Abstract
The use of microRNAs as biomarkers has been proposed for many diseases including the diagnosis of melanoma. Although hundreds of microRNAs have been identified as differentially expressed in melanomas as compared to benign melanocytic lesions, limited consensus has been achieved across studies, constraining the effective use of these potentially useful markers. In this study we applied a machine learning-based pipeline to a dataset consisting of genetic features, clinical features and next-generation microRNA sequencing from micro-dissected formalin fixed paraffin embedded melanomas and their adjacent benign precursor nevi. We identified patient age and tumor cellularity as variables that frequently confound the measured expression of potentially diagnostic microRNAs. By employing the ratios of microRNAs that were either enriched or depleted in melanoma compared to nevi as a normalization strategy, we developed a model that classified all available published cohorts with an area under the receiver operating characteristic curve of 0.98. External validation on an independent cohort classified lesions with 81% sensitivity and 88% specificity, and was uninfluenced by tumor content of the sample or patient age.
Keywords: microRNA, melanoma, feature selection
Introduction
Misdiagnosis of cutaneous melanoma is among the most significant contributors to medical malpractice lawsuits in the United States (Wallace et al. 2013). The advanced stages of melanoma are associated with five-year survival rates less than 20% and are responsible for over 10,000 deaths in the U.S. each year (Gershenwald et al. 2017; Key Statistics for Melanoma Skin Cancer). Although the disease is curable when detected early, the process of differentiating between malignant lesions and the more prevalent benign lesions, such as melanocytic nevi, is challenging. The clinical standard for diagnosing concerning lesions is histopathologic assessment of formalin fixed paraffin embedded (FFPE) biopsy specimens. However, a considerable rate of discordance in diagnoses has been established even among expert pathologists (Boiko et al. 1994; Brochez et al. 2002; Corona et al. 1996; Elder et al. 2018; Elmore et al. 2017; Farmer et al. 1996; Gaudi et al. 2013; Heenan et al. 1984; Niebling et al. 2014; Shoo et al. 2010). A large-scale study published by Elmore and colleagues in 2017 reported interobserver discordance rates as high as 57–75% and intraobserver discordance rates as 37–65% (Elmore et al. 2017). Together, these observations highlight the complexity of histopathologic assessment and emphasize the need for quantitative molecular methods for distinguishing malignant from benign lesions to augment current practices.
Molecular biomarkers can provide robust, objective and quantitative measurements of disease state (Buchbinder and Flaherty 2016; Leachman et al. 2017; Rodriguez-Cerdeira et al.). One class of candidate biomarkers are small non-coding microRNAs (miRNAs). Discovered twenty-five years ago (Lee et al. 1993), miRNAs are appreciated as potentially valuable candidate biomarkers for many conditions and diseases for several reasons (Jung et al. 2010; Sheinerman and Umansky 2013). First, as with other transcripts, miRNAs exhibit tissue- and cell-specific expression patterns during mammalian development and these patterns are mis-regulated in disease (D’Amato et al. 2013; Kosik 2010; Parchem et al. 2014; Reddy 2015). Second, due its relatively small size and simplicity, the microRNA transcriptome is more efficiently profiled with next generation sequencing than the mRNA transcriptome (Creighton et al. 2009). Third, the integrity of miRNAs is retained in FFPE samples, whereas long RNAs are most often degraded (Liu et al. 2009). Thus, if miRNA expression levels provided reliable insight into the progression state of a FFPE melanocytic neoplasm biopsy, they could assist in the diagnosis of difficult to diagnose cases where material is limiting.
However, despite these useful properties, few miRNA biomarkers have emerged in the clinical setting due to a frequent lack of reproducibility between differential expression studies (Hawkes et al. 2016; Mumford et al. 2018; Nair et al. 2012; Pogribny 2018; Raya et al. 2012; Witwer and Halushka 2016). This poor reproducibility of mis-regulated miRNA signatures is exemplified in studies that have explored the use of miRNAs as biomarkers for melanoma (reviewed in (Jarry et al. 2014; Jayawardana et al. 2016; Margue et al. 2013; Raya et al. 2012)). Among the most reproducible miRNA biomarker candidates for this application include three enriched in benign melanocytic neoplasms (MIR211, MIR205 and MIR125B) and two enriched in melanomas (MIR21 and MIR150) (Latchana et al. 2016). However, even these miRNAs do not consistently provide predictive value across cohorts. Discrepancies in differential expression signatures across comparable studies have been attributed to a variety of causes (Mumford et al. 2018; Raya et al. 2012; Witwer and Halushka 2016). For example, FFPE biopsy sections routinely include normal adjacent skin, with the concerning melanocytic component constituting only a fraction of the biopsy. Sample to sample variation in the type and amount of infiltrating or adjacent non-tumor cells could alter the small RNA profile. Similarly, variability could arise from differences in the malignant content of each section – the percent of the tumor that contains malignant cells, as opposed to adjacent benign cells from which the melanoma arose. Alternatively, with histopathologic diagnosis discordance rates of over 50%, the cohorts of samples used for each independent study are likely assembled with different degrees of diagnostic accuracy. Another common explanation for poor reproducibility are biases inherent to the different platforms for quantifying miRNA expression levels – small RNA next generation sequencing, hybridization arrays, and RT-qPCR based arrays (Witwer and Halushka 2016). Further investigations into the causes of these discrepancies are needed, and greater consistency across independently derived datasets are required before miRNA expression can be a useful molecular diagnostic.
In this study, we sought to determine the common confounding variables that influenced quantification of miRNA expression from nevus and melanoma FFPE biopsies. We developed a score based upon miRNA expression ratios that was both uninfluenced by these confounding variables and predictive of malignancy in an independent validation cohort.
Results
Variation in tumor cell content of FFPE samples confounds miRNA expression analyses
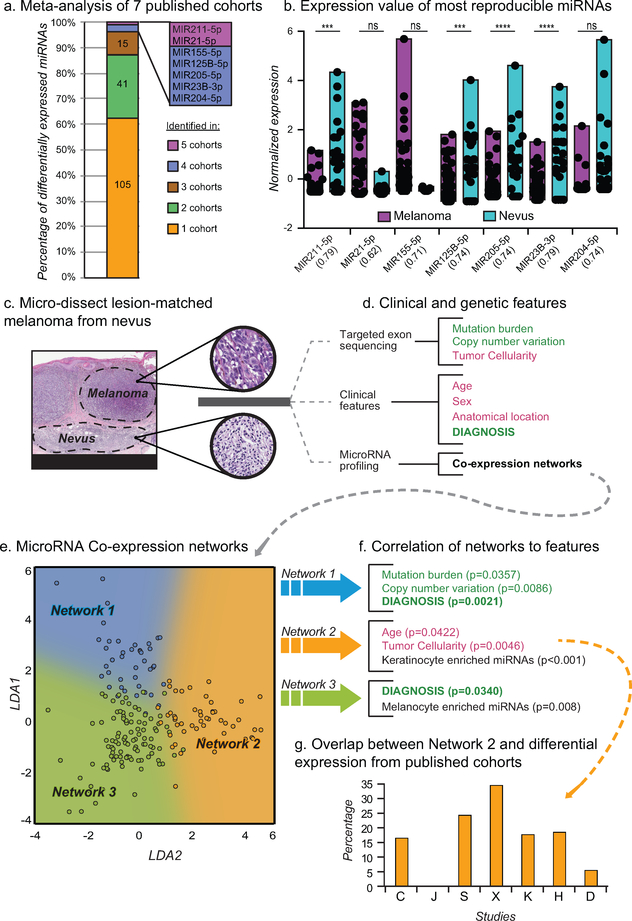
To identify miRNAs that were consistently mis-regulated across independently assembled cohorts, we performed a meta-analysis of seven publicly available miRNA expression datasets. Each dataset contained both melanoma and nevus cases (Table S1). When each cohort was considered independently, 168 unique miRNAs were identified as differentially expressed between melanoma and nevus cases with an FDR cutoff of 0.05. As discussed by previous studies, no miRNA was differentially expressed in every cohort, and only seven of these miRNAs showed reproducible expression differences in at least half of the cohorts (Fig. 1a & Table S2) (Jayawardana et al. 2016). When cohorts were considered in aggregate four of the seven miRNAs – MIR211–5p, MIR125B-5p, MIR205–5p, and MIR23B-3p – retained significance (Fig. 1b). However, the variance in expression of these top candidates was considerable, such that the use of miRNA expression as a classifier for identifying malignant cases achieved a maximum area under (AUC) the receiver operating characteristic (ROC) curve of 0.79 (Fig. 1b & S1a, MIR211–5p and MIR23B-3p). This analysis demonstrates that the melanoma-associated and nevus-associated miRNA signatures derived from profiling FFPE biopsies undergo substantial variation when sampling from independent cohorts.
Figure 1: Tumor cellularity and age confound miRNA profiling of melanoma samples.
a) Differential miRNA expression signatures between nevi and melanoma from 7 studies (Table S1–2). b) Normalized expression of common differentially expressed miRNAs. AUC of classification using each microRNA in parenthesis. c) Micro-dissected melanomas with adjacent precursor nevus regions (scale bars, black = 4mm, grey = 300μm). d) Features obtained for each region using targeted exon sequencing (genetic features) or the pathology requisition form (clinical features). Potentially confounding features (red) and target features (green) are highlighted. e) Scatter plot of expressed miRNAs separated by LDA trained on three coexpression networks (blue, orange, and green). F) Correlation between miRNA coexpression networks and features from (d). p values calculated from correlation coefficient. g) Percent of differentially expressed miRNAs from (a) in Network 2.
To identify covariates that could cause these variations in differential miRNA expression analyses, we took advantage of a cohort of primary melanomas with intact adjacent benign nevi, from which they arose (Sham et al. 2018) (Fig. 1c). To control for the discordance of diagnosis, the progression stages for each sample were diagnostically classified by a panel of at least five dermatopathologists. To minimize the mixture of malignant and benign tissue, each section was micro-dissected. To account for variability in the amount of infiltrating tumor cells, we genotyped the microdissected tissue for over five hundred cancer-related genes and estimated the tumor cell content (referred to here as tumor cellularity) using allele frequencies and magnitudes of copy number changes (Shain et al. 2018). Consequently, the dataset was annotated with both clinical features (e.g. patient age, sex, anatomical location of the lesion) as well as genomic information (e.g. mutation burden, copy number variation, tumor cellularity) for each matched pair of nevus and melanoma regions (Fig. 1d and Table S3).
To investigate the influence of each genomic and clinical feature on the miRNA expression pattern, we conducted miRNA sequencing on fifteen of the regions from seven cases (Table S3). In order to first identify potential systemic confounding features, we first employed co-expression analyses for identification of networks of miRNAs sharing similar expression patterns across all regions and identified three co-expression networks (Fig. S 1b & Table S4) that were effectively separated via Linear Discriminate Analysis (LDA) (Fig. 1e) (Langfelder and Horvath 2008). Each network consists of miRNAs with read counts that are positively correlated across all samples, regardless of level of expression. We next determined whether the expression patterns of these networks correlated with the clinical or genomic annotations of the samples, including not only diagnosis but also potentially confounding features, such as patient age. We summarized the miRNA expression matrix for each network by its first principal component and compared these to the sample covariates (Fig. 1f & Fig. S1c–d). Two of the networks (Network 1 and Network 3) were significantly correlated with a diagnosis of melanoma and were not influenced by tumor cellularity or other clinical features. Network 1 was also correlated with mutation burden and copy number variation, both measurements of genome damage that increase during progression from nevus to melanoma (Shain et al. 2015).
In contrast to the two melanoma-associated networks, Network 2 was positively correlated with tumor cellularity and, to a lesser extent, patient age. This observation suggests that although miRNAs within Network 2 were differentially expressed in melanoma and nevus samples, variation in their observed abundance may reflect the extent of contamination with non-tumor cells rather than different progression stages. Consistent with this interpretation, we observed that miRNAs known to be expressed in cultured primary human keratinocytes were enriched in Network 2 as would be expected if keratinocytes were a significant fraction of contaminating non-tumor cells (Fig. 1f, Fig. S2d). Conversely, miRNAs known to be expressed in cultured primary human melanocytes were enriched in Network 3 consistent with changes in Network 3 reflecting melanocyte biology. Together, these data suggest that miRNA profiling datasets derived from micro-dissected FFPE samples can contain sufficient levels of contaminating non-tumor cells to influence the overall miRNA expression profile.
Contamination by non-tumor cells is expected to vary among samples dependent on their size, histologic type (predominantly junctional versus intradermal), and preparation (e.g. precision of microdissection). If not controlled for, variation in tumor cellularity is expected to degrade the reproducibility of miRNA signatures across studies. Indeed, miRNAs from Network 2 constituted up to thirty percent of the miRNAs in expression signatures reported from the seven previously reported datasets (Fig. 1g). This result highlights the need to control for tumor cellularity and patient age when identifying miRNA signatures predictive of melanoma diagnosis from FFPE derived samples.
Classification of nevus from melanoma samples with miRNA ratios
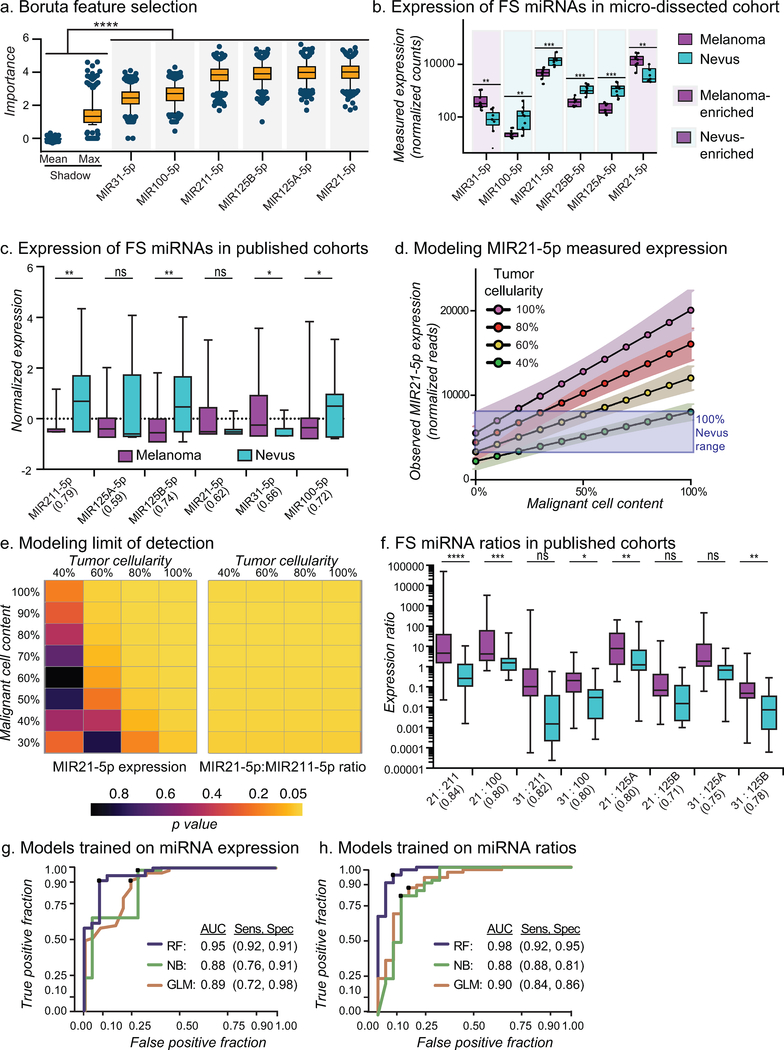
To identify miRNAs that are predictors of the diagnosis of a lesion, we employed a machine-learning analytical method called Boruta to obtain an initial list of those miRNAs that were most important for differentiating melanoma from nevus samples across 1000 random forest iterations (Kursa 2014; Li et al. 2016). All miRNAs with more than five total reads were considered, resulting in 341 unique miRNAs. In addition, for each unique miRNA, a second unique artificial feature was generated through randomized re-distribution of the read counts across samples (Fig. S2a). These ‘shadow miRNAs” provided an equal number of negative control miRNAs for which to compare each experimental feature. By requiring a miRNA to significantly outperform all shadow miRNAs, the likelihood of false positive identification is reduced (Kursa and Rudnicki 2010). We conducted Boruta with the combined 682 experimental and negative control miRNAs, ranking the importance of each for the accurate classification of nevus samples from melanoma samples with each iteration. We identified 38 miRNAs that ranked higher than the maximum-performing shadow miRNA with a p-value of less than 0.001 (Fig. S2b). To enable comparison with previous studies we also required that the expression of each miRNA was assessed in all published datasets (Table S2). The final list of feature-selected miRNAs contained two miRNAs with increased expression in melanomas (MIR31–5p, MIR21–5p) and four miRNAs (MIR211–5p, MIR125A-5p, MIR125B-5p, MIR100–5p) with decreased expression in melanomas (Fig. 2a–b). These miRNAs are referred to as melanoma-enriched (ME) and melanoma-depleted (MD) miRNAs, respectively.
Figure 2: MicroRNA ratio-trained model classifies melanocytic lesions.
a) MiRNAs that classify micro-dissected sections identified by Boruta feature selection (FS) compared to shadow max (X) and mean (M) features. b) Normalized miRNA-Seq counts from micro-dissected regions. Boxes indicate mean, first and third quartiles. c) Normalized expression of FS-miRNAs in published studies. AUC of classification using each miRNA in parenthesis. d) Model of the expected observed expression of MIR21–5p with variation in tumor content compared to expected observed expression from pure nevus samples (purple box). e) Heatplot of p values when comparing expected observed MIR21–5p expression (left) and expected observed MIR21–5p:MIR2n-5p ratio (right) of melanoma samples with variation in tumor content to nevus. f) ME-miRNA:MD-miRNA ratios from published studies. AUC of classification using each ratio in parenthesis. g-h) ROC curves for cross-validation of miRNA expression (g) and ratio (h) classification models trained on aggregate published studies. Naïve Bayes (NB), Random Forest (RF), Logistic Regression (GLM).
We then sought to determine the accuracy by which ME- and MD-miRNAs classified melanomas from nevi across published datasets. Similar to the miRNA signatures identified from the meta-analysis (Fig. 1b), classifiers trained with expression levels alone performed poorly, with a maximum area under the ROC curve of 0.79 (MIR211–5p, Fig. 2c). However, the tumor cellularity and malignant content of each sample within the external cohorts are unknown, but expected to be variable (Fig. 1g). Using the normalized reads from our micro-dissected cohort, we modeled the expected observed expression of the most consistent ME-miRNA, MIR21–5p, across a range of tumor cellularity and malignant cell content (Fig. 1d). Our model predicted that as tumor cellularity and/or malignant cell content dropped below 60%, the observed expression level of MIR21–5p would cease to be a reliable indicator of diagnosis (Fig. 1e). Similar results were obtained for the other feature selected miRNAs (Fig. S3). The use of transcript ratios has been demonstrated to both strengthen the prediction accuracy and simply feature-sets (Avissar et al. 2009; Reddy et al. 2015). Standard normalization techniques frequently consider the ratio of a candidate marker to a “housekeeping” transcript to control for RNA input. Our model already considers input normalization, but this strategy does not account for tumor cellularity. We reasoned that by considering the ratios of miRNAs highly expressed in melanomas to miRNAs highly expressed in nevi we would control for the fraction of input RNA that originated from melanocytic cells, as opposed to total RNA. Our model predicted that by using expression ratios of ME-miRNAs to MD-miRNAs, we would control for both tumor cellularity and the malignant content in each sample (Fig. 2e, Fig. S3).
To develop a diagnostic score using all ratios of ME-miRNAs to MD-miRNAs, we first divided expression levels of each of the two ME-miRNAs by each of the four MD-miRNAs, producing eight miRNA ratios (MIR31–5p/MIR211–5p, MIR31–5p/MIR125A-5p, MIR31–5p/MIR125B-5p, MIR31–5p/MIR100–5p, MIR21 −5p/MIR211–5p, MIR21–5p/MIR125A-5p, MIR21–5p/MIR125B-5p, and MIR21–5p/MIR100–5p) (Fig. 2f). Classifiers trained with individual ratios performed better than expression alone with area under ROC curves ranging from 0.71–0.84 (Fig. 2f). To consider all eight ratios together, we trained a series of classifiers using either the six expression values or the eight expression ratios of the feature-selected miRNAs using the aggregated public datasets. Crossvalidation of the final miRNA expression trained model resulted in an area under the ROC curve of 0.95 (Fig. 2g). Similarly, crossvalidation of the final miRNA Ratio Trained Model (MiRTM) resulted in an area under the ROC curve of 0.98 (Fig. 2h). We next sought to challenge each model with an external dataset representative of the range of tumor cellularity, malignant content and patient age encountered in standard clinical practice.
Validation of the MiRTM on randomly selected cases
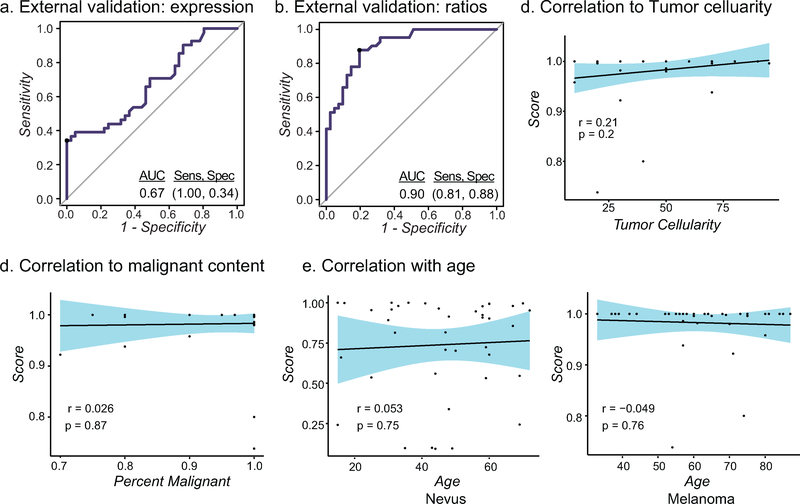
Discovery phase cohorts are often selected for unambiguous and homogenous cases. By design, we expected the MiRTM to perform equally as well on cohorts constructed with a high degree of variance in tumor cellularity, malignant content and patient age. To validate our model on cases with greater diversity in these features, we randomly retrieved 82 biopsied melanocytic lesions - 41 neoplasms diagnosed as nevi and 41 diagnosed as melanoma - from the archives of the UCSF Dermatopathology Section. All diagnoses were reviewed and confirmed by an independent dermatopathologist. This cohort contained a range of tumor cellularity and subtypes of melanocytic neoplasms (Table 1, Table S5). To test the robustness of our model against this noise, sections were not micro-dissected, but rather all tissue was harvested to obtain bulk RNA, and the abundances of the six miRNAs were assessed by RT-qPCR and applied to the two models (Table S6). The performance of the miRNA expression trained model dropped from crossvalidation to external validation, resulting in an area under ROC curve of 0.67 and sensitivity and specificity for the test set were 1.00 and 0.34, respectively (Fig. 3a). In contrast, the MiRTM performed with an area under the ROC curve of 0.90, and sensitivity and specificity of 0.81 and 0.88 (Fig. 3b). We found no correlation between the MiRTM scores and tumor cellularity, malignant content, patient age, or other clinical features, suggesting that the MiRTM score was unaffected by these variables (Fig. 3d–e, Fig. S4).
Table 1:
Sample information for unfiltered cohort
| Features | Melanoma | Nevus | |
|---|---|---|---|
| n(%) | n(%) | ||
| Totals | 41 (100) | 41 (100) | |
| Age | <30 | 0 (0) | 6 (15) |
| 30–60 | 21 (51) | 28 (68) | |
| >60 | 20 (49) | 6 (15) | |
| Stage | pT1a/b | 35 (85) | - |
| pT2a/b | 3 (7) | - | |
| pT3–4 a/b | 2 (5) | - | |
| Melanoma Subtype | SSM | 33 (80) | - |
| Nodular | 4 (10) | - | |
| NoS | 3 (7) | - | |
| Nevus Subtypes | Congenital | - | 15 (37) |
| Compound | - | 32 (78) | |
| Lentiginous | - | 26 (63) | |
| Dysplastic | - | 7(17) | |
| Percent Tumor Cell | <40 | 13 (32) | 21 (51) |
| 40–70 | 19 (46) | 15 (37) | |
| >70 | 13 (32) | 5 (12) | |
| Percent Melanoma vs Nevus | <80 | 3 (7) | - |
| 80–100 | 10 (24) | - | |
| 100 | 28 (69) | - | |
Figure 3: MiRTM validation.
a-b) ROC curve for external validation cohort using model trained with miRNA expression (a) or miRNA ratios (b, MiRTM) from the combined published cohort with the random forest classifier. c-e) Correlation of MiRTM score with percent tumor cell content (c), malignant content (d) or age (e). 95% CI is shown in blue.
Discussion
Numerous studies have analyzed miRNA expression at different stages of melanoma progression, collectively identifying over 500 miRNAs enriched in nevi or melanomas, most of which have not reproduced with external validation sets (Babapoor et al. 2017; Chen et al. 2011; Hanniford et al. 2015; Jukic et al. 2010; Komina et al. 2016; Kozubek et al. 2013; Latchana et al. 2017; Sand et al. 2013; Satzger et al. 2012; Xu et al. 2012). Our analyses have refined this expansive list to eight miRNA expression ratios that reproducibly distinguish nevi from melanoma across independent datasets and profiling platforms.
We identified this signature by controlling two important variables, interobserver variability of diagnosis and variability in tumor cell content. Our strategy was first to meticulously assemble and annotate an initial limited-sized cohort of lesions and conduct next generation small RNA sequencing. We used this cohort to first perform a feature selection step to identify miRNAs that are predictive of diagnosis, but unaffected by other confounding variables. By limiting our feature set to six miRNAs, we reduced the risk of over-training our predictive model on our training set of 25 nevi and 57 melanomas samples. The model classified benign from malignant melanocytic lesions with a crossvalidation AUC of 0.95 and an external validation AUC of 0.90 (41 nevi and 41 melanomas). The sensitivity and specificity of the MiRTM for external validation was 0.81 and 0.88. This performance is comparable to other molecular tests for distinguishing benign melanocytic nevi from melanomas, including chromosomal analysis by fluorescence in situ hybridization (sensitivity 0.72–1.00, specificity 0.90–1.00) (Ferrara and De Vanna 2016; Gerami et al. 2010) and myPath Melanoma gene expression profiling (sensitivity 0.63–0.90, specificity 0.88–0.93)(Clarke et al. 2017; Minca et al. 2016). The MiRTM does not perform as well as chromosomal analysis by array comparative genomic hybridization (aCGH, sensitivity 0.92–0.96, specificity 0.87–1.00) or proteomic analysis by mass spectrometry imaging (sensitivity 0.97, specificity 0.90) (Bastian et al. 2003; Lazova et al. 2012; Wang et al. 2013). However, assessment by the MiRTM requires only a single section of FFPE material, does not require microdissection, and RT-qPCR is a quick and affordable assay making this approach a candidate for lesions where tissue availability is limited. Future studies are required to determine the accuracy of the MiRTM in classifying ambiguous cases and to further determine whether the MiRTM is a predictor of metastasis or overall survival.
Of the six miRNAs in our signature, three (MIR211–5p, MIR21–5p, and MIR125B-5p) have been linked to melanoma previously, and changes in their expression validated by in situ hybridization (Babapoor et al. 2016; Latchana et al. 2016; Wandler et al. 2017). MIR21 is an established oncomir and regulates genes involved in increased proliferation and invasion (Satzger et al. 2012). It is upregulated in many cancers and its expression correlates with progression from nevi to primary melanomas and then to metastatic melanomas (Jiang et al. 2012; Satzger et al. 2012). Conversely, MIR125B is often downregulated in cancers, including advanced melanomas, where its loss results in increased expression of cJUN and MLK3 and its gain induces senescence in melanoma cell lines (Kappelmann et al. 2013; Nyholm et al. 2014; Zhang et al. 2014). MIR211 is among the most well-established functional miRNAs in melanocytes and is downstream of the melanocyte lineage transcription factor MITF (Mazar et al. 2010). It is often downregulated during melanoma progression and has been linked to invasion through regulation of BRN2, NFAT5 and TGFPR2 (Boyle et al. 2011; Levy et al. 2010). Interestingly, MIR211 expression elicits paradoxical cell behaviors dependent on the stage and type of melanoma. In primary human melanocytes and in amelanotic melanoma cell lines, MIR211 acts as a tumor suppressor through inhibition of invasion and proliferation (Bell et al. 2014; Levy et al. 2010; Mazar et al. 2010; Xu et al. 2012). However, in pigmented melanoma cell lines, MIR211 expression confers resistance to targeted therapy (Vitiello et al. 2018; Vitiello et al. 2017). The other miRNAs in the signature are less well characterized in melanocytic systems. As another MIR125 family member, MIR125A is expected to target a similar set of genes as MIR125B. MIR31 is upregulated in some cancers, but its role as an obligate oncomir is controversial as it is transcribed from a commonly deleted or methylated genomic region in many cancers (Asangani et al. 2012; Valastyan and Weinberg 2010). Similarly, MIR100 has also been described as both a tumor suppressor and an oncomir depending on the context (Li et al. 2015). Given these context-specific behaviors, future studies should dissect the transcriptional programs affected by each of these miRNAs specifically during the transition of growth-restricted nevus melanocytes to early stage melanomas.
We speculate that the observed increase in classifier stability when considering expression ratios is due to specific expression of the MD-miRNAs in melanocytes, with little contribution from infiltrating or adjacent non-melanocytic cells. If true, we would expect this strategy to normalize to the amount of input RNA of melanocytic origin, as opposed to total RNA, and therefore be less influenced by changes in biopsy content. Previous reports that have visualized the expression patterns of MIR211, MIR125B and MIR21 in melanocytic lesions support this proposition, but further analyses of the expression pattern of all six miRNAs are required for confirmation (Babapoor et al. 2016; Wandler et al. 2017). Regardless of their precise functional role or their local cell of origin in the context of melanocytic neoplasia, our analyses demonstrate that the relative expression ratios of these six miRNAs can assist in distinguishing benign melanocytic nevi from malignant melanoma in FFPE samples.
Materials and Methods
Clinical specimens and histopathologic assessment
A feature-selection cohort was generated from 15 microdissected regions of seven cases selected from a larger published cohort (Shain et al. 2018). All cases were retrieved from the UCSF Dermatopathology archive as FFPE tissue blocks. Histopathologically distinct areas had been independently evaluated by a panel of 5–8 dermatopathologists for staging (Shain et al. 2018). A training cohort of 25 nevi and 57 melanomas was generated by combining all samples from previously published microRNA array profiling datasets(Chen et al. 2011; Jukic et al. 2010; Komina et al. 2016; Sand et al. 2013) (Meta-analysis described in Supplemental Methods). An external validation cohort was generated by retrieving 82 diagnosed melanomas (41 cases) or nevi (41 cases). Cases were reevaluated by a separate dermatopathologist to confirm diagnosis and obtain histopathological features, but were not excluded for any reason. For more detailed methods see Supplementary Methods.
RNA quantification
MicroRNA sequencing libraries were constructed with the TailorMix Small RNA Library Preparation Kit (SeqMatic, Freemont, CA). Sequencing was performed on the Illumina HiSeq2500 platform at single-end 50bp. After adaptor sequences were removed, reads were aligned to a human reference (hg37) with Bowtie (Langmead et al. 2009) and then small RNA reference groups (miRBase21) were counted. Differential expression analysis was performed from feature counts using DeSeq2 (Love et al. 2014) with p-values adjusted for multiple testing with the Benjamin-Hochberg method (p-adj). Co-expression analysis was conducted as previously described (Horvath and Dong 2008) (see Supplementary Methods). Quantitative PCR for specific miRNA detection was conducted with TaqMan Advanced miRNA Assays (Thermo Fisher) (see Supplemental Methods).
Training and validation of classifier
A feature subset was selected using the Boruta R package (Kursa and Rudnicki 2010) to determine a minimal set of miRNAs for classifier predictive accuracy from the FFPE miRNA-seq data set. Using log fold-change information from differential expression analysis, each miRNA was associated as melanoma-enriched (ME) or melanoma-depleted (MD) and miRNA ratios were created from each combination of the 2 ME-miRNAs (MIR31–5p and MIR21–5p) and 4 MD-miRNAs (MIR211–5p, MIR125A-5p, 125B-5p, MIR100–5p). The feature subset determined from our feature selection set was then used to train several classifiers on the selected combined outside published cohort (Table S1) using this transformed (ratio) or not transformed (expression) minimal signature set. Performance was determined by 5-fold repeated cross-validation over 100 repeats with the top model from each built using a random forest model used to create a final miRNA ratio trained model (MiRTM) or expression model. These models were then used to classify each case in our validation cohort from ratio or expression transformed data with sensitivities and specificities determined using the Youden index and overall performance visualized by the area under a ROC. More details are described in Supplementary Methods.
Statistical Analysis
Statistical significance was set to 0.05 with p-values adjusted for multiple testing with the Benjamin-Hochberg method. Pearson correlation coefficients were obtained between all continuous features with the equivalent point biserial correlation coefficient for binary variables. Correlation matrices were plotted with the corrplot R Package and correlation plots with the ggpubr R package with 95% confidence intervals calculated for the curves. Sensitivities and specificities were calculated from classification models built using the caret R package. ROC curves were generated using the pROC R package. Confidence intervals (CI) were calculated from the 95% CI of 2000 bootstrap replicates for sensitivity and specificity or the ‘Delong’ method for AUCs using pROC R package. All data was processed in R (3.3.2)
Data Availability Statement
Datasets related to this article can be found at [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001550.v2.p1], hosted by dbGaP (phs001550.v2.p1).
Supplementary Material
Acknowledgements
This research was supported by NIH DP5OD019787 and the Sandler Foundation Program for Breakthrough Biomedical Research Fellowship to R.L.J., and the Marcus Program in Precision Medicine Innovation Fund Seeding Big Ideas Award and the Helen Diller Family Comprehensive Cancer Center Impact Award to R.L.J and M.L.W. We would like to thank Tim H. McCalmont for assistance in identifying cases and Hilary Faith Hickman for assistance in visual design.
Financial Support: This research was funded in part by NIH DP5OD019787, the Sandler Foundation Program for Breakthrough Biomedical Research Fellowship, the UCSF Marcus Program in Precision Medicine Innovation Fund and the UCSF Helen Diller Impact Award.
Footnotes
Conflict of Interest Statement
BB: Consulting for Lilly Inc.. Unrelated to the subject of this manuscript. The other authors state no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asangani IA, Harms PW, Dodson L, Pandhi M, Kunju LP, Maher CA, et al. Genetic and epigenetic loss of microRNA-31 leads to feed-forward expression of EZH2 in melanoma. Oncotarget. 2012;3(9):1011–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avissar M, Christensen BC, Kelsey KT, Marsit CJ. A MicroRNA Expression Ratio is Predictive of Head and Neck Squamous Cell Carcinoma. Clin. Cancer Res 2009;15(8):2850–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babapoor S, Horwich M, Wu R, Levinson S, Gandhi M, Makkar H, et al. microRNA in situ hybridization for miR-211 detection as an ancillary test in melanoma diagnosis. Mod. Pathol 2016;29(5):461–75 [DOI] [PubMed] [Google Scholar]
- Babapoor S, Wu R, Kozubek J, Auidi D, Grant-Kels JM, Dadras SS. Identification of microRNAs associated with invasive and aggressive phenotype in cutaneous melanoma by next-generation sequencing. Lab. Invest 2017;97(6):636–48 [DOI] [PubMed] [Google Scholar]
- Bastian BC, Olshen AB, LeBoit PE, Pinkel D. Classifying melanocytic tumors based on DNA copy number changes. Am. J. Pathol American Society for Investigative Pathology; 2003;163(5): 1765–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell RE, Khaled M, Netanely D, Schubert S, Golan T, Buxbaum A, et al. Transcription Factor/microRNA Axis Blocks Melanoma Invasion Program by miR-211 Targeting NUAK1. J. Invest. Dermatol Elsevier; 2014;134(2):441–51 [DOI] [PubMed] [Google Scholar]
- Boiko PE, of the Board PMW, Piepkorn MW. Reliability of skin biopsy pathology. J. Am. Board Fam. Pract American Board of Family Medicine; 1994;7(5):371–4 [PubMed] [Google Scholar]
- Boyle GM, Woods SL, Bonazzi VF, Stark MS, Hacker E, Aoude LG, et al. Melanoma cell invasiveness is regulated by miR-211 suppression of the BRN2 transcription factor. Pigment Cell Melanoma Res. 2011;24(3):525–37 [DOI] [PubMed] [Google Scholar]
- Brochez L, Verhaeghe E, Grosshans E, Haneke E, Piérard G, Ruiter D, et al. Intern-observer variation in the histopathological diagnosis of clinically suspicious pigmented skin lesions. J Pathol. 2002;196(4):459–66 [DOI] [PubMed] [Google Scholar]
- Buchbinder EI, Flaherty KT. Biomarkers in Melanoma: Lessons from Translational Medicine. Trends Cancer. 2016;2(6):305–12 [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang X, Lentz C, Abi-Daoud M, Paré GC, Yang X, et al. miR-193b Regulates Mcl-1 in Melanoma. Am. J. Pathol 2011;179(5):2162–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke LE, Flake DD, Busam K, Cockerell C, Helm K, McNiff J, et al. An independent validation of a gene expression signature to differentiate malignant melanoma from benign melanocytic nevi. Cancer. 2017;123(4):617–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corona R, Mele A, Amini M, Rosa DG, Coppola G, Piccardi P, et al. Interobserver variability on the histopathologic diagnosis of cutaneous melanoma and other pigmented skin lesions. J Clin Oncol Off. J Am Soc Clin Oncol 1996;14(4): 1218–23 [DOI] [PubMed] [Google Scholar]
- Creighton CJ, Reid JG, Gunaratne PH. Expression profiling of microRNAs by deep sequencing. Brief. Bioinform 2009;10(5):490–7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Amato NC, Howe EN, Richer JK. MicroRNA regulation of epithelial plasticity in cancer. Cancer Lett. 2013;341(1):46–55 [DOI] [PubMed] [Google Scholar]
- Elder DE, Piepkorn M, Barnhill RL, Longton GM, Nelson HD, Knezevich SR, et al. Pathologist Characteristics Associated with Accuracy and Reproducibility of Melanocytic Skin Lesion Interpretation. J Am Acad Dermatol. 2018;79(1):52–59.e5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmore JG, Barnhill RL, Elder DE, Longton GM, Pepe MS, Reisch LM, et al. Pathologists’ diagnosis of invasive melanoma and melanocytic proliferations: observer accuracy and reproducibility study. BMJ. 2017;357:j2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer ER, Gonin R, Hanna MP. Discordance in the histopathologic diagnosis of melanoma and melanocytic nevi between expert pathologists. Hum Pathol. 1996;27(6):528–31 [DOI] [PubMed] [Google Scholar]
- Ferrara G, De Vanna AC. Fluorescence In Situ Hybridization for Melanoma Diagnosis. Am. J. Dermatopathol 2016;38(4):253–69 [DOI] [PubMed] [Google Scholar]
- Gaudi S, Zarandona JM, Raab SS, English JC, Jukic DM. Discrepancies in dermatopathology diagnoses: The role of second review policies and dermatopathology fellowship training. J. Am. Acad. Dermatol Mosby; 2013;68(1): 119–28 [DOI] [PubMed] [Google Scholar]
- Gerami P, Mafee M, Lurtsbarapa T, Guitart J, Haghighat Z, Newman M. Sensitivity of Fluorescence In Situ Hybridization for Melanoma Diagnosis Using RREB1, MYB, Cep6, and 11q13 Probes in Melanoma Subtypes. Arch. Dermatol 2010;146(3):273–8 [DOI] [PubMed] [Google Scholar]
- Gershenwald JE, Scolyer RA, Hess KR, Sondak VK, Long GV, Ross MI, et al. Melanoma staging: Evidence-based changes in the American Joint Committee on Cancer eighth edition cancer staging manual. CA. Cancer J. Clin 2017;67(6):472–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanniford D, Segura MF, Zhong J, Philips E, Jirau-Serrano X, Darvishian F, et al. Identification of metastasis-suppressive microRNAs in primary melanoma. J. Natl. Cancer Inst 2015;107(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkes JE, Nguyen GH, Fujita M, Florell SR, Callis Duffin K, Krueger GG, et al. microRNAs in Psoriasis. J. Invest. Dermatol Elsevier; 2016;136(2):365–71 [DOI] [PubMed] [Google Scholar]
- Heenan PJ, Matz LR, Blackwell JB, Kelsall GR, Singh A, ten Seldam RE, et al. Inter-observer variation between pathologists in the classification of cutaneous malignant melanoma in Western Australia. Histopathology. 1984;8(5):717–29 [DOI] [PubMed] [Google Scholar]
- Horvath S, Dong J. Geometric interpretation of gene coexpression network analysis. PLoS Comput. Biol 2008;4(8):e1000117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarry J, Schadendorf D, Greenwood C, Spatz A, van Kempen LC. The validity of circulating microRNAs in oncology: five years of challenges and contradictions. Mol. Oncol 2014;8(4):819–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jayawardana K, Schramm S-J, Tembe V, Mueller S, Thompson JF, Scolyer RA, et al. Identification, Review, and Systematic Cross-Validation of microRNA Prognostic Signatures in Metastatic Melanoma. J. Invest. Dermatol 2016;136(1):245–54 [DOI] [PubMed] [Google Scholar]
- Jiang L, Lv X, Li J, Li J, Li X, Li W, et al. The status of microRNA-21 expression and its clinical significance in human cutaneous malignant melanoma. Acta Histochem. 2012;114(6):582–8 [DOI] [PubMed] [Google Scholar]
- Jukic DM, Rao UNM, Kelly L, Skaf JS, Drogowski LM, Kirkwood JM, et al. Microrna profiling analysis of differences between the melanoma of young adults and older adults. J. Transl. Med 2010;8:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung M, Schaefer A, Steiner I, Kempkensteffen C, Stephan C, Erbersdobler A, et al. Robust microRNA stability in degraded RNA preparations from human tissue and cell samples. Clin. Chem 2010;56(6):998–1006 [DOI] [PubMed] [Google Scholar]
- Kappelmann M, Kuphal S, Meister G, Vardimon L, Bosserhoff A-K. MicroRNA miR-125b controls melanoma progression by direct regulation of c-Jun protein expression. Oncogene. 2013;32(24): 2984–91 [DOI] [PubMed] [Google Scholar]
- Komina A, Palkina N, Aksenenko M, Tsyrenzhapova S, Ruksha T. Antiproliferative and Pro-Apoptotic Effects of MiR-4286 Inhibition in Melanoma Cells. PLoS One. 2016;11(12):e0168229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosik KS. MicroRNAs and cellular phenotypy. Cell. 2010;143(1):21–6 [DOI] [PubMed] [Google Scholar]
- Kozubek J, Ma Z, Fleming E, Duggan T, Wu R, Shin D-G, et al. In-Depth Characterization of microRNA Transcriptome in Melanoma. PLoS One. 2013;8(9):e72699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursa MB. Robustness of Random Forest-based gene selection methods. BMC Bioinformatics. 2014; 15:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kursa MB, Rudnicki WR. Feature Selection with the Boruta Package. J. Stat. Softw 2010;36(11): 1–13 [Google Scholar]
- Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latchana N, Del Campo SEM, Grignol VP, Clark JR, Albert SP, Zhang J, et al. Classification of Indeterminate Melanocytic Lesions by MicroRNA Profiling. Ann. Surg. Oncol 2017;24(2):347–54 [DOI] [PubMed] [Google Scholar]
- Latchana N, Ganju A, Howard JH, Carson WE. MicroRNA dysregulation in melanoma. Surg. Oncol 2016;25(3):184–9 [DOI] [PubMed] [Google Scholar]
- Lazova R, Seeley EH, Keenan M, Gueorguieva R, Caprioli RM. Imaging Mass Spectrometry—A New and Promising Method to Differentiate Spitz Nevi From Spitzoid Malignant Melanomas. Am. J. Dermatopathol 2012;34(1):82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leachman SA, Koon SM, Korcheva VB, White KP. Assessing Genetic Expression Profiles in Melanoma Diagnosis. Dermatol. Clin. 2017;35(4):537–44 [DOI] [PubMed] [Google Scholar]
- Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75(5):843–54 [DOI] [PubMed] [Google Scholar]
- Levy C, Khaled M, Iliopoulos D, Janas MM, Schubert S, Pinner S, et al. Intronic miR-211 assumes the tumor suppressive function of its host gene in melanoma. Mol. Cell 2010;40(5):841–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Gao Y, Zhang K, Chen J, Han S, Feng B, et al. Multiple Roles of MicroRNA-100 in Human Cancer and its Therapeutic Potential. Cell. Physiol. Biochem 2015;37(6):2143–59 [DOI] [PubMed] [Google Scholar]
- Li J, Tran M, Siwabessy J. Selecting Optimal Random Forest Predictive Models: A Case Study on Predicting the Spatial Distribution of Seabed Hardness. PLoS One. 2016;11(2):e0149089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A, Tetzlaff MT, Vanbelle P, Elder D, Feldman M, Tobias JW, et al. MicroRNA expression profiling outperforms mRNA expression profiling in formalin-fixed paraffin-embedded tissues. Int. J. Clin. Exp. Pathol 2009;2(6):519–27 [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margue C, Philippidou D, Reinsbach SE, Schmitt M, Behrmann I, Kreis S. New target genes of MITF-induced microRNA-211 contribute to melanoma cell invasion. PLoS One. 2013;8(9):e73473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazar J, DeYoung K, Khaitan D, Meister E, Almodovar A, Goydos J, et al. The Regulation of miRNA-211 Expression and Its Role in Melanoma Cell Invasiveness. Cheriyath V, editor. PLoS One. 2010;5(11):e13779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minca EC, Al-Rohil RN, Wang M, Harms PW, Ko JS, Collie AM, et al. Comparison between melanoma gene expression score and fluorescence in situ hybridization for the classification of melanocytic lesions. Mod. Pathol 2016;29(8):832–43 [DOI] [PubMed] [Google Scholar]
- Mumford SL, Towler BP, Pashler AL, Gilleard O, Martin Y, Newbury SF. Circulating MicroRNA Biomarkers in Melanoma: Tools and Challenges in Personalised Medicine. Biomolecules. 2018;8(2):21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair VS, Maeda LS, Ioannidis JPA. Clinical Outcome Prediction by MicroRNAs in Human Cancer: A Systematic Review. JNCI J. Natl. Cancer Inst 2012;104(7):528–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niebling MG, Haydu LE, Karim RZ, Thompson JF, Scolyer RA. Pathology Review Significantly Affects Diagnosis and Treatment of Melanoma Patients: An Analysis of 5011 Patients Treated at a Melanoma Treatment Center. Ann. Surg. Oncol 2014;21(7):2245–51 [DOI] [PubMed] [Google Scholar]
- Nyholm AM, Lerche CM, Manfé V, Biskup E, Johansen P, Morling N, et al. miR-125b induces cellular senescence in malignant melanoma. BMC Dermatol. 2014; 14(1):8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parchem RJ, Ye J, Judson RL, LaRussa MF, Krishnakumar R, Blelloch A, et al. Two miRNA clusters reveal alternative paths in late-stage reprogramming. Cell Stem Cell. 2014; 14(5):617–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogribny IP. MicroRNAs as biomarkers for clinical studies. Exp. Biol. Med 2018;243(3):283–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raya L, Sidi Y, Avni D. Aberrations in the micro-RNA biogenesis machinery and the emerging roles of micro-RNAs in the pathogenesis of cutaneous malignant melanoma. Pigment Cell Melanoma Res. 2012;25(6):740–57 [DOI] [PubMed] [Google Scholar]
- Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015; 15:38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy A, Growney JD, Wilson NS, Emery CM, Johnson JA, Ward R, et al. Gene Expression Ratios Lead to Accurate and Translatable Predictors of DR5 Agonism across Multiple Tumor Lineages. PLoS One. 2015;10(9):e0138486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez-Cerdeira C, Molares-Vila A, Carnero-Gregorio M, Corbalan-Rivas A. Recent advances in melanoma research via “omics” platforms. J. Proteomics. : 10.1016/j.jprot.2017.11.005 [DOI] [PubMed] [Google Scholar]
- Sand M, Skrygan M, Sand D, Georgas D, Gambichler T, Hahn SA, et al. Comparative microarray analysis of microRNA expression profiles in primary cutaneous malignant melanoma, cutaneous malignant melanoma metastases, and benign melanocytic nevi. Cell Tissue Res. 2013;351(1):85–98 [DOI] [PubMed] [Google Scholar]
- Satzger I, Mattern A, Kuettler U, Weinspach D, Niebuhr M, Kapp A, et al. microRNA-21 is upregulated in malignant melanoma and influences apoptosis of melanocytic cells. Exp. Dermatol 2012;21(7):509–14 [DOI] [PubMed] [Google Scholar]
- Shain AH, Joseph NM, Yu R, Benhamida J, Liu S, Prow T, et al. Genomic and Transcriptomic Analysis Reveals Incremental Disruption of Key Signaling Pathways during Melanoma Evolution. Cancer Cell. 2018;34(1):45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shain AH, Yeh I, Kovalyshyn I, Sriharan A, Talevich E, Gagnon A, et al. The Genetic Evolution of Melanoma from Precursor Lesions. N. Engl. J. Med 2015;373(20): 1926–36 [DOI] [PubMed] [Google Scholar]
- Sheinerman KS, Umansky SR. Circulating cell-free microRNA as biomarkers for screening, diagnosis and monitoring of neurodegenerative diseases and other neurologic pathologies. Front. Cell. Neurosci 2013;7:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoo AB, Sagebiel RW, Mohammed K-S. Discordance in the histopathologic diagnosis of melanoma at a melanoma referral center. J Am Acad Dermatol. 2010;62(5):751–6 [DOI] [PubMed] [Google Scholar]
- Valastyan S, Weinberg RA. miR-31: A crucial overseer of tumor metastasis and other emerging roles. Cell Cycle. 2010;9(11):2124–9 [DOI] [PubMed] [Google Scholar]
- Vitiello M, D’Aurizio R, Poliseno L. Biological role of miR-204 and miR-211 in melanoma. Oncoscience. Impact Journals, LLC; 2018;5(7–8):248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitiello M, Tuccoli A, D’Aurizio R, Sarti S, Giannecchini L, Lubrano S, et al. Context-dependent miR-204 and miR-211 affect the biological properties of amelanotic and melanotic melanoma cells. Oncotarget. 2017;8(15) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace E, Lowry J, Smith SM, Fahey T. The epidemiology of malpractice claims in primary care: a systematic review. BMJ Open. 2013;3(7):e002929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wandler A, Riber-Hansen R, Hager H, Hamilton-Dutoit SJ, Schmidt H, Nielsen BS, et al. Quantification of microRNA-21 and microRNA-125b in melanoma tissue. Melanoma Res. 2017;27(5):417–28 [DOI] [PubMed] [Google Scholar]
- Wang L, Rao M, Fang Y, Hameed M, Viale A, Busam K, et al. A genome-wide high-resolution array-cgh analysis of cutaneous melanoma and comparison of array-cgh to fish in diagnostic evaluation. J. Mol. Diagnostics. 2013;15(5):581–91 [DOI] [PubMed] [Google Scholar]
- Witwer KW, Halushka MK. Towards the Promise of microRNAs - Enhancing reproducibility and rigor in microRNA research. RNA Biol. 2016;13(11):1103–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, Brenn T, Brown ERS, Doherty V, Melton DW. Differential expression of microRNAs during melanoma progression: miR-200c, miR-205 and miR-211 are downregulated in melanoma and act as tumour suppressors. Br. J. Cancer. 2012;106(3):553–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Lu L, Xiong Y, Qin W, Zhang Y, Qian Y, et al. MLK3 promotes melanoma proliferation and invasion and is a target of microRNA-125b. Clin. Exp. Dermatol 2014;39(3):376–84 [DOI] [PubMed] [Google Scholar]
- Key Statistics for Melanoma Skin Cancer [Internet]. [cited 2018 Dec 19]. Available from: https://www.cancer.org/cancer/melanoma-skin-cancer/about/key-statistics.html
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Datasets related to this article can be found at [https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs001550.v2.p1], hosted by dbGaP (phs001550.v2.p1).