Abstract
Fusion of host and viral membranes is a crucial step during infection by enveloped viruses. In the structurally-defined “class I″ viral glycoproteins, the formation of a highly stable α-helical bundle by the ectodomain of the fusion subunit (e.g., GP2 for Marburg virus, MARV) is postulated to provide the energetic driving force to overcome barriers associated with membrane fusion. Upon cell binding, the fusion subunit is proposed to form an extended intermediate that bridges both the viral and host membranes, and collapse of this extended intermediate brings the two membranes into proximity. While there is much high-resolution structural data available for prefusion and post-fusion structures of viral glycoproteins, little information is available about intermediate conformations especially in the context of the fusion loop/peptide (FL or FP) and membrane-proximal external region (MPER)/transmembrane (TM) segments. We present structural and functional studies on segments of MARV GP2 that encompass the FL and MPER/TM in detergent micelles and lipid bicelles. A protein that contains most elements of GP2 (“MGP2-full”) is α-helical in membrane-mimicking environments and has pH-dependent membrane lytic activity. MGP2-full is monomeric under such conditions, contrasting with the trimeric species that has been described previously for MARV GP2 ectodomain in aqueous buffer. Variants of MARV GP2 containing the N- and C-terminal halves (“MGP2-FNL” and “MGP2-CMT”, respectively) have similar properties. This work provides novel insight into conformational and membrane-perturbing properties of the MARV fusion subunit and how they may relate to viral membrane fusion.
Keywords: Viral protein, Protein engineering, Protein folding, Lipid bilayer, Virology, Molecular biology, Viral membrane fusion, Membrane protein, Marburg virus, Ebola virus
Viral protein; Protein engineering; Protein folding; Lipid bilayer; Virology; Molecular biology; Viral membrane fusion; Membrane protein; Marburg virus; Ebola virus
1. Introduction
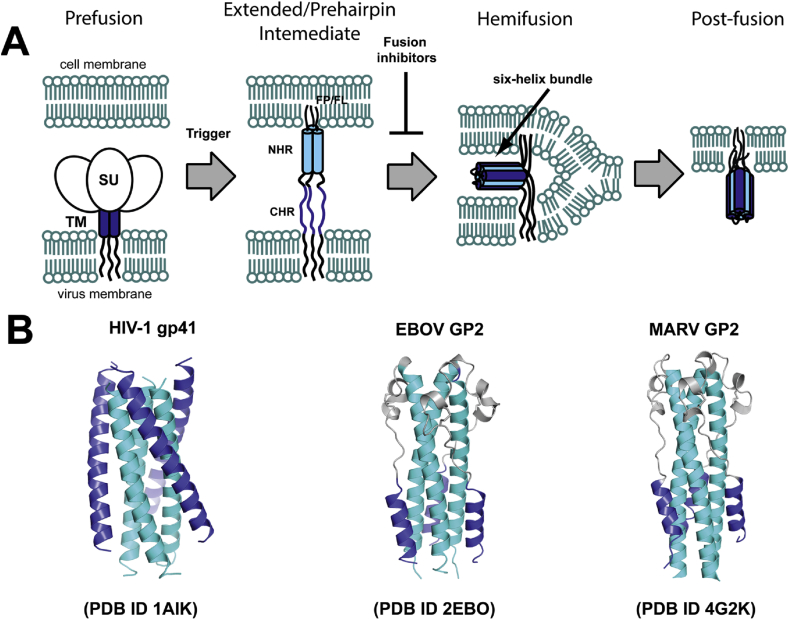
Fusion of the viral and host membranes is critical first step for infection by membrane-bound viruses, and this process is facilitated by viral envelope glycoproteins. The fusion of two membranes is a thermodynamically favorable process, but there is a high kinetic barrier (40–50 kcal/mol) associated with bringing the two phospholipid bilayers into proximity and introducing local membrane deformations required for initial lipid mixing events [1, 2, 3, 4, 5]. In general, viral glycoproteins adopt at least three distinct conformations during fusion: (i) prefusion; (ii) extended intermediate, or prehairpin; and (iii) post-fusion; various factors, host or environmental, induce the transition from one conformation to the next (Figure 1a). The prefusion conformations have been visualized in atomic detail with soluble versions of the glycoprotein for many viruses, and in lower resolution with virus- or membrane-bound glycoproteins for a few [6, 7, 8]. Many post-fusion X-ray structures have been reported for the ectodomain segments (e.g., Figure 1b), but none are available for full-length membrane- or detergent-bound glycoproteins. Several studies have provided rough geometric or topological features of the extended intermediate [9, 10, 11], but neither low- or high-resolution structures exist of the extended intermediate for any virus. Agents (small molecules, peptides, or antibodies) that bind to the prefusion and extended intermediate conformations inhibit viral entry (Figure 1a) [3, 12], and therefore atomic details of membrane fusion-associated conformations are of both basic and applied significance.
Figure 1.
General mechanism of viral membrane fusion and its inhibition for “class I” (α-helical) glycoproteins. The NHR and CHR are indicated and colored cyan or dark blue in both panels. (A) SU, surface subunit (GP1 in MARV); TM transmembrane subunit (GP2 in MARV), FP/FL, fusion peptide/fusion loop. (B) Post-fusion structures of ectodomains from HIV-1 gp41, EBOV GP2, and MARV GP2.
Marburg virus (MARV) is a member of the family Filovirdae of negative-stranded RNA viruses (“filoviruses”) that includes the ebolaviruses [13]. Infection by filoviruses causes a severe and rapidly progressing hemorrhagic fever with human case fatality rates of 30–90%. The envelope glycoproteins of filoviruses belong to the structurally defined “class I” category because the fusion subunit ectodomains contain a high α-helical content [14, 15, 16, 17, 18]. The prefusion glycoprotein (GP) assembly consists of three copies each of the surface subunit (GP1) and the fusion (transmembrane) subunit (GP2) that anchors the prefusion spike into the viral membrane [19, 20]. The proposed fusion pathway for filoviruses is as follows. The first step is attachment and uptake of the viral particle into host endosomal compartments, where host proteases remove the variable and glycosylated segments (mucin-like domain and glycan cap) of GP1 to expose a highly conserved receptor binding region (RBR) [21]. The RBR engages the lumenal C-domain of the host receptor Neimann Pick C1 (NPC1) to activate the fusion pathway [22]. NPC1 is both necessary and sufficient for viral infection, but it is possible that other host receptors are required [23]. Next, the fusion loop (FL) of GP2 is thought to extend into the host endosomal membrane, creating the extended intermediate that spans both the virus and host. This exposes the GP2 ectodomain N- and C-heptad repeat regions (NHR and CHR), which fold into a six-helix bundle that is characteristic of class I fusion proteins [14, 15].
It is postulated for MARV and other class I viruses that the energy supplied by folding of the six-helix bundle provides the driving force for overcoming initial barriers associated with membrane fusion [1, 2, 3, 4, 5]. Indeed, our group has previously determined that the MARV GP2 six-helix bundle folding stability can be as high as ~33.5 kcal/mol, and that this high stability is pH-dependent [14, 15]. Similar features have been described for the GP2 six-helix bundles of EBOV, CAS Virus (CASV), and avian leukosis sarcoma virus (ASLV) [24, 25, 26]. At physiological pH, the MARV GP2 six-helix bundle unfolds noncooperatively with a TM of 56 °C, but at pH 4–5 this TM increases to 98 °C [14]. We and others have demonstrated that the filovirus FLs also undergo pH-dependent conformational changes that induce lipid perturbing activity [27, 28, 29]. Overall, these results suggest that inherent pH-dependent structural preferences of the GP2 FL and ectodomain act as a mechanism to control fusion-active conformations until the virion is embedded deep within the endocytic pathway.
Although the structural characterizations of the GP2 FL and ectodomain have provided novel information about fusion-associated conformational rearrangements, a fundamental limitation of such studies is that they do not consider how the ectodomains or membrane-associated segments may behave in concert with one another. Recently, Tamm and coworkers reported NMR studies in detergent micelles and lipid bicelles for the EBOV GP2 FL and MPER/TM regions that indicated direct interaction between the FL and MPER in lipid environments, suggesting interplay of N- and C-terminal segments within the membrane is required for fusion events [30]. Furthermore, recent studies with HIV-1 gp41-based constructs containing the fusion peptide and transmembrane domain in membrane-mimicking environments have suggested that the fusion subunit is not exclusively trimeric, and that monomeric conformations that span both host and viral membranes may lie on the fusion pathway [31, 32, 33, 34, 35]. Single particle fusion assays with influenza virus-like particles (VLPs) indicate that there is a significant time delay (potentially on the order of ~10 s) between hemifusion, a stalk-like membrane state where the two outer leaflets are merged but the inner two leaflets are not, and formation of a full fusion pore [36]. Likely, the six-helix bundle promotes hemifusion by bringing the viral and host membranes together, but it is hypothesized that the membrane-associated segments are involved in development of the fusion pore. The decoupled kinetic parameters of hemifusion and formation of the fusion pore suggest that another set of conformational changes, perhaps involving interactions among the fusion peptide or loop with the transmembrane segment, may be required for late-stage fusion events.
Here we describe studies on the full-length MARV GP2 (containing both the FL and MPER/TM regions), and on the N- and C-terminal fragments in membrane-mimicking environments. We find that the full-length GP2 subunit exhibits pH-dependent membrane perturbing activity, and that the structure of the N-terminal fragment differs alone or in the context of the other GP2 segments. Furthermore, similar to recent reports on HIV-1 gp41, we find that the full-length GP2 as well as the ectodomain dissociates into a monomeric state in membrane-like environments. These results provide new information about the properties of the membrane-associated segments from GP2 and how they may be involved in the fusion cascade.
2. Materials and methods
2.1. Protein expression and purification
Synthetic DNA fragments encoding MARV full length GP2 with N′-terminal His-tag (MGP2-full), FL-NHR-loop (MGP2-FNL), and CHR-MPER-TM (MGP2-CMT) with N′-terminal His-tag were obtained from a commercial supplier (Genewiz, South Plainfield, NJ). The genes were cloned into pET-22b(+) vectors (Novagen, Madison, WI) using NdeI and XhoI restriction sites, so that an extra His tag was encoded at the C-terminus of the coding sequence. The correct sequences of the constructed plasmids were confirmed by DNA sequencing (Genewiz, South Plainfield, NJ).
All MARV GP2 proteins (including MGP2-full, MGP2-FNL and MGP2-CMT) were expressed in E. coli strain BL21(DE3). The cells were induced with 0.2mM isopropyl-β-D-thiogalactopyranoside (IPTG) at an A600 of 1.0 and harvested by centrifugation after shaking for 14–18 h at 22 °C. The cell pellet was lysed with Bugbuster (Merck Millipore) supplemented with Deoxyribonuclease I (Invitrogen) in PBS buffer, pH 7.0 and the insoluble fraction, which contains MARV GP2 proteins as inclusion bodies, was isolated by centrifugation (11,000rpm for 20min) and washed twice with PBS buffer. The insoluble fraction was then solubilized with 8M urea/0.2% SDS in PBS. The solubilized proteins were purified via Ni-NTA (Ni-Nitrilotriacetic acid agarose, QIAGEN) column chromatography, and refolded on-resin by exchanging with 20 mM sodium phosphate buffer with 0.05% n-dodecylphosphocholine (DPC, Avanti Polar Lipids) at pH 7.0, and then eluted with 20 mM sodium phosphate buffer with 0.05% DPC, 250 mM imidazole at pH 7.0 or in 10 mM sodium acetate buffer with 0.05% DPC, 250 mM imidazole at pH 4.0. Final purification was performed by gel filtration chromatography on PD-10 column (GE Healthcare) equilibrated with DPC to remove imidazole. The purified proteins were then reconstituted into 10 mM sodium acetate, pH 4 with 1% isotropic bicelles (q = 0.33) 1,2-dimyristoyl-sn-glycero-3-phosphocholine/1,2-dihexanoyl-sn-glycero-3-phosphocholine (DMPC/DHPC, Avanti Polar Lipids) with a PD MiniTrap G-25column (GE Healthcare).
2.2. Circular dichroism
Circular dichroism (CD) spectra of MARV GP2 constructs were recorded at room temperature on a Jasco J-815 spectrometer with a 1 cmpath-length cuvette. Protein concentrations were determined by the absorbance at 280 nm. Full wavelength spectra were obtained with a 0.5 nm step size and represent the average of two scans. The signal was converted to mean molar ellipticity (θ) using the equation: θ (in deg cm2 dmol−1) = millidegrees/(pathlength in millimeters × the molar protein concentration × the number of residues).
2.3. Sedimentation velocity-analytical ultracentrifugation (SV-AUC)
A sedimentation velocity study of MGP2-full was conducted using the absorption optics of a Beckman Optima XL-I analytical ultracentrifuge with samples loaded into two-sector cell assemblies run in the AN-60Ti rotor. Boundary movement was followed at 280 nm or 295 nm during centrifugation at 58,000 rpm and 20 °C in buffer containing 10 mM sodium acetate, 150 mM NaCl with 0.5% DPC or 1% bicelles at pH4. D2O was used to density match the DPC or bicelles present in the buffer [37, 38, 39]. Sixty to 70 scans were collected over the course of the sedimentation runs of which a subset, beginning with those where a clear plateau is evident between the meniscus and the boundary, was selected for time-derivative analysis using DCDT + version 2.4.2 [40,41]. The data presented were fit to either single or two non-interacting species models floating S (sedimentation coefficient), Mw (molecular weight, or S/D where D is the diffusion coefficient), protein concentration, and a baseline offset. The 68.3% joint confidence limits are reported. The MGP2-full was analyzed at a concentration of 22 μM in micelle, and at a concentration of 11–110 μM in bicelles. The corresponding buffer was used to blank each sample. Values of the buffer density and viscosity were calculated from the composition (including D2O but neglecting detergent) using Sedenterp version 20120828 Beta (University of New Hampshire). The partial specific volume of MGP2-full was calculated from its sequence using Sedenterp. The presented sedimentation parameters were corrected to standard conditions (20,w) using these values.
2.4. Liposome release assay
To prepare large unilamellar vesicles (LUVs) with encapsulated fluorescent dye, lipid dispersions of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine/1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPC/POPG, 4:1, molar ratio) were resuspended with HEPES buffer containing fluorescent dye 8-aminonapthalene-1,3,6 trisulfonic acid (ANTS) and its quencher p-xylene-bis-pyridinium bromide (DPX). Ten cycles of freeze/thaw and 11 rounds of extrusion with 100 nm membrane were performed to generate homogenous large unilamellar vesicles. Next, a 10 mL column Sepharose CL-2B resin was used to isolated liposomes with ANTS/DPX for the following assays. Lipid concentration was determined using an organic phosphate assay [42]. An Infinite M1000 PRO plate reader was used to monitor liposomal content release induced by fusion loop constructs. Excitation was at 355 nm with an 8 nm slit width, and emission was monitored at 520 nm with a 12 nm slit width. Varying concentrations of MGP2 (MGP2-full, MGP2-FNL and MGP2-CMT) were added to ANTS/DPX-encapsulated liposomes to test for their fusion activity. Buffer with 0.005% DPC was used as a baseline (0% content release, in the absence of protein), while buffer with 0.3% triton was used as 100% content release.
3. Results
3.1. Protein design, expression, and purification
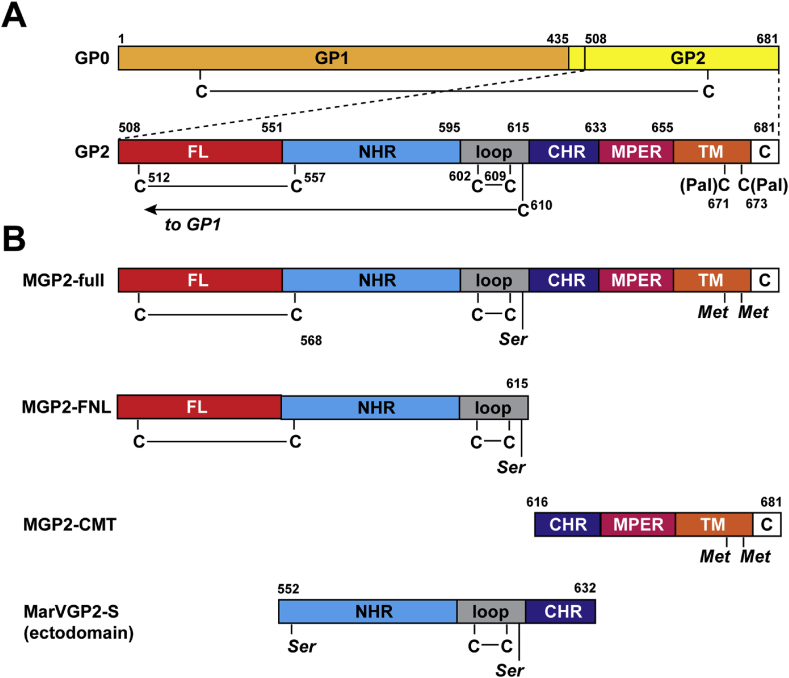
The constructs studied here are shown in Figure 2. The prefusion MARV GP spike is produced as a precursor, GP0, that is cleaved by furin upon maturation to produce the two subunits, GP1 (residues 1–435) and GP2 (436–681) (Figure 2a) [43]. In the prefusion form, GP2 contains an internal disulfide bond that stabilizes the fusion loop, which encompasses residues 508–551. Another disulfide bond is found in the loop region between the NHR and CHR. A third disulfide bond is formed with the GP1 subunit. The MPER is defined from residues 634 to 655, and the TM domain and short (4-residue) C-terminal tail encompasses residues 656–681.
Figure 2.
(A) Schematic of the MARV GP0 precursor and GP2 subunit. Cysteines are indicated by a ‘C’ with disulfide bonding or palmitoylation patterns indicated. (B) Construct design for MGP2-full, MGP2-FNL, MGP2-CMT, and MarVGP2-S (ectodomain).
The protein “MGP2-full” consists of MARV GP2 from the beginning of the FL through the ectodomain MPER, TM, and C-terminal tail. Two palmitoylated cysteines at the border of the TM and C-terminal tail have been mutated to methionine. Previous work has demonstrated that these palmitoylation sites are important for trafficking during production of the glycoprotein precursor but are expendable for cell entry in both the authentic MARV virus and virus-like particles (VLPs) containing EBOV GP [44, 45]. In addition, C610 of the ectodomain, which normally forms an intramolecular disulfide bond to the GP1 subunit, is mutated to Ser, as was done for the post-fusion X-ray structure of the isolated ectodomain [15]. Two fragments corresponding to the N- and C-terminal fragments of GP2 including the membrane segments were also studied: “MGP2-FNL” contains the FL, NHR and the loop preceding the CHR (including the C602–C609 disulfide bond), and “MGP2-CMT” contains the CHR, MPER, TM regions as well as the C-terminal tail (“MGP2-CMT”). All constructs expressed readily in E. coli but were rescued from inclusion andrequired addition of detergent micelles or lipid bicelles to stabilize against aggregation.
3.2. Characterization of MGP2-full
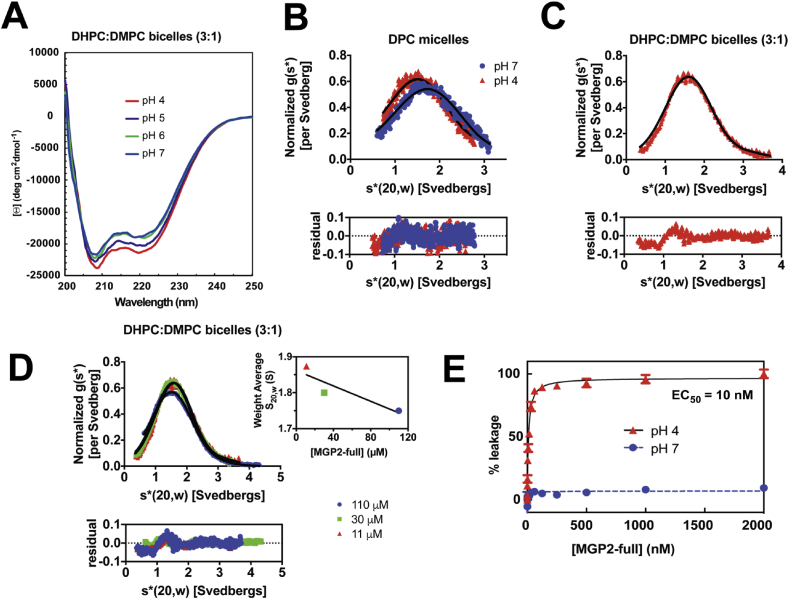
To explore the structural and functional characteristics of the MARV GP2 membrane-associated segments in the context of the fusion subunit, studies were undertaken in 3:1 DHPC:DMPC solution bicelles or DPC micelles. Circular dichroism (CD) spectra indicate that MGP2-full is α-helical in bicelles as indicated by double minima at 208 nm and 222 nm (Figure 3a). Furthermore, the MGP2-full CD signatures are invariant across pH range 4–7, in contrast to previously reported behavior of both the isolated FL (“MGP2-FL”) and ectodomain (“MarVGP2-S″) [14, 15, 29]. Ellman's test on MGP2-full indicated there were no unbounded cysteine sulfhydryl groups, confirming disulfide bond formation within the fusion loop and ectodomain (not shown).
Figure 3.
Characterization of MGP2-full. (A) CD spectra of 1.7μM MGP2-full at pHs ranging from 4 to 7, in10 mM sodium acetate (pH 4 and 5), or 20 mM sodium phosphate (pH 6 and 7) containing 0.5 % DHPC:DMPC (3:1) bicelles. B) Density-matched SV- AUC of MGP2-full at 11 μM at pH 4 and 7 in 10 mM sodium acetate, 150mM NaCl containing 0.5 % DPC. All experiments were fit to a single component model with the baseline offset floated (fit shown as solid line). (C) Density-matched SV-AUC of MGP2-full at 11 μM at pH 4 in 10 mM sodium acetate, 150mM NaCl containing DHPC:DMPC (3:1) bicelles. (D) Similar SV-AUC studies and analysis of MGP2-full at multiple concentrations (11 μM, 30 μM and 110 μM) in at pH 4 10 mM sodium acetate, 150 mM NaCl containing DHPC:DMPC (3:1) bicelles. The insert plots the protein concentration dependence of the sedimentation coefficient. (E) Liposomal release assay of MGP2-full in 10 mM sodium acetate, 100mM NaCl, Zn2+ at pH 4 or 10 mM sodium phosphate, 100mM NaCl, Zn2+ at pH 7.
SV-AUC experiments were conducted at 22 μM MGP2-full at pH 4 and pH 7 in DPC micelles. Both data sets are described as a single species (Figure 3b). S20,W and Mw values of 1.62 (1.61, 1.62) S and 15.6 (15.1, 16.1) kDa, and 1.86 (1.85,1.87) S and 14.0 (13.5–14.5) kDa were determined at pH 4 and 7, respectively. These values are less than the Mw calculated from sequence (22.7 kDa) and thus consistent with an absence of oligomerization. SV-AUC experiments conducted in lipid bicelles at pH 4 were best fit to a two-component non-interacting monomer-trimer model (Figure 3c). The trimer comprised less than 5% of the total protein. The best fit values of S20,W and Mw for the monomeric component are 1.68 (1.67, 1.69) S and 22.4 (21.9–22.9) kDa at 11 μM, 1.65 (1.65, 1.66) S and 20.8 (20.5, 21.1) kDa at 30 μM, and 1.63 (1.62, 1.64) S and 21.8 (21.0–22.0) kDa at 110 μM. The fact that S decreases with protein concentration (Figure 3d, insert) and the close agreement of the resolved values of Mw with that calculated from the peptide sequence show that MGP2-full is monomeric in lipid bicelles and not undergoing reversible assembly at the protein concentrations analyzed.
The monomeric nature of the full fusion subunit in DPC micelles or solution bicelles is perhaps surprising given that the ectodomain of MARV, EBOV, and all other class I viral glycoproteins have been found to form trimers in solutionv [3, 14, 15, 17, 18, 25, 46, 47]. However, we and others have reported that HIV-1 gp41 constructs containing the membrane-embedded segments and portions of the ectodomain tend to dissociate from trimeric states to monomers in lipid or detergent environments [31, 32, 33, 34, 35]. Furthermore, Bax and coworkers have demonstrated that the HIV-1 gp41 ectodomain forms a monomeric state in the presence of DPC micelles [31]. We found similarly that the MARV GP2 ectodomain (“MarVGP2-S″), previously shown to be trimer in solution, also dissociated to a monomer in the presence of DPC micelles (Figure 4). These data are described as a single species with S20,W = 1.08 (1.07, 1.09) S and Mw = 12.3 (11.9, 12.7) kDa. The Mw calculated from the peptide sequence is 11.1 kDa. The results obtained herein with MARV GP2 are consistent with recent data in HIV-1 systems suggesting that the fusion subunit may dissociate from the trimeric form in lipid environments.
Figure 4.
Sedimentation velocity analysis of MarVGP2-S, corresponding to the ectodomain, in 10 mM sodium acetate (pH 4), 150 m M NaCl containing 0.5 % DPC. Data were fit to a single component model (solid line).
The ability of MGP2-full to disrupt membranes was explored with a liposomal release assay (Figure 3e). Disruption of vesicles consisting of 4:1 POPC:POPG by MGP2-full was found to be concentration- and pH-dependent, with an EC50 of 10 nM at pH 4 and no activity at pH 7. We previously reported similar behavior for a construct corresponding to the MGP2-FL alone [29].
3.3. Characterization of MGP2-FNL and MGP2-CMT
The conformational and lytic properties of the N- and C-terminal fragments (MGP2-FNL and MGP2-CMT) were also explored. Both segments were found to be α-helical over a range of pH conditions in DPC micelles, and less structured in lipid bicelles (Figure 5). The degree of α-helicity for both peptides was stronger in DPC micelles than lipid bicelles, and some pH effects on CD spectra were observed in lipid bicelles. Similar to MGP2-FL and MGP2-full, both MGP2-FNL and MGP2-CMT exhibited pH-dependent liposomal content release (Figure 6). The relative activity of each half segment was attenuated in terms of EC50 values (110 nM for MGP2-FNL, and 700 nM for MGP2-CMT) in comparison to MGP2-full. Moreover, MGP2-CMT maximally resulted in ~70 % liposomal release at the highest concentration tested (2000 nM). When the two half segments (MGP2-FNL and MGP2-CMT) were mixed in a 1:1 ratio, the overall liposomal release was similar to MGP2-FNL with an EC50 of 160 nM.
Figure 5.
CD spectra of 1 μM MGP2-FNL (A) and 2 μM MGP2-CMT (B) in 10 mM sodium acetate at pH 4 and 5, and 20 mM sodium phosphate at pH 6 and pH7with 0.5 % DPC micelles or 0.5 % DHPC:DMPC (3:1) bicelles.
Figure 6.
Liposomal release assay of MGP2-FNL and MGP2-CMT alone (A and B, respectively) or as a 1:1 mixture (C) in 10 mM sodium acetate, 100mM NaCl, Zn2+ at pH 4 or 10 mM sodium phosphate, 100mM NaCl, Zn2+ at pH 7.
4. Discussion
The high-resolution X-ray structures of numerous class I viral fusion proteins in their prefusion and/or post-fusion conformations have been described, but relatively little structural information is available for intermediary conformations along the fusion pathway. The studies presented here on MGP2-full, which contains elements of the full-length GP2 subunit including the membrane-associated FL and MPER/TM, suggest that MARV GP2 is primarily α-helical but monomeric in lipid-mimicking environments. Furthermore, MGP2-full remains highly α-helical from pH 4 to 7 with no change in the CD spectra over this range, in contrast to our previous reports on the ectodomain where there were differences in the intensity ratio of the 208 nm and 222 nm peaks, at pH 5.2 and pH 7, suggesting differences in interhelical packing arrangements over this range. Despite the invariant CD spectra over this range, MGP2-full does exhibit pH-dependent membrane-perturbing characteristics in the liposomal release assay. Similar characteristics were observed with MGP2-FNL and MGP2-CMT, although the liposomal release was somewhat attenuated relative the MGP2-full, suggesting that each N- and C-terminal segment on its own has the capacity to form α-helical structure with membrane lytic activity alone.
The observation that MGP2-full as well as the MARV GP2 ectodomain sediment as monomers by sedimentation velocity analytical ultracentrifugation in lipid-mimicking environments is consistent with parallel studies on segments of HIV-1 gp41 ectodomain and on gp41 variants that contain the fusion peptide (FP) and FP proximal region (FPPR) as well as the MPER/TM. In multiple reports, such constructs have exhibited tendencies form monomers upon addition of detergent or lipids, despite the exceptional stability of trimeric α-helical bundles containing NHR/CHR segments in aqueous solutions [31, 32, 33, 34, 35]. It has been postulated that such monomeric forms of the fusion subunit (gp41 in HIV-1 and, here, GP2 in MARV) may represent intermediates along the fusion pathway that span both the host and viral membranes. However, such observations may also be dependent on the detergent or lipid mimics, as DPC micelles in particular have been shown to elicit drastically different conformations than other lipid mimicking environments in several systems [48].
We previously reported that the MARV GP2 ectodomain 6-helix bundle exhibits pH-dependent stability and proposed that this pH-dependent stability reflects the endosomal pathway for membrane fusion [14, 15]. Since the 6-helix bundle is reflective of the post-fusion conformation, where the N- and C-terminal membrane segments are in proximity, this α-helical bundle is promoted under conditions of appropriately matured cellular compartments where fusion is known to occur. Consistent with this hypothesis, we demonstrated previously that the MARV GP2 FL segment exhibits pH-dependent lytic activity and here we report similar activity for MGP2-full, MGP2-FNL, and MGP2-CMT [29]. Arguably, this confers an advantage to the virus for immune evasion, in that the most critical regions of the fusion protein required for membrane fusion are exposed only in host endosomal compartments, where they would be sheltered from recognition by antibodies or other immune molecules. pH-dependent conformational changes have been described for a number of other class I viruses, most notably influenza HA2, but perhaps a unique aspect of the filovirus and related fusion subunits is the plurality of endosomal requirements [5, 16]. The host cysteine proteases that are required to expose the RBR are resident in the endosome and have stronger activity at low pH; and the receptor itself (NPC1) is sequestered into late endosomes. The low pH dependence of the fusion subunit is also distributed in nature, because it affects both the ectodomain and the FL. However, the fact that the CD spectra for MGP2-full are highly α-helical and invariant over a broad pH range suggests that secondary structural components are less sensitive to pH in lipid environments. How precisely the pH-dependence of the ectodomain 6-helix bundle, and lytic activities of the membrane-associated segments (FL and MPER/TM) are interconnected to promote fusion under the optimal conditions remains the subject of further study.
Single particle studies on the influenza HA fusion machinery suggests there is a significant time lag between hemifusion and formation of a full fusion pore [36]. While formation of the 6-helix bundle by the ectodomain NHR and CHR segments may bring the N- and C-termini into proximity to allow initial lipid mixing events between the outer leaflets of the host and virus. The precise events that lead to formation of a full fusion pore have not been visualized in atomic detail, but, as shown here for MARV GP2 and in other systems, the N-terminal fusion loop/peptide and MPER/TM segments of many class I systems typically have membrane-perturbing properties [29, 35, 49, 50, 51]. Furthermore, it has recently been reported that the FL and MPER of EBOV GP2 interact directly in membrane environments, and that the side chains involved in this direct interaction are required for entry of VLPs [30]. Potentially, the lytic activities of either the FL and MPER/TM segments in concert promotes mixture of the inner leaflets from host and viral membranes, a key step for initiation of a fusion pore. While a direct interaction between the FL and MPER/TM in membranes is critical for EBOV GP2, additional high resolution studies would be required to determine if a similar interaction occurs in MARV GP2.
In summary, we have found that segments of the MARV GP2 fusion subunit containing the FL and MPER/TM are α-helical and monomeric in lipid-mimicking environments. Furthermore, all components exhibit pH-dependent membrane perturbing properties. These studies provide novel insights into the mechanism of viral membrane fusion by MARV GP2.
Declarations
Author contribution statement
J. Lai: Conceived and designed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
N. Liu and M. Brenowitz: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
M. Girvin: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the National Institutes of Health (R01-AI125462) and the Albert Einstein College of Medicine.J. Lai was partially supported by an Irma T. Hirschl/Monique Weill-Caulier Career Scientist Award.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- 1.Harrison S.C. Mechanism of membrane fusion by viral envelope proteins. Adv. Virus Res. 2005;64:231–261. doi: 10.1016/S0065-3527(05)64007-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison S.C. Viral membrane fusion. Nat. Struct. Mol. Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eckert D.M., Kim P.S. Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 2001;70:777–810. doi: 10.1146/annurev.biochem.70.1.777. [DOI] [PubMed] [Google Scholar]
- 4.White J.M., Delos S.E., Brecher M., Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit. Rev. Biochem. Mol. Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harrison J.S., Higgins C.D., O’Meara M.J., Koellhoffer J.F., Kuhlman B.A., Lai J.R. Role of electrostatic repulsion in controlling pH-dependent conformational changes of viral fusion proteins. Structure. 2013;21:1085–1096. doi: 10.1016/j.str.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhu P., Winkler H., Chertova E., Taylor K.A., Roux K.H. Cryoelectron tomography of HIV-1 envelope spikes: further evidence for tripod-like legs. PLoS Pathog. 2008;4 doi: 10.1371/journal.ppat.1000203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zanetti G., Briggs J., Grunewald K., Sattentau Q.J., Fuller S.D. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog. 2006;2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cao S., Zhang W. Characterization of an early-stage fusion intermediate of Sindbis virus using cryoelectron microscopy. Proc. Natl. Acad. Sci. 2013;110:13362–13367. doi: 10.1073/pnas.1301911110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y.H., Donald J.E., Grigoryan G., Leser G.P., Fadeev A.Y., Lamb R.A., DeGrado W.F. Capture and imaging of a prehairpin fusion intermediate of the paramyxovirus PIV5. Proc. Natl. Acad. Sci. 2011;108:20992–20997. doi: 10.1073/pnas.1116034108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cardone G., Brecher M., Fontana J., Winkler D.C., Butan C., White J.M., Steven A.C. Visualization of the two-step fusion process of the retrovirus avian sarcoma/leukosis virus by cryo-electron tomography. J. Virol. 2012;86:12129–12137. doi: 10.1128/JVI.01880-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matsuyama S., Delos S.E., White J.M. Sequential roles of receptor binding and low pH in forming prehairpin and hairpin conformations of a retroviral envelope glycoprotein. J. Virol. 2004;78:8201–8209. doi: 10.1128/JVI.78.15.8201-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Porotto M., Yokoyama C.C., Palermo L.M., Mungall B., Aljofan M., Cortese R., Pessi A., Moscona A. Viral entry inhibitors targeted to the membrane site of action. J. Virol. 2010;84:6760–6768. doi: 10.1128/JVI.00135-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emanuel J., Marzi A., Feldmann H. Filoviruses: ecology, molecular biology, and evolution. Adv. Virus Res. 2018;100:189–221. doi: 10.1016/bs.aivir.2017.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harrison J.S., Koellhoffer J.F., Chandran K., Lai J.R. Marburg virus glycoprotein GP2: pH-dependent stability of the ectodomain α-helical bundle. Biochemistry. 2012;51:2515–2525. doi: 10.1021/bi3000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koellhoffer J.F., Malashkevich V.N., Harrison J.S., Toro R., Bhosle R.C., Chandran K., Almo S.C., Lai J.R. Crystal structure of the Marburg virus GP2 core domain in its postfusion conformation. Biochemistry. 2012;51:7665–7675. doi: 10.1021/bi300976m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller E.H., Chandran K. Filovirus entry into cells–new insights. Current opinion in virology. 2012;2:206–214. doi: 10.1016/j.coviro.2012.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weissenhorn W., Carfí A., Lee K.-H., Skehel J.J., Wiley D.C. Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell. 1998;2:605–616. doi: 10.1016/s1097-2765(00)80159-8. [DOI] [PubMed] [Google Scholar]
- 18.Malashkevich V.N., Schneider B.J., McNally M.L., Milhollen M.A., Pang J.X., Kim P.S. Core structure of the envelope glycoprotein GP2 from Ebola virus at 1.9-Å resolution. Proc. Natl. Acad. Sci. 1999;96:2662–2667. doi: 10.1073/pnas.96.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee J.E., Fusco M.L., Hessell A.J., Oswald W.B., Burton D.R., Saphire E.O. Structure of the Ebola virus glycoprotein bound to an antibody from a human survivor. Nature. 2008;454:177–182. doi: 10.1038/nature07082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J.E., Saphire E.O. Ebolavirus glycoprotein structure and mechanism of entry. Future Virol. 2009;4:621–635. doi: 10.2217/fvl.09.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandran K., Sullivan N.J., Felbor U., Whelan S.P., Cunningham J.M. Endosomal proteolysis of the Ebola virus glycoprotein is necessary for infection. Science (New York, N.Y.) 2005;308:1643–1645. doi: 10.1126/science.1110656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Carette J.E., Raaben M., Wong A.C., Herbert A.S., Obernosterer G., Mulherkar N., Kuehne A.I., Kranzusch P.J., Griffin A.M., Ruthel G., Dal Cin P., Dye J.M., Whelan S.P., Chandran K., Brummelkamp T.R. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller E.H., Obernosterer G., Raaben M., Herbert A.S., Deffieu M.S., Krishnan A., Ndungo E., Sandesara R.G., Carette J.E., Kuehne A.I., Ruthel G., Pfeffer S.R., Dye J.M., Whelan S.P., Brummelkamp T.R., Chandran K. Ebola Virus Entry Requires the Host-programmed Recognition of an Intracellular Receptor. EMBO J. 2012 doi: 10.1038/emboj.2012.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrison J.S., Higgins C.D., Chandran K., Lai J.R. Designed protein mimics of the Ebola virus glycoprotein GP2 α-helical bundle: stability and pH effects. Protein Sci. 2011;20:1587–1596. doi: 10.1002/pro.688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koellhoffer J.F., Dai Z., Malashkevich V.N., Stenglein M.D., Liu Y., Toro R., J S.H., Chandran K., DeRisi J.L., Almo S.C., Lai J.R. Structural characterization of the glycoprotein GP2 core domain from the CAS virus, a novel arenavirus-like species. J. Mol. Biol. 2014;426:1452–1468. doi: 10.1016/j.jmb.2013.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aydin H., Smrke B.M., Lee J.E. Structural characterization of a fusion glycoprotein from a retrovirus that undergoes a hybrid 2-step entry mechanism. FASEB J. : off. publ of the Fed Am Soc. Exp Biol. 2013;27:5059–5071. doi: 10.1096/fj.13-232371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gregory S.M., Harada E., Liang B., Delos S.E., White J.M., Tamm L.K. Structure and function of the complete internal fusion loop from Ebolavirus glycoprotein 2. Proc. Natl. Acad. Sci. 2011;108:11211–11216. doi: 10.1073/pnas.1104760108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gregory S.M., Larsson P., Nelson E.A., Kasson P.M., White J.M., Tamm L.K. Ebolavirus entry requires a compact hydrophobic fist at the tip of the fusion loop. J. Virol. 2014;88:6636–6649. doi: 10.1128/JVI.00396-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu N., Tao Y., Brenowitz M.D., Girvin M.E., Lai J.R. Structural and functional studies on the Marburg virus GP2 fusion loop. JID (J. Infect. Dis.) 2015 doi: 10.1093/infdis/jiv030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee J., Nyenhuis D.A., Nelson E.A., Cafiso D.S., White J.M., Tamm L.K. Structure of the Ebola virus envelope protein MPER/TM domain and its interaction with the fusion loop explains their fusion activity. Proc. Natl. Acad. Sci. U. S. A. 2017;114:E7987–e7996. doi: 10.1073/pnas.1708052114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roche J., Louis J.M., Grishaev A., Ying J., Bax A. Dissociation of the trimeric gp41 ectodomain at the lipid–water interface suggests an active role in HIV-1 Env-mediated membrane fusion. Proc. Natl. Acad. Sci. 2014;111:3425–3430. doi: 10.1073/pnas.1401397111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Louis J.M., Baber J.L., Ghirlando R., Aniana A., Bax A., Roche J. Insights into the conformation of the membrane proximal regions critical to the trimerization of the HIV-1 gp41 ectodomain bound to dodecyl phosphocholine micelles. PLoS One. 2016;11 doi: 10.1371/journal.pone.0160597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dai Z., Tao Y., Liu N., Brenowitz M.D., Girvin M.E., Lai J.R. Conditional trimerization and lytic activity of HIV-1 gp41 variants containing the membrane-associated segments. Biochemistry. 2015;54:1589–1599. doi: 10.1021/bi501376f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Roche J., Louis J.M., Aniana A., Ghirlando R., Bax A. Complete dissociation of the HIV-1 gp41 ectodomain and membrane proximal regions upon phospholipid binding. J. Biomol. NMR. 2015;61:235–248. doi: 10.1007/s10858-015-9900-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banerjee K., Weliky D.P. Folded monomers and hexamers of the ectodomain of the HIV gp41 membrane fusion protein: potential roles in fusion and synergy between the fusion peptide, hairpin, and membrane-proximal external region. Biochemistry. 2014;53:7184–7198. doi: 10.1021/bi501159w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Floyd D.L., Ragains J.R., Skehel J.J., Harrison S.C., van Oijen A.M. Single-particle kinetics of influenza virus membrane fusion. Proc. Natl. Acad. Sci. 2008;105:15382–15387. doi: 10.1073/pnas.0807771105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanford C., Nozaki Y., Reynolds J.A., Makino S. Molecular characterization of proteins in detergent solutions. Biochemistry. 1974;13:2369–2376. doi: 10.1021/bi00708a021. [DOI] [PubMed] [Google Scholar]
- 38.Tanford C., Reynolds J.A. Characterization of membrane proteins in detergent solutions. Biochim. Biophys. Acta. 1976;457:133–170. doi: 10.1016/0304-4157(76)90009-5. [DOI] [PubMed] [Google Scholar]
- 39.Korendovych I.V., Senes A., Kim Y.H., Lear J.D., Fry H.C., Therien M.J., Blasie J.K., Walker F.A., Degrado W.F. De novo design and molecular assembly of a transmembrane diporphyrin-binding protein complex. J. Am. Chem. Soc. 2010;132:15516–15518. doi: 10.1021/ja107487b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Philo J.S. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal. Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 41.Stafford W.F. Boundary analysis in sedimentation transport experiments: a procedure for obtaining sedimentation coefficient distributions using the time derivative of the concentration profile. Anal. Biochem. 1992;203:295–301. doi: 10.1016/0003-2697(92)90316-y. [DOI] [PubMed] [Google Scholar]
- 42.Ames B.N. Assay of inorganic phosphate, total phosphate and phosphatases. Methods Enzymol. 1966;8:115–118. [Google Scholar]
- 43.Flyak A.I., Ilinykh P.A., Murin C.D., Garron T., Shen X., Fusco M.L., Hashiguchi T., Bornholdt Z.A., Slaughter J.C., Sapparapu G., Klages C., Ksiazek T.G., Ward A.B., Saphire E.O., Bukreyev A., Crowe J.E., Jr. Mechanism of human antibody-mediated neutralization of Marburg virus. Cell. 2015;160:893–903. doi: 10.1016/j.cell.2015.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Funke C., Becker S., Dartsch H., Klenk H.-D., Mühlberger E. Acylation of the Marburg virus glycoprotein. Virology. 1995;208:289–297. doi: 10.1006/viro.1995.1151. [DOI] [PubMed] [Google Scholar]
- 45.Ito H., Watanabe S., Takada A., Kawaoka Y. Ebola virus glycoprotein: proteolytic processing, acylation, cell tropism, and detection of neutralizing antibodies. J. Virol. 2001;75:1576–1580. doi: 10.1128/JVI.75.3.1576-1580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weissenhorn W., Dessen A., Harrison S., Skehel J., Wiley D. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 47.Chan D.C., Fass D., Berger J.M., Kim P.S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 48.Chipot C., Dehez F., Schnell J.R., Zitzmann N., Pebay-Peyroula E., Catoire L.J., Miroux B., Kunji E.R.S., Veglia G., Cross T.A., Schanda P. Perturbations of native membrane protein structure in alkyl phosphocholine detergents: a critical assessment of NMR and biophysical studies. Chem. Rev. 2018;118:3559–3607. doi: 10.1021/acs.chemrev.7b00570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Regula L.K., Harris R., Wang F., Higgins C.D., Koellhoffer J.F., Zhao Y., Chandran K., Gao J., Girvin M.E., Lai J.R. Conformational properties of peptides corresponding to the ebolavirus GP2 membrane-proximal external region in the presence of micelle-forming surfactants and lipids. Biochemistry. 2013;52 doi: 10.1021/bi400040v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yao H., Hong M. Membrane-dependent conformation, dynamics, and lipid interactions of the fusion peptide of the paramyxovirus PIV5 from solid-state NMR. J. Mol. Biol. 2013;425:563–576. doi: 10.1016/j.jmb.2012.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tristram-Nagle S., Chan R., Kooijman E., Uppamoochikkal P., Qiang W., Weliky D.P., Nagle J.F. HIV fusion peptide penetrates, disorders, and softens T-cell membrane mimics. J. Mol. Biol. 2010;402:139–153. doi: 10.1016/j.jmb.2010.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]