Abstract
CDKN2A and CDK4 are well-established melanoma susceptibility genes, but their effect on tumor location/distribution is unknown. We used a case-case study design to assess for differences in tumor location between mutation carriers (CDKN2A =141 patients, 348 melanomas; CDK4 =15 patients, 54 melanomas) and non-carriers (104 patients, 157 melanomas) in U.S. melanoma-prone families. Associations between groups were assessed with chi-square tests. Odds ratios (ORs) for tumor location were adjusted for diagnosis age, gender, and superficial spreading subtype. Models included random effects to account for within individual/family correlations. Compared to having a truncal melanoma, CDK4 (vs. non-carriers: lower extremities OR=14.5, 95% CI, 5.02−42.0, P<.001; upper extremities OR=6.88, 95% CI, 2.37−19.9, P<.001; head/neck OR=18.6, 95% CI, 4.04−85.2, P<.001) and CDKN2A (vs. non-carriers: lower extremities OR=3.01, 95% CI, 1.56−5.82, P<.05; upper extremities OR=1.91, 95% CI, 1.03−3.52, P<.05; head/neck OR=5.40, 95% CI, 2.10−13.9, P<.001) carriers had higher odds of developing melanoma at all other sites. Similar findings were observed for analyses stratified by gender, age, and first vs. subsequent melanoma diagnoses. Further studies are needed to understand the biology underlying these genotype-associated patterns of tumor development, which could provide new insights into melanoma treatment and prevention.
Keywords: melanoma, tumor location, cyclin-dependent kinase inhibitor 2A (CDKN2A), cyclin-dependent kinase 4 (CDK4), cancer screening, family research
INTRODUCTION
Approximately 90,000 cases of melanoma are diagnosed annually in the United States. Between 5 and 10% of new diagnoses will occur in patients with a family history of melanoma. Further, among melanoma-prone families, germline mutations of CDKN2A will be identified in 20–40% of families, with the highest likelihood of a mutation occurring in families with 3 or more members with melanoma.(Florell et al., 2005, Goldstein et al., 2007, Hayward, 2003, SEER, 2018)
The CDKN2A locus encodes two proteins, p16 and p14(ARF), which function as tumor suppressors in the Rb/E2F and HDM2/p53 pathways respectively (OMIM: 600160). Mutations in CDKN2A are associated with a highly elevated risk for melanoma, and to a lesser degree, an increased susceptibility to pancreatic cancer. A subset of alterations including large deletions or mutations affecting both the pl6 and pl4 proteins may occasionally predispose patients to multiple cancer types in addition to melanoma and pancreatic cancer including malignant peripheral nerve sheath tumors, gliomas, breast cancer, and colon cancer.(Goldstein et al., 2006, Goldstein et al., 1995, Hussussian et al., 1994, Prowse et al., 2003, Randerson-Moor et al., 2001, Sargen et al., 2016, Vanneste et al., 2013)
Germline mutations of CDK4 are also associated with an increased risk for melanoma, but are less common and have been reported in less than 20 melanoma-prone families.(Puntervoll et al., 2013, Soufir et al., 1998, Zuo et al., 1996) Pathogenic mutations in CDK4 affect the p16 binding site of this protein, thereby allowing CDK4 to remain in an activated state, which promotes cell division.(Zuo et al., 1996)
Germline mutations of CDKN2A and CDK4 are associated with a significantly increased lifetime risk for melanoma.(Bishop et al., 2002, Puntervoll et al., 2013, Soufir et al., 1998, Zuo et al., 1996) In this study, we assessed differences in the anatomic distribution of melanomas between mutation carriers (CDKN2A, CDK4) and those without a mutation in either gene (non-carriers) from U.S. melanoma-prone families. If present, associations between genotype and tumor distribution could potentially further our understanding of melanoma tumor development among individuals with a familial predisposition to melanoma.
RESULTS
Clinical Characteristics of Carriers and Non-carriers in Melanoma-prone Families
We evaluated the anatomic location for 559 tumors (CDKN2A carriers having 348 tumors; CDK4 carriers having 54 tumors; non-carriers having 157 tumors) obtained from 260 individuals with melanoma (141 CDKN2A carriers; 15 CDK4 carriers; 104 non-carriers) (Figure 1) (Table 1). All three groups were similar with respect to the gender composition (Table 1). Compared to individuals without a known mutation (median age=45 years), mutation carriers were younger (median age: CDKN2A carrier, 31 years, PWMW <.001; CDK4 carrier, 35 years, PWMW =0.003) and more likely to develop superficial spreading melanomas (Chi-square P<.001 for CDKN2A and CDK4 carriers) than other histologic subtypes (Table 1).
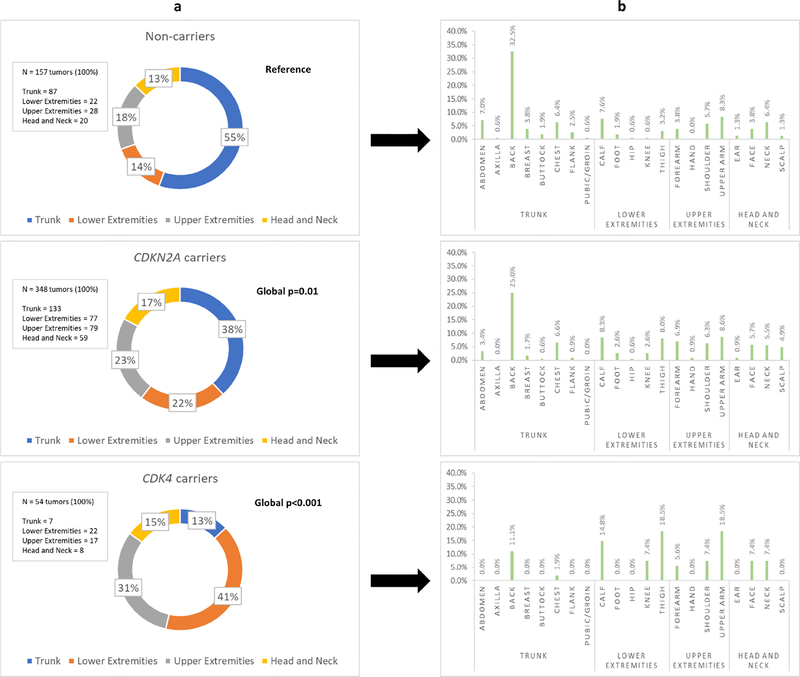
Figure 1: Cutaneous patterns of melanomagenesis in U.S. melanoma-prone families with and without germline CDKN2A and CDK4 mutations.
The above figure illustrates the regional distribution (column “a”) and specific biopsy sites (column “b”) for melanomas diagnosed in individuals from U.S. melanoma-prone families according to genotype. Global p-values comparing differences in melanoma distribution between mutation carriers (CDKN2A or CDK4) and non-carriers (individuals without a CDKN2A or CDK4 mutation) with adjustments for within individual and within family correlations, age at diagnosis, gender, and superficial spreading melanoma subtype are provided in column “a”.
Table 1:
Clinical and histologic features of U.S. melanoma-prone families
| Non-carriers | CDKN2A carriers | CDK4 carriers | CDKN2A carriers vs. Non-carriers | CDK4 carriers vs. Non-carriers | CDK4 carriers vs. CDKN2A carriers | ||||
|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | P-value | P-value | P-value | |
| Total patients | 104 | 100% | 141 | 100% | 15 | 100% | |||
| Female patients | 49 | 47% | 70 | 50% | 8 | 53% | 0.701 | 0.651 | 0.791 |
| Median age of first melanoma diagnosis (range) | 45 (18–95) | 31 (10–69) | 35 (24–64) | <0.0012 | 0.0032 | 0.212 | |||
| Average number of melanomas per patient (range) | 1.5 (1–6) | 2.5 (1–30) | 3.6 (1–13) | ||||||
| Total melanomas | 157 | 100% | 348 | 100% | 54 | 100% | |||
| Histologic Subtype | <0.0013 | <0.0013 | 0.113 | ||||||
| Acral Lentiginous Melanoma |
1 | 0.6% | 1 | 0.3% | 0 | 0% | |||
| Lentigo Maligna Melanoma |
17 | 11% | 9 | 3% | 0 | 0% | |||
| Nodular Melanoma | 12 | 8% | 14 | 4% | 0 | 0% | |||
| Superficial Spreading Melanoma |
94 | 60% | 275 | 79% | 51 | 94% | |||
| Not Otherwise Specified |
33 | 21% | 49 | 14% | 3 | 6% | |||
Chi-square test comparing differences in gender (female vs. male) between groups
Wilcoxon-Mann-Whitney test comparing differences in median age of first melanoma diagnosis between groups
Fisher exact test comparing overall differences in histologic subtype between groups
Tumor Distribution in Melanoma-prone Families
Among melanoma-prone families, tumor occurrence at non-truncal sites was strongly associated with the presence of either a CDKN2A (CDKN2A carriers vs. non-carriers: 62% vs. 45%, Global P=.01) or CDK4 (CDK4 vs. non-carriers: 87% vs. 45%, Global P<.001) mutation after adjustment for age at diagnosis, gender, and superficial spreading melanoma subtype (Figure 1). Compared to having a melanoma of the trunk, mutation carriers had higher odds of developing melanoma at any non-truncal site (CDKN2A carriers vs. non-carriers, OR=2.77, 95% CI, 1.60 to 4.80, P<.001; CDK4 carriers vs. non-carriers, OR=l1.6, 95% CI, 4.09 to 32.7, .P<.001) (Table 2). Statistically significant associations were also observed for each non-truncal anatomic region (lower extremities, upper extremities, head and neck) (Table 2). Similar findings were also observed for gender-specific (Table S1) and age-stratified (Table S2) comparisons.
Table 2:
Adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for tumor development at specific anatomic site compared to trunk
| Anatomic Site | CDKN2A carriers vs. Non-carriers | CDK4 carriers vs. Noncarriers | CDK4 carriers vs. CDKN2A carriers | |||
|---|---|---|---|---|---|---|
| OR1 | 95% CI | OR1 | 95% CI | OR1 | 95% CI | |
| Trunk | Reference | Reference | Reference | Reference | Reference | Reference |
| Non-T runcal | 2.77** | 1.60–4.80 | 11.6** | 4.09–32.7 | 4.18* | 1.59–11.0 |
| Lower extremities | 3.01* | 1.56–5.82 | 14.5** | 5.02–42.0 | 4.84* | 1.89–12.4 |
| Upper extremities | 1.91* | 1.03–3.52 | 6.88** | 2.37–19.9 | 3.62* | 1.36–9.65 |
| Head and neck | 5.40** | 2.10–13.9 | 18.6** | 4.04–85.2 | 3.45 | 0.93–12.8 |
P<. 05
P<.001
Multinomial random effects model adjusting for within individual and within family correlations, age at diagnosis, gender, and superficial spreading melanoma subtype
First and Subsequent Tumor Diagnoses in Melanoma-prone Families
Among first melanoma diagnoses in melanoma-prone families, non-truncal melanomas were more common for CDKN2A (CDKN2A carriers vs. non-carriers: 57% vs. 44%, Global P=0.04) and CDK4 (CDK4 carriers vs. non-carriers: 79% vs. 44%, Global P=.03) carriers in adjusted models (Table S3). Compared to having a melanoma of the trunk, mutation carriers had higher odds of developing their first primary melanoma at any non-truncal site (CDKN2A carriers vs. non-carriers, OR=1.98, 95% CI, 1.07 to 3.67, P<.05; CDK4 carriers vs. non-carriers, OR=6.17, 95% CI, 1.97 to 19.3, P<.05) (Table S3). Comparable associations and trends for anatomic distribution were observed for analyses of subsequent melanomas, that is, melanomas diagnosed after the patient’s first primary melanoma (Table S3). Within each group (CDKN2A carriers, CDK4 carriers, and non-carriers), anatomic distributions of tumors were similar (Chi-square P>.05) for the first and subsequent melanoma diagnoses (Table S3).
Body Surface Area and Tumor Distribution in Melanoma-prone Families
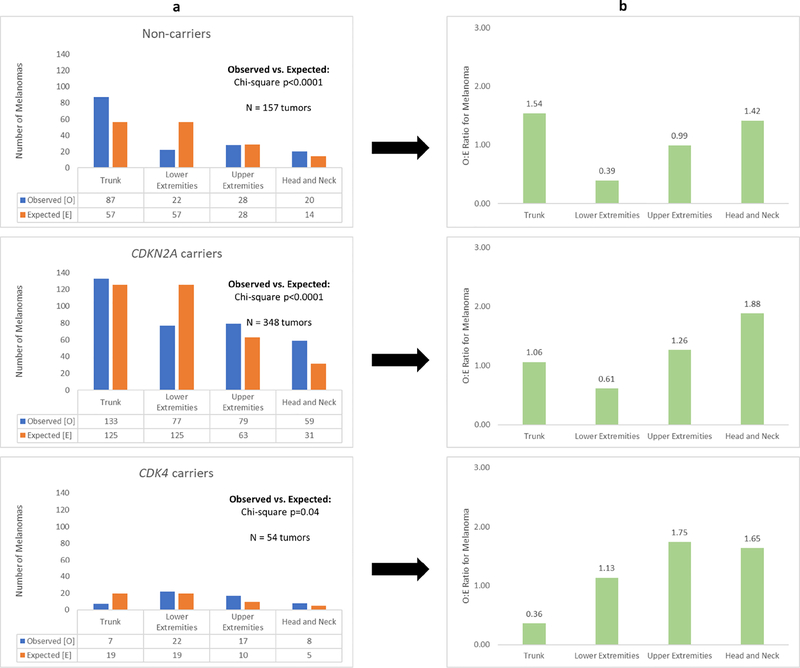
For each of the three groups (CDKN2A carriers, CDK4 carriers, and non-carriers), there was a statistically significant difference (P<.05) in the observed and expected distribution of melanomas (Figure 2). Although based on smaller numbers of tumors, the overall O/E (observed/expected) patterns for CDK4 (truncal melanomas, O/E = 0.37) carriers differed more from the patterns seen for the two other groups (non-carriers, truncal melanomas, O/E = 1.52; CDKN2A carriers, truncal melanomas, O/E = 1.06).
Figure 2: Body surface area and melanoma distribution.
The above figure illustrates the observed and expected number of melanomas for each anatomic region according to genotype. In column “a”, the total number of melanomas (N) for each group (non-carriers, CDKN2A carriers, CDK4 carriers) was multiplied by the body surface area of each anatomic region to calculate the expected number of melanomas. Differences between the observed and expected melanoma distribution were assessed by chi-square test (column “a”). The ratio of observed to expected number of melanomas for each anatomic region is presented in column “b”.
CDKN2A versus CDK4 Carriers in Melanoma-prone Families
While mutation carriers (CDKN2A and CDK4) both demonstrated increased odds of developing non-truncal tumors compared to non-carriers in U.S. melanoma-prone families, CDK4 carriers were also more likely to develop non-truncal tumors than CDKN2A carriers (CDK4 carriers vs. CDKN2A carriers: 87% vs 62%, Global P<.001). Further, the likelihood of developing a non-truncal tumor (CDK4 carriers vs. CDKN2A carriers: OR=4.18, 95% CI, 1.59 to 11.0, P<.05) was strongest for the lower extremities (CDK4 carriers vs. CDKN2A carriers: OR=4.84, 95% CI, 1.89−12.4, P<.05) and upper extremities (CDK4 carriers vs. CDKN2A carriers: OR=3.62, 95% CI, 1.36 to 9.65, P<.05) (Table 2). Similar associations and trends were observed for first and subsequent melanoma diagnoses (Table S3) as well as comparisons stratified by gender (Table S1) and age (Table S2).
Familial versus Non-Familial Cases from SEER
More than half of all melanomas diagnosed in non-carriers from melanoma-prone families were located on the trunk, which was significantly higher than the frequency of truncal tumors observed in predominantly non-familial SEER cases (non-carriers vs. SEER cases: 56% vs. 36%, Global P=.04) or in the familial mutation carrier groups (Table 3). CDK4 carriers were less likely to develop truncal tumors than was observed in SEER, but these differences were not statistically significant (CDK4 carriers vs. SEER cases: 21% vs. 36%, Global P=0.10). In contrast, the tumor distribution patterns for CDKN2A carriers and SEER cases were similar (global P=0.24).
Table 3:
Location of first melanoma diagnosis: Comparison of U.S. melanoma-prone families to Surveillance, Epidemiology, and End Results (SEER) data
| Non-carriers |
CDKN2A carriers |
CDK4 carriers | SEER cases (1973–2015) | SEER cases vs. Non–Carriers | SEER cases vs. CDKN2A carriers | SEER cases vs. CDK4 Carriers | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N | % | N | % | N | % | N | % | P-value | P-value | P-value | |
| Total patients | 104 | 100% | 141 | 100% | 15 | 100% | 313,237 | 100% | |||
|
Female patients |
49 | 47% | 70 | 50% | 8 | 53% | 137,931 | 44% | 0.521 | 0.331 | 0.561 |
|
Median age of diagnosis (range) |
45 (18–95) |
31 (10–69) |
35 (24–64) |
62 (1–87) |
<0.00012 | <0.00012 | <0.00012 | ||||
|
Total melanomas4 |
117 | 100% | 161 | 100% | 24 | 100% | 313,237 | 100% | |||
| Anatomic Site | 0.043 | 0.243 | 0.103 | ||||||||
| Trunk | 65 | 56% | 70 | 44% | 5 | 21% | 111,660 | 36% | |||
| Lower extremities |
16 | 14% | 31 | 19% | 6 | 25% | 61,229 | 20% | |||
| Upper extremities |
22 | 19% | 33 | 21% | 9 | 38% | 83,336 | 27% | |||
| Head and neck | 14 | 12% | 27 | 17% | 4 | 17% | 57,012 | 18% | |||
Chi-square test comparing differences in gender (female vs. male) between groups
Wilcoxon-Mann-Whitney test comparing differences in median age of first melanoma diagnosis between groups
Global p-value adjusting for age at diagnosis, gender, and superficial spreading melanoma subtype
Analysis restricted to first primary melanomas for non-carriers, CDKN2A carriers, and CDK4 carriers. Some noncarriers (N=8), CDKN2A carriers (N=15), and CDK4 carriers (N=4) were diagnosed with multiple melanomas at the time of their first melanoma diagnosis.
DISCUSSION
In our analysis of U.S. melanoma-prone families, the anatomic distribution of melanomas differed substantially depending upon mutation status. CDKN2A and CDK4 carriers were significantly more likely to have non-truncal tumors than individuals without a mutation.
Ascertainment for all three familial groups was similar. Mutation status was unknown at study entry for almost all families, except for a few recently ascertained kindreds (with known CDKN2A mutations). Tumor distributions according to genotype also showed consistent results even when analyses were stratified according to age at diagnosis, gender, and first versus subsequent melanoma diagnoses. Further, the study population was geographically diverse for all familial groups with individuals living across the United States (Northeast, Southeast, Midwest, Southwest, West Coast, Alaska, Hawaii).
Non-truncal sites are exposed to either chronic (head/neck) or intermittent (upper and lower extremities) ultraviolet (UV) radiation in contrast to the trunk, which is usually covered by clothing. Therefore, our results suggest that UV exposure is an important modifier of melanoma risk among individuals with germline CDKN2A and CDK4 mutations, which is consistent with prior studies demonstrating increased melanoma penetrance among CDKN2A carriers living closer to the equator where the ultraviolet index is higher.(Bishop et al., 2002) Consistent site-specific UV exposure data (UVA exposure, UVB exposure, number of blistering sunburns, tanning bed use, etc.) were not available for our analysis, and therefore, further investigation is needed to understand the potential modifying effect of UV radiation on melanoma tumor development among individuals with germline CDKN2A and CDK4 mutations.
CDKN2A and CDK4 carriers in melanoma-prone families also demonstrated a high-frequency of superficial spreading melanomas, which is consistent with prior studies.(Sargen et al., 2015, Taylor et al., 2016) This association suggests that alterations of the CDKN2A(p16)/CDK4 pathway are important in the development of tumors with this histologic subtype. Among CDKN2A carriers, BRAF mutations occur at a lower frequency than observed for sporadic melanomas.(Jovanovic et al., 2010) Further, BRAF-wild type melanomas often occur on the extremities.(Maldonado et al., 2003) Therefore, CDKN2A carriers may be at risk for melanomagenesis at non-truncal sites given their increased susceptibility to BRAF-wild type tumors. To assess this hypothesis, tumor BRAF mutational status, which was not available for our analysis, should be assessed in follow-up studies evaluating tumor distribution patterns in melanoma-prone families.
Interestingly, CDK4 carriers were more likely to develop non-truncal tumors than CDKN2A carriers and non-carriers in melanoma-prone families. Overall, 87% (47 of 54 tumors) of all melanomas diagnosed among CDK4 carriers were non-truncal. The highest number of tumors were observed on the lower extremities followed by the upper extremities. These patterns were consistent across all subset and sensitivity analyses. While these observations suggest that CDK4 mutations are strongly associated with non-truncal tumor development, our sample size (54 tumors, 15 patients, 2 families) was relatively small, and therefore additional studies are necessary to confirm these findings.
Our results for U.S. melanoma-prone families are consistent with two prior studies that reported the phenotypic characteristics for CDKN2A (Taylor et al, N=670 individuals)(Taylor et al., 2016) and CDK4 (Puntervoll et al, N=140)(Puntervoll et al., 2013) carriers from North America, Europe, and Australia. Both studies included less detailed patient/tumor data from a subset of U.S. melanoma-prone families that were analyzed in the current study. Interestingly, non-carriers (N=l, 258 individuals) reported by Taylor et al. were less likely to have melanoma occur on the trunk (38%) compared to our U.S. non-carriers (55%). However, country-specific data were not reported by Taylor et al. to assess whether non-carriers in specific countries/regions develop truncal melanomas at an increased frequency similar to what was observed for non-carriers in our U.S. melanoma-prone families. Melanoma occurrence on the trunk for U.S. non-carriers was also considerably higher than that observed for non-familial cases from SEER, 1973–2015 = 36%. Further studies are needed to examine whether a high occurrence of truncal melanomas is restricted to U.S. non-carrier melanoma-prone families.
Non-carrier patients/families, who are at increased risk for melanoma, may carry mutations in other melanoma susceptibility genes. In the vast majority of cases (n=96/104 patients), WES and WGS data were available, and single gene mutations in known high-penetrance melanoma susceptibility genes (e.g. BAP1, POT1) were not identified, suggesting that melanoma in these families may be caused by mutations in novel high-penetrance genes or multiple genes with moderate/low penetrance and these unknown genes may interact with UV exposure in different ways compared with CDKN2A/CDK4.
Despite the tendency to develop non-truncal tumors, mutation carriers also developed a substantial number of melanomas on the trunk (CDKN2A carriers = 38%; CDK4 carriers = 13%). The wide anatomic distribution of tumor sites among mutation carriers highlights the importance of performing full-body skin exams to screen for melanoma, which is the current standard of care for melanoma-prone families.(Johnson et al., 2017)
In conclusion, this study observed significant differences in the melanoma distribution patterns between mutation carriers (CDKN2A, CDK4) and individuals without a known mutation in U.S. melanoma-prone families with mutation carriers developing the majority of their tumors at non-truncal sites. Given the relatively small sample size of our study, it will be important for future studies to validate our findings before recommending any changes to melanoma screening for members of melanoma-prone families. Further investigation is also necessary to understand the biology underlying these genotype-associated patterns of tumor development, which could provide new insights into melanoma treatment and prevention.
MATERIALS AND METHODS
Study Population
Data for this study came from a non-population-based family study from the Division of Cancer Epidemiology and Genetics at the National Cancer Institute (NCI). Families with and without germline CDKN2A and CDK4 mutations were ascertained through self or health professional referrals, and have been followed prospectively for up to 40+ years, starting in 1976. All patients self-identified as non-Hispanic white. Tumor diagnosis and location were ascertained from medical records (pathology reports, physician notes). Given the possibility of phenocopies (individuals who express a particular phenotype (eg. melanoma) without the associated genotype (eg. CDKN2A or CDK4 mutation)) in melanoma-prone families, we included only individuals whose germline mutation status was known in our analyses.(Helgadottir et al., 2018) Phenocopies with known mutation status (i.e. CDKN2A or CDK4 negative) were included in the non-carrier group. Additional details of the study population are available online and have been described in prior publications.(Goldstein et al., 2018, Goldstein et al., 2000, Goldstein et al., 2017, Tucker et al., 2018) Written informed consent was obtained from all participants or their legal guardians prior to participating in this NCI institutional review board-approved protocol (NCI 002-0211; ClinicalTrials.gov identifier ).
CDKN2A and CDK4 Mutation Status
During this longitudinal study multiple techniques, including single-strand conformation polymorphism (SSCP), Sanger sequencing, array comparative genomic hybridization (aCGH), whole exome sequencing (WES), and whole genome sequencing (WGS), have been used to screen for CDKN2A and CDK4 mutations depending upon the technologies that were available at the time of patients’ enrollment or mutation testing. Testing for mutations in CDKN2A/CDK4 was performed on stored blood specimens. For the current study, we grouped individuals into “carriers” (CDKN2A and CDK4 carriers) and “non-carriers” (no mutation identified).
Statistical Analysis
The primary objective of this study was to determine if tumor distribution patterns are associated with genotype for individuals in U.S. melanoma-prone families. We used chi-square and Wilcoxon-Mann-Whitney (WMW) tests to assess differences in characteristics between groups. We modeled the anatomical location of each melanoma as a function of carrier status of the patients and other attributes (age at diagnosis, gender, and whether or not tumors were superficial spreading subtype) using multinomial models with random effects to account for correlations of multiple melanomas within individuals and correlations of family members (PROC GLIMMIX SAS 9.4). Tumor locations were grouped into 4 regional categories (lower extremities; upper extremities; trunk; head/neck). The probabilities of having a tumor at a specific location were modeled using polytomous logistic regression to obtain odds ratios (ORs) for association of tumor location with carrier status, adjusted for age, gender, and superficial spreading subtype. Adjusted ORs are presented unless specified otherwise. Models were also stratified by gender and overall median age of diagnosis for all three groups (≤39 or >39 years at diagnosis). Since patients developed different numbers of melanomas, we conducted sensitivity analyses in which we restricted models to the first diagnosed melanoma for each patient.
The effect of body surface area on melanoma distribution was evaluated by assessing differences in the observed and expected number of melanomas (see equation below) for each anatomic region using a chi-square test. The Wallace Rule of Nines was used to estimate body surface area for each anatomic region.(Orgill, 2009)
For this evaluation, our null hypothesis was that no difference would exist in the observed and expected distributions of melanoma if body surface area were an important determinant of melanoma distribution.
Two-sided p-value cut-offs of P<.05 defined statistical significance. Statistical analyses were performed using SAS 9.4 (Cary, NC) and STATA 15 (College Station, TX).
Familial vs. Non-Familial Cases
In a secondary analysis, we used chi-square and WMW tests to compare differences in attributes (gender, median age of diagnosis) and tumor location between our familial groups (CDKN2A carriers, CDK4 carriers, non-carriers) and predominantly sporadic cases (Surveillance, Epidemiology, and End Results (SEER) database).(SEER*Stat) For the familial groups, our analysis of tumor location was restricted to an individual’s first melanoma diagnosis in order to be consistent with SEER data, which is composed of first primary tumors. SEER cases diagnosed in the time period of 1973 to 2015 were analyzed.(SEER*Stat)
Supplementary Material
ACKNOWLEDGMENTS
We would like to thank the families who agreed to participate in this study (Clinicaltrials.gov ID: ) as well as the nurses and research assistants who have contributed to this study since its inception. Preliminary data from this manuscript was presented at annual meetings for GenoMel (April 8 – April 10, 2019, Athens, Greece) and the Society for Investigative Dermatology (May 8 – May 11, 2019, Chicago, IL).
Funding
This research was supported entirely by the Intramural Research Program of the Division of Cancer Epidemiology and Genetics, National Cancer Institute, National Institutes of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DATA AVAILABILITY
No datasets were generated or analyzed during the current study.
CONFLICTS OF INTEREST
The authors have no relevant conflicts of interest, financial or other, related to the contents of this article.
REFERENCES:
- Aspinwall LG, Taber JM, Kohlmann W, Leaf SL, Leachman SA. Unaffected family members report improvements in daily routine sun protection 2 years following melanoma genetic testing. Genet Med 2014;16(11):846–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop DT, Demenais F, Goldstein AM, Bergman W, Bishop JN, Bressac-de Paillerets B, et al. Geographical variation in the penetrance of CDKN2A mutations for melanoma. J Natl Cancer Inst 2002;94(12):894–903. [DOI] [PubMed] [Google Scholar]
- Floreil SR, Boucher KM, Garibotti G, Astle J, Kerber R, Mineau G, et al. Population-based analysis of prognostic factors and survival in familial melanoma. J Clin Oncol 2005;23(28):7168–77. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Gillanders EM, Hayward NK, Avril MF, et al. High-risk melanoma susceptibility genes and pancreatic cancer, neural system tumors, and uveal melanoma across GenoMEL. Cancer Res 2006;66(20):9818–28. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Chan M, Harland M, Hayward NK, Demenais F, Bishop DT, et al. Features associated with germline CDKN2A mutations: a GenoMEL study of melanoma-prone families from three continents. J Med Genet 2007;44(2):99–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with pl6INK4 mutations. N Engl J Med 1995;333(15):970–4. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Stidd KC, Yang XR, Fraser MC, Tucker MA. Pediatric melanoma in melanoma-prone families. Cancer 2018; 124(18):3715–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldstein AM, Struewing JP, Chidambaram A, Fraser MC, Tucker MA. Genotype-phenotype relationships in U.S. melanoma-prone families with CDKN2A and CDK4 mutations. J Natl Cancer Inst 2000;92(12):1006–10. [DOI] [PubMed] [Google Scholar]
- Goldstein AM, Xiao Y, Sampson J, Zhu B, Rotunno M, Bennett H, et al. Rare germline variants in known melanoma susceptibility genes in familial melanoma. Hum Mol Genet 2017; 26(24):4886–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayward NK. Genetics of melanoma predisposition. Oncogene 2003;22(20):3053–62. [DOI] [PubMed] [Google Scholar]
- Helgadottir H, Olsson H, Tucker MA, Yang XR, Hoiom V, Goldstein AM. Phenocopies in melanoma-prone families with germ-line CDKN2A mutations. Genet Med 2018;20(9): 1087–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussussian CJ, Struewing JP, Goldstein AM, Higgins PA, Ally DS, Sheahan MD, et al. Germline pl6 mutations in familial melanoma. Nat Genet 1994;8(1): 15–21. [DOI] [PubMed] [Google Scholar]
- Johnson MM, Leachman SA, Aspinwall LG, Cranmer LD, Curiel-Lewandrowski C, Sondak VK, et al. Skin cancer screening: recommendations for data-driven screening guidelines and a review of the US Preventive Services Task Force controversy. Melanoma Manag 2017;4(1): 13–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovic B, Egyhazi S, Eskandarpour M, Ghiorzo P, Palmer JM, Bianchi Scarra G, et al. Coexisting NRAS and BRAF mutations in primary familial melanomas with specific CDKN2A germline alterations. J Invest Dermatol 2010;130(2):618–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maldonado JL, Fridlyand J, Patel H, Jain AN, Busam K, Kageshita T, et al. Determinants of BRAF mutations in primary melanomas. J Natl Cancer Inst 2003;95(24): 1878–90. [DOI] [PubMed] [Google Scholar]
- Orgill DP. Excision and skin grafting of thermal bums. N Engl J Med 2009;360(9):893–901. [DOI] [PubMed] [Google Scholar]
- Prowse AH, Schultz DC, Guo S, Vanderveer L, Dangel J, Bove B, et al. Identification of a splice acceptor site mutation in p16INK4A/pl4ARF within a breast cancer, melanoma, neurofibroma prone kindred. J Med Genet 2003;40(8):el02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puntervoll HE, Yang XR, Vetti HH, Bachmann IM, Avril MF, Benfodda M, et al. Melanoma prone families with CDK4 germline mutation: phenotypic profile and associations with MC1R variants. J Med Genet 2013;50(4):264–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Randerson-Moor JA, Harland M, Williams S, Cuthbert-Heavens D, Sheridan E, Aveyard J, et al. A germline deletion of pl4(ARF) but not CDKN2A in a melanoma-neural system tumour syndrome family. Hum Mol Genet 2001;10(l):55–62. [DOI] [PubMed] [Google Scholar]
- Sargen MR, Kanetsky PA, Newton-Bishop J, Hayward NK, Mann GJ, Gruis NA, et al. Histologic features of melanoma associated with CDKN2A genotype. J Am Acad Dermatol 2015;72(3):496–507 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargen MR, Merrill SL, Chu EY, Nathanson KL. CDKN2A mutations with pl4 loss predisposing to multiple nerve sheath tumours, melanoma, dysplastic naevi and internal malignancies: a case series and review of the literature. Br J Dermatol 2016;175(4):785–9. [DOI] [PubMed] [Google Scholar]
- SEER. Cancer Stat Facts: Melanoma of the Skin, https://seer.cancer.gov/statfacts/html/melan.html; 2018. [accessed 12/7/2018.2018].
- SEER* Stat. Surveillance, Epidemiology, and End Results (SEER) Program (www.seer.cancer.gov) SEER*Stat Database: Incidence - SEER 18 Regs Research Data + Hurricane Katrina Impacted Louisiana Cases, Nov 2017 Sub (1973–2015 varying) -Linked To County Attributes - Total U.S., 1969–2016 Counties, National Cancer Institute, DCCPS, Surveillance Research Program, released April 2018, based on the November 2017 submission.
- Soufir N, Avril MF, Chompret A, Demenais F, Bombled J, Spatz A, et al. Prevalence of pl6 and CDK4 germline mutations in 48 melanoma-prone families in France. The French Familial Melanoma Study Group. Hum Mol Genet 1998;7(2):209–16. [DOI] [PubMed] [Google Scholar]
- Taylor NJ, Handorf EA, Mitra N, Avril MF, Azizi E, Bergman W, et al. Phenotypic and Histopathological Tumor Characteristics According to CDKN2A Mutation Status among Affected Members of Melanoma Families. J Invest Dermatol 2016; 136(5): 1066–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker MA, Elder DE, Curry M, Fraser MC, Pichler V, Zametkin D, et al. Risks of Melanoma and Other Cancers in Melanoma-Prone Families over 4 Decades. J Invest Dermatol 2018; 138(7): 1620–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste R, Smith E, Graham G. Multiple neurofibromas as the presenting feature of familial atypical multiple malignant melanoma (FAMMM) syndrome. Am J Med Genet A 2013; 161 A(6): 1425–31. [DOI] [PubMed] [Google Scholar]
- Zuo L, Weger J, Yang Q, Goldstein AM, Tucker MA, Walker GJ, et al. Germline mutations in the pl6INK4a binding domain of CDK4 in familial melanoma. Nat Genet 1996; 12(1):97–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.