Abstract
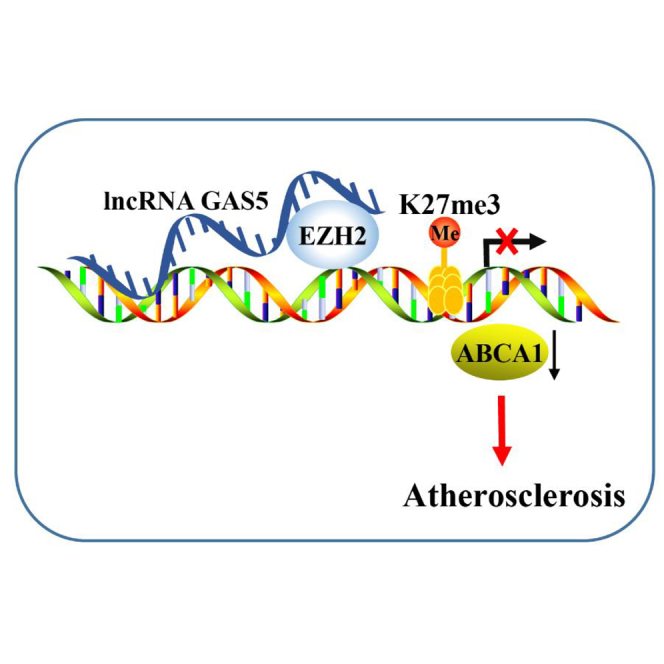
Atherosclerosis is a disorder occurring in the large arteries and the primary cause of heart diseases. Accumulating evidence has implicated long non-coding RNAs (lncRNAs) in atherosclerosis. This study aims to clarify the potential effects of lncRNA growth arrest-specific 5 (GAS5) on cholesterol reverse-transport and intracellular lipid accumulation in atherosclerosis. GAS5 was mainly localized in the nucleus and highly expressed in the human monocytic leukemia cell line (THP-1) macrophage-derived foam cells in coronary heart disease. Overexpressed GAS5 increased THP-1 macrophage lipid accumulation. Of note, GAS5 can inhibit the expression of ATP-binding cassette transporter A1 (ABCA1) by binding to enhancer of zeste homolog 2 (EZH2). Overexpression of EZH2 reduced cholesterol efflux and ABCA1 expression. EZH2 promoted triple methylation of lysine 27 (H3K27) in the ABCA1 promoter region. Subjected to overexpressed GAS5, overexpressed EZH2, or downregulated ABCA1, the Apolipoprotein E (ApoE)−/− mice with atherosclerosis showed increased total cholesterol (TC), free cholesterol (FC), cholesterol ester (CE), low-density lipoprotein (LDL) levels, aortic plaque, and lipid accumulation, accompanied by reduced high-density lipoprotein (HDL) level and cholesterol outflow. Altogether, knockdown of GAS5 can potentially promote reverse-transportation of cholesterol and inhibit intracellular lipid accumulation, ultimately preventing the progression of atherosclerosis via reducing EZH2-mediated transcriptional inhibition of ABCA1 by histone methylation.
Keywords: atherosclerosis, long non-coding RNA, GAS5, ABCA1, EZH2, THP-1 macrophage, cholesterol, epigenetics
Graphical Abstract
Introduction
Atherosclerosis is a chronic vascular disease and associates with lipid accumulation, uptake of low density lipoprotein (LDL) by macrophages, and the subsequent conversion to foam cells.1 Atherosclerosis is characterized by inflammatory response, lipid accumulation, and fibrosis of the arterial wall, all of which form the pathological basis of coronary heart disease.2 The system of reverse-transportation of cholesterol, a process of mobilizing cholesterol on high-density lipoprotein (HDL) particles to plasma from extravascular tissues,3 plays key roles in the progression of atherogenesis.4 Moreover, excessive lipid accumulation in macrophages has been shown to trigger an increase in the formation of foam cells, which is a crucial process of atherosclerosis.5 At present, the exact pathology of atherosclerosis is not fully understood, largely due to the numerous factors at play. Thus, it is necessary to elucidate the finer molecular mechanism underlying atherosclerosis and to explore a treatment regimen to promote reverse-transportation of cholesterol and inhibit intracellular lipid accumulation.
Long non-coding RNAs (lncRNAs) with a length of more than 200 nucleotides are clarified to play an important role in cardiovascular diseases and various tumors.6 Widely expressed in various tissues and cells of human and mice, lncRNA growth arrest-specific 5 (GAS5) represents a major regulator of cell growth in the pancreatic β cells in mice.7 A previous study flagged increases in the expression of GAS5 in patients with atherosclerosis, and its downregulation can inhibit apoptosis of the human monocytic leukemia cell line (THP-1) cells induced by oxidized low density lipoprotein (ox-LDL).8 Several studies have concluded that lncRNAs can inhibit the transcription of its target genes by recruiting enhancer of zeste homolog 2 (EZH2) to the promoter region of target genes.9,10 As one of the enzymatic factors of the polycomb repressive complex 2, EZH2 is often upregulated in various types of tumors, including breast, prostate, and colorectal cancers.11 EZH2 has been reported to be regulated by lncRNA SPRY4-IT1 in lung adenocarcinoma,12 by lncRNA ANCR in osteoblast differentiation,13 and by lncRNA HOTAIR in epithelial-to-mesenchymal transition.14 EZH2 has been demonstrated to function in an epigenetic manner. A previous study revealed that following treatment with an antibody against EZH2, the number of nuclei presenting with the mark of triple methylation of lysine 27 in histone H3 (H3K27me3) in tissues with atherosclerotic plaques decreased.15 EZH2 can promote the progression of atherosclerosis by regulating ATP-binding cassette transporter A1 (ABCA1) transcription in atherosclerosis.16 ABCA1 is a protein that has been implicated in the pathology of atherosclerosis, capable of regulating lipid efflux.17 ABCA1 is also a key transporter in reverse-transportation of cholesterol, and overexpression of ABCA1 can inhibit the progression of atherosclerosis.18 On the basis of aforementioned literature, we proposed the hypothesis that there may be interactions between GAS5, EZH2, and ABCA1 in the progression of atherosclerosis. Hence, the current study aimed to determine the ability of GAS5 to influence the progression of atherosclerosis in connection with the involvement of EZH2 and ABCA1.
Results
GAS5 Is Highly Expressed in Atherosclerosis
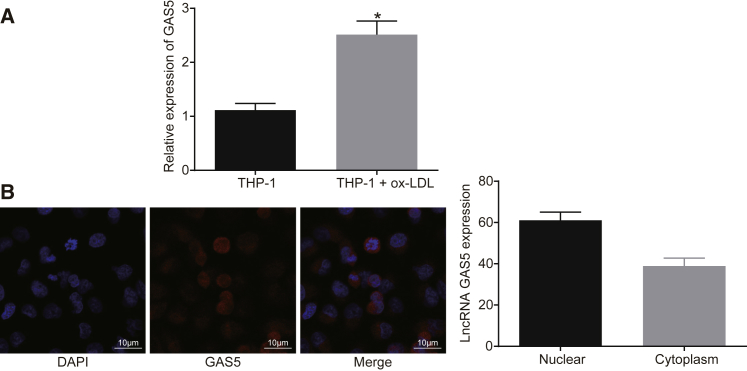
It has been reported that GAS5 was highly expressed in the atherosclerotic plaque and downregulated GAS5 could suppress the ox-LDL-induced THP-1 cell apoptosis.19 In order to identify the expression and role of GAS5 in atherosclerosis, the expression of GAS5 on a cellular level was determined. The localization of GAS5 in THP-1 macrophage-derived foam cells was determined by fluorescence in situ hybridization (FISH). qRT-PCR results demonstrated that the expression of GAS5 in THP-1 macrophage-derived foam cells was much higher than that in THP-1 macrophages (Figure 1A) (p < 0.05). It was shown that GAS5 was significantly enriched in THP-1 macrophage-derived foam cells after ox-LDL stimulation, and GAS5 expression was upregulated in the THP-1 macrophage-derived foam cells (p < 0.05), indicating that ox-LDL influenced the expression of GAS5. FISH results demonstrated that GAS5 was predominantly localized in the nucleus of the THP-1 macrophage-derived foam cells, which may be related to the mechanism by which GAS5 functions in atherosclerosis (Figure 1B).
Figure 1.
GAS5 Is Expressed at a High Level in THP-1 Macrophage-Derived Foam Cells
(A) Relative expression of GAS5 in THP-1 macrophage-derived foam cells and THP-1 macrophages detected using qRT-PCR. *p < 0.05 versus the THP-1 group; data were compared using unpaired t test. (B) Subcellular localization of GAS5 in THP-1 macrophage-derived foam cells detected using FISH assay (×1,000). Cellular experiments were repeated 3 times.
GAS5 Promotes Lipid Accumulation and Inhibits Cholesterol Efflux in THP-1 Macrophage-Derived Foam Cells
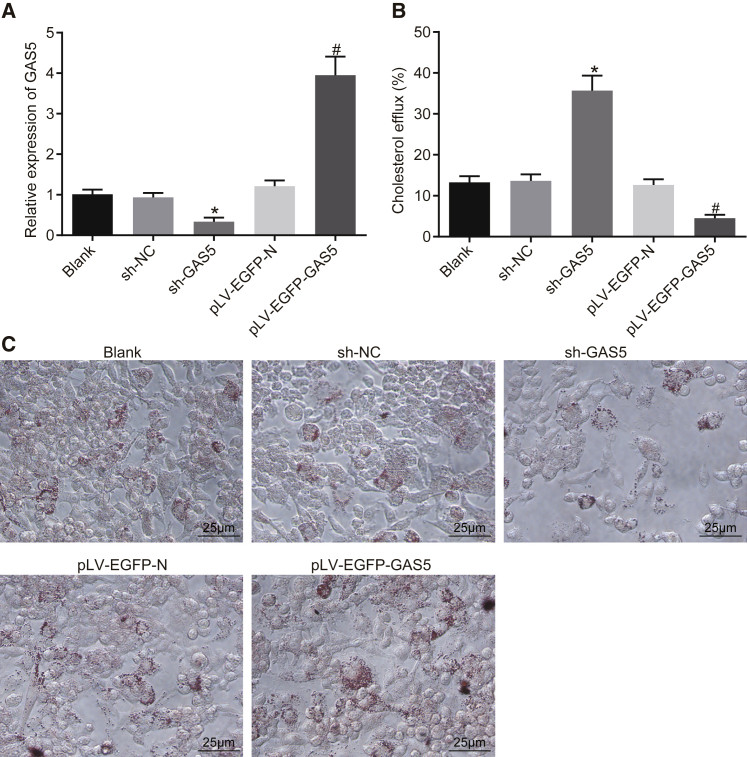
qRT-PCR was performed to measure the expression efficiency of GAS5 in each group. After knockdown or overexpression of GAS5, the expression of GAS5 in THP-1 macrophage-derived foam cells was changed as compared with the control group (Figure 2A) (p < 0.05), and there was no difference in the expression of GAS5 between the short hairpin RNA-negative control (sh-NC) and lentivirus gene overexpression vector (pLV-EGFP-N) groups (Figure 2A) (p > 0.05). The results were suggestive of the successful knockdown and overexpression of GAS5.
Figure 2.
Overexpression of GAS5 Inhibits Cholesterol Efflux but Promotes Lipid Accumulation in THP-1 Macrophage-Derived Foam Cells
(A) Transfection efficiency of GAS5 in each group determined using qRT-PCR. (B) Effect of GAS5 on the cholesterol efflux of cells detected by liquid scintillation counter. (C) Effect of GAS5 on intracellular lipid accumulation examined using oil red O staining (×400). *p < 0.05 versus the sh-NC group; #p < 0.05 versus the pLV-EGFP-N group. All cellular experiments were repeated 3 times. Comparison among multiple groups was analyzed using one-way ANOVA.
The effects of knockdown and overexpression of GAS5 on cholesterol efflux from THP-1 macrophage-derived foam cells were subsequently examined using a liquid scintillation counter. After GAS5 knockdown, the cell cholesterol efflux rate was even higher than that in the sh-NC group; following the overexpression of GAS5, the cell cholesterol efflux rate was lower, when compared to the pLV-EGFP-N group (Figure 2B) (p < 0.05), highlighting the ability of GAS5 to inhibit cholesterol efflux in THP-1 macrophage-derived foam cells. Next, oil red O staining was performed in order to detect the effects associated with the knockdown and overexpression of GAS5 on intracellular lipid accumulation in THP-1 macrophage-derived foam cells. As shown in Figure 2C, when compared with the sh-NC group, knockdown of GAS5 inhibited intracellular lipid accumulation; in comparison with the pLV-EGFP-N group, the overexpression of GAS5 increased intracellular lipid accumulation (p < 0.05). Consistent with the oil red O staining results, the results obtained from the high performance liquid chromatography (HPLC) revealed that the intracellular total cholesterol (TC), free cholesterol (FC), and cholesterol ester (CE) levels were much lower than those in the sh-NC group following the knocking down of GAS5; the intracellular TC, FC, and CE levels were higher than those in the pLV-EGFP-N group after overexpression of GAS5 (Table 1) (p < 0.05). The data collected indicated that GAS5 can promote lipid accumulation and inhibit cholesterol efflux in THP-1 macrophage-derived foam cells.
Table 1.
HPLC Detection of GAS5 on Cholesterol Content in THP-1 Macrophage-Derived Foam Cells
| Group | TC (μg/mg) | FC (μg/mg) | CE (μg/mg) | CE/TC (%) |
|---|---|---|---|---|
| Blank | 502.15 ± 50.25 | 210.32 ± 21.54 | 291.83 ± 29.28 | 58.57 |
| sh-NC | 520.31 ± 51.15 | 220.15 ± 22.16 | 300.16 ± 31.02 | 57.88 |
| sh-GAS5 | 369.11 ± 37.21∗ | 156.24 ± 16.94∗ | 212.87 ± 20.27∗ | 57.72 |
| pLV-EGFP-N | 529.16 ± 52.99 | 220.31 ± 22.64 | 308.85 ± 30.97 | 59.06 |
| pLV-EGFP-GAS5 | 710.15 ± 71.08# | 301.54 ± 30.19# | 408.61 ± 40.89# | 57.87 |
All cellular experiments were repeated 3 times, and one-way ANOVA was used for data analysis between groups. HPLC, high performance liquid chromatography; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; NC, negative control; THP-1, the human monocytic leukemia cell line.
p < 0.05 versus the sh-NC group
p < 0.05 versus the pLV-EGFP-N group.
GAS5 Inhibits Transcriptional Expression of ABCA1 by Binding to EZH2
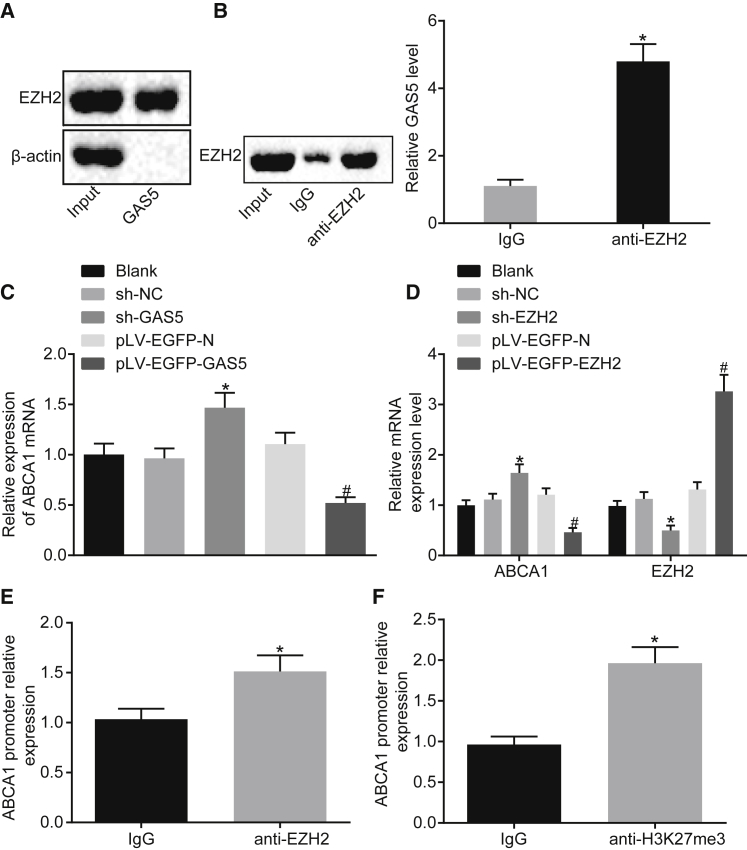
According to the prediction results from bioinformatics website available at http://pridb.gdcb.iastate.edu/RPISeq/, GAS5 may directly bind to EZH2. Besides, EZH2 was demonstrated to function together with lncRNA previously.20,21 Hence, an RNA pull-down experiment was conducted in order to determine whether GAS5 can bind to EZH2. Western blot analysis revealed the presence of the EZH2 protein, indicating that EZH2 can bind to GAS5 (Figure 3A). The results of RNA binding protein immunoprecipitation (RIP) experiment revealed that, after RNA was extracted from the pull-down protein solution by the EZH2 antibody, GAS5 could be detected using qRT-PCR (Figure 3B) (p < 0.05), further verifying the mutual interaction between GAS5 and EZH2 proteins.
Figure 3.
GAS5 Inhibits the Transcriptional Expression of ABCA1 by Binding to EZH2
(A) Interaction between GAS5 and EZH2 verified by RNA pull-down assay. (B) The enrichment of GAS5 by EZH2 assessed using RIP assay. (C) The effect of GAS5 on ABCA1 gene transcriptional expression assessed using qRT-PCR. (D) Transfection efficiency of sh-EZH2 and the effect of EZH2 on the transcriptional expression of ABCA1 gene determined using qRT-PCR. (E) Expression of EZH2 enriched by ABCA1 detected using ChIP. (F) ABCA1 expression enriched by H3K27me3 detected using ChIP assay. *p < 0.05 versus the IgG group or the sh-NC group; #p < 0.05 versus the pLV-FGFP-N group. Cellular experiment was repeated 3 times. Comparison between two groups was analyzed using t test and comparison among multiple groups was analyzed using one-way ANOVA.
Meanwhile, the qRT-PCR results revealed that the mRNA expression of ABCA1 was downregulated after the overexpression of GAS5 and increased following GAS5 knockdown treatment (Figure 3C) (p < 0.05). This indicated that GAS5 can inhibit the transcriptional expression of ABCA1. As shown in Figure 3D, after overexpression of EZH2, the mRNA expression of ABCA1 was downregulated and after knocking down EZH2, the mRNA expression of ABCA1 was increased (p < 0.05). This indicated that EZH2 can inhibit the transcriptional expression of ABCA1. Therefore, it was concluded that EZH2 can bind to the promoter region of ABCA1 to regulate transcription of ABCA1.
Next, we performed chromatin immunoprecipitation (ChIP) assay using EZH2 antibody, the results of which revealed that the expression of the ABCA1 promoter region was increased in the EZH2 antibody group when compared with the immunoglobulin G (IgG) group (Figure 3E) (p < 0.05), indicating that the ABCA1 promoter region interacted with EZH2. Meanwhile, qRT-PCR results revealed that EZH2 could inhibit the transcriptional expression of ABCA1. The results suggested that H3K27me3 modification in the promoter region of ABCA1 gene was associated with EZH2.
Subsequently, ChIP assay was employed to evaluate the expression of H3K27me3 binding to ABCA1 gene. The results showed that the expression of ABCA1 promoter region was increased in the H3K27me3 antibody group, when compared with the IgG group (Figure 3F) (p < 0.05), indicating that the ABCA1 promoter region directly binds to H3K27me3 and histone H3K27me3 modification was present in ABCA1 gene.
GAS5 and EZH2 Play a Role in THP-1 Macrophage-Derived Foam Cells via ABCA1
Previous experiments have demonstrated that stable overexpression of GAS5 in THP-1 macrophage-derived foam cells inhibited cholesterol efflux from THP-1 macrophage-derived foam cells and promoted intracellular lipid accumulation. Moreover, GAS5 may be able to regulate the transcriptional expression of ABCA1 in THP-1 macrophage-derived foam cells through EZH2. Hence, we hypothesized that the role of GAS5 in atherosclerosis may be related to ABCA1.
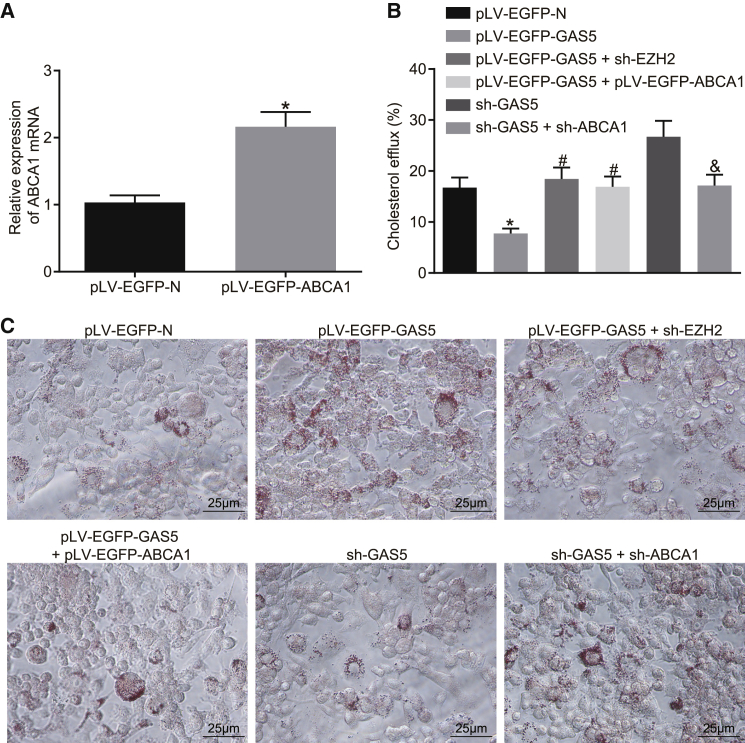
The expression efficiency of pLV-EGFP-ABCA1 was determined by qRT-PCR. Following the overexpression of ABCA1, the relative expression of ABCA1 in THP-1 macrophage-derived foam cells was confirmed to be higher than that in the control group (Figure 4A) (p < 0.05). The results of cholesterol efflux detection indicated that the cholesterol efflux rate of THP-1 macrophage-derived foam cells in the pLV-EGFP-GAS5 group was lower than that in the pLV-EGFP-N group (p < 0.05). After knockdown of EZH2 expression or overexpression of ABCA1, the cholesterol efflux rate of THP-1 macrophage-derived foam cells was much higher than that of the pLV-EGFP-GAS5 control group, while the cholesterol efflux rate of THP-1 macrophage-derived foam cells was lowered with knockdown of ABCA1, as compared with that of the sh-GAS5 control group (Figure 4B) (p < 0.05). Subsequent oil red O staining results demonstrated that after knocking down EZH2 or overexpressing ABCA1, lipid accumulation in THP-1 macrophage-derived foam cells was reduced, when compared to the pLV-EGFP-GAS5 control group, whereas with knockdown of ABCA1 expression, the accumulation of lipids in THP-1 macrophage-derived foam cells was higher than that in the sh-GAS5 control group (Figure 4C) (p < 0.05). The HPLC results illustrated that after knocking down EZH2 or overexpressing ABCA1, TC, FC, and CE in THP-1 macrophage-derived foam cells were lower than those in the pLV-EGFP-GAS5 control group, while knockdown of ABCA1 elevated levels of TC, FC, and CE in THP-1 macrophage-derived foam cells in comparison to sh-GAS5 treatment (Table 2) (p < 0.05).
Figure 4.
GAS5 Plays a Role in THP-1 Macrophage-Derived Foam Cells by Recruiting EZH2 to Mediate ABCA1 Histone Trimethylation
(A) Transfection efficiency of ABCA1 detected by qRT-PCR; the data were compared using unpaired t test. (B) The effects of GAS5, EZH2, and ABCA1 on reverse cholesterol transport function of THP-1 macrophage-derived foam cells detected by cholesterol efflux assay; one-way ANOVA was used for data analysis among multiple groups. (C) The effects of GAS5, EZH2, and ABCA1 on lipid accumulation in THP-1 macrophage-derived foam cells examined by oil red O staining (×400). *p < 0.05 versus the pLV-EGFP-N group; #p < 0.05 versus the pLV-EGFP-GAS5 group; &p < 0.05 versus the sh-GAS5 group.
Table 2.
Determination of Cholesterol in THP-1 Macrophage-Derived Foam Cells by HPLC
| Group | TC (μg/mg) | FC (μg/mg) | CE (μg/mg) | CE/TC (%) |
|---|---|---|---|---|
| pLV-EGFP-N | 479.12 ± 53.34 | 181.29 ± 20.49 | 297.83 ± 33.84 | 62.66 |
| pLV-EGFP-GAS5 | 634.13 ± 70.83∗ | 248.19 ± 27.59∗ | 385.94 ± 43.19∗ | 60.92 |
| pLV-EGFP-GAS5 + sh-EZH2 | 479.16 ± 53.73# | 183.32 ± 20.68# | 295.84 ± 33.19# | 61.80 |
| pLV-EGFP-GAS5 + pLV-EGFP-ABCA1 | 436.14 ± 49.31# | 167.56 ± 18.67# | 268.58 ± 29.93# | 62.56 |
| sh-GAS5 | 300.16 ± 34.83 | 120.08 ± 13.47 | 180.08 ± 20.42 | 60.66 |
| sh-GAS5 + sh-ABCA1 | 481.64 ± 54.25& | 186.97 ± 21.67& | 294.67 ± 33.49& | 62.13 |
All cellular experiments were repeated 3 times, and one-way ANOVA was used for data analysis. HPLC, high performance liquid chromatography; TC, total cholesterol; FC, free cholesterol; CE, cholesterol ester; EZH2, enhancer of zeste homolog 2; THP-1, the human monocytic leukemia cell line; GAS5, growth arrest-specific transcript 5; ABCA1, ATP-binding cassette transporter A1.
p < 0.05 versus the pLV-EGFP-N group
p < 0.05 versus the pLV-EGFP-GAS5 group
p < 0.05 versus the sh-GAS5 group.
These results indicated that knockdown of EZH2 or overexpression of ABCA1 suppressed the inhibitory effect of overexpression of GAS5 on reverse-transportation of cholesterol of THP-1 macrophage-derived foam cells while promoting an effect on intracellular lipid accumulation. When ABCA1 expression was knocked down, the effects of knockdown of GAS5 on cholesterol efflux and intracellular lipid accumulation could be attenuated. This suggested that GAS5 plays a role in regulating THP-1 macrophage-derived foam cells by recruiting EZH2 to mediate ABCA1 promoter region histone trimethylation.
GAS5 Promotes Progression of Atherosclerosis In Vivo
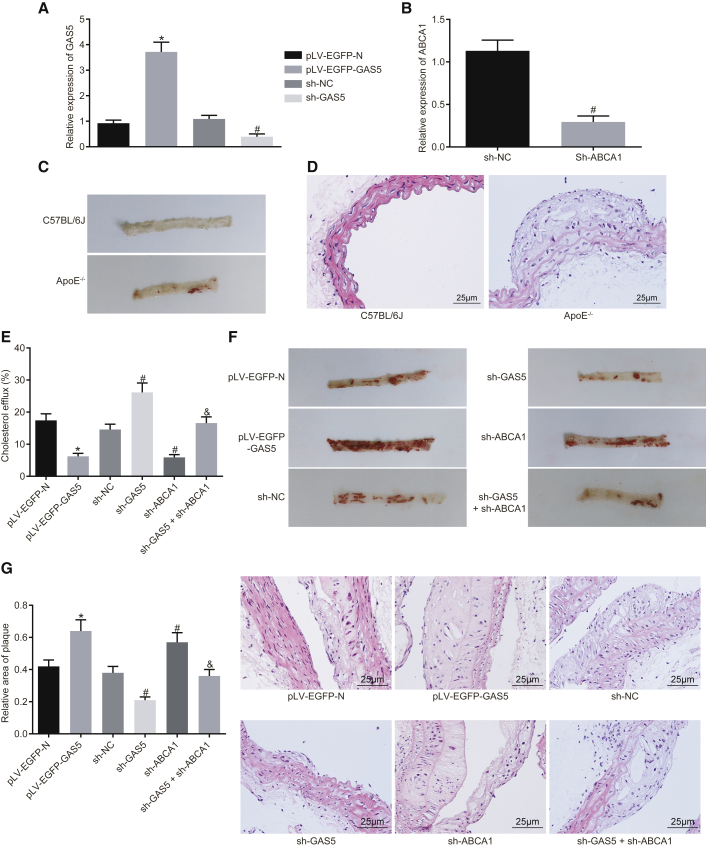
To further explore the effect of GAS5 on the progression of atherosclerosis in vivo, we fed ApoE knockout mice with a high-fat diet to establish animal models of atherosclerosis. The results of qRT-PCR demonstrated that knockdown and overexpression of GAS5 were effective in RAW264.7 macrophage-derived foam cells (Figure 5A) (p < 0.05), and ABCA1 knockdown was effective (Figure 5B) (p < 0.05). Next, the models were further evaluated. The blood lipid levels of the control group and the model group revealed that the levels of TC, triglyceride (TG), HDL, and LDL in the model group induced by high-fat diet were higher than those in the C57BL/6J mice (p < 0.05) (Table S1). Oil red O staining of aortic results showed that after a high-fat diet, the ApoE−/− mice showed obvious atherosclerotic plaques in the aortic arch, abdominal aorta, thoracic aorta, and common iliac artery branches, and the lipids in the plaque were stained with oil red O, while C57BL/6J mice had no obvious atherosclerotic plaque formation (Figure 5C). The results of H&E staining in the aortic root were similar to those of the whole aortic staining. In the mice induced by a high-fat diet, the blood vessel wall of the aortic root was thickened, foam cells were accumulated, partially ruptured, and the atherosclerotic lesions were obvious. There was no atherosclerotic change in the aortic root of C57BL/6J mice, and the cells were normal and arranged closely (Figure 5D). In conclusion, the high-fat diet-induced ApoE−/− mice exhibited typical pathological features of atherosclerosis, and the mouse models can be used for subsequent experiments.
Figure 5.
GAS5 Downregulation Prevents Atherosclerosis Progression in ApoE−/− Mice
(A) Transfection efficiency of GAS5 in each group detected by qRT-PCR and (B) transfection efficiency of sh-ABCA1 in each group detected by RT-qPCR. (C) Oil red O staining analysis of aortic plaque formation in C57BL/6J and ApoE−/−mice. (D) H&E staining analysis of aortic plaque formation in C57BL/6J and ApoE−/− mice (×400). (E) Serum cholesterol efflux in ApoE−/− mice with atherosclerosis after oe-GAS5, sh-GAS5, or sh-ABCA1 treatment. (F) Oil red O staining analysis of aortic plaque formation in ApoE−/− mice with atherosclerosis after oe-GAS5, sh-GAS5, or sh-ABCA1 treatment. (G) H&E staining analysis of aortic plaque formation in ApoE−/− mice with atherosclerosis after oe-GAS5, sh-GAS5, or sh-ABCA1 treatment (×400). *p < 0.05 versus the pLV-EGFP-N group; #p < 0.05 versus the sh-NC group; &p < 0.05 versus sh-ABCA1 group. The experiment was repeated three times. Comparison between two groups was analyzed using t test and comparison among multiple groups was analyzed using one-way ANOVA.
Measurement of the in vivo cholesterol efflux rate revealed that the radioactivity of the pLV-EGFP-GAS5 group was decreased, as compared with the pLV-EGFP-N group (p < 0.05); compared with the sh-NC group, the radioactivity of the sh-GAS5 group was increased (p < 0.05), while the radioactivity of sh-ABCA1 group was decreased (p < 0.05). Compared with the sh-ABCA1 group, the radioactivity of the sh-GAS5 + sh-ABCA1 group was elevated (p < 0.05) (Figure 5E). The aforementioned findings indicated that GAS5 could regulate the reverse translocation process of macrophage cholesterol in vivo by regulating the expression of ABCA1. Enzyme oxidation method was used to evaluate blood lipid levels. The results revealed that after overexpression of GAS5, levels of TG, TC, and LDL were increased to different extents, while HDL level was decreased (p < 0.05). After GAS5 expression was knocked down, levels of TG, TC, and LDL were downregulated and the level of HDL was increased (p < 0.05). Instead, after knocking down the expression of ABCA1, the levels of TG, TC, and LDL were increased to different degrees, while the level of HDL was decreased (p < 0.05). Furthermore, knockdown of GAS5 expression inhibited the effect of knockdown of ABCA1 expression on blood lipid levels in ApoE−/− mice (p < 0.05) (Table 3), suggesting that GAS5 regulated blood lipid levels in ApoE−/− mice with atherosclerosis by mediating reverse cholesterol transport. Oil red O staining analysis of the aorta revealed that overexpression of GAS5 promoted atherosclerosis, while knockdown of GAS5 attenuated atherosclerosis; knockdown of ABCA1 was found to increase atherosclerosis, while the knockdown of GAS5 expression inhibited the effect of knockdown of ABCA1 on atherosclerotic lesions in ApoE−/− mice (Figure 5F). H&E staining of arterial root showed intimal hyperplasia and notable plaque formation in the valve of each group, but the area of plaque in the mice overexpressing GAS5 was much higher than that in mice expressing pLV-EGFP-N when the plaque area of each mice was measured (p < 0.05); the area of plaque in the mice was reduced by knockdown of GAS5 (p < 0.05); the area of plaque in the mice was enlarged following the knockdown of ABCA1 (p < 0.05), suggesting that knockdown of GAS5 inhibited the effect of knockdown of ABCA1 on atherosclerotic lesions in ApoE−/− mice (Figure 5G). Taken together, the results indicated that GAS5 could promote atherosclerotic lesions in ApoE−/− mice.
Table 3.
Blood Lipid Levels of Each Group of ApoE−/− Mice (n = 8, mmol/L)
| Group | TC | TGs | HDL | LDL |
|---|---|---|---|---|
| pLV-EGFP-N | 18.49 ± 1.96 | 1.83 ± 0.19 | 1.45 ± 0.16 | 9.16 ± 0.98 |
| pLV-EGFP-GAS5 | 24.36 ± 2.59∗ | 2.64 ± 0.29∗ | 0.92 ± 0.09∗ | 16.76 ± 1.78∗ |
| sh-NC | 17.09 ± 1.71 | 1.68 ± 0.19 | 1.53 ± 0.16 | 7.61 ± 0.81 |
| sh-GAS5 | 15.13 ± 1.59# | 1.34 ± 0.15# | 1.74 ± 0.18# | 5.84 ± 0.59# |
| sh-ABCA1 | 23.49 ± 2.43# | 2.57 ± 0.26# | 0.96 ± 0.11# | 15.96 ± 1.67# |
| sh-GAS5 + sh-ABCA1 | 17.49 ± 1.83& | 1.76 ± 0.19& | 1.49 ± 0.15& | 7.64 ± 0.79& |
TC, total cholesterol; TGs, triglycerides; HDL, high density lipoprotein; LDL, low density lipoprotein; GAS5, growth arrest-specific transcript 5; ABCA1, ATP-binding cassette transporter A1.
p < 0.05 versus the pLV-EGFP-N group
p < 0.05 versus the sh-NC group
p < 0.05 versus the sh-ABCA1 group; data analysis was performed by one-way ANOVA.
Discussion
Atherosclerosis is a chronic inflammatory and lipid-depository disease that eventually leads to acute cardiovascular events.22 Emerging evidence supports that epigenetic processes such as DNA methylation, histone modification, and noncoding RNAs play an important role in plaque progression and vulnerability.23, 24, 25 Based on the results observed during the present study, it became clear that downregulation of GAS5 could promote reverse-transportation of cholesterol and inhibit intracellular lipid accumulation via decreasing EZH2 and inhibiting the histone methylation of ABCA1, ultimately alleviating the progression of atherosclerosis.
Atherosclerosis is a chronic inflammatory disease of the vessel wall.26 Although significant progress has been made in the research and development of atherosclerosis treatment, the drug therapies available at present still have numerous side effects particularly for those on long-term treatment plans.27 Thus, it is of great importance to find more effective and long-term treatment for patients with atherosclerosis. The results of our study showed that GAS5 was highly expressed in atherosclerotic plaque tissues in ApoE−/− mice with atherosclerosis. In line with our study, a previous study confirmed GAS5 to be upregulated not only in patients with atherosclerosis but also in atherosclerotic rats.28 Another study also provided evidence highlighting the elevation of GAS5 in the plaque of atherosclerosis from both human and animal models.19 All of the aforementioned findings support our results that GAS5 was overexpressed in atherosclerotic plaque.
Furthermore, this study found that downregulation of GAS5 can increase the transcription of ABCA1 in atherosclerotic plaque via binding to EZH2. lncRNAs could inhibit the transcription of its target genes by recruiting EZH2 to the promoter region of target genes. Specifically, lncRNA SYISL recruits EZH2 protein to the promoters of the cell-cycle inhibitor gene p21 and muscle-specific genes such as myogenin (MyoG) and muscle creatine kinase (MCK), leading to H3K27me3 and epigenetic silencing of target genes.10 Besides, lncRNA PCAT6 overexpression might decrease LATS2 promoter activity via binding to EZH2, resulting in H3K27me3 in the LATS2 promoter region.9 A previous study revealed that knockdown of lncRNA PVT1 can downregulate EZH2,29 which highlights the potential regulation of EZH2 by lncRNAs. As histone methyltransferase, EZH2 is a repressive epigenetic mark.30 Notably, EZH2 can act as a regulator of cell process in various diseases including atherosclerosis.31 More specifically, elevation of EZH2 has been observed in atherosclerotic plaque tissues.32 In our study, the results of RNA pull-down, RIP, and ChIP assays strictly proved that GAS5 can bind to EZH2 and downregulation of GAS5 decreased the expression of EZH2.
The present study also proved that downregulation of GAS5 could promote reverse-transportation of cholesterol and inhibit lipid accumulation via EZH2-mediated ABCA1, which was indicated by the decreased TC, TG, and LDL levels upon knockdown of GAS5 or EZH2 and overexpressed ABCA1. The combination of increased lipid influx, elevated production and reduced catabolism of lipids, and impaired HDL-mediated reverse cholesterol transport can promote atherosclerosis.33 Recent evidence has indicated that lncRNAs are able to control the function of the vessel wall, lipid metabolism, activation of macrophages, and inflammatory response.34 Liang et al. concluded that GAS5 knockdown could alleviate ox-LDL-stimulated apoptosis and recover ox-LDL-impaired autophagy flux in human aortic endothelial cells.35 Moreover, the reduction of GAS5 has been proved to downregulate the expression of pro-inflammatory cytokines (interleukin-6 [IL-6], IL-1β, tumor necrosis factor alpha [TNF-α]) induced by ox-LDL in macrophages.28 As shown in a previous study, the silkworm protein 30Kc6 dramatically improves the conditions of the atherosclerosis by decreasing serum levels of TG, HDL-C, LDL-C, and TC.36 Another study further suggested that TC, TG, and LDL were upregulated in atherosclerosis, and elevated ABCA1 expression induced by apigenin can inhibit the development of atherosclerosis.37 Cellular cholesterol transporter ABCA1 might play vital roles in the prevention of atherosclerosis cellular cholesterol transporters.38 Moreover, leonurine has been shown to inhibit atherosclerosis by upregulating ABCA1.39 It has been proved that atherosclerosis in ApoE–/– mice can be suppressed via increasing ABCA1 expression by botulin.40 Furthermore, it has been also detected that the expression of ABCA1 in atherosclerotic plaque tissues can be decreased by EZH2,16 which was consistent with the findings of our study: ABCA1 expression has been shown to be stimulated following the downregulation of GAS5 and EZH2. Based on these results, our study found that downregulation of GAS5 can increase the expression of ABCA1 via binding to EZH2, thus inhibiting the progression of atherosclerosis.
The in vivo results showed that the combined inhibition of GAS5 and ABCA1 completely negated the effects of inhibition of GAS5 on cholesterol efflux and lipid accumulation. However, that did not emerge in in vitro experimental results. The different environment of in vitro and in vivo cells may cause the difference between in vitro and in vivo experimental results. This is also what we will continue to explore in the future study.
The key findings from the present study provided evidence that knockdown of GAS5 could potentially stimulate reverse-transportation of cholesterol and attenuate intracellular lipid accumulation via reducing EZH2-mediated transcriptional inhibition of ABCA1, consequently preventing the progression of atherosclerosis. These findings highlighted the potential of GAS5 as a promising therapeutic target to treat atherosclerosis. Considering all the experiments in the present study were conducted in mice, we will try to develop clinical values in clinical trials in future studies.
Materials and Methods
Ethics Statement
This study was conducted under the approval of the Animal Ethics Committee of Shanghai General Hospital and in strict accordance with the Guide for the Care and Use of Laboratory Animals published by the NIH.
Cell Culture and Transfection
The human monocytic leukemia cell line THP-1 was purchased from the Basic Medical Cell Center of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China; http://www.crcpumc.com/pr.jsp?keyword = THP1&_pp = 0_312). THP-1 monocytes were cultured using RPMI 1640 medium (GIBCO, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS). The THP-1 cells were incubated in a 6-well plate with 20 μL of 100 nmol/L phorbol myristic acetate (Sigma-Aldrich Chemical Company, St. Louis, MO, USA) in each well for 72 h at 37°C with 5% CO2 to make THP-1 cells grow in a single layer and convert from monocytes to macrophages. Next, 50 μg/mL ox-LDL was added to serum-free RPMI 1640 medium containing 0.3% BSA (Hyclone-Pierce, Logan, UT, USA), and the cells were cultured in the medium for 48 h to induce macrophages to be transformed into foam cells for cell transfection.
A mouse mononuclear macrophage cell line RAW264.7 was purchased from the Basic Medical Cell Center of Institute of Basic Medical Sciences, Chinese Academy of Medical Sciences (Beijing, China; http://www.crcpumc.com/pr.jsp?keyword = RAW264.7&_pp = 0_312). RAW264.7 cells were cultured in DMEM medium (GIBCO, Carlsbad, CA, USA) containing 10% FBS with 5% CO2 at 37°C. After the cell density reached approximately 2 × 108 cells/L, the medium was replaced with DMEM medium containing 1% FBS, and the culture medium was replaced every 2 days. The final concentration of ox-LDL was 50 μg/mL, and the culture was continued for 24 h to induce macrophages to transform into foam cells for cell transfection.
The vector used to construct the lentiviral vector was pLV-enhanced GFP (EGFP)-N (lentivirus gene overexpression vector), pSIH1-H1-copGFP (lentivirus gene silencing vector of shRNA fluorescent expression). The lentiviral vectors of sh-NC, sh-GAS5, sh-EZH2, and sh-ABCA1 were constructed by Shanghai GenePharma (Shanghai, China). Lentiviral packaging was performed using 293T cells. The cells were cultured in RPMI 1640 complete medium containing 10% FBS and sub-cultured every other day. The virus was collected and THP-1 macrophage-derived foam cells were classified into the following groups according to different transfections: blank control group (transfected with no sequence); pLV-EGFP-N group (transfected with empty vector); pLV-EGFP-GAS5 group (oe-GAS5, transfected with GAS5 overexpression lentiviral vector); pSIH1-H1-copGFP-sh-NC transfection group (sh-NC, transfected with irrelevant shRNA sequence lentiviral vector); sh-GAS5 group (transfected with GAS5 shRNA lentiviral vector); pSIH1-H1-copGFP-sh-EZH2 (sh-EZH2, transfected with EZH2 shRNA lentiviral vector); pLV-EGFP-N-EZH2 (oe-EZH2, transfected with EZH2 overexpression lentiviral vector); pLV-EGFP-N-GAS5 + pLV-EGFP-N-EZH2 (oe-GAS5 + oe-EZH2, co-transfected with GAS5 overexpression and EZH2 overexpression lentiviral vector); pLV-EGFP-N-GAS5 + pLV-EGFP-N-ABCA1 (oe-GAS5 + oe-ABCA1, co-transfected with GAS5 overexpression and ABCA1 overexpression lentiviral vector); sh-GAS5 + pSIH1-H1-copGFP-sh-ABCA1 (sh-GAS5 + sh-ABCA1, co-transfected with GAS5 shRNA and ABCA1 shRNA lentiviral vector); and pSIH1-H1-copGFP-sh-ABCA1 (sh-ABCA1, transfected with ABCA1 shRNA lentiviral vector). The THP-1 macrophage-derived foam cells in the logarithmic growth phase were treated with trypsin, triturated, and made into a cell suspension of 5 × 104 cells/mL. Next, the cell suspension was then seeded in a 6-well plate with 2 mL per well and incubated overnight at 37°C. The virus (1 × 108 U/mL) was added to the cells for infection, whereas the cells with stable inheritance were collected for subsequent experimentation.
RNA Isolation and Quantitation
The total RNA from the cells was extracted by Trizol (Sigma-Aldrich Chemical Company, St. Louis, MO, USA). The RNA quality and concentration were determined using an UV-visible spectrophotometry. The extracted RNA was subsequently reversely transcribed into cDNA using the PrimeScript RT Reagent Kit (Takara Holdings, Kyoto, Japan). The cDNA was utilized as a template, with the target gene subjected to a fluorescent quantitative PCR operation with reference to the SYBR Premix Ex TaqTM II (Tli RNaseH Plus) kit (Takara Holdings, Kyoto, Japan). Quantitative real-time fluorescence PCR was performed on a Cycler Dice Real Time System (TP800, Takara Holdings, Kyoto, Japan). Next, 3 replicates were set up for each group. Primer sequences for qRT-PCR are shown in Table S2. The primers for the amplification template were designed by the Primer Express 2.0 software (synthesized by Guangzhou RiboBio, Guangzhou, Guangdong, China), with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) regarded as the internal reference gene. 2-ΔΔCT was considered to be reflective of the expression of the gene to be assessed: ΔCT = CT (target gene) – CT (internal reference).
Western Blot Analysis
The cells were collected and washed with PBS, followed by incubation with protein lysis buffer containing protease and alkaline phosphatase inhibitor at 4°C for 30 min. The protein was then separated by 10% SDS-PAGE and then transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was then blocked using 5% skimmed milk powder for 1 h and incubated overnight at 4°C with Tris-buffered saline Tween-20 (TBST) diluted primary rabbit antibody to EZH2 (#5246, 1:1,000, Cell Signaling Technologies, Beverly, MA, USA), mouse antibodies to ABCA1 (ab18180, 1:500, Abcam, Cambridge, MA, USA), β-actin (ab6276, 1:1,000, Abcam, Cambridge, MA, USA), and H3K27me3 (ab6002, 1:1,000, Abcam, Cambridge MA, USA). After 3 TBST washes, the membrane was incubated with horseradish peroxidase (HRP)-labeled secondary antibody at room temperature for 1 h, followed by an additional 6 washes with TBST. The immunocomplexes on the membrane were then visualized using enhanced chemiluminescence (ECL) with the band intensities quantified using ImageJ software. The ratio of the gray value of the target band to β-actin was considered to be representative of the relative protein expression.
FISH Assay
FISH was performed to detect the subcellular localization of GAS5 in THP-1 macrophage-derived foam cells. The cover glasses were placed in the bottom of a 24-well plate at a density of 5 × 103 cells/well. After a 24 h period of culture, the supernatant was removed, after which the cells were washed with 1 × PBS. After fixation with 4% paraformaldehyde, PBS containing 0.5% Triton X-100 was added to the cells, followed by blocking with pre-hybridization solution at 37°C. Next, GAS5 probe was hybridized overnight at 37°C. The cells were then washed with a hybridization solution at 42°C under conditions void of light. The hybridized area of the cover glasses was subsequently stained with 4’,6-diamidino-2-phenylindole (DAPI), with the cover glasses fixed on the slide using a sealing agent under conditions void of light. The experimental specimens were analyzed under a laser confocal microscope.
Cholesterol Efflux Detection
THP-1 macrophage-derived foam cells were incubated in RPMI 1640 medium containing 10% FBS with 0.2 μCi/mL [3H] cholesterol. Upon reaching 85% confluence, the cells were washed with PBS. Next, the cells were transfected in fresh culture medium and assigned into blank, sh-NC, sh-GAS5, pLV-EGFP-N, pLV-EGFP-GAS5, sh-GAS5 + sh-ABCA1, pLV-EGFP-GAS5 + pLV-EGFP-ABCA1, and pLV-EGFP-GAS5 + sh-EZH2 groups. The cells were then cultured in serum-free RPMI 1640 medium containing lipoprotein for 24 h. After PBS rinsing, the cells were cultured in fresh medium containing 25 μg/mL apoA-I for 6 h and lysed with the scintillation solution. The [3H] cholesterol radioactivity of each sample was measured using a liquid scintillation counter. The cholesterol efflux rate = radiation intensity of culture solution (count per minute, cpm) ÷ total radiation intensity (culture liquid radiation intensity [cpm] + cell lysate radiation intensity [cpm]) × 100%.
Oil Red O Staining
The THP-1 macrophage-derived foam cells were cultured in 6-well culture plates with sterile coverslips. The cells were treated, washed 3 times with PBS, fixed with 50% isopropanol for 1 min, and stained with oil red O staining solution for 10 min. After hematoxylin staining for 5 min, the cells were differentiated with 1% HCl, returned to blue, and sealed using a sealing agent. Under a microscope, the intracellular lipids were observed to be stained with red and the nuclei were stained with blue. The image analysis system was employed for images collection after which a microscope was used for photography.
HPLC
The THP-1 macrophage-derived foam cells were collected after transfection, washed 3 times with PBS, and disrupted by sonication in an ice bath. The protein content was determined through the application of the bicinchoninic acid (BCA) method. The cell lysate was divided into two portions, one portion was added to an equal volume of freshly prepared 15% KOH alcohol solution and stirred at room temperature until the cell lysate was clear to obtain TC extraction; the other portion was added to an equal volume of freshly prepared 8.9 mmol/L KOH alcohol solution for a water bath at 80°C for 1 h to obtain FC. The protein was removed following the addition of 6% trichloroacetic acid. The sample was added with an equal volume of a mixture of n-hexane and isopropanol at the ratio of 4:1, mixed and stirred for 5 min, followed by centrifugation at a speed of 1,500 rpm for 5 min at 15°C. The upper organic phase was collected and dried in a vacuum dryer at 65°C. After cooling, the upper organic phase was added with 100 μL of mixture solution of isopropanol, n-heptane, and acetonitrile at the ratio of 35:13:52 (V/V) to dissolve the sample, followed by centrifugation at 1,500 rpm for 5 min for supernatant collection. Next, 10 μL of the supernatant was collected for HPLC. A non-gradient elution was performed using a C-18 column with isopropanol:n-heptane:ethylidene as the mobile phase at a flow rate of 1 mL/min, and the column temperature was maintained at 4°C. Detection was carried out at 216 nm for 10 min. Cholesterol was quantified by peak area and CE was hydrolyzed by cholesterol esterase. CE was calculated based on the following formula: CE = TC − FC.
RNA Pull-Down Assay
GAS5 RNA fragment was in vitro transcribed using T7 RNA polymerase (Ambion, Company, Austin, TX, USA), then treated with RNeasy Plus Mini Kit (QIAGEN Company, Hilden, Germany) and DNase I (QIAGEN Company, Hilden, Germany), and purified by the RNeasy Mini Kit. Then 1 μg of labeled RNA was heated in RNA structure buffer containing 10 mmol/L Tris (pH = 7), 0.1 mol/L KCl, and 10 mmol/L MgCl2 to 95°C. After 2 min, the labeled RNA was incubated on ice for 3 min and then allowed to stand at room temperature for 30 min in order to allow the RNA to form a suitable secondary structure. Next, 3 μg of THP-1 macrophage-derived foam cells were added to the cell lysis (Sigma-Aldrich Chemical Company, St. Louis, MO, USA) for 1 h at 4°C. The lysate was centrifuged at 12,000 × g and 4°C for 10 min, followed by supernatant collection and transfer into an RNase-free centrifuge tube. Next, 400 ng of biotinylated RNA was added to 500 μL of RIP buffer and incubated with cell lysate for 1 h at room temperature while some of the cell lysate was regarded as the input. Streptavidin magnetic beads were then added to each binding reaction and incubated for 1 h at room temperature. Finally, the cells were washed 5 times with RIP buffer and added with 5 × loading buffer. After incubation at 95°C for 5 min, the eluted EZH2 protein was determined by western blot analysis.
RNA Binding Protein Immunoprecipitation
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) was used for measurement purposes. THP-1 macrophage-derived foam cells at 90% confluence were lysed in RIP lysis buffer, with the lysate supernatant collected and divided into 2 portions. Two inputs were used to determine the endogenous protein content in western blot analysis and content of RNA in quantitative real-time PCR. Afterward, 100 μL of cell extraction was incubated with 900 μL of RIP buffer containing magnetic beads coated with primary rabbit antibody to EZH2 (#5246, 1: 300, Cell Signaling Technology, Beverly, MA, USA) or NC normal mouse IgG for 3 h at 4°C overnight. After instantaneous centrifugation, the tube was placed on a magnetic separator with the supernatant then discarded. The tube was subsequently washed 6 times with 500 μL of RIP wash buffer. At the last wash, 50 μL of the wash buffer was collected with the supernatant discarded and the loading buffer added. Western blot analysis was employed to determine whether the magnetic beads were coated with the antibody. The sample was incubated with proteinase K at 55°C for 30 min and shaken continuously to detach the protein. After the detachment, the immunoprecipitated RNA was isolated with Trizol-chloroform and subjected to qRT-PCR for GAS5 enrichment detection.
ChIP
ChIP was performed using EZ-Magna ChIP A kit (17-408, Upstate Millipore, Billerica, MA, USA). In brief, 2 mL of THP-1 macrophage-derived foam cells in logarithmic growth phase were washed with pre-cooled PBS. The cells were then ice-bathed in an equal volume of PBS containing Protease Inhibitor Cocktail II for 5 min. The cell extract was centrifuged at 800 × g for 5 min at 4°C followed by discarding of the supernatant. The cells were resuspended in 2.0 mL of cell lysis buffer containing Protease Inhibitor Cocktail II, lysed on ice, and centrifuged at 800 × g for 5 min at 4°C with 0.5 μL of Protease Inhibitor Cocktail II. After the supernatant was discarded, the cells were added with 2.0 mL of nuclear lysis buffer. The mixture was lysed on ice and sonicated to a fragment of 200 to 1,000 bases in length. After ultrasonication, the product was subjected to gel electrophoresis to determine the length of the DNA fragment. After ultrasound was performed, the cells were centrifuged at 1,200 × g for 10 min at 4°C, and 50 μL of the supernatant (DNA fragment) was harvested from each group. The DNA fragment was co-immunoprecipitated with the addition of 450 μL of dilution buffer and 2.25 μL of Protease Inhibitor Cocktail II. The grouping was divided into NC, H3K27me3, and EZH2 groups. Next, 5.0 μg of rabbit IgG was added to the NC group, and 5.0 μg of mouse monoclonal antibody to H3K27me3 and rabbit monoclonal antibody to EZH2 were added to the H3K27me3 and EZH2 groups, respectively. The magnetic bead-antibody complex was washed and resuspended in 500 μL of ChIP wash buffer and added with 100 μL of ChIP elution buffer and Proteinase K complex. The sample was placed on a magnetic stand in order to collect the magnetic bead-protein complex followed by removal of the supernatant, and two rinses with ChIP wash buffer. DNA was extracted again and qRT-RCR was conducted to quantify the DNA promoter of ABCA1. The primer sequences of ABCA1 were: forward, 5′-CTCGGTGCAGCCGAATCTAT-3′; reverse, 5′-CACTC ACTC TCGCTCGCAAT-3′.
Establishment and Identification of Atherosclerosis Model in ApoE−/− Mice
A total of 60 healthy specific pathogen-free (SPF) male ApoE−/− mice (aged 6–8 weeks and weighing 20–30 g) purchased from Nanjing Better Biotechnology (Nanjing, Jiangsu, China) were included in the study to establish mouse models of atherosclerosis. 10 healthy C57BL/6J SPF male mice (aged 8−10 weeks and weighing 20–30 g) were purchased from Nanjing Better Biotechnology (Nanjing, Jiangsu, China) as a normal control group. The experimental mice were housed in a SPF animal laboratory with a humidity of 60% to 65% and a temperature of 22°C to 25°C. After 1-week period of acclimatization, the mice in the model group were fed with a high-fat diet (21% fat and 0.15% cholesterol) for 10 weeks, and C57BL/6J mice in the control group were fed with a high-fat diet. By observing the physiological state and tissue structure of the mice, it was judged whether the model establishment was successful based on the following aspects: (1) physiological state: mice with dark fur, pale and dull claws showed tiredness, sluggish appetite, and slow actions; (2) pathological examination: three modeled mice were selected, with a large number of atheromatous plaques detected in the arteries of these mice, a large number of foam cells had formed and accumulated with the thickened aortic intima, disordered tissue arrangement, as well as obvious plaque instability; and (3) abnormal levels of TC, TG, HDL, and LDL in blood lipid were detected.41 The mice in the normal group were lively and active with smooth fur and normal appetite and mental condition, and their tissues were neatly arranged without plaque. After the successful modeling of atherosclerosis in ApoE−/− mice, the mice were fed with the normal diet for 4 weeks. In the modeling process, 3 mice died, blood lipid levels of 4 mice did not meet the blood lipid level, and the modeling success rate was 88.33% (53/60). Then ApoE−/− mice were assigned into 6 groups (8 in each group): pLV-EGFP-N, pLV-EGFP-GAS5, sh-NC, sh-GAS5, sh-ABCA1, and sh-GAS5 + sh-ABCA1 groups. The RAW264.7 macrophage-derived foam cells in the logarithmic growth phase were trypsinized and pipetted to prepare a cell suspension of 5 × 104 cells/mL, which was cultured at 37°C overnight after seeding in a 6-well plate (2 mL per well). Then the virus (1 × 108 plaque-forming unit [PFU]/100 μL) infected with stably transfected cells was injected into mice of each group. In the first week, the mice were injected (80 mL/kg in 0.2 mL normal saline) twice per week, followed by one injection per week for the next 3 weeks, a total of 4 weeks. After that, the mice were euthanized, followed by tissue sample collection.
Collection of Experimental Specimens of Mice
On the night prior to blood collection, the mice in each group were fasted for 12 h. After anesthesia with 3% pentobarbital sodium, the blood was collected from the ocular venous plexus of the mice. The serum samples were obtained by centrifugation at 3,000 rpm for 15 min at 4°C. The mice were euthanized after blood collection, and the ventral midline incision was made to separate the sternum and diaphragm bluntly. The hearts and kidneys were then isolated, followed by exposure of the thoracic and abdominal aortas. The vascular adventitia was dissected, tissues were isolated, and the whole aorta was taken. After PBS rinsing, the aorta was fixed with 4% paraformaldehyde solution for 24 h for subsequent experiments
Detection of Blood Lipid
The serum levels of TC, TG, HDL, and LDL were evaluated by an automatic biochemical analyzer (AD-VIA-2400, Siemens, Erlangen, Germany) with biochemical detection reagent (01218LH, Beijing Leadman, Beijing, China).
Oil Red O Staining of Aorta
The mice were fasted 1 day prior to being anesthetized and euthanized with a subcutaneous injection of 3% sodium pentobarbital at a dose of 50 mg/kg. The aortic arch to the branch of the abdominal aorta was removed and the aorta was longitudinally dissected. The aorta was spread, stained with oil red O staining solution for 15 min and soaked for 20 min in 70% alcohol, followed by rinsing with distilled water for 30 min. The arteries were spread on a white plate, photographed, and analyzed quantitatively using Image-Pro Plus 6.0 (IPP 6.0) software (Media Cybernetics, Rockville, MD, USA) for plaque area. The red staining presented with atherosclerotic lesions.
H&E Staining
The formaldehyde-fixed specimens were rinsed with distilled water for 1 h and then completely soaked in the following reagents: 70% alcohol for 24 h; 80% alcohol for 24 h; 95% alcohol (I) for 30 min; 95% alcohol (II) for 30 min; 100% alcohol (I) for 30 min; 100% alcohol (II) for 30 min; the mixture of 100% alcohol and xylene at the ratio of 1:1 for 20 min; xylene (I) for 20 min; xylene (II) for 20 min; soft wax for 10 min; and hard wax for 10 min. The samples were subsequently embedded in paraffin blocks, after which they were lightly sliced to 5 μm-thick slices. The slices were then gently spread on a glass slide, baked at 60°C for 1 h, and stored at room temperature. Paraffin-embedded sections were routinely dewaxed with xylene, stained with hematoxylin for 5–10 min, rinsed under tap water, and differentiated using 1% aqueous hydrochloric acid. The sections were then stained with eosin solution for 1–2 min. Finally, the sections were dehydrated with ethanol, cleared, and sealed with neutral resin. An ordinary light fluoroscope was employed to analyze the morphological structure of blood vessels and plaques, while ImageJ image-processing software was employed for plaque area measurement and images collection.
In Vivo Cholesterol Efflux Detection
Ox-LDL with a final concentration of 50 μg/mL was added into a 15 mL centrifuge tube and water-bathed with [3H] cholesterol at a concentration of 5 μCi/mL at 37°C for 30 min. After the aforementioned mixture had been added into the DMEM medium, the RAW 264.7 macrophage-derived foam cells were transfected with pLV-EGFP-N, sh-NC, pLV-EGFP-GAS5, sh-GAS5, sh-ABCA1, sh-GAS5 + sh-ABCA1, followed by incubation at 37°C for 48 h. After centrifugation and resuspension processes, the cell number was adjusted to 1 × 1010 cells/L, and the radioactivity of the injection suspension was 6.2 × 106 cpm/mL. The 0.5 mL of isotope-labeled suspension was injected into the mice through the abdominal cavity. After 48 h, the serum of mice was collected. The blood samples were placed on ice for 1 h, centrifuged at 1,200 rpm for 5 min, and 20 μL of serum fluid was taken to calculate the radioactivity using a liquid scintillation counter (FJ22107P, state-owned 262 nuclear instrument factory, Xi’an, Shaanxi, China).
Statistical Analysis
All data were analyzed by SPSS 21.0 statistical software (IBM, Armonk, NY, USA). The data were all subjected to normal distribution and homogeneity of variance tests. Measurement data conforming to normal distribution were expressed as mean ± SD. Data with skewed distribution or uneven variance were expressed by the interquartile range. Comparisons between two groups were analyzed by unpaired t test, while comparisons among multiple groups were assessed by one-way ANOVA, followed by a post hoc test. Data at different time points were compared using repeated-measures ANOVA. Data with skewed distribution were evaluated using a nonparametric rank sum test. A p value <0.05 was considered to be statistically significant.
Author Contributions
X.-D.M. and H.-H.Y. conceived the project and wrote the paper; L.-M.W., S.S., and M.Y. designed experiments, performed the experiments, analyzed the data, and wrote the paper; Z.-X.Y. performed the experiments; J.L. analyzed the data; M.Y. supervised the study. All authors read and approved the final manuscript.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
The authors thank the reviewers for their helpful comments. This study was conducted under the approval of the Animal Ethics Committee of Shanghai General Hospital and in strict accordance with the Guide for the Care and Use of Laboratory Animals published by the NIH.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.10.034.
Supplemental Information
References
- 1.Fang S., Xu Y., Zhang Y., Tian J., Li J., Li Z., He Z., Chai R., Liu F., Zhang T. Irgm1 promotes M1 but not M2 macrophage polarization in atherosclerosis pathogenesis and development. Atherosclerosis. 2016;251:282–290. doi: 10.1016/j.atherosclerosis.2016.07.011. [DOI] [PubMed] [Google Scholar]
- 2.Wang H.H., Garruti G., Liu M., Portincasa P., Wang D.Q. Cholesterol and Lipoprotein Metabolism and Atherosclerosis: Recent Advances In reverse Cholesterol Transport. Ann. Hepatol. 2017;16:s27–s42. doi: 10.5604/01.3001.0010.5495. [DOI] [PubMed] [Google Scholar]
- 3.Martel C., Li W., Fulp B., Platt A.M., Gautier E.L., Westerterp M., Bittman R., Tall A.R., Chen S.H., Thomas M.J. Lymphatic vasculature mediates macrophage reverse cholesterol transport in mice. J. Clin. Invest. 2013;123:1571–1579. doi: 10.1172/JCI63685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sugamura K., Sugiyama S., Fujiwara Y., Matsubara J., Akiyama E., Maeda H., Ohba K., Matsuzawa Y., Konishi M., Nozaki T. Cannabinoid 1 receptor blockade reduces atherosclerosis with enhances reverse cholesterol transport. J. Atheroscler. Thromb. 2010;17:141–147. doi: 10.5551/jat.2865. [DOI] [PubMed] [Google Scholar]
- 5.Gu J.Q., Wang D.F., Yan X.G., Zhong W.L., Zhang J., Fan B., Ikuyama S. A Toll-like receptor 9-mediated pathway stimulates perilipin 3 (TIP47) expression and induces lipid accumulation in macrophages. Am. J. Physiol. Endocrinol. Metab. 2010;299:E593–E600. doi: 10.1152/ajpendo.00159.2010. [DOI] [PubMed] [Google Scholar]
- 6.Bitarafan S., Yari M., Broumand M.A., Ghaderian S.M.H., Rahimi M., Mirfakhraie R., Azizi F., Omrani M.D. Association of Increased Levels of lncRNA H19 in PBMCs with Risk of Coronary Artery Disease. Cell J. 2019;20:564–568. doi: 10.22074/cellj.2019.5544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jin F., Wang N., Zhu Y., You L., Wang L., De W., Tang W. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell. Physiol. Biochem. 2017;43:2062–2073. doi: 10.1159/000484191. [DOI] [PubMed] [Google Scholar]
- 8.Chen L., Yao H., Hui J.Y., Ding S.H., Fan Y.L., Pan Y.H., Chen K.H., Wan J.Q., Jiang J.Y. Global transcriptomic study of atherosclerosis development in rats. Gene. 2016;592:43–48. doi: 10.1016/j.gene.2016.07.023. [DOI] [PubMed] [Google Scholar]
- 9.Shi X., Liu Z., Liu Z., Feng X., Hua F., Hu X., Wang B., Lu K., Nie F. Long noncoding RNA PCAT6 functions as an oncogene by binding to EZH2 and suppressing LATS2 in non-small-cell lung cancer. EBioMedicine. 2018;37:177–187. doi: 10.1016/j.ebiom.2018.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jin J.J., Lv W., Xia P., Xu Z.Y., Zheng A.D., Wang X.J., Wang S.S., Zeng R., Luo H.M., Li G.L., Zuo B. Long noncoding RNA SYISL regulates myogenesis by interacting with polycomb repressive complex 2. Proc. Natl. Acad. Sci. USA. 2018;115:E9802–E9811. doi: 10.1073/pnas.1801471115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lu J., Ji H., Tang H., Xu Z. microRNA-124a suppresses PHF19 over-expression, EZH2 hyper-activation, and aberrant cell proliferation in human glioma. Biochem. Biophys. Res. Commun. 2018;503:1610–1617. doi: 10.1016/j.bbrc.2018.07.089. [DOI] [PubMed] [Google Scholar]
- 12.Wen X., Han X.R., Wang Y.J., Fan S.H., Zhuang J., Zhang Z.F., Shan Q., Li M.Q., Hu B., Sun C.H. Effects of long noncoding RNA SPRY4-IT1-mediated EZH2 on the invasion and migration of lung adenocarcinoma. J. Cell. Biochem. 2018;119:1827–1840. doi: 10.1002/jcb.26344. [DOI] [PubMed] [Google Scholar]
- 13.Zhu L., Xu P.C. Downregulated LncRNA-ANCR promotes osteoblast differentiation by targeting EZH2 and regulating Runx2 expression. Biochem. Biophys. Res. Commun. 2013;432:612–617. doi: 10.1016/j.bbrc.2013.02.036. [DOI] [PubMed] [Google Scholar]
- 14.Battistelli C., Cicchini C., Santangelo L., Tramontano A., Grassi L., Gonzalez F.J., de Nonno V., Grassi G., Amicone L., Tripodi M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene. 2017;36:942–955. doi: 10.1038/onc.2016.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wierda R.J., Rietveld I.M., van Eggermond M.C., Belien J.A., van Zwet E.W., Lindeman J.H., van den Elsen P.J. Global histone H3 lysine 27 triple methylation levels are reduced in vessels with advanced atherosclerotic plaques. Life Sci. 2015;129:3–9. doi: 10.1016/j.lfs.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 16.Lv Y.C., Tang Y.Y., Zhang P., Wan W., Yao F., He P.P., Xie W., Mo Z.C., Shi J.F., Wu J.F. Histone methyltransferase enhancer of zeste homolog 2-mediated abca1 promoter DNA methylation contributes to the progression of atherosclerosis. PLoS ONE. 2016;11:e0157265. doi: 10.1371/journal.pone.0157265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sato S., Imachi H., Lyu J., Miyai Y., Fukunaga K., Dong T., Ibata T., Kobayashi T., Yoshimoto T., Kikuchi F. Effect of TNF-α on the expression of ABCA1 in pancreatic β-cells. J. Mol. Endocrinol. 2018;61:185–193. doi: 10.1530/JME-18-0167. [DOI] [PubMed] [Google Scholar]
- 18.Xu Y., Liu Q., Xu Y., Liu C., Wang X., He X., Zhu N., Liu J., Wu Y., Li Y. Rutaecarpine suppresses atherosclerosis in ApoE-/- mice through upregulating ABCA1 and SR-BI within RCT. J. Lipid Res. 2014;55:1634–1647. doi: 10.1194/jlr.M044198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L., Yang W., Guo Y., Chen W., Zheng P., Zeng J., Tong W. Exosomal lncRNA GAS5 regulates the apoptosis of macrophages and vascular endothelial cells in atherosclerosis. PLoS ONE. 2017;12:e0185406. doi: 10.1371/journal.pone.0185406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu M., Chen X., Lin K., Zeng K., Liu X., Pan B., Xu X., Xu T., Hu X., Sun L. The long noncoding RNA SNHG1 regulates colorectal cancer cell growth through interactions with EZH2 and miR-154-5p. Mol. Cancer. 2018;17:141. doi: 10.1186/s12943-018-0894-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Song Z., Wu W., Chen M., Cheng W., Yu J., Fang J., Xu L., Yasunaga J.I., Matsuoka M., Zhao T. Long noncoding rna anril supports proliferation of adult t-cell leukemia cells through cooperation with ezh2. J. Virol. 2018;92 doi: 10.1128/JVI.00909-18. e00909-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu S., Pelisek J., Jin Z.G. Atherosclerosis is an epigenetic disease. Trends Endocrinol. Metab. 2018;29:739–742. doi: 10.1016/j.tem.2018.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang W., Agrawal D.K., Boosani C.S. Cell-specific histone modifications in atherosclerosis (Review) Mol. Med. Rep. 2018;18:1215–1224. doi: 10.3892/mmr.2018.9142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zaina S. Unraveling the DNA methylome of atherosclerosis. Curr. Opin. Lipidol. 2014;25:148–153. doi: 10.1097/MOL.0000000000000059. [DOI] [PubMed] [Google Scholar]
- 25.Abd-Elmawla M.A., Fawzy M.W., Rizk S.M., Shaheen A.A. Role of long non-coding RNAs expression (ANRIL, NOS3-AS, and APOA1-AS) in development of atherosclerosis in Egyptian systemic lupus erythematosus patients. Clin. Rheumatol. 2018;37:3319–3328. doi: 10.1007/s10067-018-4269-x. [DOI] [PubMed] [Google Scholar]
- 26.van der Valk F.M., van Wijk D.F., Stroes E.S. Novel anti-inflammatory strategies in atherosclerosis. Curr. Opin. Lipidol. 2012;23:532–539. doi: 10.1097/MOL.0b013e3283587543. [DOI] [PubMed] [Google Scholar]
- 27.Dou Y., Chen Y., Zhang X., Xu X., Chen Y., Guo J., Zhang D., Wang R., Li X., Zhang J. Non-proinflammatory and responsive nanoplatforms for targeted treatment of atherosclerosis. Biomaterials. 2017;143:93–108. doi: 10.1016/j.biomaterials.2017.07.035. [DOI] [PubMed] [Google Scholar]
- 28.Ye J., Wang C., Wang D., Yuan H. LncRBA GSA5, up-regulated by ox-LDL, aggravates inflammatory response and MMP expression in THP-1 macrophages by acting like a sponge for miR-221. Exp. Cell Res. 2018;369:348–355. doi: 10.1016/j.yexcr.2018.05.039. [DOI] [PubMed] [Google Scholar]
- 29.Zhou Q., Chen J., Feng J., Wang J. Long noncoding RNA PVT1 modulates thyroid cancer cell proliferation by recruiting EZH2 and regulating thyroid-stimulating hormone receptor (TSHR) Tumour Biol. 2016;37:3105–3113. doi: 10.1007/s13277-015-4149-9. [DOI] [PubMed] [Google Scholar]
- 30.Lue J.K., Amengual J.E. Emerging EZH2 Inhibitors and Their Application in Lymphoma. Curr. Hematol. Malig. Rep. 2018;13:369–382. doi: 10.1007/s11899-018-0466-6. [DOI] [PubMed] [Google Scholar]
- 31.Xiaoling Y., Li Z., ShuQiang L., Shengchao M., Anning Y., Ning D., Nan L., Yuexia J., Xiaoming Y., Guizhong L., Yideng J. Hyperhomocysteinemia in ApoE-/- Mice Leads to Overexpression of Enhancer of Zeste Homolog 2 via miR-92a Regulation. PLoS ONE. 2016;11:e0167744. doi: 10.1371/journal.pone.0167744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu S., Xu Y., Yin M., Zhang S., Liu P., Koroleva M., Si S., Little P.J., Pelisek J., Jin Z.G. Flow-dependent epigenetic regulation of IGFBP5 expression by H3K27me3 contributes to endothelial anti-inflammatory effects. Theranostics. 2018;8:3007–3021. doi: 10.7150/thno.21966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vaziri N.D. Lipotoxicity and impaired high density lipoprotein-mediated reverse cholesterol transport in chronic kidney disease. J. Ren. Nutr. 2010;20(5, Suppl):S35–S43. doi: 10.1053/j.jrn.2010.05.010. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y., Zheng L., Wang Q., Hu Y.W. Emerging roles and mechanisms of long noncoding RNAs in atherosclerosis. Int. J. Cardiol. 2017;228:570–582. doi: 10.1016/j.ijcard.2016.11.182. [DOI] [PubMed] [Google Scholar]
- 35.Liang W., Fan T., Liu L., Zhang L. Knockdown of growth-arrest specific transcript 5 restores oxidized low-density lipoprotein-induced impaired autophagy flux via upregulating miR-26a in human endothelial cells. Eur. J. Pharmacol. 2019;843:154–161. doi: 10.1016/j.ejphar.2018.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Yu W., Ying H., Tong F., Zhang C., Quan Y., Zhang Y. Protective effect of the silkworm protein 30Kc6 on human vascular endothelial cells damaged by oxidized low density lipoprotein (Ox-LDL) PLoS ONE. 2013;8:e68746. doi: 10.1371/journal.pone.0068746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ren K., Jiang T., Zhou H.F., Liang Y., Zhao G.J. Apigenin Retards Atherogenesis by Promoting ABCA1-Mediated Cholesterol Efflux and Suppressing Inflammation. Cell. Physiol. Biochem. 2018;47:2170–2184. doi: 10.1159/000491528. [DOI] [PubMed] [Google Scholar]
- 38.Li C., Guo R., Lou J., Zhou H. The transcription levels of ABCA1, ABCG1 and SR-BI are negatively associated with plasma CRP in Chinese populations with various risk factors for atherosclerosis. Inflammation. 2012;35:1641–1648. doi: 10.1007/s10753-012-9479-9. [DOI] [PubMed] [Google Scholar]
- 39.Jiang T., Ren K., Chen Q., Li H., Yao R., Hu H., Lv Y.C., Zhao G.J. Leonurine Prevents Atherosclerosis Via Promoting the Expression of ABCA1 and ABCG1 in a Pparγ/Lxrα Signaling Pathway-Dependent Manner. Cell. Physiol. Biochem. 2017;43:1703–1717. doi: 10.1159/000484031. [DOI] [PubMed] [Google Scholar]
- 40.Gui Y.Z., Yan H., Gao F., Xi C., Li H.H., Wang Y.P. Betulin attenuates atherosclerosis in apoE-/- mice by up-regulating ABCA1 and ABCG1. Acta Pharmacol. Sin. 2016;37:1337–1348. doi: 10.1038/aps.2016.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chao M.L., Guo J., Cheng W.L., Zhu X.Y., She Z.G., Huang Z., Ji Y., Li H. Loss of Caspase-Activated DNase Protects Against Atherosclerosis in Apolipoprotein E-Deficient Mice. J. Am. Heart Assoc. 2016;5:e004362. doi: 10.1161/JAHA.116.004362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.