Commun 2008, 371: 79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Doucleff M Clore GM: Global jumping and domain-specific intersegment transfer between DNA cognate sites of the multidomain transcription factor Oct-1. Proc Nat Acad Sci USA 2008, 105: 13871–13876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sidorova NY, Scott T, Rau DC: DNA concentration–dependent dissociation of EcoRI: Direct transfer or reaction during hopping. Biophys J 2013, 104: 1296–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Esadze A Iwahara J: Stopped-flow fluorescence kinetic study of protein sliding and intersegment transfer in the target DNA search process. J Mol Bio 2014, 426: 230–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Lee KS, Marciel AB, Kozlov AG, Schroeder CM, Lohman TM, Ha T: Ul-trafast redistribution of E. coli SSB along long single-stranded DNA via intersegment transfer. J Mol Bio 2014, 426: 2413–2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Itoh Y, Murata A, Takahashi S, Kamagata K: Intrinsically disordered domain of tumor suppressor p53 facilitates target search by ultrafast transfer between different DNA strands. Nucl Acids Res 2018, 46: 7261–7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Giuntoli RD, Linzer NB, Banigan EJ, Sing CE, de la Cruz MO, Graham JS, Johnson RC, Marko JF: DNA-segment-facilitated dissociation of Fis and NHP6A from DNA detected via single-molecule mechanical response. J Mol Biol 2015, 427: 3123–3136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Kunzelmann S, Morris C, Chavda AP, Eccleston JF, Webb MR: Mechanism of interaction between single-stranded DNA binding protein and DNA. Biochem 2010, 49: 843–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Gibb B, Ye LF, Gergoudis SC, Kwon Y, Niu H, Sung P, Greene EC: Concentration-dependent exchange of replication protein A on single-stranded DNA revealed by single-molecule imaging. PLOS ONE 2014, 9: e87922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Paramanathan T, Reeves D, Friedman LJ, Kondev J, Gelles J: A general mechanism for competitor-induced dissociation of molecular complexes. Nat Comm 2014, 5: 5207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Tsai MY, Zhang B, Zheng W, Wolynes PG: Molecular mechanism of facilitated dissociation of Fis protein from DNA. J Am Chem Soc 2016, : 13497–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Cocco SMJF Monasson R: Stochastic ratchet mechanisms for replacement of proteins bound to DNA. . Phys Rev Lett 2014, 112: 238101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kim S, Broströmer E, Xing D, Jin J, Chong S, Ge H, Wang S, Gu C, Yang L, Gao YQ, et al. : Probing allostery through DNA. Science 2013, 339: 816–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Tuttle LM, Pacheco D, Warfield L, Luo J, Ranish J, Hahn S, Klevit RE: Gcn4-mediator specificity is mediated by a large and dynamic fuzzy Protein–Protein complex. Cell Rep 2018, 22: 3251–3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Record MT, Lohman TM, De Haseth P: Ion effects on ligand-nucleic acid interactions. J Mol Biol 1976, . [DOI] [PubMed] [Google Scholar]
- [48].Mascotti DP Lohman TM: Thermodynamic extent of counterion release upon binding oligolysines to single-stranded nucleic acids. . Proc Natl Acad Sci USA 1990, 87: 3142–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Tummino PJ Copeland RA: Residence time of receptor–ligand complexes and its effect on biological function. Biochemistry 2008, 47: 5481–5492. [DOI] [PubMed] [Google Scholar]
- [50].Corzo J: Time, the forgotten dimension of ligand binding teaching. Biochem Mol Biol Ed 2006, 34: 413–416. [DOI] [PubMed] [Google Scholar]
- [51].Tanner NA, Tolun G, Loparo JJ, Jergic S, Griffith JD, Dixon NE, van Oijen AM: E. coli DNA replication in the absence of free β clamps. EMBO J 2011, 30: 1830–1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Fonseca JP, Steffen PA, Müller S, Lu J, Sawicka A, Seiser C, Ringrose L: In vivo Polycomb kinetics and mitotic chromatin binding distinguish stem cells from differentiated cells. Genes Dev 2012, 26: 857–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Steffen PA, Fonseca JP, Gänger C, Dworschak E, Kockmann T, Beisel C, Ringrose L: Quantitative in vivo analysis of chromatin binding of Polycomb and Trithorax group proteins reveals retention of ASH1 on mitotic chromatin. Nucl Acids Res 2013, 41: 5235–5250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Kim B, Eggel A, Tarchevskaya SS, Vogel M, Prinz H, Jardetzky TS: Accelerated disassembly of IgE–receptor complexes by a disruptive macromolecular inhibitor. Nature 2012, 491: 613–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Goodrich CP, Brenner MP, Ribbeck K: Enhanced diffusion by binding to the crosslinks of a polymer gel. Nat Comm 2018, 9: 4348. [DOI] [PMC free article] [PubMed] [Google Scholar]