Abstract
Genetic code expansion, which enables the site-specific incorporation of unnatural amino acids into proteins, has emerged as a new and powerful tool for protein engineering. Currently, it is mainly utilized inside living cells for a myriad of applications. However, the utilization of this technology in a cell-free, reconstituted platform has several advantages over living systems. The typical limitations to the employment of these systems are the laborious and complex nature of its preparation and utilization. Herein, we describe a simplified method for the preparation of this system from Escherichia coli cells, which is specifically adapted for the expression of the components needed for cell-free genetic code expansion. Besides, we propose and demonstrate a modular approach to its utilization. By this approach, it is possible to prepare and store different extracts, harboring various translational components, and mix and match them as needed for more than four years retaining its high efficiency. We demonstrate this with the simultaneous incorporation of two different unnatural amino acids into a reporter protein. Finally, we demonstrate the advantage of cell-free systems over living cells for the incorporation of δ-thio-boc-lysine into ubiquitin by using the methanosarcina mazei wild-type pyrrolysyl tRNACUA and tRNA-synthetase pair, which could not be achieved in a living cell.
Keywords: Cell free system, Genetic code expansion, Thio-lysine, Simplified extract preparation
Introduction
In nature, a native protein is limited to the 20 canonical amino acids, in addition to selenocysteine [1] and pyrrolysine [2] in some selected organisms, and to their specific chemical and physical properties. Genetic code expansion (henceforth GCE) expands this limit and offers the ability to site-specifically incorporate hundreds of new unnatural moieties by stop codon suppression. Thereby enabling the rational enhancement of the chemical and physical properties of proteins. This methodology was established in the early years of the current century in E. coli [3] and gradually was adapted to many other organisms in both eukaryotes [[4], [5], [6], [7], [8], [9]] and prokaryotes [10,11].
Moreover, synthetic E. coli strains were generated to remove all UAG stop codons and their cognate release factors [12]. More recently, in a remarkable technical feat, the total synthesis of an E. coli genome was achieved. This was explicitly done to afford a better application of this technology, freeing rare and stop codons for the incorporation of unnatural amino acids (Uaas) [13]. The ability to produce genetically expanded proteins inside living cells resulted in many applications driven by hundreds of Uaas [14]. However, some of these amino acids are hard to synthesize and are expensive, thereby limiting their use in the relatively large volumes of bacterial growth media. These amino acids have low cellular permeability and sometimes are toxic to the host organism, thus limiting their use in living cells. Cell-free protein synthesis with GCE capabilities alleviates these specific limitations. Another important advantage of genetically expanded cell-free protein synthesis is the absence of the host genome.In the presence of a host genome, which leads to limitations of toxicity and off-target suppression may occur by suppression of the host genome endogenous stop codons. This is even more pronounced when two different Uaas are utilized at the same time; an exploit that is possible in living cells but is more feasible in cell-free platforms [15].
The first genetically expanded cell-free protein synthesis system was, in fact, a precursor for the GCE systems in living cells, it was realized in the late 1980s by Peter Schultz's team, using yeast Phe-tRNAs which were mutated to suppress the UAG stop codon in E. coli and were chemically aminoacylated by a Uaa. These tRNAs were added to E. coli cell extracts and successfully suppressed the designated stop codons [16]. After the successful in-vivo adaptation of GCE in the early 2000s, again by the Schultz team [17], the field has mostly moved to live cells as chassis. At the same time, Cell-free transcription-translation methodologies advanced to achieve higher yields [[18], [19], [20]], simplified and improved preparation protocols [21,22], and better understanding and control of the methodology itself [23,24]. As a result, GCE in cell-free protein synthesis gradually proliferated and improved: The PURE system was introduced and achieved ca. 80 mg/mL yields of Uaa containing proteins [25]. The first systems which utilized both orthogonal tRNA and a tRNA-synthetase, exogenously added to E. coli extracts, were introduced by the Nishikawa team [26] with no reported yields but with 50% suppression efficiency, and by the Swartz team, which also improved its yields and its suppression efficiency [27]. This methodology was further enhanced in recent years by the Jewett team, which implemented it in RF1-deficient cells [28,29]. However, the main problem of an approach where the orthogonal tRNA and synthetase are added exogenously is that it requires the purification of both components. Orthogonal components purification is both laborious and may be challenging with insoluble synthetases, as in the case with pyrrolysyl tRNA synthetase.
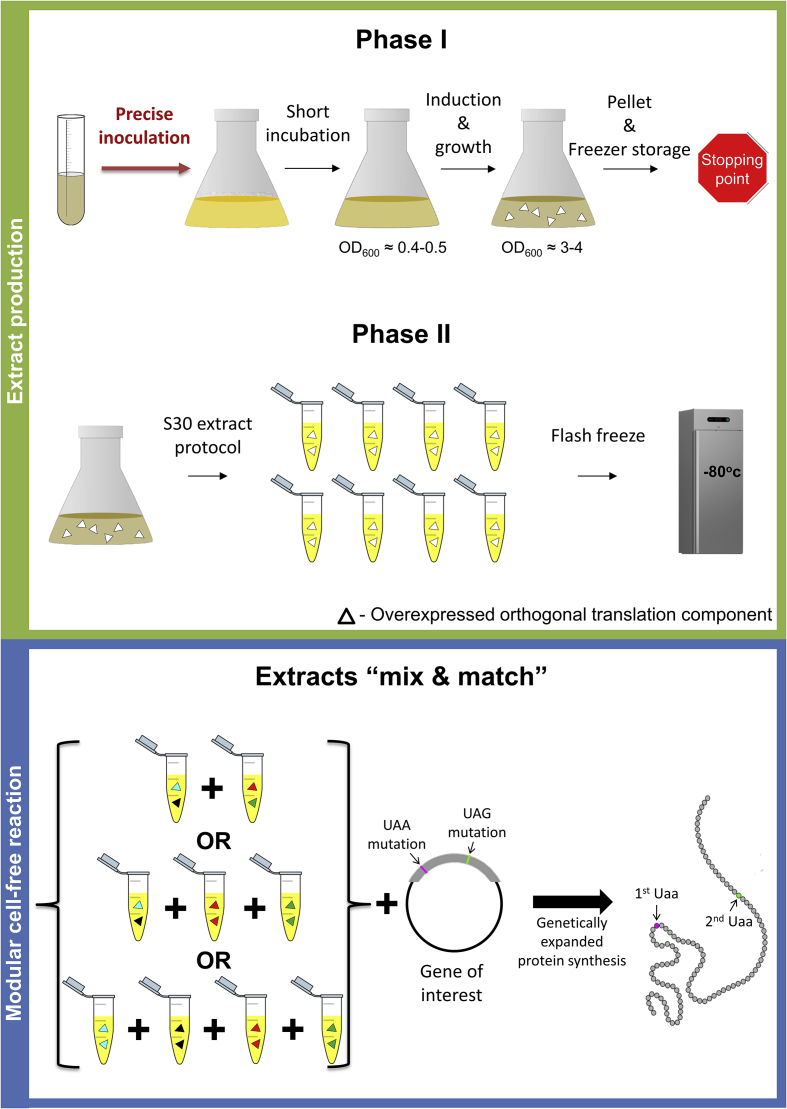
To overcome these limitations, another approach has been developed by both the Bundy team and by us, where the orthogonal tRNA and tRNA-synthetase were transformed and expressed endogenously in the host cell prior to extract preparation [30,31]. This approach completely circumvents the otherwise needed purification steps and leads to reasonable yields of up to 300 mg/L with high suppression efficiency. As we recognize that the complex, laborious nature of the preparation and use of GCE in cell-free protein synthesis has been a barrier to the more widespread use of this technology, this study will expound the extract preparation method of this approach in a straightforward manner. Moreover, we present here an adaptation of the protocol, which reduces its complexity and difficulty, without compromising its quality. Lastly, we promote a modular approach that was recently introduced [15], where different endogenous components could be expressed in separate bacterial extracts, stored for long periods of time and combined in a desired combinatorial fashion when needed (Fig. 1).
Fig. 1.
General scheme of the simplified preparation protocol and the modular approach for the cell-free reaction presented in this study. The entire protocol is divided into two phases: step, tools Phase one: bacterial growth is scaled up and induced to produce the desired components. Next, the cells are harvested, pelleted, and stored in the freezer. Phase two, the extract is prepared under the S30 protocol (vide infra) with small variations. Once several extracts are made, each containing different translational component pairs, the cell-free protein synthesis can be performed, where the modular reaction can produce one or more products while varying the components simply by mixing and matching extracts.
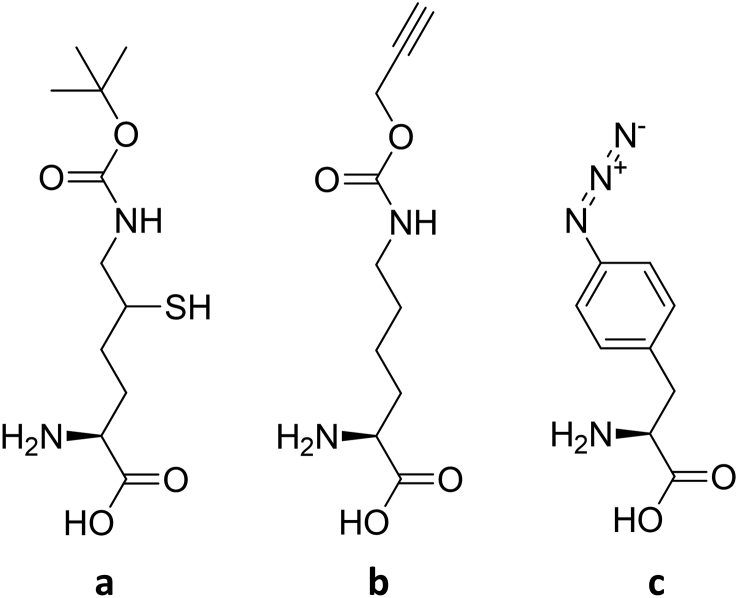
Herein, we present the utilization of this approach to simultaneously incorporate two different Uaas into the same protein. Finally, we demonstrate a new use of this approach, and cell-free GCE in general, by the incorporation of δ-thio-N-boc-lysine (TBK, Fig. 2a) to reporter proteins and yeast ubiquitin, which is both challenging and expensive to accomplish inside living cells. We further demonstrate the ability to use this Uaa as a bio-orthogonal chemical handle for future native chemical ligation of ubiquitin and polyubiquitin.
Fig. 2.
Chemical structures of Uaas used in the present research. a) δ-thio-N-boc-lysine (TBK). b) N-propargyl-l-lysine (PrK). c) p-azido-l-phenylalanine (AzF).
Results and discussion
Simplified extract preparation and cell-free reaction for genetically expanded protein synthesis
Bacterial extract preparation is the most challenging phase in cell-free protein synthesis; if GCE is not necessary, it could be purchased commercially. However, if GCE is required, the extract must be prepared by the user as the orthogonal tRNA synthetase, and a tRNA pair are not commercially available. As we realize that this phase is a significant limitation for the employment of this approach, the first goal of this study is to simplify the process and improve its accessibility to potential users.
One of the most widely used protocols for extract preparation is the S30 protocol [15,21,28,[31], [32], [33], [34], [35], [36]], which was modified in this study to create the GCE cell-free protein synthesis system. When using orthogonal translation systems (OTS) to achieve GCE harbored on a replicating bacterial vector, such as the pEVOL vector [37], the extract preparation protocol becomes longer and more difficult as the bacterial growth becomes slower and an induction step becomes necessary. The original S30 protocol starts with bacterial plating; it then requires three consecutive inoculation and growth rounds. In each round, the culture volume is increased, and the rounds should be precisely timed to re-inoculate the bacteria in the late-log-phase. This leads to difficulties in the timing of inoculations when working with bacteria with slow growth rates, like the strain which was used in this study; E. coli C321ΔA [12] transformed with the pEVOL Pyl OTS vector, encoding for the Methanosarcina mazei (Mm) pyrrolysyl orthogonal pair (Pyl-OTS). This strain has an average doubling time of ca. 50 min, which leads to difficulties in following the original S30 protocol and the required consecutive inoculations. The problem is enhanced when induction is required, as it further reduces growth rates and adds another un-predictable growth rate change caused by the addition of the inducer. To address this problem, we have divided the protocol into three stand-alone steps, which could be performed separately: The required preparations for the protocol, will not be elaborated detailed herein as it follows the S30 preparations without significant alterations, which were previously detailed by Sun et al. [38].
Phase 1: In this phase, bacterial scale-up, induction, protein over-expression, and preparation for lysis are performed. For this phase to fit into a standard workday, the S30 protocol had to be adjusted; To achieve that, we have designed four different protocol groups with variations in the inoculation method: Group (1) was prepared under the original S30 protocol [31], with its three required consecutive inoculations. In group (2) The first and second inoculations were removed, and the final inoculation step (to 1 L of growth media) was done directly from the plates, with very low initial bacterial concentrations, which then were incubated overnight. In group (3) The second inoculation step was skipped, which means that bacteria were first inoculated from plates to a volume of 50 mL of growth media and incubated overnight, during which the culture reached the stationary phase (instead of the required log phase). The final inoculation step was done by transferring 10 mL of stationary-phase bacteria to 1 L of growth medium (this volume corresponds to a 1/100 dilution, the dilution factor required by the original protocol). Lastly, group (4) is similar to (3), but the final inoculation step was done by transferring 40 mL of stationary-phase bacteria (a dilution factor of 1/25).
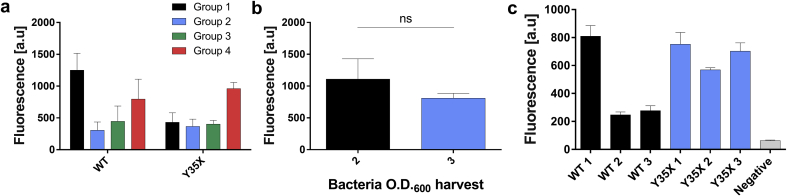
The three altered inoculation groups (2–4), represent a significant improvement to the original protocol as they enable the induction early on the workday (when the final culture reaches OD600 of ~0.5). Even if induction is not needed, these altered protocols still sidestep timing limitations and reduce the duration of inoculation time from three days to only two days. After induction, bacterial growth until harvesting OD600 is being reached as well as OTS expression can be completed by noontime. In respect to the harvesting OD600, we have found that the requirement in the original S30 protocol of cell harvest OD600 of 1.5–2.0 could be increased to ~3–3.5, thereby doubling the biomass and the resulting extract without any apparent compromise of efficiency in the system (Fig. 3b). For this reason, protocol groups (2–4) were harvested at OD600 ~3.0, which represent ~66%–77% of their measured maximal growth OD600, of ~4.5. This value is close to a previously reported optimal value of 58.5% [20].
Fig. 3.
Comparison of performance using different extract preparation protocols. a) Cell-free production of WT GFP and genetically expanded Y35PrK GFP. Fluorescence intensity of produced WT and mutant GFP for four protocols: 1) S30 protocol 2) No intermediate inoculation, log phase maintained 3) No intermediate inoculation, stationary phase instead of log-phase added to final culture in small volume 4) Similar to 3, but added in large volume to final culture to obtain near-induction OD b) Cell-free production of WT GFP with extracts harvested at different OD600. Original S30 protocol suggested bacterial harvest at OD== 2, compared to higher bacterial harvest at OD =3. The results are not significantly different (two-sided T-test, t = 1.57, df = 4, p-val = 0.19). c) Cell-free production of GFP for a comparison between different plasmids, using Pyl-OTS for UAG suppression. Fluorescence intensity of expressed protein from three different batches of WT GFP and Y35X plasmid preparations. Each plasmid type encoded had the same sequence but was purified independently.
Next, we set out to find a stopping point for the extract preparation protocol, which is not available in the published S30 protocols. We have reasoned that it could be stopped after the culture washing steps (see protocol), for a prolonged period, as a bacterial pellet before lysis. Protocol groups (2–4), were subjected to this stopping point. This roughly divided the protocol midway, which spreads the intensive work of one day to two more convenient days of work. We have tested the resulting extracts from all protocol combinations with the cell-free expression of wildtype (WT) GFP and a GFP gene mutated at position 35 from tyrosine (UAC) to amber stop codon (UAG), named GFPY35X. The reaction mixture of GFPY35X was supplemented with 1 mM of PrK (Fig. 2b), which could be incorporated through UAG suppression by the Pyl-OTS (Fig. 3a). For the GFP Y35PrK protein, we found that all the new protocol groups (2–4), which include the inoculation and the stopping point alterations, do not compromise the yields nor the fidelity of the protein with an incorporated Uaa, while significantly reducing the workload (Fig. 3a). It appears, however, that group (4) yielded the best results under these conditions.
Phase 2: In this phase, the rest of the S30 protocol is performed, including lysis (using a bead-beater), pellet separation by centrifugation of the lysate, nuclease digestion by incubation and dialysis. In this phase we have found two notable adjustments to simplify and improve the protocol for GCE, i) The dialysis should not be continued for more than 3–4 h in 4 °C as the efficiency of the genetically expanded system decreases after prolonged dialysis, presumably due to the lower stability of the orthogonal tRNA. ii) The final extract protein concentration measurement and calibration steps, in the original protocol, are both time-consuming and do not significantly affect the efficiency of the extract. Therefore, this phase should be considered optional and should be applied only when the complete optimization of the extract is needed.
Cell-free transcription/translation reaction: The genetically expanded protein synthesis reaction requires further considerations. First, the GCE cell-free buffer calibration is an important step that affects the yields of protein synthesis. In some S30 protocols, it is advised to calibrate the concentrations of K-glutamate and Mg-glutamate and the concentration of DTT. However, in GCE cell-free protein synthesis, we have found that concentrations usually could be fixed to 100 mM and 2.5 mM for K-glutamate and Mg-glutamate, respectively, without DTT, with no significant effect on the system yields. Second, the Uaa concentration was found to achieve the best yield in a concentration range of between 0.5 mM (for N-Boc-Lysine) [31] and up to 5.4 mM (for TBK, in this study), an optimal concentration should be calibrated for each specific Uaa while the starting concentration should be 1 mM. Finally, we note that cell-free reaction yield-variability is relatively high; this warrants separate attention and further investigations as to its sources. However, it is important to discuss two sources of this variability, which could be a common impediment in the employment of this system. First, the context and location of the site chosen for mutated to facilitate Uaa incorporation have a significant effect on the efficiency of the system [39,40], but notably, from our experience, the context, and locations could be different between in-vivo and cell-free systems [data not shown]. This subject warrants a systematic investigation that is outside the scope of this study. However, it will be wise to screen several sites of the protein to achieve successful cell-free Uaa incorporation. Second, the expression vector, which is added to the reaction mixture carries significant variability. We have noticed, on numerous occasions, that batches of the same plasmid, from unknown reasons, could vary considerably when added to reactions, and in extreme cases, there could be no expression at all. To demonstrate this point, we have tested three batches of two plasmids, pBEST GFP WT, and pBEST GFP Y35X, in an otherwise identical reaction conditions (Fig. 3c). The results indicated the extent of the variability that arises solely from differences in plasmid batches. Therefore, it is beneficial to test several plasmid batches, sometimes even by using different prep kits, before discarding an extract batch. We were not able to determine the source of variability by the following quality controls: accurate plasmid concentration measurements (using nano drop and standard absorbance measurements) and by plasmids re-purification from extracts.
A modular approach to cell-free protein synthesis with Uaas
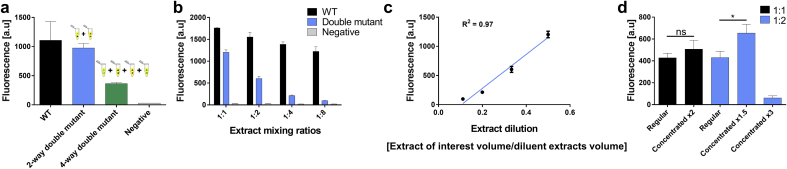
Recently, we have found that extracts from the same bacterial strain but with different genotypes and from different batches could be mixed and combined [15]; this led us to develop a modular approach for cell-free protein synthesis with GCE. In this approach, a component of choice, which could be a protein or a non-protein-coding gene like tRNAs, could be expressed separately in bacteria, which will sequentially be made into an extract that contains the said component. These separate extracts could be stored for an extended period of time (some extracts used in this study were stored for over four years), thawed upon demand and combined, mixed and matched according to the experimental design. Specifically, we have utilized this approach to express a reporter protein containing two different Uaas. A GFP gene was mutated in position 193 to UAG and position 35 to UAA: double mutant. We have tested the GFP expression in two modular combinations with and without the supplementation of the Uaas as a negative control (Fig. 4a): 1) a two-extracts system, one which contains the Pyl-OTS which incorporate PrK in response to the UAG stop codon, the other contains the p-azido-l-phenylalanine (AzF)-OTS which incorporates AzF in response to the UAA codon. Both OTSs contain a tRNA-synthetase and a tRNA pair of genes. 2) a four-extracts system each contains only one of the four above mentioned components: 1) AzF tRNA-synthetase 2) AzF tRNA for UAA suppression 3) PrK-tRNA-synthetase 4) PrK tRNA for UAG suppression. The results clearly show that this approach is viable for at least four different combinations of interacting components of tRNAs and tRNA synthetases. However, there is a noticeable decrease in the system yield between the 2-extracts system and the 4-extracts system. We reasoned that this decrease in yield is a result of the relative dilution of each orthogonal component in the final volume, as more extracts are being mixed. To test this, we have diluted the AzF-OTS with increasing concentrations of Pyl-OTS and quantified the double mutant production rates (Fig. 4b). The results show a significant decrease in the expression level of the double mutant, while only a slight decrease is observed in the level of WT protein. These results correlate with the WT protein production, which does not depend on the orthogonal translational components. In contrast with the double mutant GFP that depends on increasingly diluted orthogonal components.
Fig. 4.
Cell-free production of double mutant GFP protein (Y35UAAD193UAG), using extracts mixtures of different ratios. Fluorescence intensity of double mutant production was compared to WT protein or double mutant without any Uaas (negative control). a) mixtures of OTS pairs and mixtures of OTS single components extract comparison for double mutant GFP production. A two-extracts mixture composed of AzF-OTS for UAA suppression and Pyl-OTS for UAG suppression, while a four-extracts mixture composed of the following extracts: 1) AzF synthetase 2) AzF tRNA for UAA suppression 3) PrK synthetase 4) PrK tRNA for UAG suppression. b) OTS pairs extract mixture, between AzF-OTS for UAA suppression and increasing quantities of Pyl-OTS for UAG suppression, at different ratios. c) Linear regression of extract dilution for double mutant GFP expression measured by GFP fluorescence. d) Lyophilized extracts and mixing ratios of AzF-OTS for UAA supression and Pyl-OTS for UAG supression: 1:1, black; 1:2, blue Extracts were rehydrated with varying volumes of water (t-test, ns represents p-val>0.05, * represents p-val = 0.02).
Mixing several different batches of extracts with different orthogonal components will inevitably result in drastic OTSs dilution and therefore result in reduced productivity. However, this method still presents significant advantages when mixing a small number of extracts. It allows much-needed modularity as well as not having a single extract carry the “burden” of synthesizing all of the components. We have fitted the mixing results with a linear regression (Fig. 4c), so it explains ca. 97% of the variability in protein's levels that we observe, the two-extracts, and the four-extracts modular systems results are in agreement with the linear model and fall within the standard deviation of the model. Lastly, in order to reduce the dilution effect of OTSs when using different extracts, we have lyophilized the relevant extracts and sequentially re-hydrated them with lower volumes of water, thereby increased the relative concentrations of the OTSs (Fig. 4d). The result demonstrates that in a two-extract system and three-extract system, the GCE yields were increased by 1/2 and by 2/3 (significant increase [t-test, pval = 0.02]) then by using their original volumes, respectively. However, when concentrated to less than 1/2 of their original volume, the system collapsed, and the efficiency dropped, presumably due to high salt concentrations.
Incorporation of δ-thio-N-boc-lysine (TBK) into reporter proteins and ubiquitin
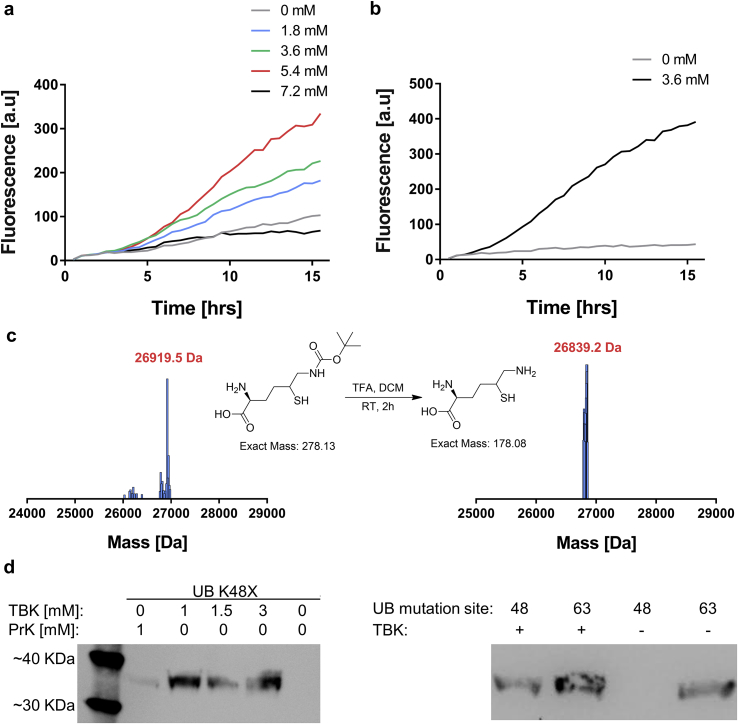
Synthesis of the Uaa, TBK was performed using a published procedure adapted from Virdee et al. [41]. The TBK amino acid was used for bio-orthogonal chemistry of native chemical ligation, which results in an isopeptide bond formation between two proteins of interest. This Uaa was shown to be incorporated exclusively in-vivo by the pyrrolysyl orthogonal translation system with an evolved pyrrolysyl tRNA synthetase [41]. In our experience, the in-vivo utilization of this Uaa was both expensive (as high concentrations in addition to large culture volumes were required) and achieved low yields, which were insufficient for downstream applications. Therefore, TBK incorporation was tested in the genetically expanded cell-free protein synthesis using the modular extract with the Pyl-OTS. Using this system, the TBK was first incorporated into GFP in position 35 (i.e., GFP Y35X) and RFP in position 15 (i.e., RFP K15X). The incorporation of the Uaa was tested using kinetic fluorescent measurements for both GFP [Fig. 5a] and RFP [Fig. 5b]. We have first tested the TBK concentration dependence in the GFP construct, which showed that 5.4 mM and 3.6 m M of TBK achieves the best results. In the RFP construct, only the 3.6 mM concentration was tested and utilized, as it represented comparatively similar results to hose with 5.4 mM TBK.
Fig. 5.
TBK incorporation into proteins, using genetically expanded cell-free protein synthesis. a) Cell-free kinetics of GFP Y35X production in the presence of different TBK concentrations. b) Cell-free kinetics of RFP K15X production in the presence and absence of TBK. c) ESI mass spectra of purified RFP K15X with TBK, before and after boc-deprotection. d) Anti-ubiquitin Western blot of genetically expanded cell-free protein synthesis of the 36.4 kDa Ub-Intein-CBD construct. In the left panel: Cell-free reactions of incorporation of PrK and TBK in different concentrations. The right panel presents a comparison between TBK incorporated into two different branching sites, K48X and K63X, in the ubiquitin construct in the presence or absence of TBK, the bend in site 63 in the absence of TBK could be a result of stop codon readthrough.
To be utilized for native chemical ligation, the TBK must first be boc-deprotected. To test this, we have expressed the RFP K15TBK variant in a large volume, purified it using nickel affinity chromatography and removed the protective group using TFA, the resulting protein RFP K15TK was validated using mass spectrometry (Fig. 5c).
To demonstrate the specific advantage of cell-free protein synthesis, we have sought to incorporate TBK to the ubiquitin gene. Ubiquitin is a highly conserved 76 amino acid protein that is present in all eukaryotes, and serves as a post-translational modification of other proteins. This system was discovered in the context of protein degradation [42] since it was found to play a role in a myriad of cellular processes. Every ubiquitin can be ubiquitinated by one or more other ubiquitins at seven different possible sites, thus creating a complex code [43]. Efficient methods for the synthetic generation of this code is a feat which is long sought after. Therefore, we have decided to utilize the cell-free GCE platform presented herein to take a step in this direction. The yeast UBI4 gene (the ubiquitin gene) was amplified from the yeast genome and fused to intein-chitin binding domain (Int-CBD) construct. This construct enables the formation of N-terminal-thioester and affinity purification [44]. The 36.4 kDa construct was cloned into the pBEST plasmid to be expressed in the genetically expanded cell-free protein synthesis system. Its expression was tested to incorporate both PrK or varying concentrations of TBK at the poly-ubiquitin branching lysine site K48 (Fig. 5d, left panel) and was also validated for a second branching site - K63 (Fig. 5d, right panel). These produced moieties represent a first step towards the synthetic synthesis of a branched poly-ubiquitin code that could be generated using native chemical ligation. This is achieved as this ubiquitin construct can act both to ubiquitinate or be ubiquitinated utilizing the boc-deprotected moiety of TBK in a combinatorial approach.
Conclusions
Herein we present a method to generate a simplified genetically expanded cell-free protein synthesis system with new capabilities. It reduces the laborious nature of the protocol by altering its inoculation steps and by adding possible stop points. Also, this protocol reduces the complexity of the S30 protocol by identifying and removing optional steps. Once a simplified protocol for extract preparation is used, it could be utilized for several genotypes of the same bacterial strain, each over-expressing a different set of components. These different genotypic extracts can contain various components that could be stored for an extended period (at least 4 years) while maintaining most of its functionality and then mixed-and-matched in any combinatorial way without the need for a new preparation. This results in a modular approach to cell-free protein synthesis with genetic code expansion. We anticipate that these results will increase the use of genetically expanded cell-free protein synthesis, in the specific cases where it presents an advantage over living cells. We acknowledge that the modular approach presented herein is not limited only to GCE, but it could directly be adapted and utilized for other applications. These applications include chaperon-required protein expression, an amalgamation of toxic or aggregating components which cannot be co-expressed in living cells, and any cell-free protein expression which requires several components which it would be beneficial to combinatorially mix and match .
Methods
Strains and plasmids
The bacterial strain used for the preparation of all cell extracts is the C321.ΔprfA strain (Addgene #48998) carrying a pEVOL plasmid, transformed before growth and extract preparation. In this paper, six types of pEVOL plasmids were used: 1) Methanosarcina mazei (Mm) orthogonal pair of Mm-PrKRS\Mm-tRNACUAPrK (Pyl-OTS) 2) Methanocaldococcus jannaschii (Mj) orthogonal pair of Mj-AzFRS\Mj-tRNAUUAAzF (AzF-OTS) 3) Mm-PrKRS (no tRNA) 4) Mm-tRNACUAPrK (no synthetase) 5) Mj-AzFRS (no tRNA) 6) Mj-tRNAUUAAzF (no synthetase) [15].
All expression genes targeted for CFPS were harbored on a pBEST plasmid [45]. pBEST deGFP [31] was expressed in three forms, WT GFP, Y35UAG GFP, Y35UAA D193UAG GFP. pBEST RFP was expressed in the form of K15UAG RFP. All mutations were introduced using standard mutagenesis protocol.
The pBEST-UB constructs were generated as follows. The S. cerevisiae UBI4 ubiquitin gene was amplified from its genome. Next, it was fused in its N terminus to intein and CBD genes from the IMPACT kit [New England Biolabs], and the entire construct was amplified and cloned to the pBEST plasmid. pBEST UB was expressed in three forms, WT UB, K48UAG UB, K63UAG UB. All mutations were introduced using standard mutagenesis protocol.
Extract preparation
We have followed Sun et al. visualized protocol [38] with some modifications: C321.ΔprfA bacteria containing a pEVOL plasmid were grown in 2xYT media +34 μg/mL chloramphenicol at 30 °C under four different conditions prior to lysis: 1) Control prepared according to the original protocol [38] using gradual bacterial cultivation. Bateria were plated overnight, first inoculation of a colony into 4 mL liquid culture (using a 12 mL tube) for 10 h, second inoculation of 400 μL of previous cultivation into a 50 mL liquid culture (using 250 mL Erlenmeyer flask) for 11 h and final inoculation of 15 mL into a 1 L liquid culture (using 5 L Erlenmeyer flask). 2) Bacteria were plated overnight. Next, an isolated colony was inoculated directly into the 1 L final culture (using a 5 L Erlenmeyer flask). Thus we have removed the first and second inoculation steps from the original protocol. The final culture was incubated for 12 h, after which its OD600 was measured to be 0.14. Note that ideally, the user should calibrate the exact incubation time to reach an induction OD600 of ~0.5 if using this condition. 3) Bacteria were plated and incubated overnight. Next, an isolated colony was inoculated directly to the second culture into 50 mL growth media (using 250 mL Erlenmeyer flask). In this case, the first inoculation step was removed from the original protocol. The final inoculation was done after overnight incubation of the previous inoculation using 10 mL of deep-stationary phase bacteria. 4) Similar to 3, but added 40 mL instead of 10 mL of stationary-phase bacterial culture. This was done to obtain near-induction OD.
For all protocol groups, induction of the pEVOL plasmid (with an ara promoter), was performed using 0.5% of l-arabinose when the OD600 of the final culture reached ~0.5. Following induction, all four protocol groups were incubated, and the bacterial culture was allowed to grow under induction for protein expression up to an OD600 of ~3–3.5 (higher than OD600 of 1.5–2 used in the original protocol). Harvest was performed on ice using chilled containers of ~450 mL each. In all four experimental conditions, the biomass was collected using large centrifugation containers at 5000 g, 4 °C for 12 min. During centrifugation, 2 L of cold S30A buffer at pH 7.7, composed of 10.88 g Mg-glutamate, 24.39 g K-glutamate, and 50 mL Tris at 2 M, was activated using 2 mL of 1 M DTT. Following centrifugation, tubes were dried on top of a paper towel, and biomass was washed and resuspended using 160 mL S30A buffer only to be centrifuged again at 5000 g, 4 °C for 12 min S30A washing step was repeated one more time.
Harvested biomass was re-suspended in 40 mL S30A buffer and moved to a pre-weighed, chilled falcon tube as a smaller container. Flacon tubes were centrifuged at 3000 g, 4 °C for 15 min, followed by supernatant decanting and a second centrifugation at 2000g, 4 °C for 2 min. The residual supernatant was removed with a pipette. Harvested biomass weight was calculated from initial Falcon tube weight followed by Falcon tubes storage in a −80 °C freezer. This is a stopping point that was added to the original protocol.
In the following day (not a requirement, could be stored for more extended periods), each Falcon tube biomass was re-suspended with chilled S30A buffer (calculated volume in mL as 0.9*pellet mass). Glass beads of 0.1 mm were added to the suspended bacteria, followed by 30 s vortex and immediately afterwards 30 s on ice (beads addition is repeated in that manner for 3 times, while beads quantity was calculated in grams as (5*pellet mass)/3). The entire mixture of bacteria and glass beads was loaded into 2 mL bead-beating tubes (a small amount in the tube's cap as well) using a sterile spatula. A bead-beater was used on each tube for 30 s. It was then placed upside-down in ice for 30 s and bead-beaten again. After bead-beating all the tubes, they were connected to a filter apparatus and a new bead-beating collection tube. The two tubes and filters were inserted into a 15 mL Falcon tube. The resulting lysate was separated from the glass beads by centrifugation at 6000g, 4 °C for 5 min. Non-turbid collection tubes had their supernatant moved to a new 1.75 mL micro-centrifugation tubes. Micro-centrifugation tubes were centrifuged for 12000 g, 4 °C for 10 min, and the supernatant was consolidated into 500 μL in new bead-beating tubes. Consolidated bead-beating tubes had their caps removed and placed in a 15 mL Falcon tube for 80 min at 37 °C, 220 rpm. During incubation, 2 L of S30B buffer at pH 8.2, composed of 10.88 g Mg-glutamate, 24.39 g K-glutamate, and 2 M Tris, was activated using 2 mL of 1 M DTT.
Following incubation at 37 °C, the extract was consolidated into 1.5 mL aliquots in a 1.75 mL micro-centrifuge tubes and centrifuged at 12000 g, 4 °C for 10 min. The collected supernatant was loaded into a 10 kDa molecular weight cutoff (MWCO) dialysis cassette, pre-soaked with activated S30B buffer. The extract was dialyzed for 3 h at 4 °C. Upon dialysis completion, the extract was separated into 1.5 mL in 1.75 mL micro-centrifuge tubes and centrifuged at 12000 g, 4 °C for 10 min. The supernatant was collected and was considered as the final extract.
The final extract was aliquoted into Eppendorf tubes in 30 μL batches, without any protein concentration measurement or calibration (in contrast to the original protocol).
All cell-free extracts used in this research including all different combinations of OTSs were produced between 2 and 4 and a half years prior to writing this manuscript. They were all stored in −80 °C in the past years and proved to be functional even up to their latest use in October 2019.
Cell-free protein synthesis reaction
The CFPS reaction volume was carried in a Nunc (black, flat transparent bottom) 384 well plate (Thermo scientific) in an incubated Synergy HT plate reader (Biotek) in temperature (29oc) for ~17 h with intervals of 30 min. GFP expression was measured with an excitation wavelength of 488 nm and an emission wavelength of 507 nm.
In each cell-free reaction, 33% of the final reaction volume was composed of bacteria extract. When a single type of extract was used, all 33% were composed of that same extract. However, when several types of extracts were used, 33% of reaction volume was being divided by the different extracts. Different extracts ratios were tested when combining two separate extracts (1:1, 1:2, 1:4, 1:8 – the ratio between extract of interest volume/dilutant extracts volume) and up to four separate extracts were combined at a 1:1:1:1 ratio.
In each cell-free reaction, 41.6% of the final reaction volume was composed of reaction buffer. The reaction buffer consists of the following compounds (concentrations are in final reaction values): 40.19 mM HEPES pH 8,1.21 mM ATP and GTP, 0.72 mM CTP and UTP, 0.16 mg/mL tRNA, 0.21 mM coenzyme A, 0.27 mM NAD, 0.60 mM cAMP, 0.055 mM folinic acid, 0.80 mM spermidine, 24.11 mM 3-phosphoglyceric acid,1.21 mM each of 20 amino acids, 1.61% PEG-8000, 96.47 mM K-glutamate, 2.41 mM Mg-glutamate (reagents were purchased from the same vendors as listed in Sun et al., 2013 [38]. The buffer amount per reaction and the expression plasmid concentration were performed as described before [15]. All plasmids were purified from E. coli DH5a cells using the Wizard Plus SV minipreps kit [Promega].
Protein expression of genetically expanded proteins with stop codons mutations was done in the presence of 1 mM final concentration per Uaa unless stated otherwise.
UB Western blot analysis
Cell-free protein synthesis samples were diluted by a factor of ten and loaded into a 4–20% SDS gels [Genscript]. After transfer, anti-ubiquitin antibodies were used, the membrane was visualized using ImageQuant LAS 4000 imager [Fujifilm].
Synthesis of δ-Thio-Boc-Lysine
The synthesis was performed as described by Virdee et al. [41]. and validated using mass-spectrometry (Finnigan Surveyor/LCQ Fleet, Thermo Scientific).
Protein purification and mass-spectrometry analysis
Proteins were fused to 6xhistag and purified using standard nickel-column affinity purification. Purified protein samples were analyzed by LC-MS (Finnigan Surveyor/LCQ Fleet, Thermo Scientific).
Author contributions
Y.C and E.O share equal contribution to this paper, Y.C conceived and performed all experimentations, synthesized the molecules and co-authored the manuscript, E.O conceived and performed all experimentations and co-authored the manuscript, M.S. performed the TBK incorporation experiments, B.Z performed the extract preparation optimization experiments, R.D synthesized the TBK, L.A supervised, designed and conceived experimentations and manuscript preparation.
Acknowledgments
We wish to thank the Azrieli (Y.C.) and Darom Ph.D. fellowships (Y.C., E.O.) for supporting this study. We would like to gratefully acknowledge an ERC grant number 260647 (L.A.) for supporting parts of the studies mentioned herein.
Footnotes
Peer review under responsibility of KeAi Communications Co., Ltd.
References
- 1.Ambrogelly A., Palioura S., Söll D. Natural expansion of the genetic code. Nat Chem Biol. 2007;3:29–35. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 2.Hao B., Gong W., Ferguson T.K., James C.M., Krzycki J a, Chan M.K. A new UAG-encoded residue in the structure of a methanogen methyltransferase. Science. 2002;296:1462–1466. doi: 10.1126/science.1069556. [DOI] [PubMed] [Google Scholar]
- 3.Wang L., Brock A., Herberich B., Schultz P.G. Expanding the genetic code of Escherichia coli. Science. 2001;292:498–500. doi: 10.1126/science.1060077. [DOI] [PubMed] [Google Scholar]
- 4.Chin J.W., Cropp T.A., Anderson J.C., Mukherji M., Zhang Z., Schultz P.G. An expanded eukaryotic genetic code. Science. 2003;301:964–967. doi: 10.1126/science.1084772. [DOI] [PubMed] [Google Scholar]
- 5.Han S., Yang A., Lee S., Lee H.-W., Park C.B., Park H.-S. Expanding the genetic code of Mus musculus. Nat Commun. 2017;8:14568. doi: 10.1038/ncomms14568. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bianco A., Townsley F.M., Greiss S., Lang K., Chin J.W. Expanding the genetic code of Drosophila melanogaster. Nat Chem Biol. 2012;8:748–750. doi: 10.1038/nchembio.1043. Nature Publishing Group. [DOI] [PubMed] [Google Scholar]
- 7.Liu W., Brock A., Chen S., Schultz P.G. Genetic incorporation of unnatural amino acids into proteins in mammalian cells. Nat Methods. 2007;4:239–244. doi: 10.1038/nmeth1016. [DOI] [PubMed] [Google Scholar]
- 8.Parrish A.R., She X., Xiang Z., Coin I., Shen Z., Briggs S.P. Expanding the genetic code of Caenorhabditis elegans using bacterial aminoacyl-tRNA synthetase/tRNA pairs. ACS Chem Biol. 2012;7:1292–1302. doi: 10.1021/cb200542j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greiss S., Chin J.W. Expanding the genetic code of an animal. J Am Chem Soc. 2011;133:14196–14199. doi: 10.1021/ja2054034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chemla Y., Friedman M., Heltberg M., Bakhrat A., Nagar E., Schwarz R. Expanding the genetic code of a photoautotrophic organism. Biochemistry. 2017;56:2161–2165. doi: 10.1021/acs.biochem.7b00131. [DOI] [PubMed] [Google Scholar]
- 11.Gan Q., Lehman B.P., Bobik T.A., Fan C. Expanding the genetic code of Salmonella with non-canonical amino acids. Sci Rep. 2016;6:39920. doi: 10.1038/srep39920. Nature Publishing Group. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lajoie M.J., Rovner A.J., Goodman D.B., Aerni H.R., Haimovich A.D., Kuznetsov G. Genomically recoded organisms expand biological functions. Science. 2013;342:357–360. doi: 10.1126/science.1241459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fredens J., Wang K., de la Torre D., Funke L.F.H., Robertson W.E., Christova Y. Total synthesis of Escherichia coli with a recoded genome. Nature. 2019 doi: 10.1038/s41586-019-1192-5. Springer US. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dumas A., Lercher L., Spicer C.D., Davis B.G. Designing logical codon reassignment – expanding the chemistry in biology. Chem Sci. 2015;6:50–69. doi: 10.1039/c4sc01534g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozer E., Chemla Y., Schlesinger O., Aviram H.Y., Riven I., Haran G. In vitro suppression of two different stop codons. Biotechnol Bioeng. 2017;114:1065–1073. doi: 10.1002/bit.26226. [DOI] [PubMed] [Google Scholar]
- 16.Noren C.J., Anthony-Cahill S.J., Noren K.A., Griffith M.C., Schultz P.G. In vitro suppression of an amber mutation by a chemically aminoacylated transfer RNA prepared by runoff transcription. Nucleic Acids Res. 1990;18:83–88. doi: 10.1093/nar/18.1.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang L., Magliery T.J., Liu D.R., Schultz P.G. 2000. A new functional suppressor tRNA/aminoacyl - tRNA synthetase pair for the in vivo incorporation of unnatural amino acids into proteins; pp. 5010–5011. [Google Scholar]
- 18.Jewett M.C., Swartz J.R. Rapid expression and purification of 100 nmol quantities of active protein using cell-free protein synthesis. Biotechnol Prog. 2004;20:102–109. doi: 10.1021/bp0341693. [DOI] [PubMed] [Google Scholar]
- 19.Caschera F., Noireaux V. Synthesis of 2.3 mg/ml of protein with an all Escherichia coli cell-free transcription-translation system. Biochimie. 2014;99:162–168. doi: 10.1016/j.biochi.2013.11.025. Elsevier Masson SAS. [DOI] [PubMed] [Google Scholar]
- 20.Dopp J.L., Reuel N.F. Process optimization for scalable E . coli extract preparation for cell-free protein synthesis. Biochem Eng J. 2018;138:21–28. Elsevier B.V. [Google Scholar]
- 21.Kigawa T., Yabuki T., Matsuda N., Matsuda T., Nakajima R., Tanaka A. Preparation of Escherichia coli cell extract for highly productive cell-free protein expression. J Struct Funct Genom. 2004;5:63–68. doi: 10.1023/B:JSFG.0000029204.57846.7d. [DOI] [PubMed] [Google Scholar]
- 22.Kwon Y.C., Jewett M.C. High-throughput preparation methods of crude extract for robust cell-free protein synthesis. Sci Rep. 2015;5:1–8. doi: 10.1038/srep08663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin J., Noireaux V. An E. coli cell-free expression toolbox: application to synthetic gene circuits and artificial cells. ACS Synth Biol. 2012;1:29–41. doi: 10.1021/sb200016s. [DOI] [PubMed] [Google Scholar]
- 24.Silverman A.D., Kelley-loughnane N., Lucks J.B., Jewett M.C. Deconstructing cell-free extract preparation for in vitro activation of transcriptional genetic circuitry. ACS Synth Biol. 2018;8:403–414. doi: 10.1021/acssynbio.8b00430. American Chemical Society. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shimizu Y., Inoue A., Tomari Y., Suzuki T., Yokogawa T., Nishikawa K. Cell-free translation reconstituted with purified components. Nat Biotechnol. 2001;19:751–755. doi: 10.1038/90802. [DOI] [PubMed] [Google Scholar]
- 26.Ohno S., Matsui M., Yokogawa T., Nakamura M., Hosoya T., Hiramatsu T. Site-selective post-translational modification of proteins using an unnatural amino acid, 3-azidotyrosine. J Biochem. 2007;141:335–343. doi: 10.1093/jb/mvm036. [DOI] [PubMed] [Google Scholar]
- 27.Goerke A.R., Swartz J.R. High-level cell-free synthesis yields of proteins containing site-specific non-natural amino acids. Biotechnol Bioeng. 2009;102:400–416. doi: 10.1002/bit.22070. [DOI] [PubMed] [Google Scholar]
- 28.Hong S.H., Ntai I., Haimovich A.D., Kelleher N.L., Isaacs F.J., Jewett M.C. Cell-free protein synthesis from a release factor 1 deficient escherichia coli activates efficient and multiple site-specific nonstandard amino acid incorporation. ACS Synth Biol. 2014;3:398–409. doi: 10.1021/sb400140t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hong S.H., Kwon Y.C., Martin R.W., Des Soye B.J., De Paz A.M., Swonger K.N. Improving cell-free protein synthesis through genome engineering of Escherichia coli lacking release factor 1. Chembiochem. 2015;16:844–853. doi: 10.1002/cbic.201402708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith M.T., Hawes A.K., Shrestha P., Rainsdon J.M., Wu J.C., Bundy B.C. vol. 49. Process Biochem. Elsevier Ltd; 2014. pp. 217–222. (Alternative fermentation conditions for improved Escherichia coli-based cell-free protein synthesis for proteins requiring supplemental components for proper synthesis). [Google Scholar]
- 31.Chemla Y., Ozer E., Schlesinger O., Noireaux V., Alfonta L. Genetically expanded cell-free protein synthesis using endogenous pyrrolysyl orthogonal translation system. Biotechnol Bioeng. 2015;112:1663–1672. doi: 10.1002/bit.25587. [DOI] [PubMed] [Google Scholar]
- 32.Zubay G. In vitro synthesis of protein in microbial systems. Annu Rev. 1973;7:267–287. doi: 10.1146/annurev.ge.07.120173.001411. [DOI] [PubMed] [Google Scholar]
- 33.Li J., Zhang C., Huang P., Kuru E., Forster-Benson E.T.C., Li T. Dissecting limiting factors of the protein synthesis using recombinant elements (PURE) system. Translation. 2017;5 doi: 10.1080/21690731.2017.1327006. e1327006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kigawa T., Yabuki T., Yoshida Y., Tsutsui M., Ito Y., Shibata T. Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 1999;442:15–19. doi: 10.1016/s0014-5793(98)01620-2. [DOI] [PubMed] [Google Scholar]
- 35.Jun S.Y., Kang S.H., Lee K.-H. Continuous-exchange cell-free protein synthesis using PCR-generated DNA and an RNase E-deficient extract. Biotechniques. 2008;44:387–391. doi: 10.2144/000112690. [DOI] [PubMed] [Google Scholar]
- 36.Liu D.V., Zawada J.F., Swartz J.R. Streamlining Escherichia coli S30 extract preparation for economical cell-free protein synthesis. Biotechnol Prog. 2005;21:460–465. doi: 10.1021/bp049789y. [DOI] [PubMed] [Google Scholar]
- 37.Young T.S., Ahmad I., Yin J.A., Schultz P.G. An enhanced system for unnatural amino acid mutagenesis in E. coli. J Mol Biol. 2010;395:361–374. doi: 10.1016/j.jmb.2009.10.030. [DOI] [PubMed] [Google Scholar]
- 38.Sun Z.Z., Hayes C a, Shin J., Caschera F., Murray R.M., Noireaux V. Protocols for implementing an escherichia coli based TX-TL cell-free expression system for synthetic biology. J Vis Exp. 2013;79:1–15. doi: 10.3791/50762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schlesinger O., Chemla Y., Heltberg M., Ozer E., Marshall R., Noireaux V. Tuning of recombinant protein expression in Escherichia coli by manipulating transcription, translation initiation rates, and incorporation of noncanonical amino acids. ACS Synth Biol. 2017;6:1076–1085. doi: 10.1021/acssynbio.7b00019. [DOI] [PubMed] [Google Scholar]
- 40.Chemla Y., Ozer E., Algov I., Alfonta L. Context effects of genetic code expansion by stop codon suppression. Curr Opin Chem Biol. 2018;46:146–155. doi: 10.1016/j.cbpa.2018.07.012. Elsevier Ltd. [DOI] [PubMed] [Google Scholar]
- 41.Virdee S., Kapadnis P.B., Elliott T., Lang K., Madrzak J., Nguyen D.P. Traceless and site-specific ubiquitination of recombinant proteins. J Am Chem Soc. 2011;133:10708–10711. doi: 10.1021/ja202799r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- 43.Komander D., Rape M. The ubiquitin code. Annu Rev Biochem. 2012;81:203–229. doi: 10.1146/annurev-biochem-060310-170328. [DOI] [PubMed] [Google Scholar]
- 44.Chong S., Montello G.E., Zhang a, Cantor E.J., Liao W., Xu M.Q. Utilizing the C-terminal cleavage activity of a protein splicing element to purify recombinant proteins in a single chromatographic step. Nucleic Acids Res. 1998;26:5109–5115. doi: 10.1093/nar/26.22.5109. doi:gkb815 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shin J., Noireaux V. Efficient cell-free expression with the endogenous E. Coli RNA polymerase and sigma factor 70. J Biol Eng. 2010;4:1–9. doi: 10.1186/1754-1611-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]