Abstract
The peptidoglycan cell wall is a unique macromolecular structure in bacteria that defines their shape and confers protection from the surrounding environment. Decades of research has focused on understanding the peptidoglycan synthesis pathway and exploiting its essentiality for antibiotic development. Recently, a new class of peptidoglycan polymerases known as the SEDS (shape, elongation, division and sporulation) proteins was identified; these polytopic membrane proteins function together with the better-known penicillin-binding proteins (PBPs) to build the cell wall. In this review, we will highlight recent developments in chemical tools and methods to label the bacterial cell wall and discuss how these developments are leading to a better understanding of peptidoglycan synthases and their cellular roles.
INTRODUCTION
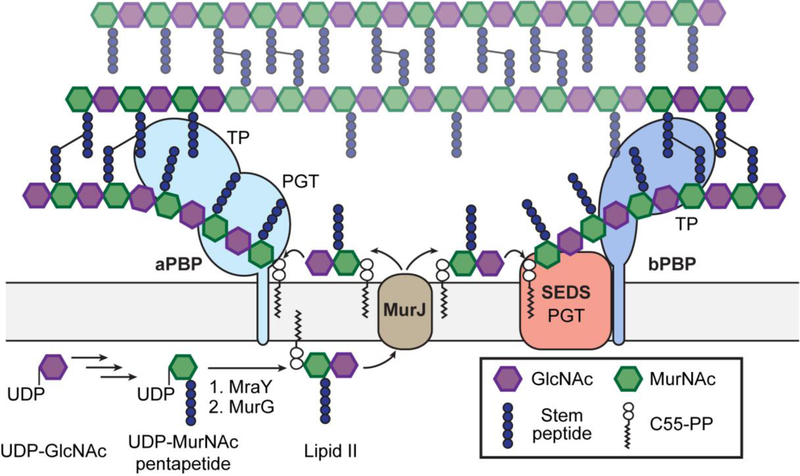
The bacterial cell envelope contains an intricate set of polymers and molecules that have evolved to promote survival and fitness in a hostile environment. Although components of the cell envelope are often present in only a subset of species, the peptidoglycan cell wall is a defining feature of prokaryotes that is present with few exceptions (e.g. Mycoplasma). The chemical steps of the peptidoglycan synthesis pathway were largely worked out by the end of the 1960s (Figure 1).1,2 In the first phase of synthesis, which is carried out by soluble enzymes in the cytoplasm, UDP-N-acetylglucosamine (UDP-GlcNAc) is converted to UDP-N-acetylmuramic acid (UDP-MurNAc)-pentapeptide via a multistep process. The basic structure of the stem pentapeptide is L-Ala-D-iGlu-mDAP (meso-2,6-diaminopimelate)/L-Lys-D-Ala-D-Ala in most species. The membrane protein MraY then catalyzes a pyrophosphate exchange reaction to couple phospho-MurNAc-pentapeptide to a polyprenyl phosphate in the membrane, and the resulting species, Lipid I, is converted by the glycosyltransferase MurG to Lipid II. Additional transformations that modify the stem pentapeptide, such as the amidation of D-iGlu at the second position and the addition of an interpeptide bridge at the third position, occur in some organisms. The complete peptidoglycan Lipid II precursor is then translocated to the extracytoplasmic side of the membrane where it is polymerized to form nascent glycan chains that are crosslinked to form cell wall peptidoglycan. Crosslinking is catalyzed by transpeptidases that are the targets of the ß-lactam antibiotics; because these enzymes were discovered based on their ability to bind penicillin, they are called penicillin-binding proteins, or PBPs. A variety of tailoring reactions, including attachment of wall teichoic acids and acetylation of the sugar backbone, can occur to diversify peptidoglycan in different organisms.3
Figure 1: Overview of peptidoglycan synthesis.
Peptidoglycan synthesis is initiated in the cytoplasm, where UDP-GlcNAc is converted to UDP-MurNAc pentapeptide, which is then converted by MraY and MurG at the cytoplasmic membrane to the peptidoglycan precursor Lipid II. After being translocated to the extracytoplasmic side, Lipid II is polymerized by peptidoglycan glycosyltransferases (PGTs) and crosslinked into the existing peptidoglycan by transpeptidases (TPs). Peptidoglycan polymerization is catalyzed by the bifunctional class A PBPs (aPBPs) or the SEDS proteins, the latter of which function together with the monofunctional class B PBPs (bPBPs).
Although the conserved chemical steps of biosynthesis have long been known, the past decade has witnessed major findings with respect to the enzymes that catalyze these steps. In part, these findings have been enabled by advances in tools to study peptidoglycan biosynthesis in vitro and in cells. This perspective will describe the recent discovery of a new family of peptidoglycan polymerases and connect this discovery to enabling advances in chemical biology.
DISCOVERY AND CHARACTERIZATION OF SEDS FAMILY PEPTIDOGLYCAN POLYMERASES
The availability of labeled beta-lactams that served as affinity-based probes for protein profiling led to the early discovery that bacteria typically contain several different PBPs having different molecular sizes.4,5 One class of high molecular weight PBPs, the class A PBPs (aPBPs), was found to contain a polymerase activity in addition to a crosslinking activity.6 The polymerase activity was found in an N-terminal domain that, for a time, served as the paradigm for what was widely believed to be the only family of peptidoglycan glycosyltransferases (PGTs). This domain was subsequently found in monofunctional glycosyltransferases (MGTs), and these were assumed to function in concert with a PBP that could catalyzing crosslinking.6,7 Although early evidence suggested that bacteria contain another, unrelated family of PGTs, this evidence was overlooked until Popham and coworkers reported in 2003 that all aPBPs can be deleted in Bacillus subtilis, which does not encode any MGTs.8 Similar observations were subsequently reported in Enterococcus faecalis and Enterococcus faecium.9,10 The requirement for aPBPs or MGTs in peptidoglycan synthesis was further questioned when peptidoglycan was detected in Chlamydia trachomatis, which was previously thought to lack a cell wall.11 The Chlamydiales family does not encode any aPBPs or MGTs, implying that an unknown PGT was responsible for making peptidoglycan polymers.12
The first clue to the identity of this mystery family of PGTs was found in the work of Matsuhashi and coworkers in 1986.13 This group reported that E. coli membranes overexpressing both RodA and PBP2 could make peptidoglycan, but this polymer was not made if only one of the proteins was overexpressed. PBP2 is a member of the class B PBPs (bPBPs), which only catalyze the transpeptidase reaction, and RodA is a core component of the Rod complex required for making side wall peptidoglycan.6 RodA belongs to the SEDS (for sporulation, elongation, division, and septation) family of polytopic membrane proteins, and a related protein, FtsW, is an essential component of the divisome that makes septal peptidoglycan.14,15 These proteins contain ten transmembrane helices, and due to the challenges of isolating membrane protein complexes and the lack of tools to reconstitute peptidoglycan synthase activity, no further studies to investigate the polymerase activity of SEDS-bPBP complexes were reported for more than thirty years. Moreover, an alternative proposal that SEDS proteins function as Lipid II flippases diverted attention from other possible functions for these essential proteins.16,17 It is now widely held that MurJ, a membrane protein that belongs to the MOP (multidrug/oligo-saccharidyl-lipid/polysaccharide) transporter family, is the primary lipid II flippase.18–20
A major breakthrough in the field was reported in 2016 when it was shown that purified B. subtilis RodA can polymerize synthetic Lipid II.21 Although the polymerase activity was weak, most likely because the bPBP partner was absent, genetic evidence was fully consistent with the role of RodA as a peptidoglycan polymerase.21–23 Homology and structural predictions identified similarities between SEDS proteins and O-antigen ligases, which catalyze the transfer of lipid-linked O-antigen to lipid A-core oligosaccharide to produce lipopolysaccharide in Gram-negative bacteria.21,24 A crystal structure of Thermus thermophilus RodA, solved using an innovative phasing strategy, revealed a conserved central cavity that was predicted to be a binding site for lipid-anchored substrates.25 At this point, however, it was still unclear whether the PGT activity observed for B. subtilis RodA was shared by other SEDS proteins. It was also unclear what role, if any, the bPBPs that form complexes with SEDS proteins played in polymerase activity.26,27
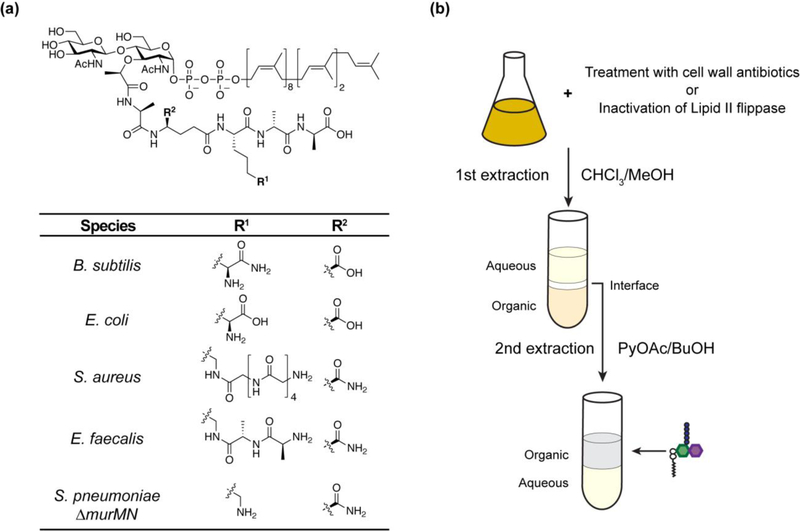
One major hurdle that for decades hindered the study of enzymes that make and modify peptidoglycan was limited access to peptidoglycan precursor. Lipid II is present in very low amounts in bacterial cells during normal growth owing to rapid turnover, and its complex structure containing an undecaprenyl (C55) pyrophosphate linked to a disaccharide-peptide moiety makes it challenging to synthesize (Figure 2a).2,28 Although chemical and enzymatic methods to prepare Lipid II were reported29–33, these required substantial effort and obtaining sufficient amounts of substrate remained one of the bottlenecks in the field for many years, particularly because structural variation in the peptide unit among different bacterial species made it necessary to tailor chemical or enzymatic routes to study key reactions (Figure 2a).3 This problem has now been solved. Following the discovery of labeling reaction that enables detection of Lipid II in cell extracts, Qiao et al. found that Lipid II accumulates in large amounts in cells treated with antibiotics or other conditions that prevent its turnover (e.g., inactivation of the Lipid II flippase).34 Subsequently, a facile two-step extraction procedure was developed to isolate Lipid II and one can now obtain any Lipid II variant in substantial quantities overnight, allowing extensive exploration of peptidoglycan assembly and processing enzymes (Figure 2b).20,35,36
Figure 2:
(a) Structure of Lipid II. Lipid II variants that have been obtained using the method described in (b) are listed.35,36 S. pneumoniae possesses a murMN operon which encodes ligases that attach L-Ser-L-Ala or L-Ala-L-Ala to the R1 position. Lipid II extracted from S. pneumoniae ∆murMN strain lacks this modification. (b) Isolation of Lipid II from bacterial cells. Accumulation of Lipid II is accomplished by treatment with vancomycin/moenomycin or conditional inactivation of the Lipid II flippase. After chloroform/methanol extraction, a white interface layer enriched with Lipid II and UDP-MurNAc-pentapeptide is observed. A second pyridinium acetate/1-butanol extraction from this interface separates Lipid II from other water-soluble molecules.
Using the newly available lipid II substrates, it has now been shown that RodA and FtsW orthologs from several different organisms have peptidoglycan polymerase activity in complex with their cognate bPBPs.37,38 It is remarkable that protein complexes with a combined eleven transmembrane helices are functional in detergent. The bPBP is required for robust polymerase activity of the SEDS proteins, but the crosslinking activity does not depend on the presence of a SEDS protein, only on the availability of a polymer substrate.38 The dependence of polymerase activity on the bPBP mirrors the initial observation from the Matsuhashi laboratory that peptidoglycan synthesis in E. coli membranes required that both RodA and the bPBP be overexpressed.13 Evidence indicates that the transmembrane helix of the cognate bPBP for each SEDS protein plays a crucial role in complex formation.24,25,38 Using state-of-the-art microscopy, it has been found that SEDS proteins and bPBPs form complexes in cells.39,40 Taken together, recent work has established these SEDS-bPBP complexes as peptidoglycan synthases. At this point, almost nothing is known about the polymerase mechanism except that divalent cations are required for activity, unlike for aPBP-mediated peptidoglycan polymerization.38
CHEMICAL PROBES FOR LABELING PEPTIDOGLYCAN
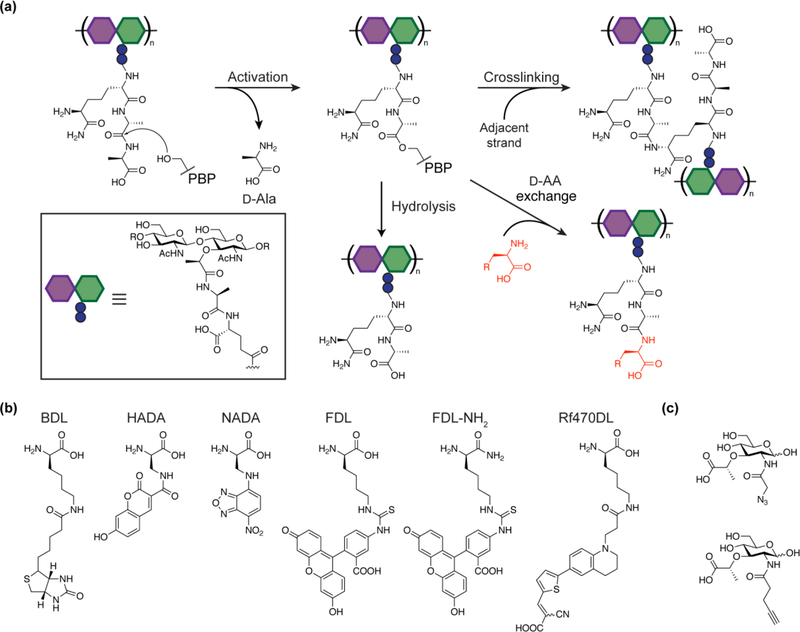
A full understanding of peptidoglycan synthesis requires not only tools that enable in vitro reconstitution, but methods to monitor peptidoglycan synthesis in cells. Seminal studies have demonstrated that noncanonical D-amino acids can be incorporated into peptidoglycan to modulate peptidoglycan structure and vancomycin resistance. While vancomycin resistance in Enterococcus was shown to be mediated by VanA, a ligase responsible for replacing D-Ala-D-Ala with D-Ala-D-lactate at the stem peptide terminus, the noncanonical D-amino acid incorporation seen in a wide range of species has been attributed to transpeptidase activities of PBPs and L,D-transpeptidases (Figure 3a).41–44 PBP-mediated D-amino acid incorporation has been confirmed in vitro for aPBPs and a family of low molecular weight PBPs that act as transpeptidases rather than carboxypeptidases.36,45 These low molecular weight PBPs can catalyze exchange of unnatural D-amino acids, including biotin-D-lysine (BDL), into Lipid II, unlike the aPBPs that only act on polymeric peptidoglycan. The ability to incorporate BDL is useful for in vitro detection of peptidoglycan (Figure 3b).
Figure 3:
(a) Schematic of PBP transpeptidase and carboxypeptidase reactions in B. subtilis. After the formation of the acyl-enzyme intermediate via the cleavage of the terminal D-Ala, transpeptidases can either accept the stem peptide from the adjacent strand or a D-amino acid. Most low molecular weight PBPs are carboxypeptidases that remove the terminal D-alanine via hydrolysis, but a subset of low molecular weight PBPs are able to catalyze the D-amino acid exchange.34,36 (b) D-amino acid probes for peptidoglycan labeling. Biotin-D-lysine (BDL)34, FDAAs (HADA, NADA & FDL)46, carboxamide FDAA (FDL-NH2)50, and rotor-fluorogenic D-amino acid (Rf470DL)52 are shown. (c) MurNAc derivatives for peptidoglycan labeling. MurNAc variants containing an azide or alkyne handle that were incorporated into the cell wall are shown.53
To detect active peptidoglycan synthesis in cells, a number of fluorescent D-amino acid (FDAA) probes have been developed (Figure 3b).46 By combining these probes with methods to fluorescently label proteins, it is possible to investigate the relationship between protein localization and peptidoglycan synthesis in cells.47 Recent studies have investigated the relationship between septal peptidoglycan synthesis and treadmilling of FtsZ, a tubulin homolog required for bacterial cell division.40,48,49 FDAAs can be tailored for each species to increase incorporation efficiency. In B. subtilis and some other organisms better peptidoglycan labeling has been observed for FDAAs in which the acid is converted to a carboxamide, perhaps because the stem peptides modified with carboxamide probes are poorer substrates for transpeptidase or hydrolysis activities that could cleave other probes.50,51 One drawback of FDAAs is that it is necessary to wash out excess probe before imaging. To circumvent this issue, Hsu et al. recently reported the design and synthesis of rotor-fluorogenic D-amino acids, which only fluoresce upon peptidoglycan incorporation and do not require any washing prior to imaging, thus providing better temporal resolution.52
In addition to stem peptide modifications, strategies to label the sugar backbone of peptidoglycan have been explored. Incorporation of MurNAc derivatives containing a bioorthogonal handle into peptidoglycan has been demonstrated in live cells, and it has been shown that the modified cell wall can be labeled with an appropriate fluorophore (Figure 3c).53 Another strategy to label the MurNAc residues takes advantage of PatB, a peptidoglycan O-acetyltransferase with promiscuous properties, to install a bioorthogonal handle at the 6-OH position of MurNAc.54 These studies have established that peptidoglycan disaccharide units are targets for installation of chemical probes and that labeling the glycan backbone can have advantages over stem peptide modification for studying the peptidoglycan architecture. Finally, fluorescent substrate-binding antibiotics that bind Lipid II and nascent peptidoglycan are also useful probes because they report on sites of new polymer synthesis.55,56
CONCLUSION
Despite being a subject of intense research for decades, the bacterial cell wall still contains many surprises. Here we have highlighted recent innovative methods in chemical biology that have made it possible to study peptidoglycan biosynthesis both in vitro and in cells. Although we focused on the recent discovery of a new family of peptidoglycan polymerases, the methods mentioned here more generally make it possible to link cell biology, genetics, and biochemistry to arrive at a comprehensive understanding of the role of cell wall assembly and modification enzymes in bacterial physiology.
With respect to SEDS-bPBP complexes, their primary function has been uncovered, but many outstanding questions remain to be answered. As noted, the biochemical mechanisms of these complexes are not understood. Moreover, how these complexes coordinate with aPBPs to build peptidoglycan in cells has only recently begun to be examined.22,57 Perhaps the most important question from the standpoint of significance is whether SEDS proteins are “druggable” antibiotic targets. SEDS proteins are found in all bacteria that have a cell wall and could thus be broad-spectrum targets like the PBPs. Moreover, their active sites are accessible from the extracytoplasmic side, which may allow targeting by compounds that cannot cross the cytoplasmic membrane. Errington and coworkers have recently described a partially purified set of natural products that appear to have an inhibitory effect on B. subtilis RodA, but the structures of these molecules have not been reported and inhibition has not been confirmed in a pure assay.23 Nevertheless, this report is promising and gives reason for optimism.
ACKNOWLEDGEMENTS
We thank Dr. Michael Welsh for critical reading of the manuscript. This work was supported by National Institutes of Health grants R01 GM076710 and CETR U19 AI109764 to D.K and S.W. A.T. is supported in part by the Funai Overseas Scholarship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CONFLICTS OF INTEREST STATEMENT
Nothing declared.
REFERENCES
- 1.Blumberg PM & Strominger JL Interaction of penicillin with the bacterial cell: penicillin-binding proteins and penicillin-sensitive enzymes. Bacteriol. Rev 38, 291–335 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Heijenoort J Lipid intermediates in the biosynthesis of bacterial peptidoglycan. Microbiol. Mol. Biol. Rev 71, 620–35 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vollmer W & Born P Bacterial Cell Envelope Peptidoglycan. Microbial Glycobiology (Elsevier Inc, 2010). doi: 10.1016/B978-0-12-374546-0.00002-X [DOI] [Google Scholar]
- 4.Blumberg PM & Strominger JL Five penicillin-binding components occur in Bacillus subtilis membranes. J. Biol. Chem 247, 8107–13 (1972). [PubMed] [Google Scholar]
- 5.Spratt BG Distinct penicillin binding proteins involved in the division, elongation, and shape of Escherichia coli K12. Proc. Natl. Acad. Sci. U. S. A 72, 2999–3003 (1975). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goffin C & Ghuysen JM Multimodular penicillin-binding proteins: an enigmatic family of orthologs and paralogs. Microbiol. Mol. Biol. Rev 62, 1079–93 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spratt BG, Zhou J, Taylor M & Merrick MJ Monofunctional biosynthetic peptidoglycan transglycosylases. Mol. Microbiol 19, 639–40 (1996). [DOI] [PubMed] [Google Scholar]
- 8.McPherson DC & Popham DL Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J. Bacteriol 185, 1423–31 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arbeloa A et al. Role of class A penicillin-binding proteins in PBP5-mediated beta-lactam resistance in Enterococcus faecalis. J. Bacteriol 186, 1221–8 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice LB et al. Role of class A penicillin-binding proteins in the expression of beta-lactam resistance in Enterococcus faecium. J. Bacteriol 191, 3649–56 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liechti GW et al. A new metabolic cell-wall labelling method reveals peptidoglycan in Chlamydia trachomatis. Nature 506, 507–10 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacquier N, Viollier PH & Greub G The role of peptidoglycan in chlamydial cell division: towards resolving the chlamydial anomaly. FEMS Microbiol. Rev 39, 262–75 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Ishino F et al. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and rodA protein. J. Biol. Chem 261, 7024–31 (1986). [PubMed] [Google Scholar]
- 14.Ikeda M et al. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J. Bacteriol 171, 6375–8 (1989). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Henriques AO, Glaser P, Piggot PJ, Moran CP & Moran CP Jr Control of cell shape and elongation by the rodA gene in Bacillus subtilis. Mol. Microbiol 28, 235–47 (1998). [DOI] [PubMed] [Google Scholar]
- 16.Höltje JV Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol. Mol. Biol. Rev 62, 181–203 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mohammadi T et al. Identification of FtsW as a transporter of lipid-linked cell wall precursors across the membrane. EMBO J 30, 1425–1432 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruiz N Bioinformatics identification of MurJ (MviN) as the peptidoglycan lipid II flippase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A 105, 15553–7 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sham L-T et al. Bacterial cell wall. MurJ is the flippase of lipid-linked precursors for peptidoglycan biogenesis. Science 345, 220–2 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rubino FA, Kumar S, Ruiz N, Walker S & Kahne DE Membrane Potential Is Required for MurJ Function. J. Am. Chem. Soc 140, 4481–4484 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meeske AJ et al. SEDS proteins are a widespread family of bacterial cell wall polymerases. Nature 537, 634–638 (2016).This paper provides genetic and biochemical evidences of B. subtilis RodA as a peptidoglycan polymerase. It challenged the wide-held assumption that aPBPs and MGTs are the only enzymes capable of polymerizing peptidoglycan in bacterial cells.
- 22.Cho H et al. Bacterial cell wall biogenesis is mediated by SEDS and PBP polymerase families functioning semi-autonomously. Nat. Microbiol 1, 1–8 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Emami K et al. RodA as the missing glycosyltransferase in Bacillus subtilis and antibiotic discovery for the peptidoglycan polymerase pathway. Nat. Microbiol 2, 1–8 (2017).This paper provides genetic evidence that B. subtilis RodA has PGT activity and reports a set of compounds that potentially inhibit RodA.
- 24.Ovchinnikov S et al. Large-scale determination of previously unsolved protein structures using evolutionary information. Elife 4, 1–25 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sjodt M et al. Structure of the peptidoglycan polymerase RodA resolved by evolutionary coupling analysis. Nature 556, 118–121 (2018).The first structure of RodA, solved using evolutionary covariance-based fold prediction, is reported.
- 26.Fay A, Meyer P & Dworkin J Interactions between late-acting proteins required for peptidoglycan synthesis during sporulation. J. Mol. Biol 399, 547–561 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fraipont C et al. The integral membrane FtsW protein and peptidoglycan synthase PBP3 form a subcomplex in Escherichia coli. Microbiology 157, 251–259 (2011). [DOI] [PubMed] [Google Scholar]
- 28.Lazar K & Walker S Substrate analogues to study cell-wall biosynthesis and its inhibition. Curr. Opin. Chem. Biol 6, 786–93 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Ye XY et al. Better substrates for bacterial transglycosylases. Journal of the American Chemical Society 123, 3155–3156 (2001). [DOI] [PubMed] [Google Scholar]
- 30.Schwartz B, Markwalder JA & Wang Y Lipid II: Total synthesis of the bacterial cell wall precursor and utilization as a substrate for glycosyltransfer and transpeptidation by penicillin binding protein (PBP) 1b of Eschericia coli. J. Am. Chem. Soc 123, 11638–11643 (2001). [DOI] [PubMed] [Google Scholar]
- 31.VanNieuwenhze MS et al. The first total synthesis of lipid II: The final monomeric intermediate in bacterial cell wall biosynthesis. J. Am. Chem. Soc 124, 3656–3660 (2002). [DOI] [PubMed] [Google Scholar]
- 32.Breukink E et al. Lipid II Is an Intrinsic Component of the Pore Induced by Nisin in Bacterial Membranes. J. Biol. Chem 278, 19898–19903 (2003). [DOI] [PubMed] [Google Scholar]
- 33.Huang LY et al. Enzymatic synthesis of lipid II and analogues. Angew. Chemie - Int. Ed 53, 8060–8065 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Qiao Y et al. Detection of lipid-linked peptidoglycan precursors by exploiting an unexpected transpeptidase reaction. J. Am. Chem. Soc 136, 14678–14681 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qiao Y et al. Lipid II overproduction allows direct assay of transpeptidase inhibition by β-lactams. Nat. Chem. Biol 13, 793–798 (2017).A facile method to obtain large amounts of Lipid II directly from bacterial cells is reported. This method enables rapid preparation of substrates for biochemical studies of peptidoglycan synthases.
- 36.Welsh MA et al. Identification of a Functionally Unique Family of Penicillin-Binding Proteins. J. Am. Chem. Soc 139, 17727–17730 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rohs PDA et al. A central role for PBP2 in the activation of peptidoglycan polymerization by the bacterial cell elongation machinery. PLOS Genet 14, e1007726 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Taguchi A et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol 4, 587–594 (2019).This paper reports the biochemical reconstitution of FtsW PGT activity from multiple species, establishing SEDS proteins as peptidoglycan polymerases.
- 39.Reichmann NT et al. SEDS-bPBP pairs direct lateral and septal peptidoglycan synthesis in Staphylococcus aureus. Nat. Microbiol (2019). doi: 10.1038/s41564-019-0437-2 [DOI] [PubMed] [Google Scholar]
- 40.Perez AJ et al. Movement dynamics of divisome proteins and PBP2x:FtsW in cells of Streptococcus pneumoniae. Proc. Natl. Acad. Sci. U. S. A 116, 3211–3220 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bugg TDH, Dutka-Malen S, Arthur M, Courvalin P & Walsh CT Identification of vancomycin resistance protein VanA as a D-alanine:D-alanine ligase of altered substrate specificity. Biochemistry 30, 2017–2021 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Caparrós M, Pisabarro AG & de Pedro MA Effect of D-amino acids on structure and synthesis of peptidoglycan in Escherichia coli. J. Bacteriol 174, 5549–59 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lam H et al. D-amino acids govern stationary phase cell wall remodeling in bacteria. Science 325, 1552–5 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cava F, de Pedro MA, Lam H, Davis BM & Waldor MK Distinct pathways for modification of the bacterial cell wall by non-canonical D-amino acids. EMBO J 30, 3442–53 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lupoli TJ et al. Transpeptidase-mediated incorporation of D-amino acids into bacterial peptidoglycan. J. Am. Chem. Soc 133, 10748–51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuru E et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem. Int. Ed. Engl 51, 12519–23 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharifzadeh S et al. Novel Electrophilic Scaffold for Imaging of Essential Penicillin-Binding Proteins in Streptococcus pneumoniae. ACS Chem. Biol 12, 2849–2857 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bisson-Filho AW et al. Treadmilling by FtsZ filaments drives peptidoglycan synthesis and bacterial cell division. Science 355, 739–743 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang X et al. GTPase activity-coupled treadmilling of the bacterial tubulin FtsZ organizes septal cell wall synthesis. Science 355, 744–747 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lebar MD et al. Reconstitution of peptidoglycan cross-linking leads to improved fluorescent probes of cell wall synthesis. J. Am. Chem. Soc 136, 10874–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pidgeon SE et al. Metabolic Profiling of Bacteria by Unnatural C-terminated D-Amino Acids. Angew. Chem. Int. Ed. Engl 54, 6158–62 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Hsu Y-P et al. Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays. Nat. Chem 11, 335–341 (2019).Novel D-amino acid probes that become fluorescent only when conjugated to peptidoglycan are reported. These probes allow real-time detection of transpeptidase activity.
- 53.Liang H et al. Metabolic labelling of the carbohydrate core in bacterial peptidoglycan and its applications. Nat. Commun 8, 15015 (2017).This paper describes the synthesis of MurNAc derivatives with chemical handles that can be incorporated into peptidoglycan of live cells.
- 54.Wang Y et al. Postsynthetic Modification of Bacterial Peptidoglycan Using Bioorthogonal N-Acetylcysteamine Analogs and Peptidoglycan O-Acetyltransferase B. J. Am. Chem. Soc 139, 13596–13599 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Daniel RA & Errington J Control of cell morphogenesis in bacteria: two distinct ways to make a rod-shaped cell. Cell 113, 767–76 (2003). [DOI] [PubMed] [Google Scholar]
- 56.Tiyanont K et al. Imaging peptidoglycan biosynthesis in Bacillus subtilis with fluorescent antibiotics. Proc. Natl. Acad. Sci. U. S. A 103, 11033–8 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dion MF et al. Bacillus subtilis cell diameter is determined by the opposing actions of two distinct cell wall synthetic systems. Nat. Microbiol (2019). doi: 10.1038/s41564-019-0439-0 [DOI] [PMC free article] [PubMed] [Google Scholar]