Abstract
Corynebacterium glutamicum ATCC 13032 is an established and industrially-relevant microbial host that has been utilized for the expression of many desirable bioproducts. Tetra-methylpyrazine (TMP) is a naturally occurring alkylpyrazine with broad applications spanning fragrances to resins. We identified an engineered strain of C. glutamicum which produces 5 g/L TMP and separately, a strain which can co-produce both TMP and the biofuel compound isopentenol. Ionic liquids also stimulate TMP production in engineered strains. Using a fed batch-mode feeding strategy, ionic liquid stimulated strains produced 2.2 g/L of tetra-methylpyrazine. We show that feedback from a specific heterologous gene pathway on host physiology leads to acetoin accumulation and the production of TMP.
Keywords: Corynebacterium glutamicum; Alkaloids; Terpene; 2,3,5,6-Tetra-methylpyrazine; Isopentenol; Bioreactor
Highlights
-
•
Evaluation of 2,3,5,6-tetra-methylpyrazine (TMP) is limited by its scarcity.
-
•
TMP can be used as a flavoring agent.
-
•
Engineered Corynebacterium glutamicum ATCC 13032 produces >5 g/L TMP.
-
•
Ionic liquids such as cholinium lysinate ([Ch][Lys]) also stimulate TMP production.
-
•
Fed-batch mode production for TMP was demonstrated.
Abbreviations
- TMP
2,3,5,6-tetra-methylpyrazine
- GC/MS
gas chromatography mass spectrometry
- GC-FID
gas chromatography flame ionization detector
- IL
ionic liquid
- [C2C1im][OAc]
1-ethyl-3-methyl imidazolium acetate
- [Ch][OAc]
cholinium acetate
- [Ch][Lys]
cholinium lysinate
- [ETA][OAc]
ethanolamine acetate
- [DEOA][OAc]
diethanolamine acetate
1. Introduction
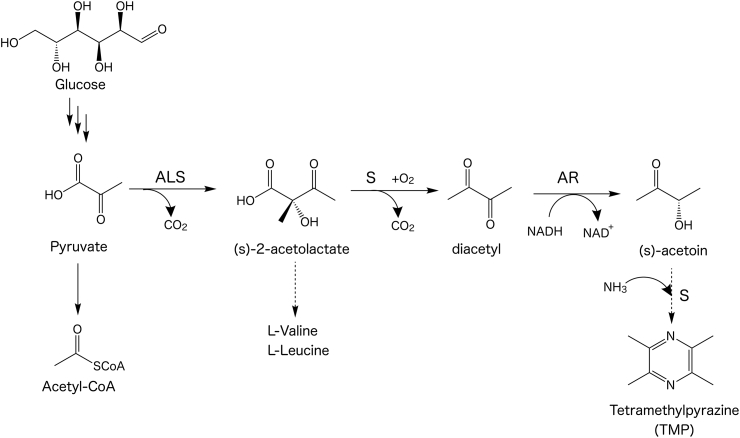
Alkylpyrazines are naturally distributed, heterocyclic aromatic compounds with a nutty or roasted flavor profile (Masuda and Mihara, 1988). Of the alkylpyrazine compounds, 2,3,5,6-tetra-methylpyrazine (also known as TMP or ligustrazine) is a biologically active alkaloid commonly found in the rhizobiome of Ligusticum wallichii (Ke-ji et al., 1983). It has been used as a flavoring agent (Zhu et al., 2010), studied as a potentially pharmacologically active molecule (Chen et al., 2018; Ferreira and Kaiser, 2012; Kao et al., 2013), or applied onto surfaces as a coating (Lee, 2014; Ng et al., 2017). While TMP has been examined for potential use in many fields, its widespread evaluation is hindered due to its limited commercial availability. Chemical synthesis routes to TMP involve multiple synthesis steps and are inefficient (Dickschat Jeroen et al., 2010; Deng et al., 2006). Commercially available TMP is often purified through an ethanol-ethyl ether extraction from plants such as Ephedra sinica (Li et al., 2001). More sustainable, biological routes towards TMP production have focused on optimizing cultivation conditions of Bacillus subtilis, which can naturally produce the TMP precursor, acetoin (Kosuge and Kamiya, 1962; Xiao et al., 2014). TMP is inefficiently generated from acetoin (26% yield) by heating the cell lysate to ~65 °C for 2 h (Xiao et al., 2014). Outside of B. subtilis, trace amounts of alkylpyrazines (such as TMP) have been detected in the headspace of Corynebacterium glutamicum (Dickschat Jeroen et al., 2010), but evidence of higher titers of TMP has only been reported from a single C. glutamicum isolate (MB-1923) which is not publicly available and has no associated genomic metadata (Demain et al., 1967). It has been postulated that TMP could be produced from pyruvate via a four-step reaction (Fig. 1). The two units of the proposed immediate precursor, acetoin (Rizzi, 1988), would spontaneously react with each other to form TMP under high nitrogen concentrations (Fig. 1).
Fig. 1.
Diagram of proposed tetra-methylpyrazine (TMP) production pathway in C. glutamicum.
Model. Pyruvate is generated from glucose using the Embden-Meyerhof-Parnas (EMP) pathway and converted to TMP in four steps as indicated. ALS, acetolactate synthase (ilvB, Cgl1271; ilvN, Cgl1272); AR, acetoin reductase (butA, Cgl2674). The upper-case letter S with arrows represents non-enzymatic spontaneous reactions. This figure was adapted and expanded upon based on Xiao et al. (2014). Pathway for the production to isopentenol is described in Kang et al. (2016).
In this communication, we examined engineered strains of C. glutamicum for the production of TMP. Specific gene substitutions in a heterologous gene pathway result in strains which produce high (>5 g/L) titers of TMP or the co-production of TMP with an acetyl-CoA derived compound, isopentenol (Sasaki et al., 2019). Other factors that enhance the production of TMP in these engineered C. glutamicum strains are also described.
2. Materials and methods
2.1. Chemicals and reagents
All chemicals and reagents were purchased from Sigma-Aldrich (St. Louis, MO) or as otherwise indicated, and were of molecular biology grade or higher. When cells were cultivated in a microtiter dish format, plates were sealed with a gas-permeable film (Sigma-Aldrich, St. Louis, MO).
2.2. Strain and plasmid construction
All strains and plasmids used in this study are listed in Table 1 and their sequences are available at http://public-registry.jbei.org. Oligo-nucleotide primers were synthesized by Integrated DNA Technologies, Inc. (San Diego, CA). Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, MA) was used for polymerase chain reaction. Isothermal DNA assembly (Gibson et al., 2009) was utilized to assemble plasmids using 40 nucleotide overhangs (NEBuilder HiFi DNA Assembly Master Mix, New England Biolabs, Ipswich, MA). Plasmids were constructed using chemically competent E. coli DH10β (New England Biolabs). Where indicated, heterologous isopentenol production pathway was modified to incorporate both hmgr and mk homologs from S. aureus or C. kroppenstedtii, to replace the existing gene from S. cerevisiae. For S. aureus, the mk homolog is encoded by mvaK1 (NCBI: WP_000197034.1) and similarly, the hmgr homolog is encoded by mvaS (NCBI: WP_045179588.1). For C. kroppenstedtii, the mk homolog is encoded by mvaK1 (NCBI: ACR16826.1) and likewise, the hmgr homolog is encoded by mvaA (NCBI: WP_012730718.1). All sequences were confirmed by colony PCR and Sanger sequencing.
Table 1.
Strains and plasmids used in this study.
| Strain | Description | Selection | Reference |
|---|---|---|---|
| JBEI-7936 | Corynebacterium glutamicum ATCC 13032/NHRI 1.1.2, biotin auxotroph | NxR | Sasaki et al. (2019) |
| JBEI-19571 | JBEI-7936 harboring p/JBEI-19559 | KanR | Sasaki et al. (2019) |
| JBEI-19652 | JBEI-7936 harboring p/JBEI-19628 | KanR | This study |
| JBEI-19658 | JBEI-7936 harboring p/JBEI-19634 | KanR | This study |
| JBEI-19566 | JBEI-7936 ΔpoxB ΔldhA | SucR, KanS | Sasaki et al. (2019) |
| E. coli DH1 | F−λ–endA1 recA1 relA1 gyrA96 thi-1 glnV44 hsdR17(rK–mK–) | Meselson and Yuan (1968) | |
|
E. coli DH10β |
F− endA1 deoR+ recA1 galE15 galK16 nupG rpsL Δ(lac)X74 ϕ80lacZΔM15 araD139 Δ(ara,leu)7697 mcrA Δ(mrr-hsdRMS-mcrBC) StrR λ– |
Invitrogen |
|
| Plasmid |
Description |
Selection |
Reference |
| JBEI-2600 | pEC-XK99E, E. coli-C. glutamicum shuttle expression vectors based on the medium-copy number plasmid including pGA1, KanR, oriV, Ptrc | KanR | Kirchner and Tauch (2003) |
| JBEI-19559 | pEC-XK99E-AK-IP-bypass | KanR | Sasaki et al. (2019) |
| JBEI-19628 | pTE221 pEC-XK99E-AK-IP-bypass-S. aureus mvaK1, mvaS (substitution) | KanR | This study |
| JBEI-19634 | pTE222 pEC-XK99E-AK-IP-bypass-C. kroppenstedtii mvaK1, mvaA (substitution) | KanR | This study |
2.3. Growth media composition
Production was analyzed in several different common growth medias. Lysogeny-Broth (LB): 10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl. Tryptone and yeast extract were purchased from BD Biosciences (Franklin Lakes, NJ). NCM media (Ruan et al., 2015): 17.4 g/L K2HPO4, 11.6 g/L NaCl, 5 g/L d-glucose, 5 g/L tryptone, 1 g/L yeast extract, 0.3 g/L trisodium citrate, 0.05 g/L MgSO4·7H2O, and 91.1 g/L sorbitol, pH 7.2. CGXII minimal medium was prepared as previously described (Keilhauer et al., 1993; Sasaki et al., 2019). d-glucose was used as a carbon source at the 4% (w/v) concentration or as otherwise indicated.
2.4. Preparation of electrocompetent C. glutamicum cells
C. glutamicum was made electrocompetent as previously described (Sasaki et al., 2019). In brief, cells were grown in NCM medium supplemented with 3% (v/v) glycine and electroporated with a Micro Pulser Electroporator (Bio-Rad Laboratories, Inc., Hercules, CA) at 10 μF, 600 Ω, and 1800 V. After electroporation cells were immediately mixed with 400 μL of BHIS broth and heat-shocked for 6 min at 46 °C. After a two-hour outgrowth at 30 °C, cells were plated on the appropriate selective agar plate.
2.5. Cultivation of C. glutamicum for isopentenol and TMP production
All cells taken from −80 °C glycerol stocks were plated on LB agar plates containing the appropriate antibiotic following standard laboratory procedures. Single colonies were inoculated and grown overnight in 5 mL LB media (with antibiotics as necessary) at 30 °C on a rotary shaker at 200 rpm. Kanamycin was added to the growth media at a final concentration of 50 μg/mL. Unless otherwise noted, all seed cultures were first inoculated for growth in culture tubes. If cells were grown in a 24-well deep well format, 2 mL of culture media was used per well. Deep well plates were incubated Infors Multitron Incubator with a 3 mm Orbital Shaking Platform shaken at 999 rpm (Bottmingen, Switzerland).
To measure production of TMP or isopentenol, the adapted cultures of C. glutamicum were first back-diluted to OD600 of 0.1 into CGXII minimal medium including 4% (w/v) d-glucose at the concentrations described above. Cells from a seed culture were sub-cultured twice to adapt cells to grow in the media, as previously described (Sasaki et al., 2019). The production pathway was then induced as before when cultures reached an OD600 of ~0.8. Exogenous ionic liquids were added to the adapted cultures at the same time of induction with IPTG.
2.6. Analytical methods for chemical identification and quantification
For metabolite quantification, 300 μL of cell culture media was combined with 300 μL of ethyl acetate containing n-butanol (10 mg/L) as an internal standard and processed as described previously (Goh et al., 2012; Kang et al., 2016; Sasaki et al., 2019). Briefly, samples were shaken at maximum speed for 15 min using an MT-400 microtube mixer (TOMY Seiko, Tokyo, Japan) and then centrifuged at 14,000g for 3 min to separate the organic phase from the aqueous phase. 60 μL of the organic layer was transferred into a GC vial and 1 μL was analyzed by Agilent GC/MS equipped with a DB-5 column (Agilent Technologies, Santa Clara, CA, USA) or Thermo GC-FID equipped with a DB-WAX column (Agilent Technologies, Santa Clara, CA, USA) for quantification of TMP, acetoin, diacetyl, and isopentenol (3-methyl-3-buten-1-ol). Analytical grade standards were purchased from Sigma-Aldrich (St. Louis, MO) and used to calculate analyte concentrations and confirm identification of peaks. To compare the extraction efficiency of TMP into dichloromethane, the same protocol as above was used where dichloromethane was used in place of ethyl acetate as the extraction solvent. Reported TMP titers were calculated using a linear curve of TMP peak areas normalized to n-butanol generated from authentic standards resuspended directly into ethyl acetate. Values were corrected for inefficient extraction from CGXII media into ethyl acetate by multiplying GC-FID values by 5.88 (refer to Supplemental Fig. 1A).
To determine the spontaneous conversion rate of acetoin or diacetyl to tetra-methylpyrazine, pure 100 mM of pure analytical grade standards were added to CGXII media supplemented with 4% d-glucose. These cultures were then incubated at 30 °C as described in section 2.5 for 48 h, after which samples were harvested for ethyl acetate extraction and quantification by GC-FID. Conversion of acetoin or diacetyl to TMP was quantified using authentic standards, and samples were tested in triplicate.
A commonly used extraction solvent, ethyl acetate, showed a 17% extraction efficiency for TMP from CGXII culture media (Supplemental Fig. 1A). While toxic and more challenging to handle, dichloromethane showed a higher extraction efficiency for TMP at 50% from culture media (Supplemental Fig. 1B).
2.7. Comparison of pathway protein abundance in E. coli and C. glutamicum
E. coli (DH1) and C. glutamicum (ATCC 13032) strains harboring plasmid pTE220 were cultivated as described in section 2.5. Cells were grown in LB media supplemented with 56 mM glucose and 50 μg/mL Kanamycin +500 μM IPTG to ensure an equitable comparison between these two hosts. Crude cell extracts containing were prepared exactly as described in (Eng et al., 2018) and analyzed with an Agilent 6550 iFunnel Q-TOF mass spectrometer (Agilent Technologies, Santa Clara, CA) coupled to an Agilent 1290 UHPLC system, with a method as described previously (Gonzalez Fernandez-Nino et al., 2015). To measure global proteomic changes between pathway variants in C. glutamicum, we analyzed the set of proteins absent in wild-type C. glutamicum but were detected in the pathway variant samples by shotgun proteomics, filtering for proteins with at least two unique total peptides detected. Endogenous proteins were functionally annotated using eggNOG-mapper (Huerta-Cepas et al., 2017) to assign COG annotations (Galperin et al., 2014). Hierarchical clustering using a one minus Pearson correlation was calculated for both proteins and strains using the Morpheus software package (https://software.broadinstitute.org/morpheus).
2.8. Fed-batch production of TMP in a 2 L bioreactor format
Fed-batch production was performed using a 2 L bioreactor equipped with a Sartorius BIOSTAT B plus control unit for regulating dissolved oxygen (DO), pH, and temperature. A seed train was used to generate the starting inoculum for the bioreactor, which was electronically controlled to a pH of 7.0±0.3 using 7.5 M ammonium hydroxide and 4 M sulfuric acid. The temperature of the bioreactor was kept constant at 30 °C throughout the production time course. Diluted 10% (v/v) PEG-PPG-PEG, Poly(ethylene glycol)-block-poly(propylene glycol)-block-poly(ethylene glycol) (Sigma-Aldrich) was added as needed to control foaming. During the initial growth phase, DO was controlled at 30% saturation by varying agitation speed from 400 to 1200 rpm, and then the air-flow rate was subsequently varied from 0.5 to 1.5 volume of air per volume of medium per minute (vvm). Cultures were induced with 500 μM IPTG after 3 h of the batch phase. After depletion of the starting d-glucose (~10 g/L), feeding was initiated in low oxygen conditions by dropping DO levels down to 0–5% of saturation by varying agitation speed from 400 to 750 rpm and gassing with 0.25 vvm, and the feeding rate was controlled to keep the d-glucose concentration above 10 g/L. The feed solution contained 500 g/L d-glucose, 5 g/L (NH4)2SO4, 1.5 g/L KH2PO4, 1.5 g/L K2HPO4, 5 g/L yeast extract, 0.5 mM IPTG, and kanamycin. At specific timepoints, 5 mL samples were collected from the bioreactor by syringe affixed to the sampling tube and used for growth, GC-FID, and HPLC analysis.
3. Results
3.1. Engineered strains harboring a heterologous gene pathway produce tetra-methylpyrazine
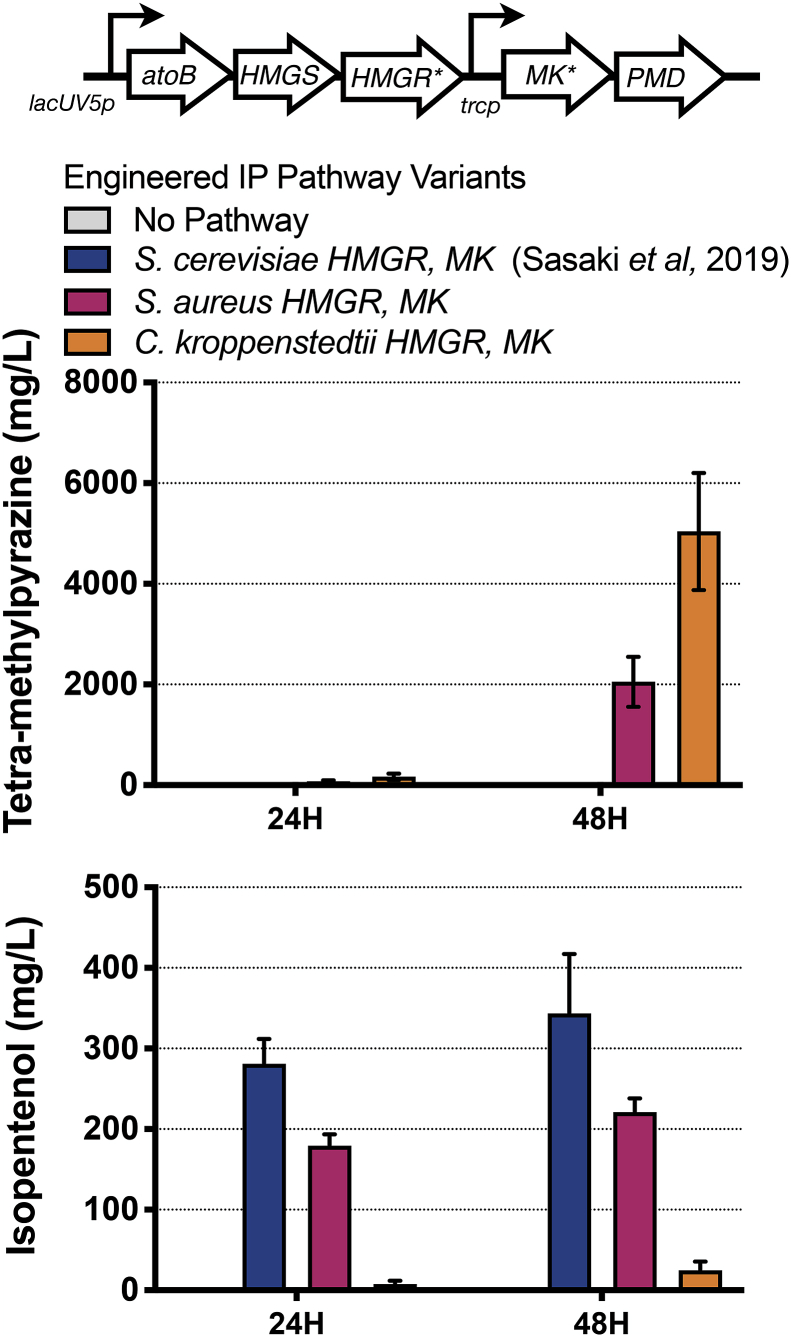
We observed the formation of tetra-methylpyrazine (TMP) while analyzing heterologous gene pathway variants expressed in engineered C. glutamicum strains. In a previous study, we had engineered C. glutamicum for the production of isopentenol using a heterologous, mevalonate-based five gene pathway, referred to as the “original” pathway (Sasaki et al., 2019). Pathway variants were constructed because two of the genes (mk and hmgR derived from Saccharomyces cerevisiae) were poorly expressed under standard laboratory growth conditions (Supplemental Table 1). These variants were constructed by substituting mk and hmgR gene homologs from either Staphylococcus aureus or Corynebacterium kroppenstedtii into the heterologous gene pathway and assayed for TMP production over a 48 h time course.
We report that these strains with two successfully expressed mk homologs (from S. aureus and C. kroppenstedtii) and hmgR from S. aureus resulted in the production of TMP as detected by GC analysis (Fig. 2). In the absence of a heterologous gene pathway, no new products were detected. The C. glutamicum strain harboring the original pathway produced 300 mg/L isopentenol and did not produce detectable TMP. In contrast to the original pathway, the S. aureus Mk, HmgR pathway variant produced 2.2 g/L of TMP and ~200 mg/L of isopentenol. Furthermore, the C. kroppenstedtii Mk, HmgR pathway variant produced 5 g/L of TMP and did not produce isopentenol. These results indicate that the choice of pathway variant expressed in C. glutamicum impacted the amount of TMP produced.
Fig. 2.
mk and hmgR Variants from bacterial species bias production towards tetra-methylpyrazine.
Top panel. Schematic of the heterologous isopentenol production pathway. The two genes selected for optimization by substitution with homologs are indicated with an asterisk (*). The proposed metabolic pathway for the conversion of glucose to isopentenol has been previously described in Sasaki et al (2019).
Bottom panels. Analysis of the engineered isopentenol production pathway in C. glutamicum using mk and hmgR homologs from S. aureus and C. kroppenstedtii in a 24 well plate format. C. glutamicum ΔpoxB ΔldhA strain harboring the original isopentenol production pathway (JBEI-19559) was compared with variants where the S. cerevisiae MK and HMGR were replaced with mk and hmgR from S. aureus (JBEI-19652) or C. kroppenstedtii (JBEI-19658). Samples were cultivated in CGXII media 4% d-glucose in a 24 well plate format. TMP or isopentenol titers were analyzed at the timepoints indicated and are an average of three biological replicates. The error bars represent standard error.
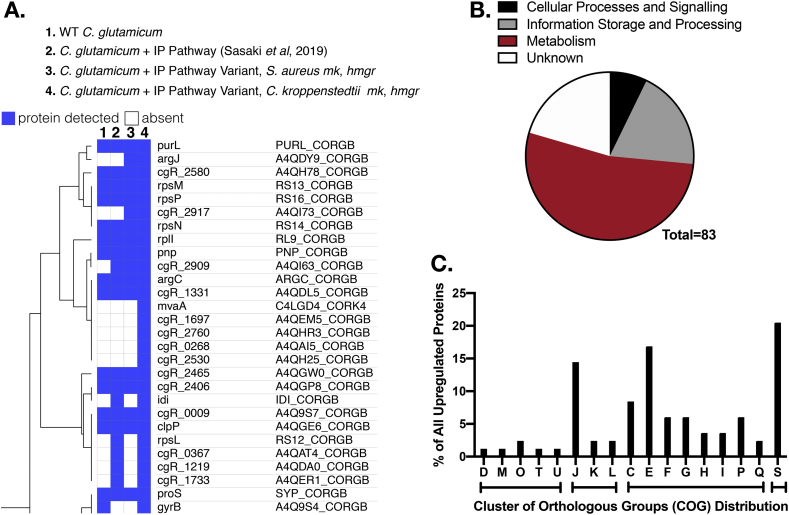
Next, we examined if candidate TMP pathway proteins (Fig. 1) had increased expression in these two new strains but did not detect significant differences in protein abundance for the three candidate pathway genes, IlvB, IlvN, or ButA among the strain variants (Supplemental Table 2). As there was no detectable change in protein abundance for the pathway genes, we searched for proteome-wide changes to cellular metabolism which could favor spontaneous conversion to TMP.
Four proteins involved in the tryptophan synthesis pathways (TrpE/CgR_2916, TrpG/CgR_2917, TrpB/CgR_2920, TrpA/CgR_2921) were newly detected in strains expressing the isopentenol pathway variants from S. aureus or C. kroppenstedtii (Fig. 3 and Supplemental Table 2). Additionally, two proteins involved in nitrogen metabolism (ArgJ/CgR_1458, CgR_1470) were also identified as enriched in these strains. There were four native proteins which were upregulated only in the C. kroppenstedtii pathway variant strain: CgR_1697 (a putative cadherin-like superfamily protein); CgR_2760 (a putative polyketide synthase); CgR_0268 (a short-chain dehydrogenase); and CgR_2530 (uncharacterized protein, no predicted homologs). A systems level analysis (Galperin et al., 2014) of the 83 upregulated proteins indicated that 53% of these proteins were involved in cellular physiology, many related to amino acid metabolism and transport (Fig. 3B). Of particular interest, ortholog groups in protein translation related activities (category J) and amino acid metabolism and transport (category E) were upregulated (Fig. 3C). Complete data of all enriched proteins from these engineered strains is plotted in Supplemental Fig. 2. As the expression of specific heterologous gene pathways favored nitrogen-accumulating processes that could enhance TMP production (Rizzi, 1988), it was reasonable that the enrichment of new compounds could be favored in these variant strain backgrounds.
Fig. 3.
Proteomic Analysis of Engineered C. glutamicum strains.
A. Hierarchical clustering of proteins enriched after heterologous gene pathway expression in engineered C. glutamicum strains. B. Gene functions of enriched proteins as modeled with eggNOG-mapper to assign genes into categories of orthologous groups (COGs). WT C. glutamicum (JBEI-7936) was compared against strains harboring the original isopentenol pathway (JBEI-19571) or the plasmid variants (JBEI-19652 and JBEI-19658). C. Distribution of enriched proteins into specific COGs. COGs falling into related categories from the top panel are grouped together in brackets below. COG definitions: D, cell division and chromosome partitioning; M, cell wall structure and biogenesis and outer membrane; O, molecular chaperones and related functions; T, signal transduction; Intracellular trafficking, secretion; J, Translation, ribosomal structure and biogenesis; K, transcription; L, replication, recombination, repair; C, energy production and conversion; E, amino acid transport and metabolism; F, nucleotide transport and metabolism; G, carbohydrate transport and metabolism; H, coenzyme transport and metabolism; I, lipid transport and metabolism; P, inorganic ion transport and metabolism; Q secondary metabolite transport and metabolism; S, unknown function.
3.2. Analysis of tetra-methylpyrazine biosynthesis and its downstream extraction
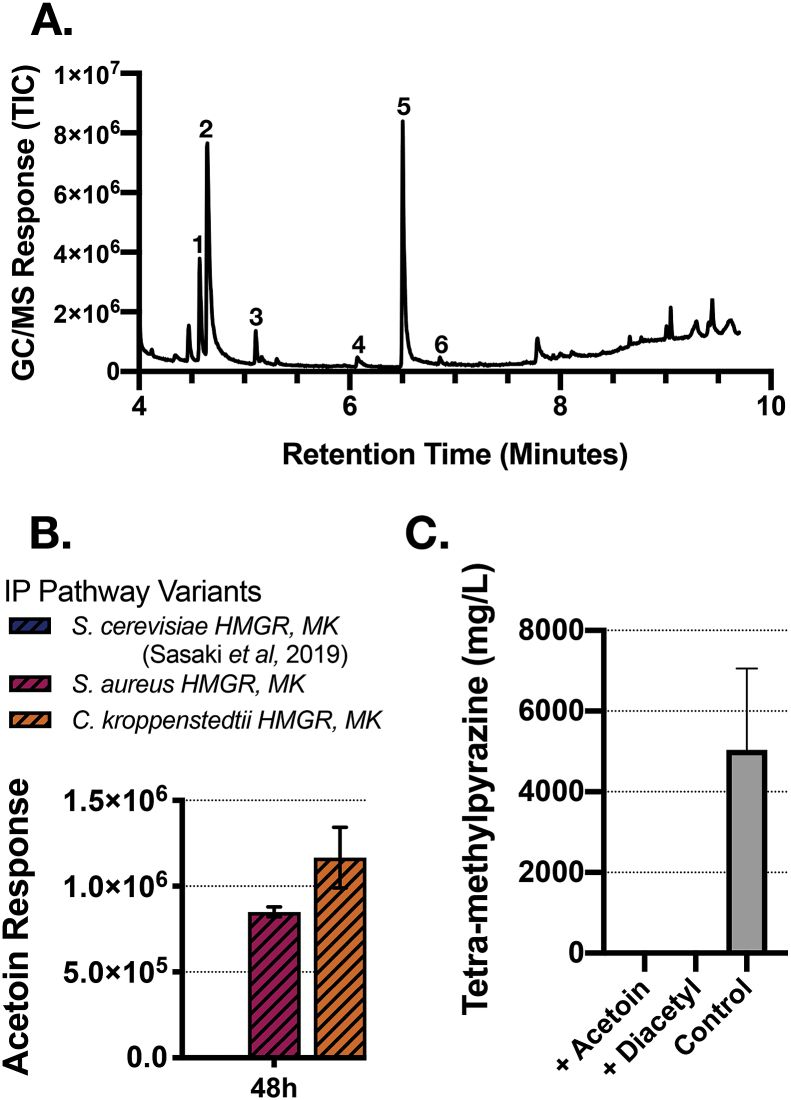
To understand how TMP might be produced in these strains, we examined the GC/MS trace files from these samples to determine if other related pathway intermediates might also be detected. GC/MS analysis of the additional analytes indicated the additional main peak to be tetra-methylpyrazine, TMP (Fig. 4A peak no. 5 and Supplemental Fig. 3). We also detected the accumulation of tri-methylpyrazine and 3,5-diethyl-2-methyl-pyrazine but did not investigate either of these products any further (Fig. 2, peak no. 4 and peak no. 6).
Fig. 4.
GC/MS analysis of tetra-methylpyrazine and S-acetoin. A. Identification of tetra-methylpyrazine and other peaks by GC/MS analysis. The genotypes of the strains used were the same as described in Fig. 2. Peak identification. 1. Acetoin (3-hydroxy-2-butanone). 2. Isopentenol (3-methyl-3-buten-1-ol). 3. 4-penten-1-ylacetate. 4. 2,3,5-tri-methyl-pyrazine. 5. 2,3,5,6-tetra-methyl-pyrazine. 6. 3,5-diethyl-2-methyl-pyrazine. B. Comparison of acetoin peak height across different engineered strains by GC/MS analysis. C. Detection of spontaneous TMP formation in CGXII media from precursors, acetoin or diacetyl, after 48 h post incubation. 100 mM of either acetoin or diacetyl was added to CGXII media. As a reference, TMP produced from the C. kroppenstedtii variant in JBEI-19658 is replotted from Fig. 2 on the same graph.
Consistent with the proposed pathway for pyrazine synthesis (Fig. 1), we also detected S-Acetoin (3-Hydroxybutanone; Fig. 4A peak no. 1 and Supplemental Fig. 3), the proposed precursor of tri/tetra-methylpyrazine (Karp et al., 2011; Xiao et al., 2014). S-Acetoin was specifically identified in samples with detectable TMP production levels and absent in samples which did not produce TMP (Fig. 4B). The accumulation of acetoin in these strains strengthens the evidence in support of the proposed biological route to produce TMP.
We next examined if TMP could form spontaneously from acetoin or diacetyl in the absence of a microbial host under biologically relevant cultivation conditions. Previous TMP synthesis routes have a post-cultivation step in which the culture broth is heated >60 °C to favor the synthesis of TMP (Rizzi, 1988; Xiao et al., 2104). However, it is unclear if a biologically relevant temperature with nitrogen concentrations equal to that in the GCXII medium would be sufficient for its spontaneous conversion. To examine this, commercially purchased authentic standards for acetoin and diacetyl were spiked into CGXII media and tested for the formation of TMP after the same cultivation time period of 48 h. We did not detect spontaneous conversion of either acetoin or diacetyl to TMP, suggesting production of the observed TMP requires the engineered strain (Fig. 4C).
3.3. Ionic liquids, a renewable pretreatment reagent, enhances production of tetra-methylpyrazine
Most sustainable production requires the use of renewable carbon sources such as plant-derived biomass and pose trade-offs due to the incompatibilities between processes (Baral et al., 2019b; Eng et al., 2018; Ouellet et al., 2011; Sasaki et al., 2019; Wang et al., 2018). We evaluated the behavior of C. glutamicum strains when cultivated with reagents used in the pretreatment of sustainable carbon streams. Specifically, we asked if trace levels of ionic liquids, a promising reagent for plant biomass deconstruction (George et al., 2015; Li et al., 2010), changed the amount of TMP produced in strains expressing the original IP pathway.
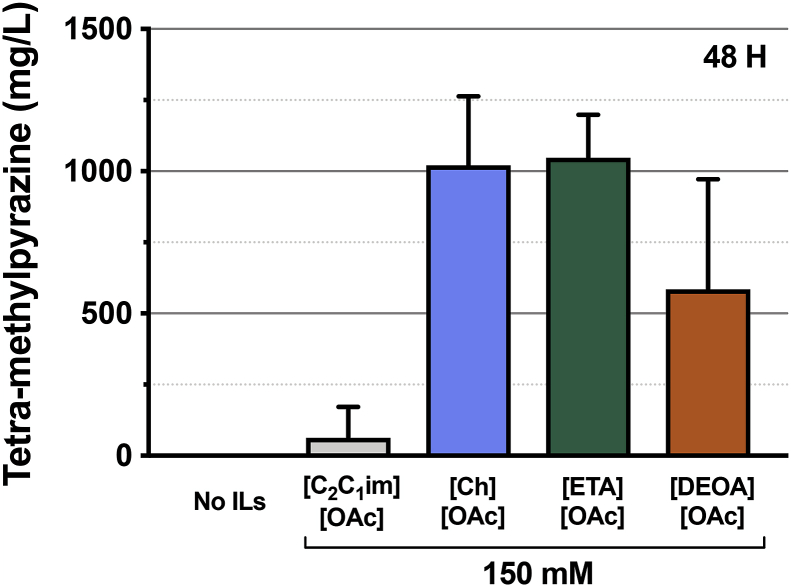
We characterized the effect of three different classes of ionic liquids on the behavior of engineered C. glutamicum strains for the production of TMP. The ionic liquids chosen were acetate salts with 1-ethyl-3-methyl imidazolium ([C2C1im]+); cholinium ([Ch]+); ethanolamine [ETA]; and diethanolamine [DEOA], [ETA] and [DEOA] cations. [C2C1im] and [Ch] are two biogenic ILs whereas [ETA] and [DEOA] are two representative protic ILs. We detected TMP at the 48 h time point under these ILs-stressed conditions (Fig. 5). In the presence of exogenous 150 mM [C2C1im][OAc], we detected 63 mg/L TMP. Interestingly, in contrast to bioproduction in other microbial hosts (Eng et al., 2018; Ouellet et al., 2011), we did not observe inhibition of production in the presence of ILs. Rather, when treated with 150 mM [Ch][OAc], we detected higher levels of TMP production, with titers reaching 1021 mg/L. Of the protic ILs, treatment with 150 mM [ETA][OAc] produced a similar titer to [Ch][OAc], with measured titers around 1050 mg/L. 150 mM [DEOA][OAc] treatment resulted in an accumulation of TMP to 585 mg/L. When expressed in C. glutamicum, the original IP pathway does not produce TMP under standard cultivation conditions, but treatment with specific ILs result in TMP production from undetectable to over 1 g/L.
Fig. 5.
Specific forms of ionic liquids induce tetra-methylpyrazine production in C. glutamicum.
The production profiles of the C. glutamicum (JBEI-19571) against three types (imidazolium, cholinium, and protic form) of ionic liquids were examined in CGXII media with 4% d-glucose in a 24 well plate format. ([C2C1im]+); cholinium ([Ch]+); ethanolamine acetate [ETA][OAc]; and diethanolamine acetate [DEOA][OAc] A. Produced titers of tetra-methylpyrazine are shown at 48 h post induction under 75 mM (A) or 150 mM (B) [C2C1im][OAc], [Ch][OAc], [ETA][OAc], and [DEOA][OAc]. The control experiment was performed without IL supplementation (No ILs). Data shown are an average of biological triplicates, and the error bars represent standard error.
Ionic liquids alone were not sufficient to induce TMP production in wild-type C. glutamicum (Supplemental Fig. 1C). The amount of TMP produced in response to the IL was more variable when treating cells with 75 mM ILs, but TMP still accumulated in the same set of ILs. These strains also co-produced TMP and ~350 mg/L isopentenol using the unmodified isopentenol production pathway (Supplemental Fig. 4). These results provide a new perspective on using ILs as pretreatment reagents. Rather than framing ILs as pretreatment contaminants that must be removed, trace ionic liquids remaining from biomass pretreatment regimes could be beneficial as inducers allowing the coproduction of TMP and isopentenol.
3.4. Production of TMP in fed batch mode cultures
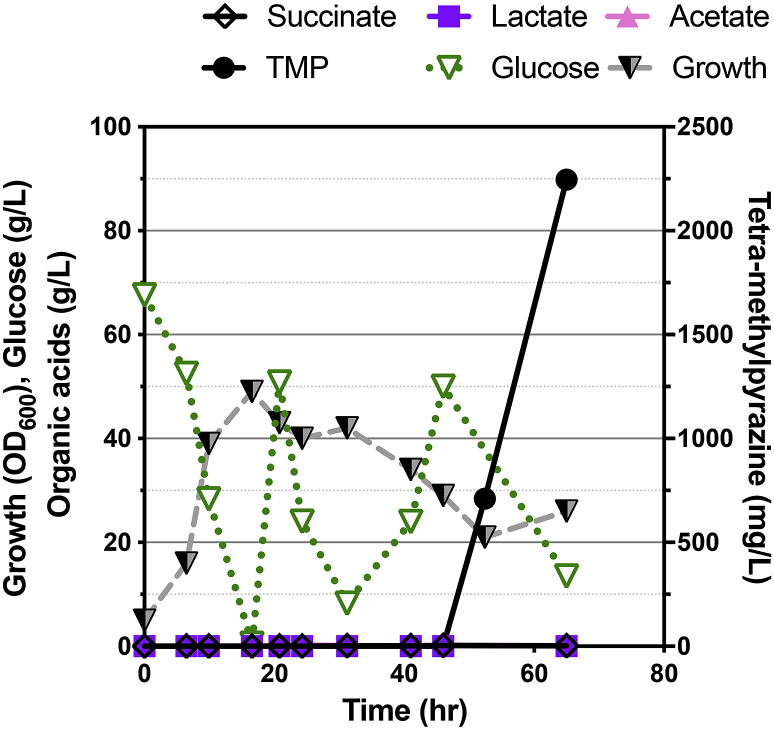
Having observed the accumulation of TMP in our engineered strains, we wanted to next determine if TMP production could be produced under more industrially relevant cultivation conditions. We thus changed our cultivation format from a 24 well deep well microtiter dish to a 2L bioreactor format in fed batch mode. To retain flexibility in the choice of the final product, we added 50 mM of the IL [Ch][Lys] to stimulate TMP production in the engineered C. glutamicum strain using the original pathway. In fed-batch mode, the rate of TMP production was slower than what was observed in batch mode, as we only detected 700 mg/L of TMP after 48 h of cultivation (Fig. 6, compare to time course in Fig. 2). It is possible that TMP is only produced after nutrient exhaustion in stationary phase, as we only detected TMP production 6 h after the final addition of the feed solution. The TMP titer after 65 h cultivation reached 2 g/L (Fig. 6), which was higher than the final titer in the smaller lab-scale format using IL-stimulated production (compare with Fig. 5). Isopentenol production under these cultivation conditions was near the detection limit (5 mg/L) potentially due to the increased aeration from the impeller driven mixing inherent to cell growth conditions in the bioreactor (Fig. 6). We attempted to phase separate isopentenol with a dodecane overlay (Peralta-Yahya et al., 2011), but dodecane inhibited growth of C. glutamicum under these conditions. These results indicate the scalability of TMP production under industrially relevant formats, but further work would be required to co-produce isopentenol.
Fig. 6.
Fed-batch mode production of TMP in engineered C. glutamicum.
C. glutamicum (JBEI-19571) was initially cultivated in batch mode using CGXII minimal media including 8% starting d-glucose and 50 mM [Ch][Lys] in a 2L Sartorius Bioreactor. Subsequently, fed-batch mode was initiated when the initial d-glucose concentration decreased below 1%. A pulse mode feeding strategy was utilized to raise the d-glucose concentration above 1%. Final products and organic acid concentrations were quantified and are indicated as labeled in the legend above.
4. Discussion
This report illustrates two parameters by which C. glutamicum can be modified to produce tetra-methylpyrazine, TMP. Our highest final titers from batch and fed-batch production modes compare favorably with published titers from Bacillus subtilis (Xiao et al., 2014; Yin et al., 2018) and do not require a post-cultivation denaturing step to produce TMP. Unlike E. coli, C. glutamicum responds to the burden of expressing a heterologous IP pathway by accumulating TMP. A previous report had indicated that redirecting central metabolism in C. glutamicum away from acetate led to the accumulation of high intracellular levels of acetoin (Mao et al., 2017), but did not report TMP in their strains. This result from Mao et al. is consistent with our hypothesis, in which the burden of a specific heterologous gene pathway can instead favor high level TMP accumulation. Moreover, the kinetics of TMP formation is dependent on the specific pathway variant analyzed. There are likely some interactions between the heterologous gene pathway and native metabolism at the metabolite level. Efficient and universal metabolite quenching for metabolic flux analysis could be utilized to examine this possibility (Wellerdiek et al., 2009; Zhang et al., 2018).
Certain ionic liquids are known to impact cellular morphology and physiology (Mehmood et al., 2015). Their impact on engineered strains can also be pathway-specific; in E. coli, the addition of an isopentenol production pathway to an IL-tolerant strain can result in acetate accumulation rather than isopentenol (Eng et al., 2018). In contrast, limonene can be produced from the same IL-tolerant strain without detectable acetate accumulation (Eng et al., 2018). While additional studies are required to understand the beneficial impact of ionic liquids on the production of TMP in engineered C. glutamicum strains, this study provides an alternate paradigm of ILs as a useful tool for improving production instead of merely being an unwanted contaminant. The added burden of ionic liquids could be a new metric for assessing how a heterologous gene pathway can impinge upon native metabolism, especially when evaluating hosts originating from different environmental niches and primary carbon sources (Wehrs et al., 2019).
The production of TMP is correlated with the accumulation of acetoin, providing additional evidence to strengthen its proposed biosynthetic pathway (Fig. 1). We do not detect abiotic accumulation of TMP from either acetoin or di-acetyl when added directly into culture media (Fig. 4C). Rather our results suggest that TMP production is strain dependent and likely involve the accumulation of acetoin. These observations highlight the importance of considering both the specific heterologous gene pathway as well as potential feedback on the chosen microbial host. With high titers of TMP, these C. glutamicum strains will facilitate rapid evaluation of TMP derivatives using candidate enzyme libraries to be expressed in a gram-positive microbial host (Hu et al., 2018; Stankevičiūtė et al., 2016; Zhang et al., 2016). Being able to produce >1 g/L quantities of many TMP derivatives will unlock the potential of this emerging molecule for a broader spectrum of applications.
While we report the exclusive production of TMP, examples of the co-production of two different final products from the same microbial host can also be of value (Liang and Qi, 2014). As these two compounds have distinct applications, such an industrial process would likely require refinement of existing purification techniques for efficient separation. Isopentenol easily partitions into organic solvents such as ethyl acetate, and TMP can be precipitated from the aqueous phase after cooling to ~4 °C (Xiao et al., 2006). This report is also the first instance in which either TMP or isopentenol has been the final product for a co-production study. The use of TMP as a possible co-produced compound with isopentenol will allow more nuance and control in the technoeconomic analysis of scaling up either product (Baral et al., 2019a, 2019b).
5. Conclusions
In summary, this report describes the successful use of the gram-positive industrial microorganism, C. glutamicum, for the high titer (>5 g/L) production of TMP. These strains can also be used to co-produce TMP as well as the biofuel candidate, isopentenol. Production of TMP was scaled-up to industrially relevant conditions in a 2L fed-batch bioreactor, where we observed 2 g/L TMP when cells were treated with exogenous ionic liquid.
Contributions
Conceptualization of the project: AM TE YS. Strain construction, molecular biology, analytical chemistry, scale-up: YS TE RH JT YC MM CJP. Interpreted results: YS TE AM YC CJP. Contributed critical reagents: TE YS JT. Drafted the manuscript: YS TE AM. All authors read, contributed feedback, and approved the manuscript for publication.
CRediT authorship contribution statement
Thomas Eng: Conceptualization, Investigation, Validation, Formal analysis, Visualization, Project administration, Writing - original draft. Yusuke Sasaki: Conceptualization, Investigation, Visualization, Writing - original draft. Robin A. Herbert: Investigation, Validation. Andrew Lau: Investigation. Jessica Trinh: Investigation. Yan Chen: Investigation, Formal analysis. Mona Mirsiaghi: Investigation. Christopher J. Petzold: Resources, Writing - review & editing. Aindrila Mukhopadhyay: Conceptualization, Supervision, Resources, Writing - original draft, Supervision, Funding acquisition.
Declaration of competing interest
The authors have no conflicts of interest to declare.
Acknowledgements
We thank James Sun (LBNL) for sharing the ionic liquids used in this study, and the Advanced Biofuels and Bioproducts Process Development Unit (ABPDU) for technical assistance, and Steve Singer (LBNL) for helpful comments on the manuscript. The work was fully funded by by the U. S. Department of Energy, Office of Science, through contract DE-AC02-05CH11231 between Lawrence. Berkeley National Laboratory and the U. S. Department of Energy. However, one of the authors (YS) was supported by the a Grant-in-Aid for JSPS Fellows (No. 18J10103) from the Japan Society for the Promotion of Science. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2019.e00115.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Supplemental Table 1. Shotgun proteomic analysis of isopentenol production pathway in E. coli DH1 and C. glutamicum.
Supplemental Table 2. Shotgun proteomic analysis of isopentenol production pathway variants in C. glutamicum.
Supplemental Figure 1. Extraction efficiency of TMP. A. Extraction efficiency of TMP using ethyl acetate as the extraction solvent. The linear range of TMP dissolved in ethyl acetate is plotted on the right. B. Extraction efficiency of TMP using dichloromethane. The linear range of TMP dissolved in dichloromethane is plotted on the right. C. Impact of exogenous ionic liquid on wild-type C. glutamicum. Without the presence of a heterologous gene pathway, no TMP was detected with any ionic liquid treatment.
Supplemental Figure 3. Mass fragmentation pattern of acetoin and TMP.
Supplemental Figure 4. Related to Fig. 5. Quantification of isopentenol production in production strains treated with exogenous ionic liquids. The data shown are an average of biological triplicates, and the error bars represent standard error.
Supplemental figure 2.

Supplemental Figure 2. Related to Fig. 3 Complete heat map of hierarchical clustered strains and proteins upregulated in engineered C. glutamicum strains.
References
- Baral N.R., Kavvada O., Mendez-Perez D., Mukhopadhyay A., Lee T.S., Simmons B.A., Scown C.D. Techno-economic analysis and life-cycle greenhouse gas mitigation cost of five routes to bio-jet fuel blendstocks. Energy Environ. Sci. 2019;12:807–824. [Google Scholar]
- Baral N.R., Sundstrom E.R., Das L., Gladden J., Eudes A., Mortimer J.C., Singer S.W., Mukhopadhyay A., Scown C.D. Approaches for more efficient biological conversion of lignocellulosic feedstocks to biofuels and bioproducts. ACS Sustain. Chem. Eng. 2019;7(10):9062–9079. [Google Scholar]
- Chen H., Cao J., Zhu Z., Zhang G., Shan L., Yu P., Wang Y., Sun Y., Zhang Z. A novel tetramethylpyrazine derivative protects against glutamate-induced cytotoxicity through PGC1α/nrf2 and PI3K/Akt signaling pathways. Front. Neurosci. 2018;12 doi: 10.3389/fnins.2018.00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng T., Tao S., Luo D., Qin P. 2006. China Patent CN1546474A. Method for Preparing Tetramethyl Pyrazine. [Google Scholar]
- Demain A., Jackson M., Trenner N. Thiamine-dependent accumulation of tetramethylpyrazine accompanying a mutation in the isoleucine-valine pathway. J. Bacteriol. 1967;94:323–326. doi: 10.1128/jb.94.2.323-326.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickschat Jeroen S., Wickel S., Bolten Christoph J., Nawrath T., Schulz S., Wittmann C. Pyrazine biosynthesis in Corynebacterium glutamicum. Eur. J. Org. Chem. 2010;2010:2687–2695. [Google Scholar]
- Eng T., Demling P., Herbert R.A., Chen Y., Benites V., Martin J., Lipzen A., Baidoo E.E., Blank L.M., Petzold C.J. Restoration of biofuel production levels and increased tolerance under ionic liquid stress is enabled by a mutation in the essential Escherichia coli gene cydC. Microb. Cell Factories. 2018;17:159. doi: 10.1186/s12934-018-1006-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira S.B., Kaiser C.R. Pyrazine derivatives: a patent review (2008–present) Expert Opin. Ther. Pat. 2012;22:1033–1051. doi: 10.1517/13543776.2012.714370. [DOI] [PubMed] [Google Scholar]
- Galperin M.Y., Makarova K.S., Wolf Y.I., Koonin E.V. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 2014;43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- George A., Brandt A., Tran K., Zahari S.M.N.S., Klein-Marcuschamer D., Sun N., Sathitsuksanoh N., Shi J., Stavila V., Parthasarathi R. Design of low-cost ionic liquids for lignocellulosic biomass pretreatment. Green Chem. 2015;17:1728–1734. [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Goh E.B., Baidoo E.E., Keasling J.D., Beller H.R. Engineering of bacterial methyl ketone synthesis for biofuels. Appl. Environ. Microbiol. 2012;78:70–80. doi: 10.1128/AEM.06785-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez Fernandez-Nino S.M., Smith-Moritz A.M., Chan L.J., Adams P.D., Heazlewood J.L., Petzold C.J. Standard flow liquid chromatography for shotgun proteomics in bioenergy research. Front. Bioeng. Biotechnol. 2015;3:44. doi: 10.3389/fbioe.2015.00044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S., Hu H., Mak S., Cui G., Lee M., Shan L., Wang Y., Lin H., Zhang Z., Han Y. A novel tetramethylpyrazine derivative prophylactically protects against glutamate-induced excitotoxicity in primary neurons through the blockage of N-Methyl-D-aspartate receptor. Front. Pharmacol. 2018;9 doi: 10.3389/fphar.2018.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J., Forslund K., Coelho L.P., Szklarczyk D., Jensen L.J., von Mering C., Bork P. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol. Biol. Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang A., George K.W., Wang G., Baidoo E., Keasling J.D., Lee T.S. Isopentenyl diphosphate (IPP)-bypass mevalonate pathways for isopentenol production. Metab. Eng. 2016;34:25–35. doi: 10.1016/j.ymben.2015.12.002. [DOI] [PubMed] [Google Scholar]
- Kao T.-K., Chang C.-Y., Ou Y.-C., Chen W.-Y., Kuan Y.-H., Pan H.-C., Liao S.-L., Li G.-Z., Chen C.-J. Tetramethylpyrazine reduces cellular inflammatory response following permanent focal cerebral ischemia in rats. Exp. Neurol. 2013;247:188–201. doi: 10.1016/j.expneurol.2013.04.010. [DOI] [PubMed] [Google Scholar]
- Karp P.D., Latendresse M., Caspi R. The pathway tools pathway prediction algorithm. Stand. Genomic. Sci. 2011;5:424–429. doi: 10.4056/sigs.1794338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke-ji C., Zhen-huai Q., Wei-Liang W., Mu-Ying Q. Tetramethylpyrazine in the treatment of cardiovascular and cerebrovascular diseases. Planta Med. 1983;47 89-89. [PubMed] [Google Scholar]
- Keilhauer C., Eggeling L., Sahm H. Isoleucine synthesis in Corynebacterium glutamicum: molecular analysis of the ilvB-ilvN-ilvC operon. J. Bacteriol. 1993;175:5595–5603. doi: 10.1128/jb.175.17.5595-5603.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchner O., Tauch A. Tools for genetic engineering in the amino acid-producing bacterium Corynebacterium glutamicum. J. Biotechnol. 2003;104:287–299. doi: 10.1016/s0168-1656(03)00148-2. [DOI] [PubMed] [Google Scholar]
- Kosuge T., Kamiya H. Discovery of a pyrazine in a natural product: tetramethylpyrazine from cultures of a strain of Bacillus subtilis. Nature. 1962;193:776. doi: 10.1038/193776a0. [DOI] [PubMed] [Google Scholar]
- Lee, J. L., Halogen-free Flame Retardant Material. US Patent US8871843B2. Apple Inc. Cupertino, CA, US., United States, 2014.
- Li C., Knierim B., Manisseri C., Arora R., Scheller H.V., Auer M., Vogel K.P., Simmons B.A., Singh S. Comparison of dilute acid and ionic liquid pretreatment of switchgrass: biomass recalcitrance, delignification and enzymatic saccharification. Bioresour. Technol. 2010;101:4900–4906. doi: 10.1016/j.biortech.2009.10.066. [DOI] [PubMed] [Google Scholar]
- Li H.-X., Ding M.-Y., Lv K., Yu J.-Y. Separation and determination of ephedrine alkaloids and tetramethylpyrazine in ephedra sinica Stapf by gas chromatography-mass spectrometry. J. Chromatogr. Sci. 2001;39:370–374. doi: 10.1093/chromsci/39.9.370. [DOI] [PubMed] [Google Scholar]
- Liang Q., Qi Q. From a co-production design to an integrated single-cell biorefinery. Biotechnol. Adv. 2014;32:1328–1335. doi: 10.1016/j.biotechadv.2014.08.004. [DOI] [PubMed] [Google Scholar]
- Mao Y., Fu J., Tao R., Huang C., Wang Z., Tang Y.-J., Chen T., Zhao X. Systematic metabolic engineering of Corynebacterium glutamicum for the industrial-level production of optically pure D-(−)-acetoin. Green Chem. 2017;19:5691–5702. [Google Scholar]
- Masuda H., Mihara S. Olfactive properties of alkylpyrazines and 3-substituted 2-alkylpyrazines. J. Agric. Food Chem. 1988;36:584–587. [Google Scholar]
- Mehmood N., Husson E., Jacquard C., Wewetzer S., Buchs J., Sarazin C., Gosselin I. Impact of two ionic liquids, 1-ethyl-3-methylimidazolium acetate and 1-ethyl-3-methylimidazolium methylphosphonate, on Saccharomyces cerevisiae: metabolic, physiologic, and morphological investigations. Biotechnol. Biofuels. 2015;8:17. doi: 10.1186/s13068-015-0206-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M., Yuan R. DNA restriction enzyme from E. coli. Nature. 1968;217(5134):1110–1114. doi: 10.1038/2171110a0. [DOI] [PubMed] [Google Scholar]
- Ng H., Seto K., Zhang W. PROMERUS, LLC; Brecksville, OH, US, United States: 2017. US Patent US9765168B2. Flame Retardant Vinyl Addition Polycycloolefinic Polymers. [Google Scholar]
- Ouellet M., Datta S., Dibble D.C., Tamrakar P.R., Benke P.I., Li C., Singh S., Sale K.L., Adams P.D., Keasling J.D., Simmons B.A., Holmes B.M., Mukhopadhyay A. Impact of ionic liquid pretreated plant biomass on Saccharomyces cerevisiae growth and biofuel production. Green Chem. 2011;13:2743–2749. [Google Scholar]
- Peralta-Yahya P.P., Ouellet M., Chan R., Mukhopadhyay A., Keasling J.D., Lee T.S. Identification and microbial production of a terpene-based advanced biofuel. Nat. Commun. 2011;2:483. doi: 10.1038/ncomms1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzi G.P. Formation of pyrazines from acyloin precursors under mild conditions. J. Agric. Food Chem. 1988;36:349–352. [Google Scholar]
- Ruan Y., Zhu L., Li Q. Improving the electro-transformation efficiency of Corynebacterium glutamicum by weakening its cell wall and increasing the cytoplasmic membrane fluidity. Biotechnol. Lett. 2015;37:2445–2452. doi: 10.1007/s10529-015-1934-x. [DOI] [PubMed] [Google Scholar]
- Sasaki Y., Eng T., Herbert R.A., Trinh J., Chen Y., Rodriguez A., Gladden J., Simmons B.A., Petzold C.J., Mukhopadhyay A. Engineering Corynebacterium glutamicum to produce the biogasoline isopentenol from plant biomass hydrolysates. Biotechnol. Biofuels. 2019;12:41. doi: 10.1186/s13068-019-1381-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankevičiūtė J., Vaitekūnas J., Petkevičius V., Gasparavičiūtė R., Tauraitė D., Meškys R. Oxyfunctionalization of pyridine derivatives using whole cells of Burkholderia sp. MAK1. Sci. Rep. 2016;6:39129. doi: 10.1038/srep39129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Cheng G., Dong J., Tian T., Lee T.S., Mukhopadhyay A., Simmons B.A., Yuan Q., Singer S.W. Tolerance characterization and isoprenol production of adapted Escherichia coli in the presence of ionic liquids. ACS Sustain. Chem. Eng. 2018;7:1457–1463. [Google Scholar]
- Wehrs M., Tanjore D., Eng T., Lievense J., Pray T.R., Mukhopadhyay A. Trends in Microbiology. 2019. Engineering robust production microbes for large-scale cultivation. [DOI] [PubMed] [Google Scholar]
- Wellerdiek M., Winterhoff D., Reule W., Brandner J., Oldiges M. Metabolic quenching of Corynebacterium glutamicum: efficiency of methods and impact of cold shock. Bioproc. Biosyst. Eng. 2009;32:581–592. doi: 10.1007/s00449-008-0280-y. [DOI] [PubMed] [Google Scholar]
- Xiao Z., Hou X., Lyu X., Xi L., Zhao J.-y. Accelerated green process of tetramethylpyrazine production from glucose and diammonium phosphate. Biotechnol. Biofuels. 2014;7:106. [Google Scholar]
- Xiao Z., Xie N., Liu P., Hua D., Xu P. Tetramethylpyrazine production from glucose by a newly isolated Bacillus mutant. Appl. Microbiol. Biotechnol. 2006;73:512–518. doi: 10.1007/s00253-006-0491-6. [DOI] [PubMed] [Google Scholar]
- Yin D., Yang M., Wang Y., Yin D., Liu H., Zhou M., Li W., Chen R., Jiang S., Ou M. High tetramethylpyrazine production by the endophytic bacterial Bacillus subtilis isolated from the traditional medicinal plant Ligusticum chuanxiong Hort. Amb. Express. 2018;8:193. doi: 10.1186/s13568-018-0721-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Zheng X., Wang Y., Yu J., Zhang Z., Dele-Osibanjo T., Zheng P., Sun J., Jia S., Ma Y. Comprehensive optimization of the metabolomic methodology for metabolite profiling of Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 2018;102:7113–7121. doi: 10.1007/s00253-018-9095-1. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhang G., Sun Y., Szeto S.S.W., Law H.C.H., Quan Q., Li G., Yu P., Sho E., Siu M.K.W., Lee S.M.Y., Chu I.K., Wang Y. Tetramethylpyrazine nitrone, a multifunctional neuroprotective agent for ischemic stroke therapy. Sci. Rep. 2016;6:37148. doi: 10.1038/srep37148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B.-F., Xu Y., Fan W.-L. High-yield fermentative preparation of tetramethylpyrazine by Bacillus sp. using an endogenous precursor approach. J. Ind. Microbiol. Biotechnol. 2010;37:179–186. doi: 10.1007/s10295-009-0661-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1. Shotgun proteomic analysis of isopentenol production pathway in E. coli DH1 and C. glutamicum.
Supplemental Table 2. Shotgun proteomic analysis of isopentenol production pathway variants in C. glutamicum.
Supplemental Figure 1. Extraction efficiency of TMP. A. Extraction efficiency of TMP using ethyl acetate as the extraction solvent. The linear range of TMP dissolved in ethyl acetate is plotted on the right. B. Extraction efficiency of TMP using dichloromethane. The linear range of TMP dissolved in dichloromethane is plotted on the right. C. Impact of exogenous ionic liquid on wild-type C. glutamicum. Without the presence of a heterologous gene pathway, no TMP was detected with any ionic liquid treatment.
Supplemental Figure 3. Mass fragmentation pattern of acetoin and TMP.
Supplemental Figure 4. Related to Fig. 5. Quantification of isopentenol production in production strains treated with exogenous ionic liquids. The data shown are an average of biological triplicates, and the error bars represent standard error.