Abstract
Non-obstructive azoospermia (NOA) is the most severe form of male infertility. However, the etiology of NOA is largely unknown, resulting in a lack of clinical treatments. Here, we performed a comparative genome-wide profiling of DNA methylation and identified SOX30 as the most notably hyper-methylated gene at promoter in testicular tissues from NOA patients. This hyper-methylation at promoter of SOX30 directly causes its silencing of expression in NOA. The reduced levels of SOX30 expression are correlated with severity of NOA disease. Deletion of Sox30 in mice uniquely impairs testis development and spermatogenesis with complete absence of spermatozoa in testes leading to male infertility, but does not influence ovary development and female fertility. The pathology and testicular size of Sox30 null mice highly simulate those of NOA patients. Re-expression of Sox30 in Sox30 null mice at adult age reverses the pathological damage of testis and restores the spermatogenesis. The re-presented spermatozoa after re-expression of Sox30 in Sox30 null mice have the ability to start a pregnancy. Moreover, the male offspring of Sox30 re-expression Sox30 null mice still can father children, and these male offspring and their children can live normally more than 1 year without significant difference of physical appearance compared with wild-type mice. In summary, methylated inactivation of SOX30 uniquely impairs spermatogenesis, probably causing NOA disease, and re-expression of SOX30 can successfully restore the spermatogenesis and actual fertility. This study advances our understanding of the pathogenesis of NOA, offering a promising therapy target for NOA disease.
Keywords: male infertility, methylation, non-obstructive azoospermia, spermatogenesis, therapy target
Introduction
Infertility is the most severe problem in reproductive health.1 Approximately half of infertility in humans is attributed to male factors, and ∼15% of these factors are azoospermia with complete absence of sperm in the semen.2, 3, 4 Azoospermia is classified into obstructive azoospermia (OA), comprising about 40%, and non-obstructive azoospermia (NOA), comprising about 60% of azoospermic cases. OA accompanied by normal spermatogenesis in testes is the consequence of physical blockage to the excurrent ducts and fails to produce mature sperm.5, 6, 7 NOA, as the severest form of human male infertility, is the consequence of abnormal spermatogenesis in testes and fails to produce testicular sperm. Up to now, more than 80% of NOA cases remain unclear in etiology.8
Although numerous genetic testing studies have been performed, genetic abnormalities have been detected in only 20% of NOA patients.4,9 It remains largely unknown whether epigenetic changes are associated with NOA disease. DNA methylation is an epigenetic modification that plays important roles in various diseases.10, 11, 12, 13, 14 Accumulating evidence has suggested that a normal methylation pattern in germ cell is highly associated with male fertility.15, 16, 17, 18 In our present study, we performed a comparative genome-wide profile of DNA methylation and gene expression in testicular biopsy specimens between OA and NOA patients. SOX30 was identified as the most notably hyper-methylated gene at promoter and a silent gene in NOA patients. SOX30 was inactivated by DNA methylation at promoter rather than genetic variation in NOA. Indeed, SOX30 deficiency was tightly correlated with NOA disease. The in vivo function of Sox30 was then explored in testis development of Sox30 knockout mice. The pathology and testicular volume of Sox30 null mice were compared with that of NOA patients. Moreover, the potential application of Sox30 to cure NOA disease by restoring Sox30 expression was evaluated. This study identified SOX30 as a key male-specific factor involved in infertility, providing a prospective target for the treatment of human NOA disease.
Results
Study Participants
The main purpose of this study is to identify novel and key methylated genes associated with germ cells or spermatogenesis in NOA disease. The OA men who exhibit normal tissue morphology with a large number of sperm and no significant reduction in spermatogenic cells in testis tissues were selected as the control tissues. Moreover, the selected OA patients underwent testicular sperm extraction (TESE) or microsurgical epididymal sperm aspiration (MESE) for assisted reproduction and could father children. The composition of cell types varies greatly in NOA patients. To eliminate other factors as much as possible, the NOA samples with clear composition of cell types and pathological morphology were selected. The NOA patients could be classified into four groups according to composition of cell types: NOA-I patients without spermatozoa, NOA-II patients without spermatids, NOA-III patients without spermatocytes, and NOA-SCO (Sertoli cell-only) patients without spermatogenic cells (Figure S1). However, the NOA-SCO patients were excluded in the present study because we would like to identify the novel and key related genes in NOA disease that are associated with germ cells or spermatogenesis. Based on the above criteria, we screened 502 cases of OA and NOA in men and selected 15 well-matched OA patients from 326 OA men and 58 NOA patients from 176 NOA men for this study. These selected NOA patients included 31 cases of NOA-I, 22 cases of NOA-II, and 5 cases of NOA-III. The detailed characteristics of the participants selected are shown in Table S2.
SOX30 Is Hyper-Methylated in Testicular Tissues of NOA Patients
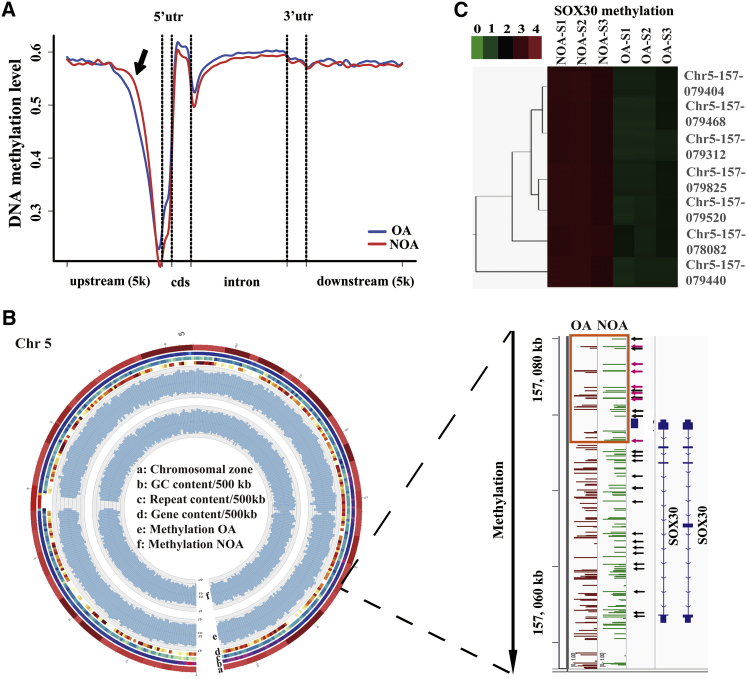
To understand the pathogenesis of human NOA disease, we analyzed by direct bisulfite sequencing the global DNA methylation in five NOA and five OA patients’ testicular tissues selected randomly from the 15 OA patients and 58 NOA patients. A total of 5,832 differentially methylated regions (DMRs; p < 0.01) were detected in NOA compared with OA. These DMRs were mainly distributed in intergenic regions (48.56%), intron regions (34.74%), upstream regions (promoter regions, 9.23%), and downstream regions (5.74%) of genes (Figure S2A). The 5,832 DMRs were located at 2,189 genes showing a different status of DNA methylation in NOA compared with OA (1,391 hyper-methylated genes and 798 hypo-methylated genes) (Table S3). The distribution of the hyper-methylated DMRs in NOA compared with OA was centered in the upstream regions of genes, and the hyper-methylated DMRs at promoter of genes preferentially existed on chromosome 3 (Chr3), chromosome 18 (Chr18), and chromosome 5 (Chr5) (Figure 1A; Figure S2B; Table S3). Among these hyper-methylated regions at promoter of genes on Chr3, Chr18, and Chr5, SOX30 was found to be one of the most notably hyper-methylated genes at promoter (p = 3.23E−6) (Table 1; Table S3). Moreover, 25 serious hyper-methylated sites of CpG island were identified at the promoter of SOX30 in NOA patients compared with OA patients (Figure 1B; Table S4). The global DNA methylation was also analyzed using microarrays in three other independent NOA and three OA patients’ testicular tissues.19,20 In the differentially methylated genes of methylation microarray, SOX30 was also significantly hyper-methylated at promoter in NOA compared with OA patients (kindly analyzed by Z.-m.L. and X. Zhuang in Xiamen University; Figure 1C). These data demonstrate that SOX30 is a remarkably hyper-methylated gene at promoter in testicular tissues of NOA patients.
Figure 1.
SOX30 Is Identified as a Hyper-Methylated Gene in NOA Testicular Tissues
(A) DNA methylation levels of different gene regions in NOA testicular tissues compared with in OA testicular tissues. The arrow represents the hyper-methylated region in NOA. (B) Identification of SOX30 as a preferentially methylated gene on chromosome 5 (Chr5) in NOA samples using genome-wide methylation screening. Circos plot shows Chr5 methylation pattern in OA and NOA samples. The chromosomal zone, GC content, repeated sequence content, gene sequence content, methylation level of OA, and methylation level of NOA are presented from the outer circle to the inner circle in the Circos figure. The arrows represent preferential methylated sites of SOX30 gene in NOA samples. Orange box represents SOX30 promoter. The hyper-methylated CpG sites that are negatively associated with SOX30 expression have been indicated with arrows in red. (C) Hierarchical clustering of differentially methylated sites of SOX30 in other independent testicular tissue samples of NOA and OA patients. The key color bar indicates methylation levels. NOA-S1, NOA-S2, NOA-S3, OA-S1, OA-S2, and OA-S3 represent NOA-Sample1, NOA-Sample2, NOA-Sample3, OA-Sample1, OA-Sample2, and OA-Sample3, respectively.
Table 1.
The Hyper-Methylated Genes of Chromosome 3, Chromosome 18, and Chromosome 5 with DMRs on the Promoter Region Were Listed between Testicular Tissues of OA and NOA Patients
| DMR Location | Methylation |
p Value (MWU-Test) | Gene Symbol and No. | Gene Region | |
|---|---|---|---|---|---|
| OA | NOA | ||||
| Chr3 | 0.17033 | 0.59108 | 0.0083 | CRTAP; NM_006371 | promoter |
| Chr3 | 0.2225 | 0.7972 | 0.00073 | C3orf84; NM_001080528 | promoter |
| Chr3 | 0.0000 | 0.2695 | 0.00013 | USP4; NM_003363 | promoter |
| Chr3 | 0.42783 | 0.86725 | 0.0005 | NICN1; NM_032316 | promoter |
| Chr3 | 0.22264 | 0.79 | 0.00014 | RPL29; NM_000992 | promoter |
| Chr3 | 0.10645 | 0.57564 | 0.00039 | DENND6A; NM_152678 | promoter |
| Chr3 | 0.07791 | 0.29973 | 0.0066 | LINC00960; NR_040005 | promoter |
| Chr3 | 0.075 | 0.8352 | 2.20E−5 | ROBO1; NM_00114584 | promoter |
| Chr3 | 0.22642 | 0.87092 | 6.40 E−6 | OR5AC2; NM_054106 | promoter |
| Chr3 | 0.31021 | 0.76793 | 0.0028 | TXNRD3; NM_001173513 | promoter |
| Chr3 | 0.118 | 0.72015 | 3.70 E−6 | TPRA1; NM_001136053 | promoter |
| Chr3 | 0.0000 | 0.4205 | 0.0011 | RBP1; NM_002899 | promoter |
| Chr3 | 0.1516 | 0.6282 | 0.0029 | LOC100128164; NR_027622 | promoter |
| Chr3 | 0.123 | 0.4503 | 0.0015 | SKIL; NM_001145097 | promoter |
| Chr3 | 0.4009 | 0.9353 | 1.10E−5 | HTR3E; NM_001256614 | promoter |
| Chr3 | 0.3481 | 0.8349 | 0.00032 | SDHAP2; NR_003265 | promoter |
| Chr18 | 0.1459 | 0.7482 | 1.10E−5 | IMPA2; NM_014214 | promoter |
| Chr18 | 0.0275 | 0.4705 | 0.0039 | OSBPL1A; NM_080597 | promoter |
| Chr18 | 0.1906 | 0.7729 | 1.10E−5 | TCEB3CL; NM_001100817 | promoter |
| Chr18 | 0.1191 | 0.7259 | 1.10E−5 | TCEB3CL2; NM_001242907 | promoter |
| Chr18 | 0.2709 | 0.9206 | 0.00049 | ELAC1; NM_018696 | promoter |
| Chr18 | 0.000 | 0.67609 | 3.80E−6 | C18orf63; NM_001174123 | promoter |
| Chr5 | 0.1871 | 0.891 | 1.10E−5 | PDZD2; NM_178140 | promoter |
| Chr5 | 0.2086 | 0.7136 | 0.0039 | SDHA; NM_004168 | promoter |
| Chr5 | 0 | 0.9022 | 6.70E−6 | MCCC2; NM_022132 | promoter |
| Chr5 | 0.2142 | 0.8657 | 1.10E−5 | SSBP2; NM_001256732 | promoter |
| Chr5 | 0.1527 | 0.6681 | 0.0015 | ZCCHC10; NM_017665 | promoter |
| Chr5 | 0.1865 | 0.4772 | 0.00049 | CDC23; NM_004661 | promoter |
| Chr5 | 0.2264 | 0.671 | 0.00027 | SNORD63; NR_002913 | promoter |
| Chr5 | 0.2774 | 0.8002 | 0.00073 | MAT2B; NM_182796 | promoter |
| Chr5 | 0.4716 | 0.6994 | 0.0014 | FAM153B; NM_001265615 | promoter |
| Chr5 | 0.2628 | 0.5196 | 0.0033 | HNRNPAB; NM_004499 | promoter |
| Chr5 | 0.4017 | 0.9711 | 3.23E−6 | SOX30; NM_178424 | promoter |
SOX30 Silenced by Hyper-Methylation at Promoter Is Highly Associated with NOA
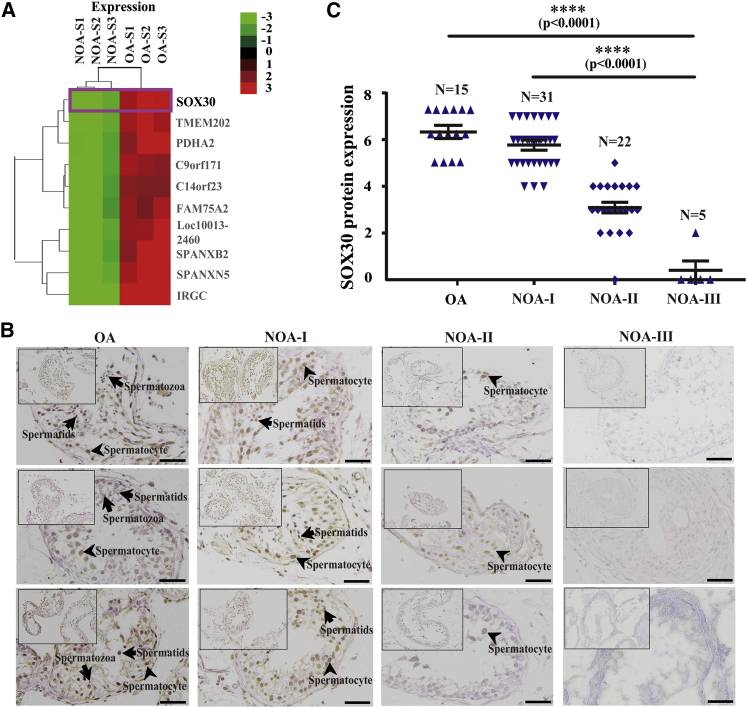
To determine the effect of methylation on gene expression, we performed a transcriptome study using microarray in the same NOA and OA patients’ testicular tissues as the methylation microarray analyzed. Over 1,900 upregulated and 2,700 downregulated genes were found comparing NOA with OA (fold change >2 or <1/2), and SOX30 was identified as a prominent silenced gene in NOA by cluster analysis of gene expression profiling (Figure 2A). By integration analyses of the data from methylation and expression microarrays, we found that multiple hyper-methylated CpG sites on SOX30 promoter are negatively associated with SOX30 expression (Figures 1B, 1C, and 2A). Immunohistochemistry (IHC) staining of testicular tissues from the OA and NOA patients confirmed that SOX30 expression was significantly decreased in testicular tissues of NOA patients compared with OA patients (p < 0.0001; Figures 2B and 2C). In the divided three groups of NOA patients, NOA-I, NOA-II, and NOA-III patients, the levels of SOX30 expression were continuously decreased from NOA-I patients (5.742 ± 0.9147), to NOA-II patients (3.091 ± 1.0405), to NOA-III patients (0.4000 ± 0.8; p < 0.0001; Figures 2B and 2C). Moreover, Sox30 is silenced by hyper-methylation and can be restored by de-methylation treatment with 5-aza-dc in mouse GC2 (spermatocyte), TM3 (Leydig cell), and TM4 (Sertoli cell) cell lines in our previous study.21 To exclude the possibility that silencing of SOX30 in NOA is caused by genetic variations, we screened mutations and deletions in testicular tissues of the five NOA patients that performed global DNA methylation using direct sequencing. DNA sequencing revealed that there was no mutation or deletion except only a known SNP site (rs35793864) of SOX30 in NOA patients (Table S5). These data indicate that the silencing of SOX30 caused by hyper-methylation at promoter is closely associated with NOA disease.
Figure 2.
SOX30 Silenced by Hyper-Methylation Is Closely Associated with NOA
(A) Hierarchical clustering of differentially expressed genes (≥2-fold) in testicular tissues of three NOA (NOA-S1, -S2, -S3) patients as compared with that of three OA (OA-S1, -S2, -S3) patients, and identification of SOX30 as a preferentially silenced gene in NOA patients. The key color bar indicates mRNA expression levels. (B) IHC staining of SOX30 was performed in the testicular biopsy specimens from OA and NOA patients. NOA-I, NOA-II, and NOA-III represent the NOA patients without spermatozoon, the NOA patients without spermatid, and the NOA patients without spermatocyte, respectively. The arrows represent spermatozoa, spermatids, and spermatocyte. Scale bars represent 20 μm. (C) IHC staining of SOX30 was quantified in the testicular biopsy specimens of OA and NOA patients. ****p < 0.0001.
Sox30 Deletion Uniquely Impairs Testis Development, Causing Infertility
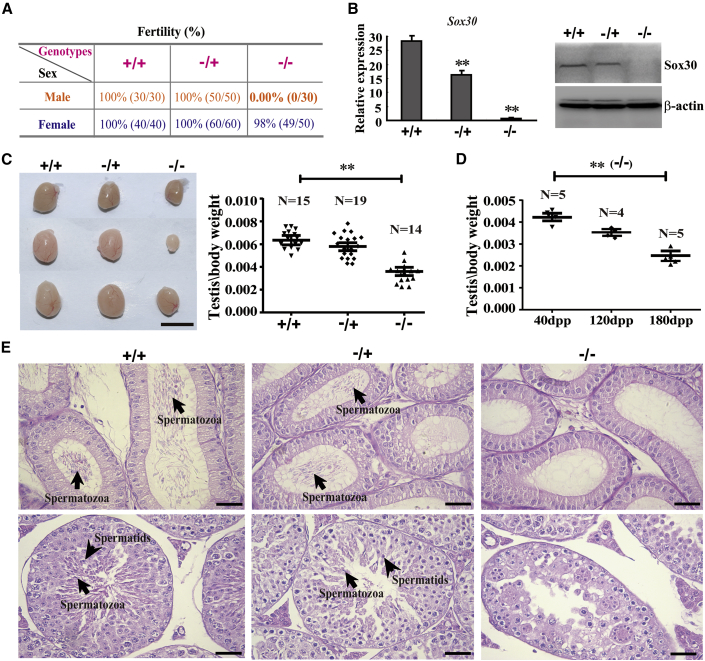
Functional network analysis of gene expression profiling demonstrated that SOX30-associated gene sets were involved in sexual reproduction, male gamete generation, and spermatogenesis processes (Figure S3). Furthermore, SOX30 expression was positively associated with testicular volume (p = 0.001) of NOA patients (Table S6). These data suggested a potential role for SOX30 in regulating testis development and spermatogenesis. To explore the biological role of Sox30 on testis development, we generated a Sox30 null mouse model (Figures S4A and S4B). Homozygous (Sox30−/−) and heterozygous (Sox30−/+) mice developed normally and lived more than 17 months without obvious differences in general physical appearance compared with their wild-type (Sox30+/+) littermates. Sox30−/− female mice were fertile, but Sox30−/− male mice were fully sterile (Figures 3A and 3B; Table S7). Sox30−/−, Sox30−/+, and Sox30+/+ female mice have similar ovaries, whereas Sox30−/− male mice showed significantly small testes compared with Sox30+/+ or Sox30−/+ mice (Figure 3C; Figures S5 and S6A–S6D). Furthermore, the loss of Sox30−/− testis weight became more severe as mice aged (Figure 3D; Figure S6E). These results demonstrate that Sox30 is essential for testis development and male fertility.
Figure 3.
Sox30 Is Required Specifically for Male Fertility and Testis Development
(A) The fertility was analyzed in different Sox30 genotypes of mice. +/+ represents wild-type mice (Sox30+/+), −/+ represents heterozygous mice (Sox30−/+), and −/− represents homozygous mice (Sox30−/−). (B) qRT-PCR and WB analyses of the expression of Sox30 in testes of mice with indicated genotypes. **p < 0.01. β-Actin was used as an internal control. (C) The morphology and weight of testes (Sox30+/+: n = 15, Sox30−/+: n = 19, and Sox30−/−: n = 14) were analyzed in Sox30+/+, Sox30−/+, and Sox30−/− mice. **p < 0.01. Scale bar represents 0.5 cm. (D) The weight of testes from Sox30−/− mice at different developmental stages (40, 120, and 180 days post-partum [dpp]) were compared. **p < 0.01. (E) H&E staining of epididymis and testis sections were shown from Sox30+/+ (n = 10), Sox30−/+ (n = 10), and Sox30−/− (n = 10) mice older than 5 months. The arrows in epididymis represent spermatozoa. The arrows in testes represent spermatozoa and spermatids. Scale bars represent 50 μm.
Sox30 Deletion Impairs Spermatogenesis, Resulting in Absence of Spermatozoa
To determine the potential causes of abnormal testis development and male infertility, we examined the gametes in epididymides and testes of Sox30 null mice. Sox30−/− mice completely lacked spermatozoa in their epididymides and exhibited disorganized testis morphology with multi-nucleated spermatogenic cells, few spermatids, and complete absence of spermatozoa in their seminiferous tubules by histological examination and analyzing semen (Figure 3E; Figures S7A and S7B). The few spermatids and complete absence of spermatozoa in testes of Sox30−/− mice were further confirmed by semithin histological section, electron-microscopy examination, and expression analysis of spermatid markers: Tnp1 and Prm2 (Figures S8A–S8C). These results reveal that deletion of Sox30 in mice impairs spermatogenesis, exhibiting few spermatids and complete absence of spermatozoa in testes.
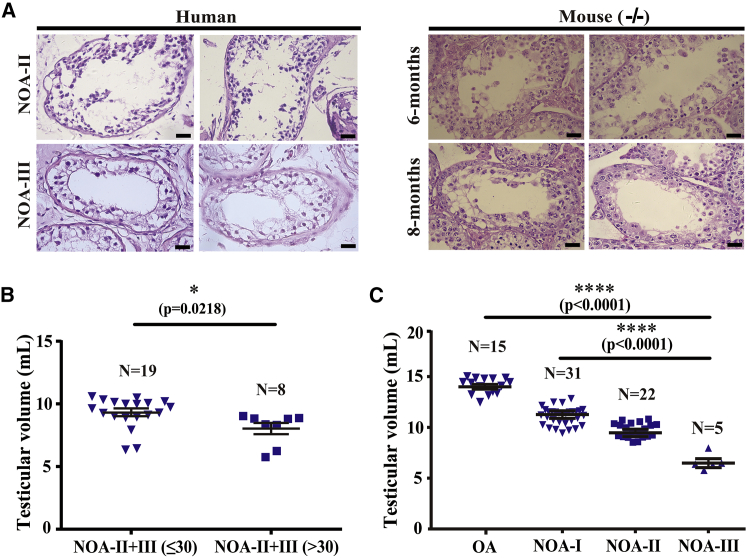
The Pathology and Testicular Size of Sox30−/− Mice Simulate Those of NOA Patients
To further determine the implication of SOX30 on NOA disease, we compared the testicular pathology of Sox30 null mice with that of NOA patients. No spermatozoa and few spermatids were observed in testes of 6-month-old Sox30-deficient mice, which are similar to the phenotypes of NOA-II patients (Figure 4A; Figure S9A). No spermatozoa, no spermatids, and a few spermatocyte-like cells were found in 8-month-old Sox30-deficient mice, which are very similar to the phenotypes of NOA-III patients (Figure 4A; Figure S9A). To determine whether the loss of testicular volume of NOA patients becomes more severe as they age, we compared the testicular volume of NOA and OA patients at different ages. The testicular volume of the NOA-II+III patients (highly similar to the phenotypes of Sox30-deficient mice) who are young (≤30 years) was significantly greater than that of the NOA-II+III patients who are older (>30 years) (p = 0.0218; Figure 4B). However, no difference was found between testicular volume of the young (≤30 years) NOA-I patients and the older (>30 years) NOA-I patients (p = 0.9895; Figure S9B), which is also applicable to the OA patients (p = 0.3943; Figure S9B). In addition, the testicular volume of OA patients is significantly greater than that of NOA patients (p < 0.0001), and the testicular volume was continuously decreased from NOA-I patients, to NOA-II patients, to NOA-III patients (p < 0.0001; Figure 4C). The data are consistent with the results of SOX30 expression in OA and NOA patients (Figures 2B and 2C) and the testis size of Sox30−/− mice (Figure 3C). These results indicate that the testicular pathology and testicular size of Sox30-deficient mice highly simulated those of NOA patients.
Figure 4.
The Pathology and Testicular Size of Sox30−/− Mice Simulate Those of NOA Patients
(A) H&E staining of testes from human NOA patients and Sox30−/− mice at 6-month (n = 10) or 8-month (n = 8) developmental stage were compared. Almost only spermatocyte-like, spermatogonia, and Sertoli cells were observed in 6-month-old Sox30-deficient mice. A few spermatocyte-like and almost only spermatogonia and Sertoli cells were found in 8-month-old Sox30-deficient mice. Scale bars, 50 μm. (B) The testicular volume of NOA patients at different ages was compared. The NOA-II+III (≤30) represents these NOA-II and NOA-III patients who are less than and equal to 30 years, and the NOA-II+III (>30) represents these NOA-II and NOA-III patients who are greater than 30 years. (C) The testicular volume of OA and NOA patients was compared. ****p < 0.0001.
The Infertility of Sox30 Deletion Can Be Cured by Restoration of Sox30
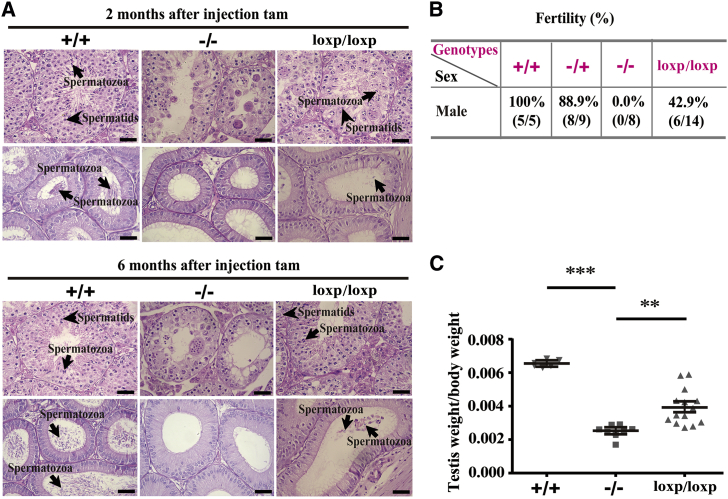
We next investigated whether the male infertility of Sox30 deficiency can be cured by re-expression of Sox30 in adult mice. Sox30 expression was restored in Sox30−/− mice by deleting the floxed cassette (Sox30loxp/loxp) using tamoxifen (tam) that activates the estrogen receptor (ER)-Cre (Figure S4A). These mice were sacrificed at 2 and 6 months after the first tam injection. The histological data showed a presence of spermatids and spermatozoa in some seminiferous tubules of testes and visible spermatozoa in epididymides from Sox30loxp/loxp mice (Figure 5A). These results reveal that re-expression of Sox30 can restore spermatogenesis in adult Sox30−/− mice. It is noteworthy that the mice analyzed at 6 months seemed to have a better restoration of spermatogenesis than those analyzed at 2 months after tam administration (Figure 5A).
Figure 5.
Infertility of Sox30−/− Mice Can Be Restored by Sox30 Re-expression
(A) H&E staining of testes and epididymides from Sox30+/+, Sox30−/−, and Sox30 re-expressed Sox30−/− (Sox30loxp/loxp) mice were shown. Spermatozoa and spermatids were observed in testes and epididymides of Sox30loxp/loxp mice, which were killed at 2 (n = 5) or 6 months (n = 5) after tam injection. The arrows with long tail indicate spermatozoa in testes and epididymides, and the arrows with short tail indicate spermatids in testes. Scale bars, 50 μm. (B) The ability to conceive of the re-presented spermatozoa was evaluated by fertility analysis after re-expression of Sox30 in Sox30−/− mice. (C) The weight of testes was analyzed in Sox30+/+ (n = 5), Sox30−/− (n = 8), and Sox30loxp/loxp (n = 14) mice. +/+, Sox30+/+ mice injected with tam; −/+, Sox30−/+ mice injected with tam; −/−, Sox30−/− mice injected with solvent; loxp/loxp, Sox30−/− mice injected with tam. ***p < 0.001; **p < 0.01.
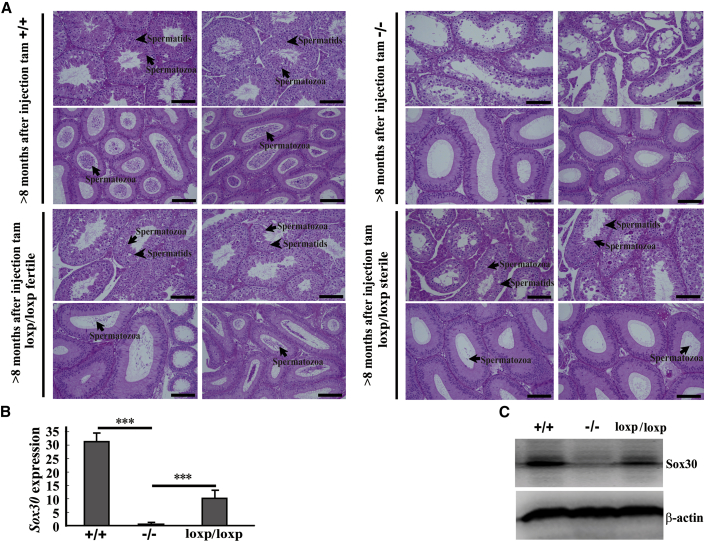
We then evaluated the ability to conceive of the re-presented spermatozoa after re-expression of Sox30 in male Sox30−/− mice by fertility analyses. The data showed that the male Sox30+/+ mice injected with tam were fertile in 100% (5/5), the male Sox30−/+ mice injected with tam were fertile in 88.9% (8/9), and the male Sox30−/− mice injected with solvent were fully sterile (0/8). Unexpectedly, the male Sox30loxp/loxp mice (Sox30−/− mice injected with tam) were fertile in 42.9% (6/14) (Figure 5B), whereas the number of descendants was relatively small (usually two to four) in the Sox30loxp/loxp mice group. Consistently, the testis weight of Sox30loxp/loxp mice was indeed restored when compared with that of Sox30−/− mice injected with solvent (Figure 5C; Figure S10). The histological results showed a presence of spermatids and spermatozoa in many seminiferous tubules of testes and a lot of spermatozoa in epididymides from fertile Sox30loxp/loxp mice (Figure 6A). There were also spermatids and spermatozoa in some seminiferous tubules of testes, but only a few spermatozoa in epididymides from sterile Sox30loxp/loxp mice (Figure 6A). To determine whether the recovery of spermatogenesis and fertility is the result of Sox30 re-expression in Sox30 null mice, we evaluated Sox30 expression in testes of these mice from the inducible system. Sox30 expression was indeed restored in testes of Sox30loxp/loxp mice (Figures 6B and 6C). These data indicate that the infertility of Sox30-deficient mice can be cured by re-expression of Sox30.
Figure 6.
The Pathological Morphology of Mice Testes with Different Genotypes Was Shown
(A) H&E staining of testes and epididymides from Sox30+/+, Sox30−/−, and Sox30 re-expressed Sox30−/− (Sox30loxp/loxp) mice was shown. Lots of spermatids were observed in testes and epididymides of Sox30+/+ mice (n = 5) injected with tam, but were not observed in testes and epididymides of Sox30−/− (n = 8) mice injected with solvent. Many spermatids were observed in testes and epididymides of fertile Sox30loxp/loxp mice (n = 6). A considerable amount of spermatids was still observed in testes of sterile Sox30loxp/loxp mice (n = 8), but only a few spermatids were found in epididymides of these mice. The arrows in epididymides represent spermatozoa; the arrows in testes represent spermatozoa and spermatids. Scale bars, 100 μm. (B and C) qRT-PCR (B) and WB (C) analyses of the expression of Sox30 in testes of Sox30+/+, Sox30−/−, and Sox30loxp/loxp mice were performed. ***p < 0.001. β-Actin was used as an internal control.
The Male Offspring of Sox30loxp/loxp Mice Can Still Father Children
To determine the fertility of offspring after re-expression of Sox30 in Sox30 null mice, we performed the fertility analyses of the male offspring. In the offspring of the 6 fertile Sox30loxp/loxp mice, 12 were male mice, and each male mouse at 2 months was mated with 2 females with known fertility for a minimum of 20 weeks. These male offspring were fertile in 91.67% (11/12). Moreover, the male offspring of the fertile Sox30loxp/loxp mice and their children can develop normally and live more than 1 year without significant difference of physical appearance compared with wild-type mice. These data show that the male offspring of Sox30loxp/loxp mice can still father children normally.
Discussion
More than 80% of NOA patients are unexplained, resulting in lack of adequate clinical treatments. Although plenty of studies have tended to explain the etiology of NOA by screening for genetic abnormality, it is still elusive. DNA methylation as an epigenetic mechanism plays key roles in various diseases.10, 11, 12, 13, 14 During spermatogenesis, the genome of germ cells generally undergoes extensive DNA demethylation followed by DNA methylation.22 The abnormality of this progress is closely associated with male infertility.15, 16, 17, 18 In the present study, SOX30 was demonstrated to be a preferential hyper-methylation gene at promoter, and this hyper-methylation of SOX30 resulted in its silence in NOA patients. The reduced levels of SOX30 expression are associated with the severity of NOA disease. Moreover, the pathology and testicular volume of Sox30-deficient mice simulate those of NOA patients. These results indicate that SOX30 silenced by hyper-methylation is likely an important contributor to the pathogenesis of NOA disease. In the morphological comparison between testis tissues of Sox30-deficient mice and NOA patients, maybe a question will be raised that there are no multinucleated cells in the phenotype images of NOA. It is likely that the silence of SOX30 in these NOA patients is too long because the multinucleated cells have also almost disappeared in testes of over 6-month-old Sox30-deficient mice. Further analysis of the phenotype of younger NOA patients will be required to determine this issue in the future. In addition, our study reveals that SOX30 level is negatively associated with NOA severity. This phenomenon may be related to the silent degree of SOX30 caused by different hyper-methylation CpG sites and/or the composition of different cell types (for example, the absence of spermatids) for NOA samples.
To determine whether the infertility of Sox30 deficiency can be cured by re-expression of Sox30, we restored Sox30 expression in Sox30−/− adult mice by inductive deletion of floxed cassette and evaluated the actual fertility of these Sox30loxp/loxp mice. The arrest of spermatogenesis in Sox30−/− mice can be restored after Sox30 re-expression. Moreover, the re-presented spermatozoa seem to be fertile, and their male offspring can also father children normally. These experimental data reveal that the infertility of Sox30 deletion can be successfully cured by restoration of Sox30 expression in adult mice. Given that DNA methylation is reversible, SOX30 may be used to develop an important therapy for NOA disease. The latest technologies developed for targeted DNA methylation editing23, 24, 25 will be useful for developing this therapy. This work will be our focus for future study, which is now underway.
In very recent works, Sox30 also has been found to be essential for spermiogenesis,26, 27, 28 which further highlights the key role of SOX30 in NOA diseases. Feng et al.26 and Zhang et al.27 reveal that germ cells arrest at the post-meiotic round spermatids in Sox30 knockout mice, and Bai et al.28 indicate that there is also an accumulation of diplotene spermatocytes, suggesting a delayed or impaired transition from meiosis to post-meiosis in Sox30 knockout mice. In our present study, we found few spermatids and complete absence of spermatozoa in testes of Sox30 null mice at 6 months. However, complete absence of spermatids and spermatozoa was found in testes of Sox30 null mice at 8 months. These data suggest that the male germ cells in Sox30 null mice may mainly arrest at an earlier stage rather than post-meiotic round spermatids. Although data of our study show that the number of spermatocytes is drastically reduced in Sox30 null mice as they aged, it seems that spermatocytes still exist in the old Sox30 null mice. This phenomenon means that the male germ cells in Sox30 null mice may arrest at an early stage of spermatocyte or arrest in differentiation from spermatogonia to spermatocyte stage. However, whether the germ cells in Sox30 null mice arrest at spermatocytes or in differentiation from spermatogonia to spermatocyte is still unclear at the moment. Further research is required to clarify the precise period of these germ cells arrest in Sox30 null mice. In addition, the phenotype observed in our study is somewhat different from that observed in previous studies,26, 27, 28 which may be caused by different strategies to create Sox30-deficient mice (for example, systemic and conditional knockout) or by differences in the degree of gene knockout. Anyway, the data of the recent reports and our present study show that Sox30 is a key participant in male infertility, which further strengthens the important contributor of inactive SOX30 to human NOA disease.
The most interesting result of our present study is that the phenotype and fertility can be reversed by restoration of Sox30 using an inducible knockout model. The presence of many spermatozoa is observed, and the actual fertility is restored following inducible Sox30 expression in adult Sox30−/− mice. However, it appears that despite analyses of several complete cycles of spermatogenesis (over 8 months), there is still a marked difference of the phenotype and fertility between Sox30+/+ and Sox30loxp/loxp mice. The possible cause may be the incomplete restoration of Sox30 in Sox30loxp/loxp mice due to efficiency of recombination. The expression analyses demonstrate that the expression of Sox30 is indeed incompletely restored in testes of Sox30loxp/loxp mice compared with Sox30+/+ mice.
A remaining question of this study is whether the effect of Sox30 on male germ cells is cell autonomous or non-cell autonomous. To clarify this issue, two experiments should be performed in the future: (1) different conditional knockout mouse lines (only germ cell knockout of Sox30 and only Sertoli cell knockout of Sox30) will be generated to compare the different effect of Sox30 on male germ cells; and (2) the spermatogonia of Sox30+/+ mice will be transplanted to testes of Sox30−/− mice to observe the changes of spermatogenesis.
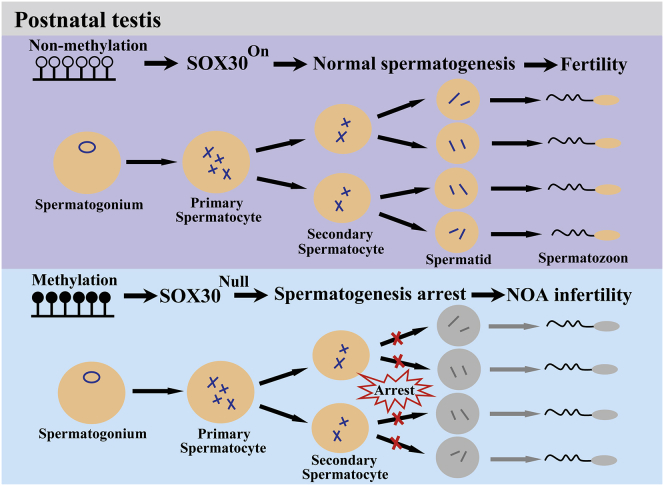
In summary, our study finds that methylated inactivation of SOX30 specifically impairs testis development and spermatogenesis probably causing NOA disease (Figure 7), and re-expression of SOX30 can successfully restore the spermatogenesis and actual fertility. This study provides an important therapy target for human NOA disease.
Figure 7.
A Schematic Illustration of Human NOA Disease Development by SOX30
In post-natal testes, when the promoter is un-methylated, SOX30 is highly expressed and the spermatogenesis is normal. However, when the promoter is hyper-methylated, SOX30 expression is silenced. The spermatogenesis is arrested, and the NOA disease is developed.
Materials and Methods
Patients
Testicular biopsy specimens were obtained from OA and NOA patients at Daping Hospital. The patients with previous testicular injury were excluded. Clinical assessment of infertile men was done as described previously.29 These infertile male patients have received a routine semen examination according to the World Health Organization (WHO) criteria. Azoospermia was confirmed for all men on two different occasions by testing centrifuged ejaculates according to WHO guidelines.30 The patients were first diagnosed as azoospermia due to no ejaculated spermatozoa in semen. Then, OA was defined based on the following characteristics: no sperm in the semen, visible sperm in testicular tissue or motile spermatozoa sampled from MESE, or number of mature spermatozoa sampled using TESE, normal testicular volume, normal blood hormone, and normal semen biochemistry. The 15 OA men aged 20–42 years who exhibit normal tissue morphology with a large number of sperm and no significant reduction of all type spermatogenic cells in testes were selected as control tissues from 326 OA patients (the ideal normal tissue control is the volunteer with known fertility, but the difficulties in acquiring testicular samples of the volunteers make this strategy impractical). These patients were identified as the NOA men according to the following characteristics: no sperm in semen, small testicular volume, abnormal blood hormone and semen biochemistry, no sperm in testicular tissue, and/or a reduction of spermatogenic cells in testicular tissue. The NOA samples were divided into different groups based on the clear composition of cell types and pathological morphology, and finally, 58 NOA men aged 20–45 years were selected from 176 NOA patients, including 31 cases of NOA patients without spermatozoa (NOA-I), 22 cases of NOA patients without spermatid (NOA-II), and 5 cases of NOA patients without spermatocyte (NOA-III), for further study. In these NOA patients, the clinical genetic testing including Y chromosome microdeletion and karyotype was performed in 84.48% (49/58) cases, and no positive findings of genetic test were observed. When defining OA, NOA-I, NOA-II, and NOA-III, we examined multiple sections from at least three separate parts of the testicular tissue sample to determine these qualifications by two independent pathologists. In the 15 OA patients and 58 NOA patients, 5 OA patients underwent MESE, 10 OA patients underwent TESE, and 7 NOA men underwent microsurgical TESE (microTESE) for assisted reproduction. These 15 OA patients selected who underwent TESE or MESE can father children by assisted reproductive technology. None of these patients were exposed to adjuvant hormonal therapy, chemotherapy, and radiation prior to biopsy. The following parameters were recorded for each patient: a detailed medical history, physical examination including the presence or absence of a varicocele, and a hormone profile. Testicular volume using a standard orchidometer was noted. Hormonal evaluation included serum follicle-stimulating hormone (FSH), luteinizing hormone (LH), and total testosterone (TT). There was no family history for the male sterility of all the patients. This study was approved by the ethics committees of Daping Hospital Affiliated to Army Medical University, and informed consent was signed by all of the patients. All experiments were carried out in accordance with the approved guidelines of Army Medical University.
DNA Extraction and Construction of Whole Genome Bisulfite Sequencing Libraries
The genomic DNA was extracted using the MagPure FFPE DNA LQ Kit (Magen) according to the manufacturer’s instructions. The fixed samples from five OA or five NOA patients who underwent testicular puncture between 2015 and 2016 (stored at 4°C) were pooled together for extraction. No significant DNA degradation was found for the extracted genomic DNA determined by 1% agarose gel electrophoresis. The concentration and integrity of genomic DNA were analyzed on a Qubit fluorometer using a Quant-iT dsDNA HS Assay kit (Invitrogen) and the Bioanalyzer analysis system (Agilent), respectively. The libraries were constructed using the Fragmented MicroMethyl DNA library kit (E-GENE) according to the manufacturer’s instructions. In brief, the genomic DNA was further fragmented by an ultrasonic instrument followed by end-repairing and dA addition to the 3′ end using end repair enzymes. After the reaction, methylation-modified adaptor was ligated to the genomic DNA. The adaptor ligated DNA was further treated with bisulfite conversion, followed by PCR amplification. The products were purified using 0.8-fold dilution purification beads and then quantified using the Bioanalyzer analysis system (Agilent) and real-time PCR assay. The qualified libraries were finally analyzed using an Illumina HiSeq.
Bisulfite-Sequencing Read Processing and Alignment
After sequencing, adapters were removed using cutadapt-1.2.1 software (https://cutadapt.readthedocs.io/en/stable/), and only reads with minimally 45-bp length were kept. After removing the adapters, low-quality reads that contained more than 10% Ns or more than 50% of the sequence with low-quality value (quality value <5) per read were filtered out. Reads mapping was done using BSMAP (version 2.73) alignment software that combines genome hashing and bitwise masking to achieve fast and accurate bisulfite mapping, allowing for maximally five mismatches or one small insertion or deletion. After mapping, PCR duplicates were removed using SAMtools software. Finally, methylation level of each cytosine in human genome was calculated on the basis of the alignments by using a Python script metyratio.py supplied with BSMAP.
Global Patterns of DNA Methylome
Circos (version 0.67) was used to visualize the global pattern of DNA methylome. The G (Guanine) and C (Cytidine) (GC) content, repeat sequence ratio, gene sequence proportion, and average methylation level of each 500-kb window on the whole genome were calculated. Chromosome name and scale are indicated on the outer rim.
Identification of DMR
The software metilene (version [v.]0.2-6) was used to screen regions in the genome that were differentially methylated between OA and NOA samples. Metilene uses a circular binary segmentation and a two-dimensional Kolmogorov-Smirnov (2D-KS) test to call DMRs. Adjusted p values are calculated using the Bonferroni correction. Only cytosine sites covered by at least five reads were used. Regions containing CpG sites ≥10, adjusted p values <0.01, q values <0.05, and changes of methylation level between two samples >0.2 were identified as DMRs.
Identification and Annotation of Sequence Variation with Bisulfite-Sequencing Data
BS-SNPer that can identify mono-allelic DNA methylation and polymorphisms in cis-regulatory sequences was used to detect SNPs in our DNA bisulfite treatment sequencing data. First, SNPs were identified in each sample according to the following filtering criteria: (1) a SNP had to be covered by at least 10 reads; (2) the minimum base quality for calling a base was 15; (3) the minimum phred-scaled confidence threshold at each variant for calling should be 20; (4) the minimum heterozygous frequency should be >0.1; and (5) the minimum homozygous frequency should be >0.85. After detection of SNPs, annovar (2017-07-17) was used to functionally annotate genetic variants detected from human genomes (hg19) and identify whether SNPs cause protein-coding changes and the amino acids that are affected.
DNA Isolation, Preparation, and Methylation Microarray Analysis
DNA isolation, preparation, and methylation microarray analysis were performed in three OA and three NOA independent patient samples (the cell types and pathological morphology of these three OA and three NOA men were very similar to the original cohort of five OA and five NOA men, respectively: one of three OA patients/two of five OA in the original cohort were slightly reduced in spermatogenic cells with many sperm in testis tissues, and two of three OA patients/three of five OA in the original cohort were not significantly reduced in all type spermatogenic cells with a lot of sperm in testis tissues; two of three NOA patients/three of five NOA in the original cohort were highly similar to the NOA-III patients without spermatocyte, and one of three NOA patients/two of five NOA in the original cohort were highly similar to the NOA-II patients without spermatids). These three OA and three NOA men were also used for the following RNA extraction and gene expression microarray as previously described.19 In brief, the frozen testicular tissue was dissolved in lysis buffer and Proteinase K from DNA Micro Kit (QIAGEN, Germany). Genomic DNA was extracted using a DNA Micro Kit based on the manufacturer’s instructions (QIAamp DNA Micro Kit; QIAGEN). For each sample, bisulfite modification of DNA (500 ng) was performed using the EZ DNA methylation kit (Zymo Research, Orange, CA, USA) based on the manufacturer’s instructions. After bisulfite conversion, bisulfite-converted DNA (4 μL) was used to hybridize on the array (Infinium Human Methylation 450 BeadChips; Illumina). Bead-bound probes were used to quantify the amount of thymine or cytosine in the hybridization process. Fluorescent signal of two bead types was obtained using BeadArray Reader. Methylation status of the CpG sites was calculated with average beta value. The beta value was the ratio of the signal intensity for the methylated probe to the sum for methylated and unmethylated probes.
RNA Extraction and Gene Expression Microarray
RNA extraction and gene expression microarray were performed in the three OA and three NOA patient samples as previously described.19 In brief, total RNAs were extracted from the frozen testicular tissues using miRNeasy Micro Kit (QIAGEN, Germany). An Agilent 2100 Bioanalyzer and RNA 6000 NanoLabChip Kit (Agilent Technologies, Waldbronn, Germany) were used to determine the RNA concentration and integrity, respectively. Total RNAs (100 ng) were prepared with Agilent’s Labeling kit (One-Color Microarray-Based Gene Expression Analysis Low Input Quick Amp Labeling kit) for microarray hybridization and were then hybridized to Agilent SurePrint G3 human gene expression microarray based on the manufacturer’s instructions. The Agilent microarray scanner (G2565CA) was used to scan the arrays. The data of the microarray were processed using Agilent Feature Extraction 10.7.3.1 Software (Agilent).
Immunohistochemical Analysis
IHC was performed using SOX30 antibody (1:50; ab26024; Abcam, Cambridge, MA, USA) as described previously.21 After IHC staining, we consolidated the results by positive percentage and intensity of staining using a scoring system. Positive percentage of staining was classified into five categories: <5% positive cells for 0 score, 5%–25% for 1 score, 26%–50% for 2 score, 51%–75% for 3 score, and ≥76% for 4 score. Intensity of staining was graded into three groups: negative scored as 0, weak staining scored as 1, moderate staining scored as 2, or strong staining scored as 3. Expression levels of SOX30 in samples were defined by the sum of category for positive percentage and grade for intensity of staining, and the range of the calculation was 0–7. All samples were independently reviewed by two blinded pathologists.
Sox30 Null Mice
Sox30 null mice were generated by Model Animal Research Center of Nanjing University. In brief, a targeted gene including homologous arms was retrieved from the BAC vector, and the LoxP-SA-IRES-GFP-NEO-STOP-PPS-LoxP cassette was introduced between Exon1 and Exon2 of Sox30 by homologous recombination. The targeting vector was confirmed by PCR, enzyme digestion, sequencing, and then linearized with AsiSI and electroporated into C57BL/6 embryonic stem cells (ESCs). The recombinants were selected by G418 and ganciclovir (Ganc); then the targeted ESCs were screened by PCR and Southern blotting. Finally, the positive clones were chosen for microinjection to generate the chimeras, which were crossed with C57BL/6 mice to produce the heterozygous mice. To restore Sox30 expression, we generated mice expressing Cre recombinase by breeding Sox30 heterozygous (Sox30+/−) mice with B6.Cg-transgenic (Tg) (CAG-cre/Esr1)5Amc/JNju tool mice (stock no. 004682; The Jackson Laboratory), to produce Sox30-KI (knockin) and Cre double-positive heterozygote mice. Tam-inducible Cre-mediated recombination will result in deletion of the floxed sequences in the offspring of these mice. The genotype of the offspring was identified by PCR. Mice were maintained in a specific pathogen-free unit under a 12-h light, 12-h dark cycle with ad libitum access to water and food. All mouse experiments were carried out with the permission of Institutional Animal Care and Use Committee of Nanjing University and Army Medical University, China.
Fertility Assays
Breeding assays with wild-type (Sox30+/+), heterozygous (Sox30+/−), and homozygous (Sox30−/−) mice were carried out. At least 30 independent mice (8–10 weeks old) of each sex for different genotypes were mated with known fertility wild-type mice for a minimum of 8 weeks. If progeny were born, the line was classed as fertile.
Epididymal Sperm Count by Sperm Class Analyzer
The epididymis was dissected from adult mice and cut into pieces in 0.5 mL of medium incubating for 10 min at 37°C. Each sample (10 μL) was transferred to a Glodcyto standard count chamber slide (Microptic, Barcelona, Spain), warmed on a microscope heating stage. Sperms on the slides were observed and counted at least in five random fields by Sperm Class Analyzer (SCA) system (Microptic, Barcelona, Spain).
RNA and Protein Extraction
Total RNA of testis tissues was extracted using TRIzol Reagent (Invitrogen) and treated with DNase I to eliminate the genomic DNA contamination. cDNA was synthesized using GoScript Reverse Transcription System (Promega) and stored at −20°C. The protein of testis tissues was extracted using lysis buffer (Beyotime, Shanghai, China) containing complete protease inhibitor (Roche, Mannheim, Germany) and stored at −80°C after centrifugation.
qRT-PCR
qRT-PCR was performed using CFX96 Real-Time PCR Detection System (Bio-Rad Laboratories) and GoTaq qPCR Master Mix (Promega, USA) as previously described.31 The expression was calculated by 2−ΔΔCT. All assays were performed in triplicate three times. The primers used in this study were listed in Table S1.
Restoration of Sox30 Expression in Sox30−/− Mice by Tam Injection
To induce Cre-mediated Sox30 expression, we injected tam (Sigma-Aldrich, St Louis, MO, USA) into mice once a day intraperitoneally (i.p.) for 5 days with 1 mg/mouse to remove the insertion cassette via activating the Cre recombinase (the control Sox30−/− mice were injected with solvent i.p following the same steps). The mice were injected with tam or solvent at 2 months old and were sacrificed at 2 or 6 months after the first injection. The histology of testes was examined by microscopy.
Fertility Analysis for Recovery Mice
Breeding assays were performed in Sox30+/+ male mice injected with tam, Sox30−/+ male mice injected with tam, Sox30−/− male mice injected with tam (Sox30loxp/loxp), and the control Sox30−/− male mice injected with solvent. Each male mouse (2 months old when injected with tam or solvent) at 6 months after tam injection was mated with two known fertility wild-type females for a minimum of 20 weeks. If progeny were born, the treated male mouse was defined as fertile.
Histology H&E Staining
Testes were dissected and immediately fixed in Bouin’s fluid. The fixed testes were dehydrated, embedded in paraffin, and cut into 5-μm-thick sections. These sections were de-waxed, rehydrated, and stained with H&E.
Transmission Electron and Optical Microscopy
Testes were dissected, cut into 1-mm cubes, and placed into fresh 2.5% (w/v) glutaraldehyde overnight at 4°C. The tissues were washed in PBS post-fixed for 1 h in 1% osmium tetroxide at room temperature, dehydrated through a graded series of ethanol and acetone, and infiltrated with and embedded in Epon 812. Sections (100 nm) were obtained with an ultramicrotome, stained with uranyl acetate and lead citrate, and observed under JEM-1400 Plus transmission electron microscope (JEOL, Japan). Semi-thin sections (1 μm) were stained with toluidine blue and examined by optical microscope (Olympus, Japan).
Western Blotting Analysis
Western blotting (WB) analysis was performed as previously described.31 The SOX30 rabbit polyclonal antibody (1:1000, ab26024; Abcam) was used. The secondary (anti-rabbit) antibody was horseradish peroxidase (HRP)-conjugated.
Statistical Analysis
Statistical analyses were performed using SPSS 15.0 software (SPSS, Chicago, IL, USA). The data were expressed as mean ± SEM. The differences between two or three groups were analyzed using Student’s t test, Tukey’s test, and one-way ANOVA. The different methylations between OA and NOA patients who underwent global DNA methylation were analyzed using 2D KS test and Mann-Whitney U (MWU) test. The p values less than 0.05 were considered statistically significant.
Author Contributions
F.H., J.-y.L., and J.C. designed the project; F.H., X.J., X.Zhang., and W.-b.L. performed the experiments; F.H., X.J., Z.-h.C., L.A., and Q.C. analyzed the experimental data; Z.-m.L., X.Zhuang, and F.G. performed the bioinformatics analysis; C.-y.M. and Y.-f.L. collected samples and performed histopathologic evaluation; F.H., J.-y.L., J.C., and W.-m.O. wrote and revised the manuscript with comments from all authors.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This work was supported by Key Program of Natural Science Funding of China (grant 81630087) and Chongqing Research Program of Basic Research and Frontier Technology (grant cstc2017jcycBX0064). The authors would like to thank all patients involved in this study and thank E-GENE Technologies (Shenzhen, China) for technical support of visualizing the global pattern of DNA methylome.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.038.
Contributor Information
Jia Cao, Email: caojia1962@126.com.
Jin-yi Liu, Email: jinyiliutmmu@163.com.
Supplemental Information
References
- 1.de Kretser D.M. Male infertility. Lancet. 1997;349:787–790. doi: 10.1016/s0140-6736(96)08341-9. [DOI] [PubMed] [Google Scholar]
- 2.Huynh T., Mollard R., Trounson A. Selected genetic factors associated with male infertility. Hum. Reprod. Update. 2002;8:183–198. doi: 10.1093/humupd/8.2.183. [DOI] [PubMed] [Google Scholar]
- 3.Maduro M.R., Lamb D.J. Understanding new genetics of male infertility. J. Urol. 2002;168:2197–2205. doi: 10.1016/S0022-5347(05)64355-8. [DOI] [PubMed] [Google Scholar]
- 4.Lee J.Y., Dada R., Sabanegh E., Carpi A., Agarwal A. Role of genetics in azoospermia. Urology. 2011;77:598–601. doi: 10.1016/j.urology.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Practice Committee of American Society for Reproductive Medicine in collaboration with Society for Male Reproduction and Urology The management of infertility due to obstructive azoospermia. Fertil. Steril. 2008;90(Suppl 5):S121–S124. doi: 10.1016/j.fertnstert.2008.08.096. [DOI] [PubMed] [Google Scholar]
- 6.Hamada A.J., Esteves S.C., Agarwal A. A comprehensive review of genetics and genetic testing in azoospermia. Clinics (São Paulo) 2013;68(Suppl 1):39–60. doi: 10.6061/clinics/2013(Sup01)06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X.N., Li Z.S., Ren Y., Jiang T., Wang Y.Q., Chen M., Zhang J., Hao J.X., Wang Y.B., Sha R.N. The Wilms tumor gene, Wt1, is critical for mouse spermatogenesis via regulation of sertoli cell polarity and is associated with non-obstructive azoospermia in humans. PLoS Genet. 2013;9:e1003645. doi: 10.1371/journal.pgen.1003645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dohle G.R., Halley D.J., Van Hemel J.O., van den Ouwel A.M., Pieters M.H., Weber R.F., Govaerts L.C. Genetic risk factors in infertile men with severe oligozoospermia and azoospermia. Hum. Reprod. 2002;17:13–16. doi: 10.1093/humrep/17.1.13. [DOI] [PubMed] [Google Scholar]
- 9.Yatsenko A.N., Georgiadis A.P., Röpke A., Berman A.J., Jaffe T., Olszewska M., Westernströer B., Sanfilippo J., Kurpisz M., Rajkovic A. X-linked TEX11 mutations, meiotic arrest, and azoospermia in infertile men. N. Engl. J. Med. 2015;372:2097–2107. doi: 10.1056/NEJMoa1406192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.American Association for Cancer Research Human Epigenome Task Force. European Union, Network of Excellence, Scientific Advisory Board Moving AHEAD with an international human epigenome project. Nature. 2008;454:711–715. doi: 10.1038/454711a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Esteller M. Epigenetics in cancer. N. Engl. J. Med. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 12.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat. Rev. Genet. 2012;13:484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 13.Tatton-Brown K., Seal S., Ruark E., Harmer J., Ramsay E., Del Vecchio Duarte S., Zachariou A., Hanks S., O’Brien E., Aksglaede L., Childhood Overgrowth Consortium Mutations in the DNA methyltransferase gene DNMT3A cause an overgrowth syndrome with intellectual disability. Nat. Genet. 2014;46:385–388. doi: 10.1038/ng.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trasler J.M. Epigenetics in spermatogenesis. Mol. Cell. Endocrinol. 2009;306:33–36. doi: 10.1016/j.mce.2008.12.018. [DOI] [PubMed] [Google Scholar]
- 15.Wu H., Hauser R., Krawetz S.A., Pilsner J.R. Environmental susceptibility of the sperm epigenome during windows of male germ cell development. Curr. Environ. Health Rep. 2015;2:356–366. doi: 10.1007/s40572-015-0067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pilsner J.R., Parker M., Sergeyev O., Suvorov A. Spermatogenesis disruption by dioxins: Epigenetic reprograming and windows of susceptibility. Reprod. Toxicol. 2017;69:221–229. doi: 10.1016/j.reprotox.2017.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Camprubí C., Salas-Huetos A., Aiese-Cigliano R., Godo A., Pons M.C., Castellano G., Grossmann M., Sanseverino W., Martin-Subero J.I., Garrido N., Blanco J. Spermatozoa from infertile patients exhibit differences of DNA methylation associated with spermatogenesis-related processes: an array-based analysis. Reprod. Biomed. Online. 2016;33:709–719. doi: 10.1016/j.rbmo.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 18.Chong S., Vickaryous N., Ashe A., Zamudio N., Youngson N., Hemley S., Stopka T., Skoultchi A., Matthews J., Scott H.S. Modifiers of epigenetic reprogramming show paternal effects in the mouse. Nat. Genet. 2007;39:614–622. doi: 10.1038/ng2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z., Zhuang X., Zeng J., Tzeng C.M. Integrated Analysis of DNA Methylation and mRNA Expression Profiles to Identify Key Genes in Severe Oligozoospermia. Front. Physiol. 2017;8:261. doi: 10.3389/fphys.2017.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhuang X., Li Z., Lin H., Gu L., Lin Q., Lu Z., Tzeng C.M. Integrated miRNA and mRNA expression profiling to identify mRNA targets of dysregulated miRNAs in non-obstructive azoospermia. Sci. Rep. 2015;5:7922. doi: 10.1038/srep07922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han F., Dong Y., Liu W., Ma X., Shi R., Chen H., Cui Z., Ao L., Zhang H., Cao J., Liu J. Epigenetic regulation of sox30 is associated with testis development in mice. PLoS ONE. 2014;9:e97203. doi: 10.1371/journal.pone.0097203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kimmins S., Sassone-Corsi P. Chromatin remodelling and epigenetic features of germ cells. Nature. 2005;434:583–589. doi: 10.1038/nature03368. [DOI] [PubMed] [Google Scholar]
- 23.Vojta A., Dobrinić P., Tadić V., Bočkor L., Korać P., Julg B., Klasić M., Zoldoš V. Repurposing the CRISPR-Cas9 system for targeted DNA methylation. Nucleic Acids Res. 2016;44:5615–5628. doi: 10.1093/nar/gkw159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu X.S., Wu H., Ji X., Stelzer Y., Wu X., Czauderna S., Shu J., Dadon D., Young R.A., Jaenisch R. Editing DNA methylation in the mammalian genome. Cell. 2016;167:233–247.e17. doi: 10.1016/j.cell.2016.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Maeder M.L., Angstman J.F., Richardson M.E., Linder S.J., Cascio V.M., Tsai S.Q., Ho Q.H., Sander J.D., Reyon D., Bernstein B.E. Targeted DNA demethylation and activation of endogenous genes using programmable TALE-TET1 fusion proteins. Nat. Biotechnol. 2013;31:1137–1142. doi: 10.1038/nbt.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Feng C.A., Spiller C., Merriner D.J., O’Bryan M.K., Bowles J., Koopman P. SOX30 is required for male fertility in mice. Sci. Rep. 2017;7:17619. doi: 10.1038/s41598-017-17854-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang D., Xie D., Lin X., Ma L., Chen J., Zhang D., Wang Y., Duo S., Feng Y., Zheng C. The transcription factor SOX30 is a key regulator of mouse spermiogenesis. Development. 2018;145:dev164723. doi: 10.1242/dev.164723. [DOI] [PubMed] [Google Scholar]
- 28.Bai S., Fu K., Yin H., Cui Y., Yue Q., Li W., Cheng L., Tan H., Liu X., Guo Y. Sox30 initiates transcription of haploid genes during late meiosis and spermiogenesis in mouse testes. Development. 2018;145:dev164855. doi: 10.1242/dev.164855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Esteves S.C., Miyaoka R., Agarwal A. An update on the clinical assessment of the infertile male. Clinics (Sao Paulo) 2011;66:691–700. doi: 10.1590/S1807-59322011000400026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.World Health Organization and Department of Reproductive Health and Research . World Health Organisation; 2010. WHO Laboratory Manual for the Examination and Processing of Human Semen, Fifth Edition. [Google Scholar]
- 31.Han F., Liu W., Jiang X., Shi X., Yin L., Ao L., Cui Z., Li Y., Huang C., Cao J., Liu J. SOX30, a novel epigenetic silenced tumor suppressor, promotes tumor cell apoptosis by transcriptional activating p53 in lung cancer. Oncogene. 2015;34:4391–4402. doi: 10.1038/onc.2014.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.