Abstract
MicroRNAs (miRNAs) have increasingly been shown to be involved in human cancer, and interest has grown about the potential use of miRNAs for cancer therapy. miRNA levels are known to be altered in cancer cells, including in non-small cell lung cancer (NSCLC), a subtype of lung cancer that is the most prevalent form of cancer worldwide and that lacks effective therapies. The let-7 miRNA is involved in the regulation of oncogene expression in cells and directly represses cancer growth in the lung. let-7 is therefore a potential molecular target for tumor therapy. However, applications of RNA interference for cancer research have been limited by a lack of simple and efficient methods to deliver oligonucleotides (ONs) to cancer cells. In this study, we have used in vitro and in vivo approaches to show that HCC827 cells internalize hydrophobically modified let-7b miRNAs (hmiRNAs) added directly to the culture medium without the need for lipid formulation. We identified functional let-7b hmiRNAs targeting the HMGA2 mRNA, one of the let-7 target genes upregulated in NSCLC, and show that direct uptake in HCC827 cells induced potent and specific gene silencing in vitro and in vivo. Thus, hmiRNAs constitute a novel class of ONs that enable functional studies of genes involved in cancer biology and are potentially therapeutic molecules.
Introduction
let-7 encodes a microRNA (miRNA) that targets HMGA2 in lung cancer.1, 2, 3, 4, 5, 6 Lung cancer is responsible for the most cancer-related deaths for both men and women in the US with an estimated 224,390 new cases and 158,080 deaths from the disease in 2016.7 Regardless of improving survival rates in the last decades, only about 18% of patients are alive 5 years after diagnosis, emphasizing the urgent need for better therapies.
MiRNAs are small non-coding RNAs that control gene expression at a post-transcriptional step and have important functions in tumorigenesis1,8,9 (Figure S1). miRNAs are known to be dysregulated in many types of cancer, and specific miRNA expression patterns characterize normal versus tumor tissue or differentiated versus poorly differentiated tumors.8,10, 11, 12. Upregulated miRNAs in cancer cells promote carcinogenesis by attenuating tumor suppressor genes and are known as oncomiRs (oncogenic miRNAs).13 Alternatively, miRNAs with low expression levels in cancer cells that typically halt cancer progression by suppression of the expression of oncogenes are called tumor suppressor miRNAs.14 Silencing highly expressed miRNA with anti-miRNA (antimiRs or antagomirs) or substituting tumor suppressor miRNAs with miRNA mimetics has been demonstrated as a valuable experimental strategy for the treatment of cancer. In particular, we have shown the let-7 has promise as a tumor suppressor in pre-clinical mouse models of lung cancer.15, 16, 17
Therapeutic oligonucleotides (ONs) are an emerging therapeutic alternative to treat diseases with known genetic origin—i.e., antisense ONs, small interfering RNA (siRNA),18 miRNAs,11 and aptamers19—and are revealing themselves as a new class of drugs. Their advantages over conventional drugs include: simple construction, logically achieved based on sequence data and straightforward screening, short time frame, leading to drug candidates, and good potency and duration of effect.11 Nonetheless, as this emerging field has developed in the last two decades it has become obvious that there are several limitations to overcome before such RNAi-based drug candidates become a reality.
Problems such as delivery-associated toxicity, poor transfection efficiency, systemic clearance, nonspecific biodistribution, and degradation in circulation have delayed the therapeutic potential of miRNAs.20,21 Perhaps the most challenging concern that is now dealt with by many academic laboratories and biotech companies is oligonucleotide drug delivery to extrahepatic tissues in the body.22,23 This major limitation is partly due to the membrane impermeability of oligonucleotide therapeutics, since there is no passive diffusion of the ON to most cells and therefore the ON is delivered at a high (potentially toxic) dose or needs some directed mechanism by which it reaches its specific target. The absence of powerful delivery vehicles to target tissues outside the liver is now a primary bottleneck in miRNA-based therapeutics.
Lipid nanoparticles (LNPs) are attractive carriers for the delivery of nucleic acids due to their protection from nucleases,24,25 and they have been used for miRNA delivery in vivo.26,27 Most LNP fabrication favors delivery of miRNAs to liver, but engineering the lipid design of LNPs has been shown to alter their distribution, granting potent delivery to other organs.28,29 Thus, different lipid elements might be used to enhance miRNA distribution. Indeed, lipid conjugates have been studied to increase miRNA bioavailability. Attachment of the first-tested lipophilic group, cholesterol (Chol), which interacts with serum components such as albumen and lipoproteins, increased circulation times and passive accumulation in the liver,30,31 but it allowed distribution to other tissues too, including muscle and placenta, where they produced RNA silencing. Other lipid-conjugated miRNAs have been explored, including fatty acids and vitamin-conjugated miRNA, but delivery and potency were only evaluated in liver.32, 33, 34
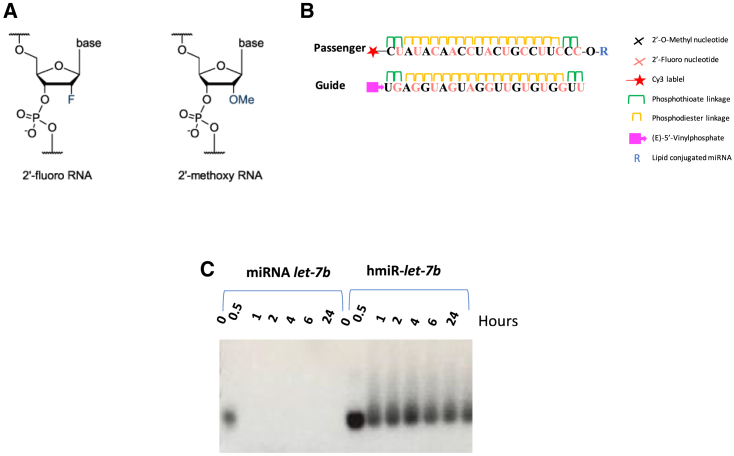
Moreover, there are potential unwanted effects that are the result of the RNA duplex itself. These include (1) the activation of the immune response; (2) the incorporation of the passenger strand (sense) into RISC (RNA-induced silencing complex); (3) the miRNA potentially silencing genes other than the intended target gene; and (4) circulating time in the tissues. Several chemical modifications such as 2′-fluoro (2′-F)35 and 2′-O-methyl (2′-OMe)1 (Figure 1A) have been shown to lower such activities, pointing to the beneficial results of using synthetic RNA molecules to not only improve biostability but also to reduce toxicity.
Figure 1.
Chemical modifications stabilize let-7b
(A) Modifications of ribose at the 2′ position. (B) General scheme of lipid-conjugated hmiRNAs. Top is the passenger strand shown 5′ to 3′. Bottom is the guide strand shown 5′ to 3′. (C) Stability of 5 μM hmiR-let-7b versus miRNA let-7b in 50% serum.
Reducing the loading of the passenger strand to RISC has also been achieved by a variety of chemical modifications, some by specific miRNA design36 and others by replacing natural bases with synthetic ones. There are a variety of nucleotide mimics where the ribose is modified to increase affinity for the target and/or increase nuclease resistance (e.g., locked nucleic acid [LNA]37) or by 5′-capping.38,39 The 2′-OMe modification has been shown to improve stability, reduce the immune response, and improve passenger strand RISC incorporation.40 Moreover, the substitution of only one non-bridging oxygen of a phosphodiester with a sulfur atom creates the phosphorothioate linkage. Not only does the PS linkage protect oligonucleotides from nucleases, but PS oligonucleotides associate with plasma proteins, resulting in longer circulation times.41 Recently, fully hydrophobically modified siRNA (hsiRNAs) were shown to improve functionality, while lipid-conjugated hsiRNAs were delivered to a variety of extra-hepatic tissues.42, 43, 44, 45
Here, we examined a novel oligonucleotide design to mimic let-7b and avoid some of the barriers of miRNA delivery mentioned above. We show fast internalization of hydrophobically modified let-7b miRNAs (hmiRNAs) in the non-small cell lung cancer (NSCLC) cell line HCC827 when added directly to the culture medium without lipid formulation. We analyzed functional let-7b hmiRNAs targeting of the mRNA of HMGA2,16,46,47 which is one of the genes upregulating during NSCLC, and show that direct uptake in HCC827 cells leads to potent and specific silencing in vitro. In addition, we systematically evaluated how lipid conjugates affect hmiRNA distribution in vivo. We synthesized hmiRNAs conjugated to three lipophilic moieties. We show that lipid conjugates enable miRNA accumulation and productive silencing in tumors after subcutaneous injection. Although most of the injected miRNA accumulates in clearance organs, we identified conjugates that enable functional miRNA delivery to lung tumor cells. The tissue concentrations of miRNA necessary for effective silencing varied depending on the tissue and lipid construct. Our findings provide proof of principle for the development of lipid-conjugated hmiRNA oligonucleotides as possible therapeutics for NSCLC.
Results
Synthesis of DCA, EPA, and Cholesterol-hmiR-let-7b Conjugates
hmiRNA is a symmetric compound composed of a 21-nt modified RNA duplex. Nucleotides in the hmiRNA are modified with 2′-OMe or 2′-F (Figure 1A, passenger and guide strands) to promote stability, and the 3′ end of the passenger strand is conjugated to eicosapentaenoic acid (EPA), docosanoic acid (DCA), or Chol lipid to promote membrane binding and association. The 5′ end of the passenger strand is conjugated to Cy3 for imaging. The strands backbone contains hydrophobic phosphorothioate linkages and promotes cellular uptake by a mechanism similar to that of antisense oligonucleotides. The presence of phosphorothioates, ribose modifications, and a lipid conjugate contributes to overall hydrophobicity and is essential for compound stabilization and efficient cellular internalization.
To evaluate how the structures of lipid conjugates affect miRNA distribution in vitro and in vivo, we synthesized three hmiRNAs-let-7b mimics conjugated with naturally occurring lipids, including EPA, the saturated fatty acid 22:0, DCA, and sterols Chol (Figure 1B). Each lipid conjugate was covalently attached to the 3′ end of the miRNA passenger strand, which tolerates a range of covalent modifications.48 The miRNA-let-7b used in these studies was fully chemically modified for maximal stability.
To test the stability of hmiR-let-7b as compared to the naked miRNA let-7b, both sequences were exposed to 50% serum, which mimic the physiological conditions. miRNA let-7b was highly unstable in the presence of serum, whereas hmiR-let-7b remained intact for up to 24 h (Figure 1C), suggesting that the modifications protect the miRNA mimic from serum nucleases.
hmiRNAs are Efficiently Internalized by HCC827 NSCLC Cells
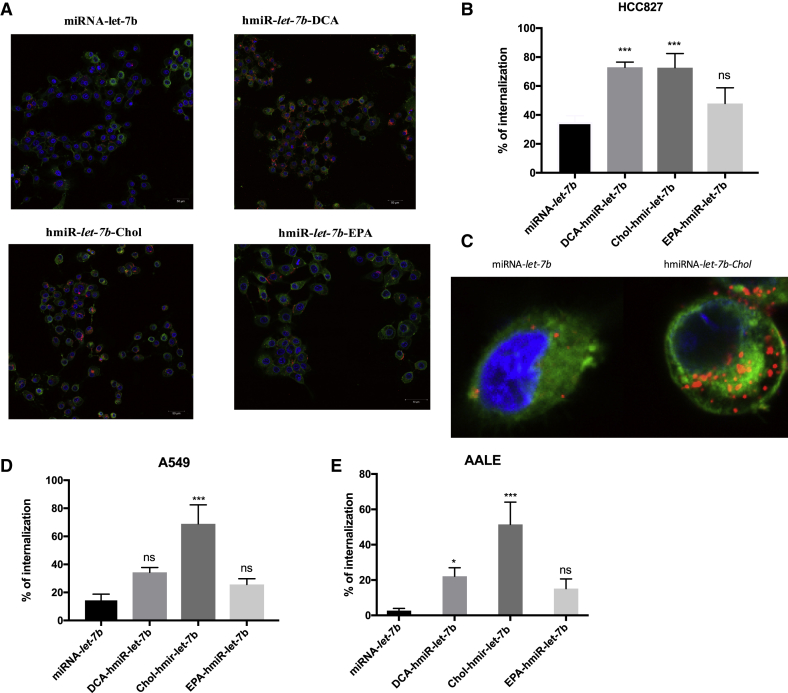
Here, we evaluated whether hydrophobically modified miRNAs are efficiently internalized by NSCLC cell lines HCC827 and A549, and by normal lung cells, AALE. We found that when added to the culture medium, the Cy3-labeled hmiRNA-cholesterol conjugate was rapidly internalized into all three cell lines (Figure 2). Cy3-labeled hmiRNA-Cholestrol was observed in almost every cell in the culture, demonstrating efficient and uniform uptake with 73%, 69%, and 51% for HCC827, A549, and AALE cells, respectively. Internalization of Cy3-labeled hmiRNA-DCA was observed frequently in the HCC827 cell line (73%), but less frequently in the normal lung cells and in A549 cells. The internalization of the EPA conjugate was more frequent than the naked miRNA-let-7b in all three cell lines, although this was not significant.
Figure 2.
hmiR-let-7b-conjugate internalization
(A) Representative images of hmiR-let-7b-conjugate in the HCC827 cell line. (B) Internalization percentage of 0.4 μM hmiR-let-7b conjugates in the HCC827 cell line after 24 h. ***p < 0.005. Statistical analysis by one-way ANOVA was adjusted for multiple comparisons. Size bar: 50 μm. (C) Magnified representative images of a miRNA-let-7b and hmiRNA-let-7b-Cholesterol-treated cell. (D) Internalization percentage of hmiR-let-7b conjugates in the A549 cell line after 24 h. (E) Internalization percentage of hmiR-let-7b conjugates in the AALE cell line after 24 h.
hmiR-let-7b-Cholesterol Increases Apoptosis in HCC827 and A549 Cells
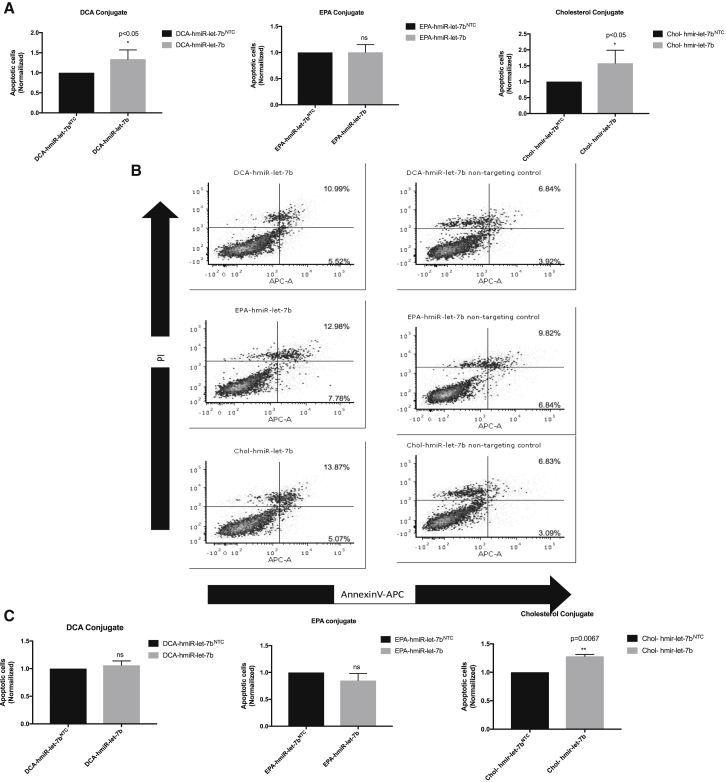
To further investigate the potential tumor suppressor role of hmiR-let-7b conjugates in HCC827 and A549 cells, we treated cells with 1 μM hmiR-let-7b conjugate and a control hmiRNA non-targeting control (NTC) for 72 h. hmiRNA-NTC consisted of a double-stranded miRNA containing a sequence without any target gene. Cells then were labeled with annexin V-allophycocyanin (APC) and propidium iodide (PI) and analyzed using flow cytometry. A significant increase in apoptosis activity was observed with the cholesterol conjugate as compared to control hmiRNA-NTC in HCC827 and A549 cell lines (Figures 3A and 3C). The DCA and EPA conjugates showed less of an increase of apoptotic cells (Figures 3A and 3C). Representative scatterplots for hmiR-let-7b conjugates in HCC827 are shown in Figure 3B.
Figure 3.
hmiR-let-7b Conjugates Increase Apoptosis In Vitro
(A) Mean of annexin V+ PI+ from at least three independent experiments at 72 h post-treatment with hmiR let-7b conjugate (DCA, EPA, and Chol) in the HCC827 cell line. *p < 0.05, analyzed using t test. (B) Representative annexin V and PI scatterplots for HCC827 cells following treatment with hmiR-let-7b conjugates and NTC. Apoptotic cells were normalized to NTC from each individual experiment. (C) Mean of annexin V+ PI+ from at least three independent experiments at 72 h post-treatment with hmiR let-7b conjugate (DCA, EPA, and Cholesterol) in A549 cell line. **p < 0.005.
Delivery of hmiR-let-7b Conjugates Promotes let-7b Engagement in RISC
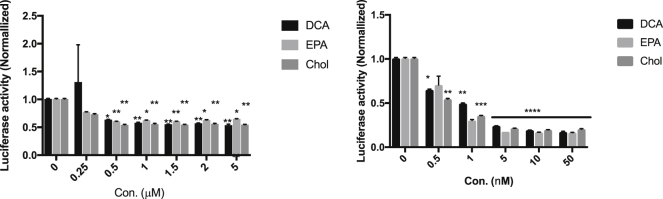
To validate the activity of hmiR-let-7b conjugates, we used a functional assay to monitor RISC/RNAi activity using a let-7 sensor. Luciferase reporter assays were performed in HCC827 cells that express a luc reporter with perfect let-7 complementary sites in its 3′ UTR. Luciferase fluorescence levels in these cells are thus inversely correlated with let-7 activity. To test whether hmiR-let-7b conjugates preserve their RNAi efficiency, HCC827 cells were co-transfected (lipid mediated) with the luc 3′ UTR let-7 sensor together with each conjugate separately using Lipofectamine 2000 in a range of concentrations of 0.5–100 nM. Following 72 h, luciferase activity was measured and showed a decrease of up to 80% and a dose response with all three conjugates (Figure 4B). This reduction in luciferase activity was not observed in untreated cells.
Figure 4.
Dose Response of HCC827 Cells to hmiR-let-7b Conjugates
Renilla values were measured 72 h after treatment. Data points were normalized to untreated cells. (A) Passive delivery. (B) Lipid-mediated (lipo2000) delivery. *p < 0.05, **p < 0.005, ****p < 0.0001. Means ± SD. Statistical analysis was by two-way ANOVA adjusted for multiple comparisons.
To further test self-delivery (passive delivery) efficacy of the three conjugates, cells were transfected with the luc 3′ UTR let-7 sensor on the first day, and the next day cells were treated with hmiR-let-7b conjugates (0.25–5 μM). Following 72 h, luciferase activity was measured and showed a decrease up to 45% and a dose response with all three conjugates (Figure 4A). Thus, our cell-based experiments indicated a rapid reduction in luciferase activity in hmiR-let-7b conjugate-treated cells 72 h after treatment regardless of the method of delivery.
Lipid Conjugates Increase Endogenous let-7 miRNA and Decrease Its Target Genes In Vitro
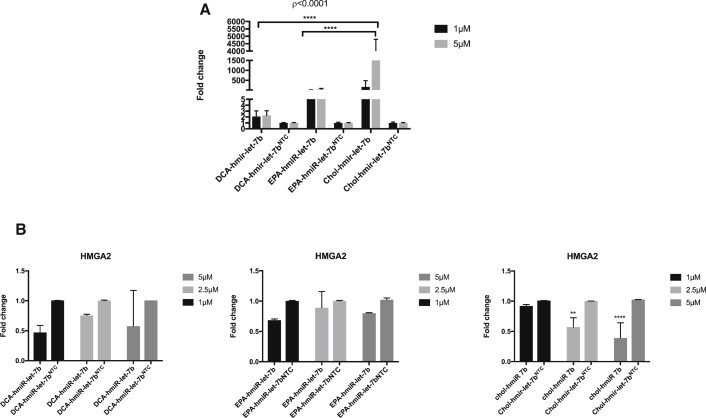
To confirm in vitro delivery of miRNA let-7b, we first treated HCC827 cells with EPA, DCA, or cholesterol hmiR-let-7b conjugates, and the levels of miR-let-7b and its target genes were quantified using quantitative real-time PCR. A significant increase in let-7b levels following 48 h of treatment was observed compared to the hmiR-NTC conjugate-treated cells, with a 2,000-fold change increase for cholesterol conjugate, 150-fold change increase for EPA, and a 2-fold change increase for DCA (Figure 5A). hmiR-let-7b conjugate delivery was active and potently downregulated the let-7 target gene HMGA2. Downregulation of this target gene ranged between 40% and 60% (Figure 5B). Treatment with a hmiR-NTC had no effect on the expression levels of the evaluated genes.
Figure 5.
Expression levels of let-7b and HMGA2
(A) The expression level of let-7b-5p in HCC827 48 h post-treatment with miRNA conjugates. Assays were performed in triplicate. miRNA let-7b levels were normalized to miRNA-423 levels. ****p < 0.001. Means ± SEM are shown. (B) Expression level of HMGA2 in HCC827 48 h post-treatment with miRNA conjugates. Assays were performed in triplicate, and HMGA2 mRNA was normalized to GAPDH mRNA levels. **p < 0.005, ****p < 0.0001. Means ± SD are shown. Statistical analysis was by two-way ANOVA adjusted for multiple comparisons
HmiR-let-7b Conjugate Distribution In Vivo after Subcutaneous Injection
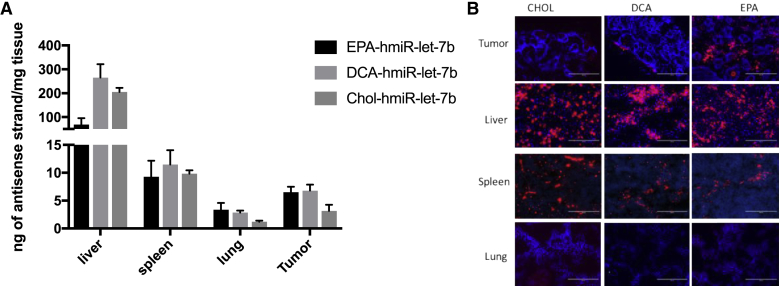
We next determined the tissue-target specificity of the hmiR-let-7b conjugate in vivo. To this end, immunodeficient (nu/nu) mice bearing HCC827 tumors were treated with a single subcutaneous injection (20 mg/kg) of EPA, Cholesterol, DCA, or PBS. Mice were then sacrificed 24 h after administration of the conjugates, and tumors/organs (lung, liver, and spleen) were harvested. The miRNA passenger/sense strands were labeled at their 5′ ends with a Cy3 fluorophore for qualitative analysis of tissue sections by fluorescence microscopy. We also quantified the absolute concentrations of let-7b guide/antisense strand in tissue using a peptide nucleic acid (PNA) hybridization assay and compared levels of miRNA in those tissues with the control group (qRT-PCR). As expected, conjugated hmiR-let-7b accumulated mostly in liver and spleen (Figure 6). We observed a clear relationship between miRNA lipophilicity and the amount of miRNA that accumulated in these tissues. High lipophilic hmiRNAs with DCA and cholesterol conjugates accumulated to higher levels in liver and spleen than did the low lipophilic miRNA-EPA conjugate, which accumulated less in the liver or spleen (PNA assay, absolute amount) (Figure 6A). Although the highest amounts of hmiR-let-7b were detected in liver and spleen, significant amounts of let-7b were detected in the tumor, with the highest amount with EPA and DCA conjugates (∼8 ng miRNA per mg of tissue). The lowest levels of let-7b were in normal lung tissues. Taken together and as compared to DCA and Cholesterol conjugates, the hmiR-let-7b-EPA conjugate accumulates less in liver, spleen, and normal lung tissue and accumulates more in tumor, making it a potential drug delivery strategy for NSCLC. Similar results were observed in fluorescence microscopy analysis (Figure 6B).
Figure 6.
MiRNA-let-7b expression in vivo after 24 hours
(A) Quantification of Let-7b antisense strand in liver, spleen, lung, and tumor after 24 h subcutaneously at 20 mg/kg (PNA hybridization assay). (B) Fluorescence images of Cy3-labeled DCA-, EPA-, and Cholesterol-conjugated miRNAs in liver, lung, spleen, and tumor. Scale bars, 200 μm.
EPA-hmiR-let-7b Conjugates Increased the Amount of Tumor let-7b miRNA and Enabled Functional Gene Silencing In Vivo
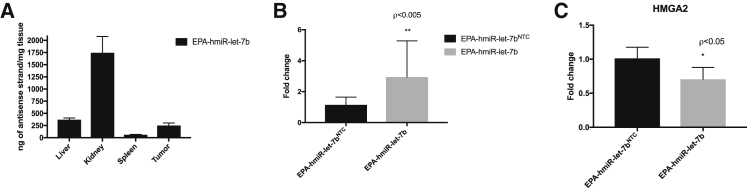
To determine whether EPA-conjugated hmiR-let-7b accumulates to levels sufficient for productive gene silencing, we injected mice subcutaneously with EPA-conjugated hmiR-let-7b. We injected the EPA conjugate at a dose of 20 mg/kg (seven mice per hmiRNA conjugate [hmiR-let-7b and non-targeting control (NTC), twice a week for 21 days and sacrificed the mice 48 h after the last injection. We measured the amount of let-7b in the tumors by PNA hybridization assay. Although most of the miRNA was found in the kidney, we found that the concentration of let-7b in tumor was 250 ng of miRNA per mg of tissue (Figure 7A). We measured a 3-fold change of miRNA let-7b levels in tumors relative to hmiRNA-NTC (Figure 7B). Furthermore, the increase of let-7b levels in tumor-bearing mice treated with EPA-hmiR-let-7b resulted in 30% downregulation of HMGA2 mRNA (GAPDH, serving as a control housekeeping gene) (Figure 7C). hmiRNA-NTC showed no significant reduction in target gene expression, indicating that the observed silencing is due to sequence-specific effects rather than the general chemical scaffold.
Figure 7.
let-7b and HMGA2 expression levels invivo
(A) Quantification of EPA-let-7b miRNA in liver, kidney, spleen, and tumor after 3.5 weeks subcutaneously at 2 × 20 mg/kg per week (PNA hybridization assay, n = 7). (B) qRT-PCR expression of EPA-miR-let-7b-5p expression. miRNA levels were normalized to miRNA-423 levels. (C) HMGA2 mRNA expression levels in tumors. mRNA levels were normalized to GAPDH mRNA levels. *p < 0.05, **p < 0.005. Statistical analysis was by two-way ANOVA adjusted for multiple comparisons
Effect of EPA-hmiR-let-7b on Tumor Growth and Ki-67 Immunoreactivity
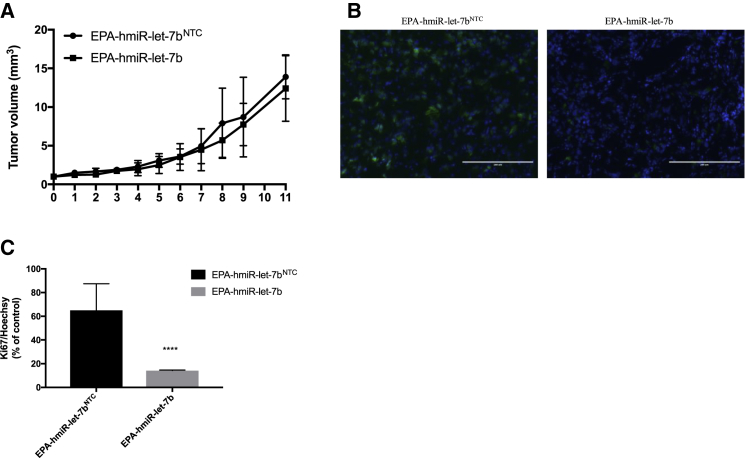
We observed a small but not significant change in tumor size between the treatment and NTC arms of the xenograft experiment (Figure 8A). In addition, tumor samples from the treatment group and NTC treatment were examined histologically using standard H&E staining after OCT (optimal cutting temperature) fixation. In both treatment and control groups, the histological findings did not identify cellular structures significantly different between the groups (Figure S2).
Figure 8.
HCC827 cell proliferaton in vivo
(A) Tumor volume. (B) The effect of EPA-hmiR-let-7b on the cell proliferation was investigated by antibody against Ki-67. Representative images from sections of the treated tumors are shown. C) Quantification of the intensity of Ki-67 and Hoechst staining. Images were captured at ×20 magnification from at least four randomly selected fields for each of six independent mice. ****p < 0.0005, analyzed using a t test. Size bar: 200 μm.
In order to determine whether EPA-hmiR-let-7b is able to reduce the proliferation in xenograft tumors at 3.5 weeks post-treatment, in this study, Ki-67 expression was used to detect alterations in cell proliferation. Tumor sections from the two groups were stained using immunocytochemistry method with Ki-67, a proliferating marker (green), and Hoechst (blue). The results (Figures 8B and 8C) showed that EPA-hmiR-let-7b reduced the proliferative condition compared to the NTC control. As shown in Figure 8B, EPA-hmiR-let-7b decreased the ratio of Ki-67/Hoechst to 14.21 ± 0.378% of the control treatment (p < 0.0001). The result revealed that EPA-hmiR-let-7b treatment decreased the expression of Ki-67 when compared to EPA-hmiR-let-7b NTC (Figures 8B and 8C). These immunohistochemistry (IHC) studies suggested that EPA-hmiR-let-7b slowed proliferation of the tumor cells, but this effect did not result in a significant effect on tumor volume at 3.5 weeks post-treatment.
Discussion
NSCLC represents about 85% of all lung cancers and is mostly diagnosed at an advanced stage.49 Overexpression of miRNA mimics in cancer cells has been known as a potential therapeutic strategy for the last 10 years, including for lung cancer.15, 16, 17,50,51 However, there are challenges in the translation of miRNAs into clinical applications, including low stability due to its degradation by endonucleases/exonucleases,52,53 and lack of potent delivery to the cancer cells due to their negative charges.54 Previous work has revealed that hydrophobic modifications to siRNAs improves their stability without a reduction in their efficacy,55 while lipids are class of molecules that can be used to improve drug-targeted delivery.56,57
Our research in the present study addresses the potential of a novel therapeutic approach combining a lipid and hydrophobically modified let-7b miRNA that can be useful to target NSCLC with silencing of HMGA2 mRNA. Indeed, the presence of high levels of HMGA2 mRNA has been observed with lung cancer.2,5,6,58,59
Here, we show that cell-targeting lipids, which are crucial for miRNA retention in the body, might be used as a carrier for therapeutic miRNAs in vivo. Lipids with different lipophilicity, Cholesterol, EPA, and DCA were covalently linked56 to let-7b miRNA, a tumor suppressor miRNA.60 To test sequence specificity, we also conjugated the same lipid to NTC miRNA.
A major limitation of RNA-based drugs in vivo is the fast degradation (few minutes) of natural ribonucleotide in serum or blood. In order to keep the RNA intact and protect it from degradation, we synthesized miRNA mimics with 2′-F and 2′-OMe modifications. We evaluated the internalization of three lipid conjugates in three cell lines, HCC827, A549, and AALE, and found that the Cholesterol conjugate was internalized in all three cell lines, while the EPA conjugate and DCA conjugate were less well internalized.
We showed that in vitro in HCC827 cells, hmiR-let-7b (the silencing moiety with lipid conjugates) preserved their activity, resulting in high levels of miR-let-7b and silencing of a well-known let-7b oncogene target, HMGA2. The Cholesteol conjugate appeared to be best for silencing in vitro. Interestingly, at 1 μM the silencing for the Cholesterol conjugate was not significant even though the miRNA levels were high. This could be explained by the known observation that the degree of silencing does not always correlate with the level of compound accumulation.44 The structure of the conjugate (EPA, cholesterol, or DCA) may define productive silencing by impacting cellular internalization and/or endosomal escape.
Moreover, we have found that overexpression of hmiR-let-7b-cholesterol in the target cell lines HCC827 and A549 was correlated with an increase in apoptotic cells and endogenous miRNA let-7b, and this compound was again found to be the best candidate in vitro. The AALE cell line was not tested because of difficulty with cell growth, but we can assume that the degree of apoptosis would be less, as the internalization in this cell line was also less than in the other two cell lines.
Our in vivo biodistribution study with the three lipid conjugates revealed a different result, and hmiR-let-7b-EPA was found to be the best candidate. Its injection in three mice showed the highest expression of miRNA let-7b in tumor tissue. With successive subcutaneous administration of the optimized lipid (EPA)-hmiR-let-7b construct, we noticed detectable uptake into target HCC827 tumors in vivo. The injected amount of the lipid-conjugated miRNAs let-7b was delivered mostly to primary clearance tissues, including liver, kidney, and spleen. Nevertheless, lipid-miRNA let-7b conjugate delivered to HCC827 tumor cells was capable of silencing HMGA2 mRNA that is overexpressed in the disease tumor tissue. Furthermore, hmiR-let-7b-EPA was able to reduce tumor cell proliferation.
Conclusion
Our study demonstrates that fully hydrophobically modified miRNAs are effective in gene silencing, and thus they might be a useful chemical compound in therapeutic studies. Direct conjugation of RNAi to a targeting agent is emerging as a promising alternative to nanoparticle-based cargoes for selective delivery, especially to the liver.61,62 However, many lipid delivery approaches attempt to target tissues other than the liver. We show lipid-targeted delivery of a miRNA with biologic activity to tumor tissue in vivo as a proof of concept for future progress of other targeted miRNA-based therapies. Although our studies were in the context of cancer, this application can be applied to different fields of RNA therapeutics by specifying a safe, effective strategy to deliver functional miRNAs to target tissues.
Materials and Methods
Cell Line
The HCC827 cell line was obtained from Dr. S. Kobayashi (Beth Israel Deaconess Medical Center/Harvard Medical School, USA). A549 cells were obtained from ATCC. AALE cells were a gift from Matthew Meyerson’s laboratory. Cells were grown in 90% DMEM supplemented with 10% fetal bovine serum (FBS) and 1× penicillin/streptomycin solution. Media and supplements were purchased from Thermo Fisher (Waltham, MA, USA). All cells were maintained at 37°C in 5% CO2.
Oligonucleotide Synthesis, Deprotection, and Purification
Oligonucleotides were synthesized following standard protocols on a MerMade 12 (BioAutomation, Irving, TX, USA). Sense strands were synthesized at 1- or 2-μmol scales on synthesized fatty acid-functionalized CPG supports.44 Antisense strands were synthesized at 10-μmol scales on CPG functionalized with Unylinker (ChemGenes, Wilmington, MA, USA). 2′-O-methyl phosphoramidites (ChemGenes, Wilmington, MA, USA), 2′-fluoro phosphoramidites (BioAutomation, Irving, TX, USA), Cy3-labeled phosphoramidites (GenePharma, Shanghai, China), and custom synthesized (E)-vinylphosphonate phosphoramidites were used.63 Oligonucleotides were removed from CPG and deprotected using 40% aqueous methylamine, purified by high-performance liquid chromatography (HPLC), and desalted by size-exclusion chromatography. Oligonucleotide purity and identity were determined by liquid chromatography-mass spectrometry (LC-MS) analysis on an Agilent 6530 Accurate-Mass Q-TOF LC/MS (Agilent Technologies, Santa Clara, CA, USA). The NTC sequence has the same modification and lipids conjugates: antisense, Pi-UPsUPsAAUCUCUUUACUGAUAUPsAPsU; sense, lipid-CPsAPsUAUAUCAGUAAAGAGAUUPsAPsA-Cy3 (X, 2′-OMe nucleotide; X, 2′-F nucleotide; Ps, phosphothioate).
Live Cell Imaging
To monitor live cell hmiRNA internalization, cells were plated at a density of 0.2 × 105 cells per well on a chambered glass-bottom dish. Imaging was performed in phenol red-free medium (Thermo Fisher, Waltham, MA, USA). Cells were treated with 0.4 μmol/L Cy3-labeled hmiRNA, and live cell imaging was performed after 24 h by a Zeiss LSM 880 confocal microscope at the Confocal Imaging Core of Beth Israel Deaconess Medical Center.
hmiR-let-7b Stability in Serum
miRNA duplex at 0.5 mM was diluted in 50% FBS with a final concentration of 5 μM and incubated at 37°C. At the desired time points, an aliquot from the assay tube was transferred to a fresh tube containing GreenGlo (5 μL, Thomas Scientific, Swedesboro, NJ, USA). Samples were run on 4%–20% PAGE and analyzed by a Bio-Rad camera.
Flow Cytometry Analysis of Apoptosis
Cultured cells were harvested by trypsinization and washed with 1× PBS. For each sample, 1 × 106 cells were stained with the annexin V‐APC/PI apoptosis detection kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s instructions, and then analyzed using fluorescence-activated cell sorting (FACS; BD FACSCanto II, BD Biosciences, CA, USA).
Activity Study in psiCHECK-2 System
The psiCHECK constructs contained a single copy of matched sequence complementary to the miRNA guide or one copy of a mismatched sequence. For testing the activity of miRNA, 1 × 105 human HCC827 cells were seeded in 24-well plates. Cells were then incubated in a 37 ± 1°C, 5% CO2 incubator for 24 h. The next day the cells were transfected with 500 ng of let-7 luciferase reporter, which carries the wild-type miR-let-7 sequence (Addgene plasmid no. 50827) using Lipofectamine 2000 reagent, according to the manufacturer’s procedure, and incubated for 24 h at 37 ± 1°C and 5% CO2. Following incubation, cells were transfected (using Lipofectamine 2000 reagent) or treated with hmiRNA conjugates at final concentrations ranging from 0.5 to 100 nM (lipid mediated) or 0.25 to 5 μM (passive delivery) in a 500-μL final volume. Cells were then incubated for 72 h at 37 ± 1°C following assessment of Renilla and firefly luciferase activities. Activities were measured in each of the miRNA-transfected or treated samples using the Dual-Luciferase assay kit (Promega, catalog no. E1960) according to the manufacturer’s procedure. Renilla luciferase activity value was divided by Firefly luciferase activity value for each sample (normalization).
RNA Isolation and miRNA Expression Analyses Using qPCR
HCC827 derived tumors from Nu/Nu mice (NU-Foxn1nu; Jackson Laboratory) were collected in tubes and stored at −80°C. Tumor and tissues were ground and placed in 400 μL of TRIzol. 100 μL of RNA was extracted using a Direct-zol RNA Minirep kit (Zymo Research) according to the manufacturer’s instructions.
qRT-PCR
Total RNA from the HCC827 cell line or tumors, lungs, livers, and spleen was isolated using the Direct-zol RNA MiniPrep Plus kit (Zymo Research) according to the vending company’s instructions. RNA quantification was estimated with a DS-11 spectrophotometer (DeNovix). For RT-PCR detection of let-7 mRNA target genes, 200–500 ng of RNA was reverse transcribed with a TaqMan miRNA assay (Applied Biosystems, Foster City, CA, USA) with the following conditions: 42°C for 60 min and 85°C for 5 min. For RT-PCR detection of the let-7b guide strand, 10 ng of purified RNA was reverse transcribed using the let-7b TaqMan miRNA assay (Applied Biosystems, Foster City, CA, USA) with the following conditions: 42°C for 15 min and 85°C for 5 min. Gene and let-7b expression levels were determined by real-time PCR using Taq polymerase reagents (Invitrogen) on the LightCycler 480 II (Roche). TaqMan gene expression assays (Applied Biosystems) were used with the following cycling conditions: 95°C for 1 min (initial denaturation), then 45 cycles of 95°C for 5 s, 60°C for 30 s. The GAPDH mRNA or miRNA 423 was amplified as an internal reference. For the in vivo biodistribution study, levels of let-7b were normalized to mice treated with PBS.
PNA Hybridization Assay
Levels of the hmiRNA guide (antisense) strand in tumor, liver, spleen, kidney, and lung were measured by a PNA hybridization assay. In this assay, a fully complementary Cy3-labeled PNA probe was used to hybridize the antisense strand of hmiRNA. Briefly, tumor, liver, spleen, kidney, and lung samples were lysed and digested in 200 μL of QG homogenizing solution (catalog no. QG0517; Affymetrix) containing 2 μL of proteinase K (20 mg/mL). SDS was precipitated using 30 μL of potassium chloride (3 mol/L) per sample and centrifuged at 4,000 × g for 15 min. Supernatants were then diluted in 150 mL of hybridization buffer (50 mM Tris, 10% Acetonitril, pH 8.8) containing 33 nM (5 pmol/150 mL of total sample) of Cy3-labeled PNA (PNABio, Thousand Oaks, CA, USA) and transferred to a PCR 96-well plate (catalog no. AB-0800/W; Thermo Scientific). Annealing was carried out on a Bio-Rad C1000 thermal cycler using the following protocol: 90°C for 15 min and 50°C for 15 min. Plates were then loaded into an Agilent Technologies 1260 Infinity Quad-pump HPLC system with a 1260 HiP ALS autosampler. Samples were run through a DNAPac PA100 anion exchange column (Thermo Fisher Scientific) and detected by a 1260 FLD fluorescent detector. The mobile phase consisted of buffer A (50% water, 50% ACN, 25 mM Tris-HCl [pH 8.5], and 1 mM EDTA) and buffer B (800 mM NaClO4 in buffer A). A steep gradient of buffer B (10%–100% within 2.5 min) was used for elution of guide strand-PNA hybrids. The Cy3 signal (550 nm excitation/570 nm emission) was monitored and recorded, and peaks were integrated to obtain the area under the curve (AUC). Final concentrations were ascertained by correlating AUCs obtained from experimental samples with a corresponding calibration curve. Calibration curves were generated by spiking known amounts of the respective hmiRNA conjugate into tissue lysates derived from an untreated animal. Spiked samples for calibration and experimental samples were processed and analyzed side by side under the same laboratorial conditions.
Tumor Histology and Imaging
Tumor tissues were fixed in OCT. Embedding and sectioning were processed by the Histology Core of Beth Israel Deaconess Medical Center (Boston, MA, USA). 4-μm tissue sections were prepared and stained with Hochest 443300 and Ki-67 antibody according to standard protocols. Phase-contrast and fluorescent images were acquired on an EVOS FL cell imaging system with a 4×, 10×, or 20× objective.
Lung Cancer Xenografts, Biodistribution, and Tumor Growth
All mice were maintained at Beth Israel Deaconess Medical Center in accordance with Institutional Animal Care and Use Committee guidelines. For single-dose studies, subcutaneous tumors were induced in female Nu/Nu mice (NU-Foxn1nu; 6 weeks; n = 12). Human HCC827 NSCLC cells were cultured in DMEM media following standard tissue culture procedures. HCC827 cells were trypsinized, counted, and subcutaneously injected into the left flank of 6-week-old nude mice (Jackson Laboratory, Bar Harbor, ME, USA) using 1 × 106 cells in 100 μL of DMEM with 50% Matrigel (BD Biosciences, San Jose, CA, USA) per injection. When tumors reached an average volume of 500 mm3 (caliper measurements were taken every 2 days, and tumor volume was calculated using the formula V = length × width2/2), 100 μL of synthetic hmiRNA-let-7b conjugates was delivered subcutaneously. Mice were sacrificed by CO2 inhalation after 24 h, and tumors, lungs, livers, and spleens were collected and prepared for histology and RNA isolation. All animal experiments were performed under an Institutional Animal Care and Use Committee-approved animal study protocol.
Statistical Analysis
Data represent the results of three independent experiments. Differences between groups were analyzed using one-way or two-way ANOVA analysis and expressed as mean ± SD from at least three separate experiments. A t test was used to compare between two groups. For all tests, p < 0.05 was considered to be statistically significant (∗p < 0.05, ∗∗p < 0.005, ∗∗∗∗p < 0.0001).
Author Contributions
M.S. conducted all in vitro and in vivo experiments. A.B. synthesized all compounds and performed PNA hybridization assay measurements. J.L. assisted with real-time qPCR experiments. M.-E.G., E.A., and R.D.C. assisted with in vivo experiment. M.S wrote the manuscript.
Conflicts of Interest
A.K. owns stock of RXi Pharmaceuticals and Advirna. F.J.S. discloses financial interests and SAB roles with Mira DX and MiRNA Therapeutics. The remaining authors declare no competing interests.
Acknowledgments
We thank A.K and A.B for critical reading of this manuscript. F.J.S. acknowledges NIH-YALE SPORE in Lung Cancer P50CA196530 and the NCI Outstanding Investigator Award (R35CA232105). Additional support to F.J.S. was provided by the P50CA196530-03S1 and support from the Ludwig Center at Harvard.
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.omtn.2019.11.008.
Supplemental Information
References
- 1.Durso M., Gaglione M., Piras L., Mercurio M.E., Terreri S., Olivieri M., Marinelli L., Novellino E., Incoronato M., Grieco P. Chemical modifications in the seed region of miRNAs 221/222 increase the silencing performances in gastrointestinal stromal tumor cells. Eur. J. Med. Chem. 2016;111:15–25. doi: 10.1016/j.ejmech.2016.01.047. [DOI] [PubMed] [Google Scholar]
- 2.Dai F.-Q., Li C.-R., Fan X.-Q., Tan L., Wang R.-T., Jin H. miR-150-5p inhibits non-small-cell lung cancer metastasis and recurrence by targeting HMGA2 and β-catenin signaling. Mol. Ther. Nucleic Acids. 2019;16:675–685. doi: 10.1016/j.omtn.2019.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Di Cello F., Hillion J., Hristov A., Wood L.J., Mukherjee M., Schuldenfrei A., Kowalski J., Bhattacharya R., Ashfaq R., Resar L.M. HMGA2 participates in transformation in human lung cancer. Mol. Cancer Res. 2008;6:743–750. doi: 10.1158/1541-7786.MCR-07-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kumar M.S., Armenteros-Monterroso E., East P., Chakravorty P., Matthews N., Winslow M.M., Downward J. Retraction. Nature. 2015;523:370. doi: 10.1038/nature14551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Naghizadeh S., Mansoori B., Mohammadi A., Kafil H.S., Mousavi Z., Sakhinia E., Baradaran B. Effects of HMGA2 gene downregulation by siRNA on lung carcinoma cell migration in A549 cell lines. J. Cell. Biochem. 2019;120:5024–5032. doi: 10.1002/jcb.27778. [DOI] [PubMed] [Google Scholar]
- 6.Sarhadi V.K., Wikman H., Salmenkivi K., Kuosma E., Sioris T., Salo J., Karjalainen A., Knuutila S., Anttila S. Increased expression of high mobility group A proteins in lung cancer. J. Pathol. 2006;209:206–212. doi: 10.1002/path.1960. [DOI] [PubMed] [Google Scholar]
- 7.Zappa C., Mousa S.A. Non-small cell lung cancer: current treatment and future advances. Transl. Lung Cancer Res. 2016;5:288–300. doi: 10.21037/tlcr.2016.06.07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 9.How C., Pintilie M., Bruce J.P., Hui A.B.Y., Clarke B.A., Wong P., Yin S., Yan R., Waggott D., Boutros P.C. Developing a prognostic micro-RNA signature for human cervical carcinoma. PLoS ONE. 2015;10:e0123946. doi: 10.1371/journal.pone.0123946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Adams B.D., Kasinski A.L., Slack F.J. Aberrant regulation and function of microRNAs in cancer. Curr. Biol. 2014;24:R762–R776. doi: 10.1016/j.cub.2014.06.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng C.J., Bahal R., Babar I.A., Pincus Z., Barrera F., Liu C., Svoronos A., Braddock D.T., Glazer P.M., Engelman D.M. MicroRNA silencing for cancer therapy targeted to the tumour microenvironment. Nature. 2015;518:107–110. doi: 10.1038/nature13905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shah M.Y., Ferrajoli A., Sood A.K., Lopez-Berestein G., Calin G.A. microRNA therapeutics in cancer—an emerging concept. EBioMedicine. 2016;12:34–42. doi: 10.1016/j.ebiom.2016.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Higgs G., Slack F. The multiple roles of microRNA-155 in oncogenesis. J. Clin. Bioinforma. 2013;3:17. doi: 10.1186/2043-9113-3-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou K., Liu M., Cao Y. New insight into microRNA functions in cancer: oncogene-microRNA-tumor suppressor gene network. Front. Mol. Biosci. 2017;4:46. doi: 10.3389/fmolb.2017.00046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esquela-Kerscher A., Trang P., Wiggins J.F., Patrawala L., Cheng A., Ford L., Weidhaas J.B., Brown D., Bader A.G., Slack F.J. The let-7 microRNA reduces tumor growth in mouse models of lung cancer. Cell Cycle. 2008;7:759–764. doi: 10.4161/cc.7.6.5834. [DOI] [PubMed] [Google Scholar]
- 16.Trang P., Medina P.P., Wiggins J.F., Ruffino L., Kelnar K., Omotola M., Homer R., Brown D., Bader A.G., Weidhaas J.B., Slack F.J. Regression of murine lung tumors by the let-7 microRNA. Oncogene. 2010;29:1580–1587. doi: 10.1038/onc.2009.445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Trang P., Wiggins J.F., Daige C.L., Cho C., Omotola M., Brown D., Weidhaas J.B., Bader A.G., Slack F.J. Systemic delivery of tumor suppressor microRNA mimics using a neutral lipid emulsion inhibits lung tumors in mice. Mol. Ther. 2011;19:1116–1122. doi: 10.1038/mt.2011.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ryther R.C.C., Flynt A.S., Phillips J.A., 3rd, Patton J.G. siRNA therapeutics: big potential from small RNAs. Gene Ther. 2005;12:5–11. doi: 10.1038/sj.gt.3302356. [DOI] [PubMed] [Google Scholar]
- 19.Zhou J., Rossi J. Aptamers as targeted therapeutics: current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esau C.C., Monia B.P. Therapeutic potential for microRNAs. Adv. Drug Deliv. Rev. 2007;59:101–114. doi: 10.1016/j.addr.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 21.McDermott A., Heneghan H., Miller N., Kerin M. The therapeutic potential of micrornas: disease modulators and drug targets. Pharm Res. 2011;28:3016–3029. doi: 10.1007/s11095-011-0550-2. [DOI] [PubMed] [Google Scholar]
- 22.Juliano R.L. The delivery of therapeutic oligonucleotides. Nucleic Acids Res. 2016;44:6518–6548. doi: 10.1093/nar/gkw236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kortylewski M., Nechaev S. How to train your dragon: targeted delivery of microRNA to cancer cells in vivo. Mol. Ther. 2014;22:1070–1071. doi: 10.1038/mt.2014.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Y., Miao L., Satterlee A., Huang L. Delivery of oligonucleotides with lipid nanoparticles. Adv. Drug Deliv. Rev. 2015;87:68–80. doi: 10.1016/j.addr.2015.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhao Y., Huang L. Lipid nanoparticles for gene delivery. Adv. Genet. 2014;88:13–36. doi: 10.1016/B978-0-12-800148-6.00002-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campani V., De Rosa G., Misso G., Zarone M.R., Grimaldi A. Lipid nanoparticles to deliver miRNA in cancer. Curr. Pharm. Biotechnol. 2016;17:741–749. doi: 10.2174/138920101708160517234941. [DOI] [PubMed] [Google Scholar]
- 27.Wang H., Liu S., Jia L., Chu F., Zhou Y., He Z., Guo M., Chen C., Xu L. Nanostructured lipid carriers for microRNA delivery in tumor gene therapy. Cancer Cell Int. 2018;18:101. doi: 10.1186/s12935-018-0596-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Akinc A., Querbes W., De S., Qin J., Frank-Kamenetsky M., Jayaprakash K.N., Jayaraman M., Rajeev K.G., Cantley W.L., Dorkin J.R. Targeted delivery of RNAi therapeutics with endogenous and exogenous ligand-based mechanisms. Mol. Ther. 2010;18:1357–1364. doi: 10.1038/mt.2010.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shi B., Keough E., Matter A., Leander K., Young S., Carlini E., Sachs A.B., Tao W., Abrams M., Howell B., Sepp-Lorenzino L. Biodistribution of small interfering RNA at the organ and cellular levels after lipid nanoparticle-mediated delivery. J. Histochem. Cytochem. 2011;59:727–740. doi: 10.1369/0022155411410885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baumann V., Winkler J. miRNA-based therapies: strategies and delivery platforms for oligonucleotide and non-oligonucleotide agents. Future Med. Chem. 2014;6:1967–1984. doi: 10.4155/fmc.14.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang Y., Wang Z., Gemeinhart R.A. Progress in microRNA delivery. J. Control. Release. 2013;172:962–974. doi: 10.1016/j.jconrel.2013.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nishina K., Unno T., Uno Y., Kubodera T., Kanouchi T., Mizusawa H., Yokota T. Efficient in vivo delivery of siRNA to the liver by conjugation of α-tocopherol. Mol. Ther. 2008;16:734–740. doi: 10.1038/mt.2008.14. [DOI] [PubMed] [Google Scholar]
- 33.Orellana E.A., Tenneti S., Rangasamy L., Lyle L.T., Low P.S., Kasinski A.L. FolamiRs: ligand-targeted, vehicle-free delivery of microRNAs for the treatment of cancer. Sci. Transl. Med. 2017;9:eaam9327. doi: 10.1126/scitranslmed.aam9327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 35.Deleavey G.F., Damha M.J. Designing chemically modified oligonucleotides for targeted gene silencing. Chem. Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 36.Bramsen J.B., Kjems J. Development of therapeutic-grade small interfering RNAs by chemical engineering. Front. Genet. 2012;3:154. doi: 10.3389/fgene.2012.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corey D.R. Chemical modification: the key to clinical application of RNA interference? J. Clin. Invest. 2007;117:3615–3622. doi: 10.1172/JCI33483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma J.-B., Yuan Y.-R., Meister G., Pei Y., Tuschl T., Patel D.J. Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature. 2005;434:666–670. doi: 10.1038/nature03514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parmar R., Willoughby J.L.S., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-vinylphosphonate: a stable phosphate mimic can improve the RNAi activity of siRNA-GalNAc ccnjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 40.Shmushkovich T., Monopoli K.R., Homsy D., Leyfer D., Betancur-Boissel M., Khvorova A., Wolfson A.D. Functional features defining the efficacy of cholesterol-conjugated, self-deliverable, chemically modified siRNAs. Nucleic Acids Res. 2018;46:10905–10916. doi: 10.1093/nar/gky745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lennox K.A., Owczarzy R., Thomas D.M., Walder J.A., Behlke M.A. Improved performance of anti-miRNA oligonucleotides using a novel non-nucleotide modifier. Mol. Ther. Nucleic Acids. 2013;2:e117. doi: 10.1038/mtna.2013.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alterman J.F., Hall L.M., Coles A.H., Hassler M.R., Didiot M.-C., Chase K., Abraham J., Sottosanti E., Johnson E., Sapp E. Hydrophobically modified siRNAs silence huntingtin mRNA in primary neurons and mouse brain. Mol. Ther. Nucleic Acids. 2015;4:e266. doi: 10.1038/mtna.2015.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Biscans A., Coles A., Echeverria D., Khvorova A. The valency of fatty acid conjugates impacts siRNA pharmacokinetics, distribution, and efficacy in vivo. J. Control. Release. 2019;302:116–125. doi: 10.1016/j.jconrel.2019.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biscans A., Coles A., Haraszti R., Echeverria D., Hassler M., Osborn M., Khvorova A. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. Nucleic Acids Res. 2019;47:1082–1096. doi: 10.1093/nar/gky1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ly S., Navaroli D.M., Didiot M.-C., Cardia J., Pandarinathan L., Alterman J.F., Fogarty K., Standley C., Lifshitz L.M., Bellve K.D. Visualization of self-delivering hydrophobically modified siRNA cellular internalization. Nucleic Acids Res. 2017;45:15–25. doi: 10.1093/nar/gkw1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee Y.S., Dutta A. The tumor suppressor microRNA let-7 represses the HMGA2 oncogene. Genes Dev. 2007;21:1025–1030. doi: 10.1101/gad.1540407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shi G., Perle M.A., Mittal K., Chen H., Zou X., Narita M., Hernando E., Lee P., Wei J.J. Let-7 repression leads to HMGA2 overexpression in uterine leiomyosarcoma. J. Cell. Mol. Med. 2009;13(9B):3898–3905. doi: 10.1111/j.1582-4934.2008.00541.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tai W. Current aspects of siRNA bioconjugate for in vitro and in vivo delivery. Molecules. 2019;24:E2211. doi: 10.3390/molecules24122211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Molina J.R., Yang P., Cassivi S.D., Schild S.E., Adjei A.A. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin. Proc. 2008;83:584–594. doi: 10.4065/83.5.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hosseinahli N., Aghapour M., Duijf P.H.G., Baradaran B. Treating cancer with microRNA replacement therapy: a literature review. J. Cell. Physiol. 2018;233:5574–5588. doi: 10.1002/jcp.26514. [DOI] [PubMed] [Google Scholar]
- 51.Jansson M.D., Lund A.H. MicroRNA and cancer. Mol. Oncol. 2012;6:590–610. doi: 10.1016/j.molonc.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bail S., Swerdel M., Liu H., Jiao X., Goff L.A., Hart R.P., Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032–1039. doi: 10.1261/rna.1851510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glinge C., Clauss S., Boddum K., Jabbari R., Jabbari J., Risgaard B., Tomsits P., Hildebrand B., Kääb S., Wakili R. Stability of circulating blood-based microRNAs—pre-analytic methodological considerations. PLoS ONE. 2017;12:e0167969. doi: 10.1371/journal.pone.0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y., Gao D.-Y., Huang L. In vivo delivery of miRNAs for cancer therapy: challenges and strategies. Adv. Drug Deliv. Rev. 2015;81:128–141. doi: 10.1016/j.addr.2014.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Selvam C., Mutisya D., Prakash S., Ranganna K., Thilagavathi R. Therapeutic potential of chemically modified siRNA: Recent trends. Chem. Biol. Drug Des. 2017;90:665–678. doi: 10.1111/cbdd.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Biscans A., Coles A., Haraszti R., Echeverria D., Hassler M., Osborn M., Khvorova A. Diverse lipid conjugates for functional extra-hepatic siRNA delivery in vivo. bioRxiv. 2018 doi: 10.1093/nar/gky1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Osborn M.F., Coles A.H., Biscans A., Haraszti R.A., Roux L., Davis S., Ly S., Echeverria D., Hassler M.R., Godinho B.M.D.C. Hydrophobicity drives the systemic distribution of lipid-conjugated siRNAs via lipid transport pathways. Nucleic Acids Res. 2019;47:1070–1081. doi: 10.1093/nar/gky1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gao X., Dai M., Li Q., Wang Z., Lu Y., Song Z. HMGA2 regulates lung cancer proliferation and metastasis. Thorac. Cancer. 2017;8:501–510. doi: 10.1111/1759-7714.12476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.MadhuKumar S., Elena Armenteros-Monterroso, East P., Probir Chakravorty, Nik Matthews, MonteWinslow M. HMGA2 acts as a ceRNA to stimulate lung cancer metastasis. Cancer Discov. 2014;4:141. [Google Scholar]
- 60.Johnson C.D., Esquela-Kerscher A., Stefani G., Byrom M., Kelnar K., Ovcharenko D., Wilson M., Wang X., Shelton J., Shingara J. The let-7 microRNA represses cell proliferation pathways in human cells. Cancer Res. 2007;67:7713–7722. doi: 10.1158/0008-5472.CAN-07-1083. [DOI] [PubMed] [Google Scholar]
- 61.Janas M.M., Schlegel M.K., Harbison C.E., Yilmaz V.O., Jiang Y., Parmar R., Zlatev I., Castoreno A., Xu H., Shulga-Morskaya S. Selection of GalNAc-conjugated siRNAs with limited off-target-driven rat hepatotoxicity. Nat. Commun. 2018;9:723. doi: 10.1038/s41467-018-02989-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Maier M.A., Jayaraman M., Matsuda S., Liu J., Barros S., Querbes W., Tam Y.K., Ansell S.M., Kumar V., Qin J. Biodegradable lipids enabling rapidly eliminated lipid nanoparticles for systemic delivery of RNAi therapeutics. Mol. Ther. 2013;21:1570–1578. doi: 10.1038/mt.2013.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haraszti R.A., Roux L., Coles A.H., Turanov A.A., Alterman J.F., Echeverria D., Godinho B.M.D.C., Aronin N., Khvorova A. 5′-Vinylphosphonate improves tissue accumulation and efficacy of conjugated siRNAs in vivo. Nucleic Acids Res. 2017;45:7581–7592. doi: 10.1093/nar/gkx507. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.