Abstract
Chemical modifications of DNA and RNA regulate genome functions or trigger mutagenesis resulting in aging or cancer. Oxidations of macromolecules, including DNA, are common reactions in biological systems and often part of regulatory circuits rather than accidental events.
DNA alterations are particularly relevant since the unique role of nuclear and mitochondrial genome is coding enduring and inheritable information. Therefore, an alteration in DNA may represent a relevant problem given its transmission to daughter cells. At the same time, the regulation of gene expression allows cells to continuously adapt to the environmental changes that occur throughout the life of the organism to ultimately maintain cellular homeostasis.
Here we review the multiple ways that lead to DNA oxidation and the regulation of mechanisms activated by cells to repair this damage. Moreover, we present the recent evidence suggesting that DNA damage caused by physiological metabolism acts as epigenetic signal for regulation of gene expression. In particular, the predisposition of guanine to oxidation might reflect an adaptation to improve the genome plasticity to redox changes.
Keywords: Guanosine oxidation, Oxidative stress, Transcription, Histone modifications
1. Introduction
8-oxo-7,8-dihydroguanosine (8-oxo-dG) is the most frequent DNA oxidation product. Its syn arrangement pairs with adenine leading to G:C-T:A transversion upon replication. Despite an efficient removal system, 8-oxo-dG adducts can be still detected and used as indicators of DNA exposure to reactive oxygen species.
In addition, the mechanisms of 8-oxo-dG formation and processing across the genome suggest that this adduct can play a role in the regulation of genomic functions.
2. Oxidized deoxyguanosine is the main product of DNA oxidation
Pro-oxidant species are relatively abundant within cells, continuously generated by endogenous metabolism or upon exposure to external factors such as different types of radiation or pollutants. They can oxidize DNA either by altering the four bases or the deoxyribose. Reactive oxygen species (ROS)-mediated DNA oxidation is a relevant phenomenon and it has been estimated to occur only at a 50% lower rate as compared to the protein oxidation one [1].
In the DNA context, guanine has the lowest oxidation potential (−1.29 mV vs. normal hydrogen electrode) with respect to other bases [2,3]. The oxidation product of the C8 of the imidazole ring of deoxyguanosine (dG) generates the 8-oxo-dG (8-oxoguanosine, tautomer known as 8-hydroxyguanosine) (Fig. 1). 8-oxo-dG can be either formed at level of free nucleotides (8-oxo-GTP) [4] or directly at the level of the DNA molecule and represents the most stable and common product of DNA oxidation. Notably, oxidation of the dG located at the 5′ end of G repeats is favored because of its redox potential and because of the migration of radical cation [5,6].
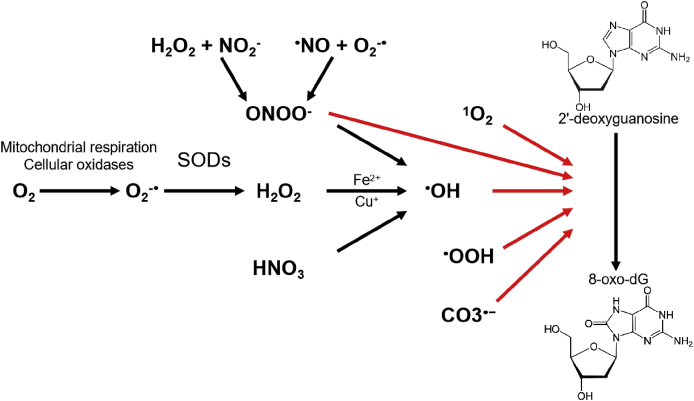
Fig. 1.
Schematic overview of 8-oxo-dG generation. Cellular 2′-deoxyguanosine undergoes an oxidative process which is mediated by a plethora of different agents. The most abundant intermediate in this process is hydrogen peroxide (H2O2) which derives from cellular metabolism and superoxide dismutases (SODs) activity. H2O2 promotes •OH release in the presence of transition metals. However, other reactive oxygen species (ONOO−, •OOH, CO3•− or 1O2) may also intervene in this process and increase the rate of 8-oxo-dG formation.
The percentage of steady-state oxidized dG within the nuclear genome has been estimated to account for approximately 104 to 106 [[7], [8], [9]] 8-oxo-dG adducts in a single nucleus.
Hydroxyl radical (•OH), hydroperoxide radical (•OOH), or the rare singlet oxygen (1O2) can attack the C8 of the imidazole ring of dG and form 8-oxo-dG. Moreover, •OH nucleophilic attack at C4, C5 and C8 positions of guanine following dehydration generates guanine radicals that decay to oxazolone in the case of C4 and C5 oxidation, or to 8-oxo-dG following hydrolysis of C8–OH. The reactions of dG with •OH and the •OOH are both highly exothermic despite the significant reaction barrier observed for •OOH-mediated C8-dG oxidation [10]. In parallel, peroxynitrite anion (ONOO−), a reactive nitrogen species (RNS) produced by the reactions of hydrogen peroxide (H2O2) with nitrite (NO2−) or superoxide (O2−•) with nitric oxide radical (NO), can also form 8-oxo-dG though at slower rate due to an unfavored reaction barrier [11]. Therefore, the major source of 8-oxo-dG is the reaction of DNA with •OH. Two-electron transfer reaction generates guanine radical and then 8-oxo-dG. Eventually, the carbonate radical (CO3•−) can oxidize guanine to guanine radical cation [12]. However, the majority of studies on the source of 8-oxo-dG focus on the reaction of DNA with •OH.
3. The origin of the •OH forming 8-oxo-dG
Within the cell, •OH is formed by the decay of H2O2, ONOO− or peroxynitrous acid (ONOOH). The presence of reduced transition metals (e.g. Fe2+ and Cu+) is required for the substantial conversion of peroxide into •OH as described first by Henry John Horstman Fenton in 1894 [13]. However, a metal-independent •OH generation mechanism has been recently evidenced: this mechanism relies on the polyhalogenated quinones-mediated H2O2 homolytic decomposition [14]. Eukaryotic cells do not display significant levels of free intracellular copper, whereas ferrous ion is relatively abundant [15,16]. On the other hand, transition metals are not required for the release of •OH by the RNS decomposition. Indeed, peroxynitrous acid decays by homolysis of its peroxo bond (k = 0.9 s−1 at 37 °C and 0.26 s−1 at 25 °C and pH 7.4) generating both nitrogen dioxide (•NO2) and hydroxyl radicals (•OH) [17]. Eventually, highly concentrated HNO3 has been shown to generate •OH radical through a homolytic reaction, releasing both •OH and •NO2 as products [18]. •OH reacts immediately with every species around the site it is formed at a rate that is restricted only by the diffusion rate of reactants in solution (diffusion-control rate reaction) [19]. Therefore, the •OH necessary to oxidize dG must be generated very close to the DNA, within an estimated diffusion range of ~5 nm, which corresponds to the width of 2–3 double helices [20].
The effective amount of ONOO− or H2O2 and Fe2+ in close proximity to DNA under physiological conditions is still undetermined. ONOO− passes across biological membranes and has an intracellular half-life of 10 ms, sufficient to diffuse across an entire cell diameter [21]. Thus, ONOO− generated inside the cytosol upon the reaction between NO and O2− might be present within the nucleus. However, the most stable and abundant ROS within cellular cytoplasm and nucleus is considered to be H2O2 [22]. Thus, the cellular system displays particular sensitivity to the lack of catalase (CAT) or glutathione peroxidase (GPX) scavenging activities [23]. Under physiological conditions, up to 2% of O2 is converted to O2−• inside cells. This production is mainly due to mitochondrial respiration [22] or to the activity of cellular oxidases [24]. Superoxide anion is then converted to hydrogen peroxide by the protein family of superoxide dismutases (SODs), acting in the cytosol, in mitochondria, and in the extracellular space [25]. At the same time, H2O2 can be originated within the nucleus. The O2−• producing NAD(P)H oxidase Nox4 has been shown to display a nuclear localization in endothelial and myocardial cells [26]. Notably, suppressing Nox4 nuclear activity affects both cysteine oxidation and histone deacetylase HDAC4 functions [27]. In hemangioendotheliomas [28] and myelodysplastic/leukemic cells [29] aberrant expression of Nox4 in the nucleus has been reported to correlate with higher levels of 8-oxo-dG critical for tumor progression. Moreover, O2−• dismutation by nuclear manganese-dependent superoxide dismutase (MnSOD) may also contribute to the nuclear production of H2O2 [30]. In addition, Lys-demethylases (LSD1 and LSD2) located on nucleosomes catalyze histone lysine (Lys) oxidative demethylation through flavin redox cycles releasing H2O2 [31]. Hundreds of enzymes containing flavin as coenzymes are expressed in mammals. Flavin coenzymes are very versatile due to their multiple redox states. They transfer one or two electrons and protons and are implicated in the divalent reduction of molecular oxygen to H2O2 [32]. Among these enzymes, LSDs are the only flavoenzymes present in the nucleus. Other Lys-demethylases such as the Jumonji C family of mono-, di- and trimethylated Lys-demethylases operate on histones, but use Fe2+ and the α-ketoglutarate-to-succinate co-reaction [33]. Intriguingly, LSD-mediated methyl group oxidation results in the release of formaldehyde, whose role in the inhibition of 8-oxo-dG removal has been reported [34]. On this basis, LSDs activity can indirectly promote the formation of 8-oxo-dG in the nucleus. Finally, Lysyl oxidase [35] and spermine oxidase [36] are other sources of nuclear H2O2, even though their contribution to DNA oxidation has never been reported.
As far as iron is concerned, direct measures of Fe2+ ions in the nucleus are scarce. X-ray fluorescence was used to detect iron pools in the nucleolus of pea plant cells [37]. Recently, Cloetens and colleagues described that iron within the nucleus of lymphocytes is mainly associated with the nuclear envelope rather than with the chromatin [38]. In addition, X-ray analysis on liver biopsies revealed a markedly lower iron abundance inside the nucleus as compared to other intracellular compartments [39]. These data suggest that iron is generally kept far from the genome thus lowering the risk of Fenton reactions.
Moreover, CAT, GPX and Glutathione transferases (GSTs) have been reported to preferentially localize in the perinuclear region or associate to the nuclear envelope of hepatocytes [40]. At the same time, the cytosolic pool of reduced glutathione (GSH) has been reported to prevent nuclear oxidative stress by scavenging cytosolic ROS [41]. Despite the abundance of iron-containing enzymes within the nucleus (e.g. replicases, helicases, primases and DNA repair glycosylases) [42], a pool of nuclear ferritin plays a crucial role in protecting DNA by quenching nuclear free-iron [43]. However the ROS-dependent regulation of transcription factors [44], replication enzymes [45] and structural proteins such as lamin [46] suggests that a critical ROS concentration has to be maintained physiologically in the nucleus.
4. The fate of 8-oxo-dG
In the B-form of DNA, the 8-oxo-dG presence impairs the “anti” angle around the glycosidic bond as the oxygen on the C8 would hinder the deoxyribose sugar, resulting in a stable “syn” conformation. In the syn conformation 8-oxo-dG mimics thymidine and pairs to “anti” adenine. This mismatch provides a way to bypass the dG alteration during DNA or RNA polymerization differently from other kinds of DNA damage [47]. Therefore, 8-oxo-dG maintenance prior to replication leads to a miscode transversion of GC to TA in the replicated strand [48]. However, dA is not always efficiently incorporated opposite to 8-oxo-dG template since DNA polymerase β (Pol β) can rotate the backbone of 8-oxo-dG for efficient pairing with dC [49]. On the contrary, DNA polymerase δ stalls in the presence of 8-oxo-dG, thus allowing the switch to other polymerases including the Pol η, which maintains high fidelity pairing with dC [50].
The specific enzymatic removal of 8-oxo-dG is catalyzed by nucleotide excision repair (NER) or by base excision repair (BER), two mechanisms that evolved from prokaryotes. In mammals the 8-oxo-dG DNA glycosylase (OGG1) recognizes the 8-oxo-dG opposite to dC [51] and cleaves the N-glycosidic bond between the base and deoxyribose. In this way, it generates an apurinic site (AP) and triggers the BER mechanism (Fig. 2).
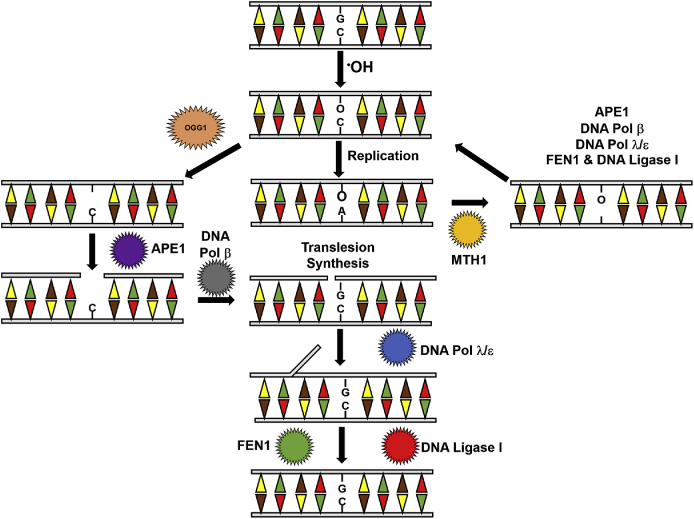
Fig. 2.
Base excision repair of oxidative DNA damage. The presence of 8-oxo-dG in double strand DNA may be repaired before replication by the concerted activity of OGG1, APE1 and translesion synthesis machinery. Alternatively, MTH1 (the homolog of E.coli mutY) recognizes the adenine introduced by replication and excises it. Upon base excision, DNA damage is repaired through the action of APE1 and translesion synthesis machinery.
If 8-oxo-dG is not removed and DNA is replicated, MTH1 (homolog of E. coli mutY) glycosylase excises 8-oxo-dG-paired adenine while promoting at the same time OGG1 activity. Upon removal, the adenine can be replaced by cytosine [52]. Thus, both MTH1 and OGG1 prevent 8-oxo-dG accumulation in DNA.
Even if OGG1 can excise the AP site at low efficiency [53], AP site is the substrate of the apurinic/apyrimidic endonuclease 1 (APE1). This enzyme hydrolyzes the phosphodiester bond at the 5′ of the AP site and produces a single-strand break (SSB) [54], which can be repaired by the replacement of a single or of few [[2], [3], [4], [5], [6], [7], [8], [9], [10]] nucleotides [51]. APE1 also removes 3′ oxidatively damaged DNA ends, including 3′ 8-oxo-dG, and 3′ unsaturated aldehyde generated by the AP endonuclease activity of DNA glycosylases [55]. This activity results in 3′-phosphate generation that can be subsequently converted to 3′-OH by the phosphatase activity of polynucleotide kinase 3′-phosphatase (PNKP). On the contrary, the 5′-deoxyribose phosphate terminus is mainly removed by the Pol β [56] that synthesizes the missing base. Then, DNA Pol λ and Pol ε with the Proliferating Cell Nuclear Antigen (PCNA) complete the DNA synthesis [57]. At the end, the flap endonuclease 1 (FEN1) removes flap ends and DNA ligase I seals the remaining nicks, thus rescuing DNA integrity.
Accessory proteins involved in the BER process include the X-ray Repair Cross-Complementing Protein 1 (XRCC1) that coordinates the assembly of BER enzymes and polyADP-ribose polymerase 1 (PARP1) that senses DNA breaks and recruits other repair proteins [58].
5. Modeling the fluxes of H2O2 and •OH forming 8-oxo-dG
The intracellular ROS levels are determined by the balance of several redox couples such as GSSG/GSH and NAD/NADH and reflects overall endogenous metabolism [22]. Exogenous challenges such as ischemia-hypoxia/reperfusion, carbon source and metal availability or macrophages exposure during inflammation affect intracellular ROS levels [[59], [60], [61], [62]].
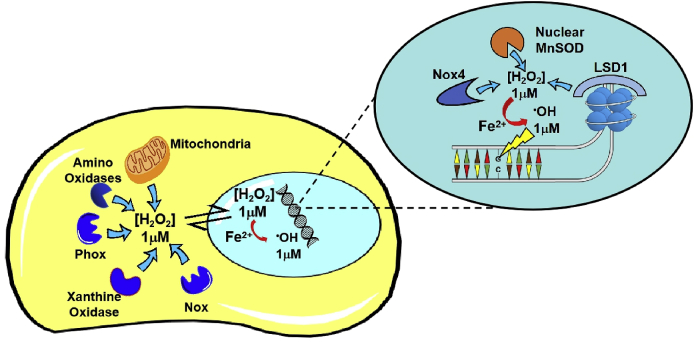
The amount of O2−• produced in cells in physiological steady state leads to an intracellular [H2O2] in the low micromolar range (1 μM, estimated in plasma cells and phagocytic cells) [[63], [64], [65], [66]]. The rate of Fenton decay of H2O2 to •OH is relatively fast in cellular conditions (20,000–30,000 M−1 s−1 at 37 °C pH 6–7) [67]. In this scenario, under the assumption of iron availability in the proximity of DNA and of not-limiting diffusion rate of H2O2 to the nucleus [68], a maximal few micromolar [•OH] could act on nuclear genome. Given the extremely low •OH range of action, the amount of nuclear volume in which the radical can exert its action, is accordingly reduced. The nuclear volume within 5 nm from the DNA double helix corresponds to about 10% of total nuclear volume, approximately 600 fL (6 × 10−13 L). Therefore, assuming a nuclear [•OH] of 1 μM, a total number of one thousand •OH molecules could constantly attack DNA (Fig. 3).
Fig. 3.
H2O2fluxes leading to dG oxidation. Cytosolic H2O2, generated from mitochondria or cytosolic oxidases, may diffuse to the nucleus. On the other hand, H2O2 may also be generated in the nucleus by LSD1 or in the nucleoplasm by nuclear oxidases. Upon conversion of H2O2 to •OH, dG may be oxidized.
Notably, OGG1-mediated 8-oxo-dG removal appears to exceed the rate of 8-oxo-dG formation as tissues from mice bearing OGG1 heterozygous deletion (OGG1+/−) show unaltered 8-oxo-dG contents in a context of reduced OGG1 activity [69]. This high repair efficiency is mainly due to DNA-binding time (in the order of subseconds), to a sliding diffusion constant equal to 5 × 106 bp/s (i.e. the theoretical upper limit for one-dimensional diffusion) and to a sliding activation barrier of 0.5 kcal/mol [70]. However, in an equilibrium situation, where 8-oxo-dG formation and removal rates are the same, the value of 105 to 106 8-oxo-dG adducts per nucleus is observed [[7], [8], [9]]. Interestingly, OGG1−/− mouse embryonic fibroblasts have been shown to display a nearly two-fold increase in 8-oxo-dG genomic accumulation when compared to wild type fibroblasts [71]. This finding suggests that completely abrogated OGG1 activity leads to a significant 8-oxo-dG accumulation in the genome.
Despite the various approximations included in this analysis, it is undeniable that the content of 8-oxo-dG is not compatible with sole or major contribution of extra-nuclear H2O2. Oxidants such as CO3•− and intra-nuclear H2O2 close to G sites of DNA may lead to further 8-oxo-dG generation. In this sense, lipid peroxides have been recently shown to induce a much higher 8-oxo-dG production in living cells when compared to H2O2 [72]. However, several evidence indicate intracellular H2O2 as the main source of the observed 8-oxo-dG in DNA. For example, CAT overexpression reduces 8-oxo-dG content in cell cultures and in vivo models [73]. Similarly, 2-mercaptoethanol depletion from embryonic cell medium increases 8-oxo-dG levels, leading to mutations in cardiac specification factor Tbx5i promoter and thus allowing cardiac-like differentiation [74]. In several mammalian cell lines glutathione depletion induces up to two-fold increase in 8-oxo-dG levels [75]. Consistently, mice supplemented with the glutathione precursor, acetyl-cysteine, display decreased 8-oxo-dG in several tissues [76]. Moreover, iron depletion has also been shown to reduce guanosine oxidation [77]. Eventually, recent works have substantiated that other stimuli affect 8-oxo-dG content independently of their redox effect, such as carbon tetrachloride [78], forms of graphene [79], diesel exhaust particles or hydroxyapatite nanoparticles [80].
6. Regulation of OGG1 activity
8-oxo-dG repair is not constitutive. The mechanism of action of the OGG1 enzyme involves 8-oxo-dG DNA recognition and removal, a process that is tightly regulated by external and internal stimuli (Fig. 4).
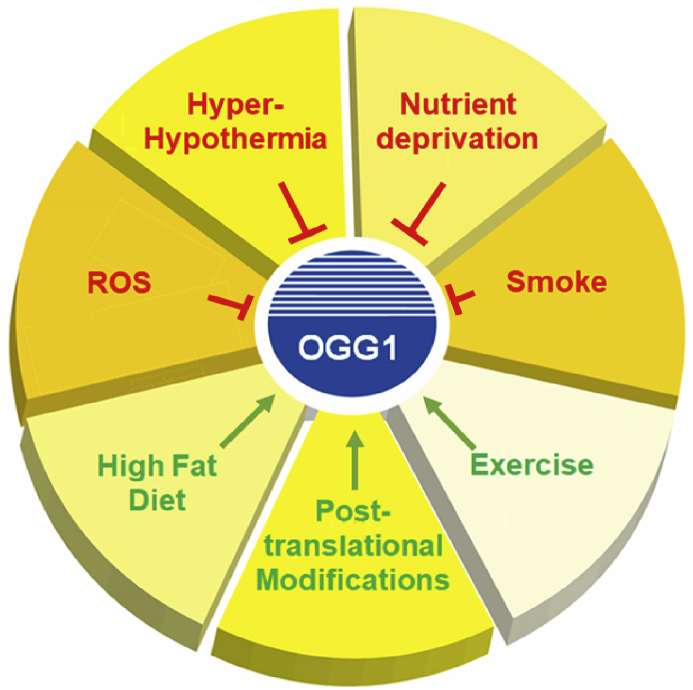
Fig. 4.
Several factors regulate OGG1 and 8-oxo-dG removal. OGG1 is regulated by a plethora of stimuli influencing both its expression, stability and eventually activity. ROS accumulation, smoke, nutrient deprivation and temperature imbalance inhibit OGG1, whereas high fat diet, exercise and several post-translational modifications may enhance its activity.
In particular, the 8-oxo-dG-repair activity of OGG1 has been shown to decrease upon oxidative stress and then to be rescued once the cellular redox state is re-established [81]. Notably, this effect cannot be ascribed to OGG1 transcription inhibition. The proposed inactivating mechanism envisions the oxidation of some OGG1 cysteine residues. Intriguingly, nitric oxide donors may mediate OGG1 inhibition [82] whereas H2O2 does not display this capability [83]. It has been proposed that OGG1 cysteine oxidation triggers the enzyme re-localization into stress vesicles in response to oxidative stress [84]. Besides its inhibitory effects on OGG1 activity, oxidative stress has been shown to upregulate APE1 gene expression. At the same, increased APE1 concentration has been linked to an increased OGG1 AP-lyase activity [85]. Moreover, Ca2+ influx-mediated neuronal membrane depolarization has been shown to induce mitochondrial ROS production and, in parallel, to increase APE1 transcription and DNA repair activity. In this context, cAMP-response element-binding protein (CREB) activation upon oxidative stress has been proposed to bind APE1 promoter, stimulate expression of the gene, and limit DNA damage upon mitochondrial ROS burst [86].
Furthermore, OGG1 activity is precisely fine-tuned by post-translational modifications. As an example,p300/CBP acetyltransferase complex catalyzes the acetylation of critical lysine residues and enhances OGG1 performance [87]. Interestingly, oxidative stress has been shown to induce an acute amplification of p300 levels through auto-acetylation and stabilization [88]. Moreover, ROS also promote the acetylation process of p300/CBP [89] thus stimulating OGG1 activity, whereas deacetylation of OGG1 by class I histone deacetylases reduces 8-oxo-dG repair [87]. On the contrary, NAD-dependent deacetylase Sirtuitin (Sirt1) significantly reduces the levels of acetylated OGG1 and therefore reduces OGG1 repair activity [90]. In an opposite way, Sirt1 induces APE1 deacetylation, thus enhancing APE1-mediated DNA repair, in a process which is further promoted by genotoxic stress [91]. In the mitochondria, instead, OGG1 is targeted by the sirtuin 3 (Sirt3), a mitochondrial NAD(+)-dependent deacetylase, that increases OGG1 stability and repair activity [92]. In addition, several serine/threonine kinases, including Protein kinase C (PKC), Cyclin-dependent kinase 4, and c-Abl phosphorylate OGG1 and increase its nicking and AP lyase activities [93]. Similarly to p300 acetyltransferase, PKC displays a significantly increased activity upon ROS [94]. On the other hand, c-Abl has been shown to be activated upon nitroxidative stress [95]. Upon phosphorylation, OGG1 associates preferentially to chromatin compartment, whereas the nuclear matrix contains un-phosphorylated OGG1 [93].
More recently, the inflammatory responses induced by endotoxin administration or autoimmune processes have been shown to promote OGG1 transcription through the Signal transducer and activator of transcription 1 (STAT1) [96]. STAT1 promotes gene expression by assembling interferon-γ complex on the interferon-γ activation motif of target promoters [97]. In parallel it induces chromatin relaxation through the acetylation and the demethylation of specific histone H3 lysine residues [96]. These results suggest a prominent role for inflammation signaling in OGG1 activity promotion and are also corroborated by recent data substantiating a significant OGG1 activation in mice lung upon pollen antigen exposition [98]. Several alternative splice variants for the OGG1 gene have been described. However, common variants of OGG1 have also been evidenced to be inhibited by the action of the inflammatory cytokine tumor necrosis factor alpha (TNF-α) [99], thus suggesting an even more complicated landscape in inflammation-mediated OGG1 fine-tuning.
The 8-oxo-dG content has also been shown to be influenced by stress conditions associated to life habits. As an example, rats exposed to ROS-inducing cigarette smoke display a significant down-regulation of OGG1 and MTH1 [100]. From a dietary point of view, mice undergoing obesogenic diet show increased OGG1 levels in different tissues [101]. Interestingly, the same result is observed in rats or humans practicing enhanced physical activities [102]. On the contrary, cardiomyocytes exposed to nutrient deprivation display a significant reduction in base excision repair due to autophagy-dependent OGG1 degradation [103]. In a similar manner, non-toxic mild hyperthermia induces OGG1 proteasomal degradation in HeLa [104]. On the other hand, OGG1 levels have been shown to be down-regulated in newborn pigs upon therapeutic hypothermia, thus suggesting a specific temperature range of action for OGG1 activity [105].
7. Distribution of 8-oxo-dG in the mammalian genome
The genomic distribution of 8-oxo-dG is not uniform and represents a controversial issue. Nakabeppu and colleagues first mapped steady state 8-oxo-dG distribution in the human genome by in situ immunodetection and found that high-density single-nucleotide polymorphisms (SNPs) areas are predominantly distributed within 8-oxo-dG-enriched regions [106]. In addition, high-resolution mapping of 8-oxo-dG in rat genome was obtained by 8-oxo-dG immunoprecipitation followed by microarray hybridization. This analysis revealed a negative correlation between 8-oxo-dG distribution and gene density [107]. Although it should be noted that gene-free regions show higher GC content than gene-rich ones, authors interpreted this negative correlation as the consequence of gene-rich regions localization within the nucleus. Indeed, gene-rich euchromatic regions occupy the inner portion of the nucleus, whereas gene-poor regions stick to the periphery and thus are more vulnerable to extra-nuclear oxidative stress. However, more recently, the sequencing of immunoprecipitated 8-oxo-dG-containing fragments (OxiDIP-Seq) showed 8-oxo-dG enrichment within the gene body and promoter regions of both human and mouse cells [108]. This result is in line with the data by Burrows and colleagues, who analyzed by high resolution 8-oxo-dG sequencing (OG-Seq) the distribution of 8-oxo-dG in mouse embryonic fibroblasts in vitro. The authors showed that intergenic regions display lower accumulation of 8-oxo-dG when compared to genic regions: in particular, promoters, 5′-UTRs, and 3′-UTRs provide the highest 8-oxo-dG enrichment in the genome [71], again suggesting a higher proneness of euchromatic regions to guanosine oxidation.
Notably, 8-oxo-dGhas been shown to map in the proximity of DNA replication origins (ORIs) within the gene body of transcribed long genes in mammalian cells [108]. Accumulation of 8-oxo-dG at the ORIs within the body of these genes is compatible with the increased sensitivity to oxidation of persistent ssDNA deriving from collisions between transcription and replication machineries, or with the block of the leading-strand replication by a G-quadruplex structure [[109], [110], [111]].
Interestingly, no association was found between 8-oxo-dG content and transcription levels. In particular, very low 8-oxo-dG signals were observed at the most highly transcribed genes, while the strongest signals were measured within the gene body of particularly long genes showing low transcription levels [108]. These data may suggest that ROS-mediated DNA damage is repaired more efficiently in the highly transcribed genes as compared to poorly transcribed ones. Traditionally OGG1 has been preferentially observed in euchromatic regions [89], where 8-oxo-dG is more efficient in recruiting OGG1 and other BER enzymes [112]. However, a more recent model proposes that OGG1 recruitment is independent of the transcription levels, but highly transcribed genes facilitate the recruitment of BER machinery [113]. On top of that, both in vivo [114] and in vitro [115] studies have shown that heterochromatic telomeric regions display 8-oxo-dG enrichment. This accumulation has been linked to telomere shortening [116] since the presence of oxidized guanosine impairs telomere maintenance and replication [[117], [118], [119]]. In addition, Opresko and colleagues showed that telomerase itself may introduce 8-oxo-dG into replicating telomeres by incorporating 8-oxo-dGTP [120], eventually leading to telomere crisis. Beside the increased amount of 8-oxo-dG in telomeric regions, the work by Liu and colleagues showed also that OGG1 repair activity is diminished in the telomeric regions due to secondary structures that may form in the human telomeres [114].
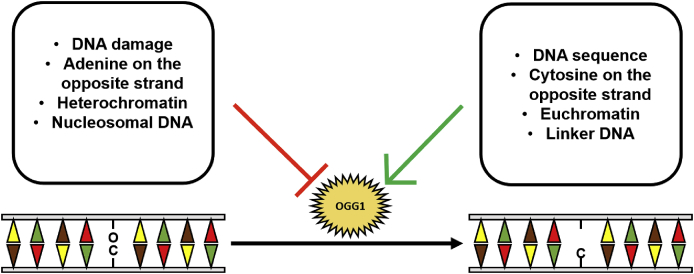
Beside the chromatin structure, OGG1 activity has been shown to be largely dependent on the DNA sequence at the genomic sites where it is recruited (Fig. 5), with some sequences being more efficiently repaired than others [121].
Fig. 5.
Factors affecting OGG1 activity in oxidative damage repair. OGG1 recruitment and repair activity may be strongly inhibited by several factors, mainly related to DNA structure compaction and alteration. On the contrary, relaxed chromatin structure and specific DNA sequences facilitate OGG1 recruitment and activity.
Indeed, OGG1-mediated 8-oxodG excision efficiency is increased by the presence of a neighboring cytosine in the opposing DNA strand, whereas a neighboring adenine reduces the efficiency of the 8-oxo-dG cleavage. Furthermore, 8-oxo-dG repair is severely inhibited by the proximity to other kinds of DNA damage (e.g. mismatches, abasic sites, single-strand breaks) [122].
Finally, oxidative damage has been reported to be more frequent in linker DNA between nucleosomes than in the octamer-embedding DNA [123]. In parallel, OGG1 activity on chromatin is also profoundly affected by 8-oxo-dG position with respect to nucleosomes: Angelov and colleagues have indeed demonstrated that 8-oxo-dG is quickly repaired when it is located in the internucleosomal linker DNA, whereas the proximity of histones decreases ten-fold the efficacy of repair [124].
8. Effect of 8-oxo-dG on gene transcription
The direct effect of 8-oxo-dG on transcription is still controversial. In vitro assays revealed that human recombinant RNA polymerase II (Pol2) partially pauses in the presence of 8-oxo-dG, slowing down the transcription rate and eventually leading to cytosine or adenine incorporation in the nascent mRNA opposite 8-oxo-dG [125,126]. Pol2 stalling on 8-oxo-dG has been suggested to induce a transcription coupled repair (TCR) of 8-oxo-dG [127] by recruiting the DNA repair machinery. Several works showed that the pausing phenomenon is largely model-dependent and suggested a role for TFIIS transcription factor in shortening the stalling time [126,128]. However, other studies claimed the absence of a role for 8-oxo-dG in promoting a transcription block [129,130]. In particular, in vivo experiments conducted on murine embryonic fibroblasts showed the absence of any significant block or delay of Pol2 at the 8-oxo-dG sites [131], suggesting a marginal role of the oxidized base in pausing transcription [132]. Furthermore, recent evidence suggests that 8-oxo-dG does not represent per se a barrier for transcription. Rather, the OGG1 recruitment and activity may induce a significant delay in Pol2 activity, thus promoting the observed Pol2 stalling [130]. Interestingly, a recent study of Vermeulen and colleagues explained the 8-oxo-dG-related Pol2 pausing as the result of 8-oxo-dG repair intermediates [113]. However, it has been recently proposed that these discrepancies may be largely due to the specific position of 8-oxo-dG in the promoter, the promoter strength, and the nucleotidic sequence surrounding the lesion [133].
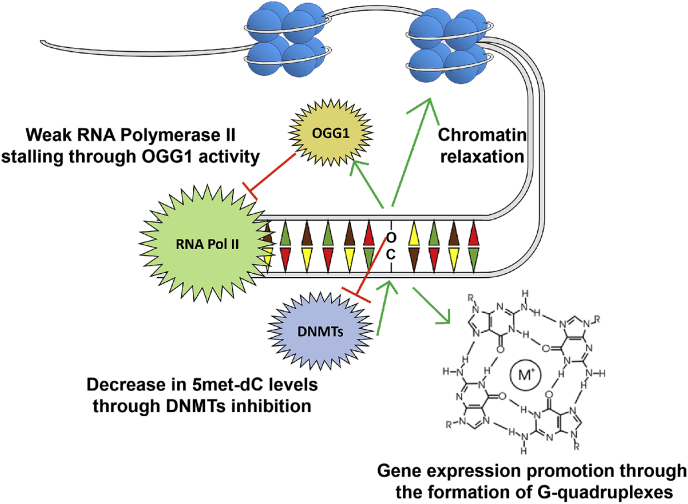
Besides being involved in Pol2 pausing, 8-oxo-dG has been shown to possess an epigenetic activity when found in gene promoter (Fig. 6).
Fig. 6.
8-oxo-dG accumulation alters gene expression in multiple ways. 8-oxo-dG-mediated recruitment on chromatin has been suggested to weakly dampen RNA polymerase transcription activity. On the contrary, oxidative damage has been demonstrated to promote gene expression through the induction of G-quadruplex formation, chromatin relaxation and reduced DNA methylation.
In particular, multiple studies have shown that guanosine oxidation in promoters leads to increased gene expression [[134], [135], [136], [137]]. The most acknowledged mechanism relies on the formation of G-quadruplex DNA secondary structures, that have been shown to promote gene expression [138,139]. Several works pointed out that 8-oxo-dG incorporation in potential G-quadruplex–forming sequences (PQSs) located in the coding strand of gene promoter leads to increased transcription. In details, OGG1 recruitment on 8-oxo-dG generates an abasic site which unmasks the PQS, leading to the formation of a G-quadruplex where APE1 binds [140] and interacts with transcription factors [141]. This mechanism seems to be particularly relevant in human DNA repair genes, which display PQS enrichment in their promoter and 5′-UTR [142]. Notably, the same mechanism has been shown to be crucial for the expression of several genes such as VEGF [143,144], PCNA [145], NTHL [141], and, more recently, to the DNA repair gene NEIL3 [146]. Interestingly, another work by Burrows and colleague showed that, when 8-oxo-dG is incorporated in PQSs located in the template strand, the formation of G-quadruplex downregulates gene expression [147]. Coherently, two recent studies by Xodo and coworkers proposed a related mechanism responsible for KRAS and HRAS transcription regulation. In their model, the formation of G-quadruplexes in the gene promoter leads to gene repression [148]. However, the recruitment of nuclear proteins essential for KRAS [149] and HRAS [150] transcription to the abasic G-quadruplex (i.e. MAZ, hnRNPA1, and PARP-1) regenerates the double helix, boosting the gene transcription. Interestingly, in both these genes the PQS regions are located in the template strand, therefore corroborating the model by Burrows and colleagues.
Accumulation of 8-oxo-dG has been reported for several loci and upon different stimuli (listed in Table 1).
Table 1.
List of genes found upregulated by 8-oxo-dG on their promoters.
| Gene name | Function | Cell type | Inducer | Ref. |
|---|---|---|---|---|
| EDN1 | Vasoconstriction | Endothelial | Hypoxia | [145] |
| HMOX1 | Heme catabolysm | Endothelial | Hypoxia | [145] |
| VEGF | Angiogenesis | Endothelial | Hypoxia | [145] |
| BCL2 | Apoptosis regulation | breast cancer | Estrogen | [131] |
| TTF1 | Secreted protein | Breast cancer | Estrogen | [151] |
| KLK3 | Serine protease | Prostate epithelial | Androgen | [152] |
| TMPRSS2 | Serine protease | Prostate epithelial | Androgen | [152] |
| MIR125B2 | Proliferation regulation | Prostate epithelial | Androgen | [152] |
| MIR133B | Apoptosis regulation | Prostate epithelial | Androgen | [152] |
| NCL | Ribosomes synthesis | Fibroblast | Myc | [153] |
| CAD | Pyrimidine biosynthesis | Fibroblast | Myc | [153] |
| CXCL2 | Immunoregulation | Lung epithelial | TNFα | [155] |
| IL1B | Immunoregulation | Lung epithelial | TNFα | [155] |
| TNF | Immunoregulation | Lung epithelial | TNFα | [155] |
Estrogen increases transcription levels and 8-oxo-dG content of Bcl-2 and pS2/TFF1 promoter regions [151]. Likewise, androgen stimulates the transcription of kallikrein 3 (Prostate Specific Antigen) and other androgen-specific target genes in prostate cells following accumulation of 8-oxo-dG in the androgen response elements of the promoters of those genes [152]. The c-Myc proto-oncogene activates transcription and induces 8-oxo-dG formation at the promoters of Nucleolin and Aspartate Transcarbamylase genes, two critical mediators of Myc-driven transformation [153]. Moreover, 8-oxo-dG plays a critical role also in the NF-κB binding site as it increases the recruitment of the p50 NF-κB transcription factor [136,154,155]. As an example, TNF-α-induced oxidative stress and increase in 8-oxo-dG content of nuclear DNA favor NF-κB-mediated expression of cytokine genes and accumulation of inflammatory cells in mouse airways [156]. Notably, hypoxia-induced genes, such as VEGF, Heme Oxigenase 1 and Endothelin 1 display increased 8-oxo-dG-content in the promoter regions upon hypoxic conditioning [145].
The specific mechanism leading to the accumulation of 8-oxo-dG in the hypoxic response element (HRE) of VEGF, or in the NF-κB binding site, is less clear. However, upon dG oxidation, the presence of OGG1 at promoter sequences of NF-κB target genes has been shown to promote gene expression [154,155]. The proposed mechanism relies on the introduction of a bend in the DNA duplex [156] and on the concomitant recruitment of TFIID, NF-κB/RelA, Sp1, and phosphorylated RNA pol II [157]. In this regard, the position of 8-oxo-dG in the DNA is relevant, since 8-oxo-dG placed 2, 3, or 5 bp (5′ or 3′) distance from the NF-κB-binding motif results in reduced binding of the transcription factor [154]. This latter outcome may involve either 8-oxoG itself or the interference of OGG1 [156]. Likewise, OGG1 and APE1 recruitment on HRE of VEGF and other hypoxia-induced genes facilitates the recruitment and the activity of the Hypoxia inducible factor α [158].
Recently, it has been suggested that 8-oxo-dG generation may favor transcription through chromatin relaxation. In particular, Avvedimento and coworkers showed that estrogen signaling activates LSD1 which in turn facilitates the transcription machinery activity [151]. LSD1 catalyzes histone demethylation by two divalent reduction of the FAD cofactor. The reconstitution of the oxidized FAD cofactor causes H2O2 production in proximity to the DNA [33] leading to 8-oxo-dG accumulation in the regulatory sites of estrogen target genes. Then, OGG1 and topoisomerase IIβ are recruited at these 8-oxo-dG sites where the OGG1-induced transient nick is used as entry points by the topoisomerase which relaxes the DNA strand to accommodate the transcription initiation complex [151]. The process involving LSD1-mediated 8-oxo-dG generation has been shown to be necessary in promoting expression of Myc target genes: LSD1 is activated by Myc and induces OGG1, APE1 and Pol2 recruitment upon guanosine oxidation [152]. Moreover, H3K9me2 demethylation by LSD1 and the subsequent local 8-oxo-dG formation are also associated to the transcription of androgen target genes [153].
In addition, 8-oxo-dG has been shown to affect dC methylation, which mainly occurs in CpG dinucleotides and generates 5 mC. Following the initial Holliday's hypothesis on the role of DNA damages on the epigenetic dC code [159], it has been extensively demonstrated that 8-oxo-dG affects C5 dC methylation by interfering with the binding of DNA methyltransferases (DNMTs) to dC [[160], [161], [162], [163]]. Consistently, a negative correlation between the content of 8-oxo-dG and 5 mC in human DNA was observed [164]. Interestingly, CuZn-SOD deficient mice display 8-oxo-dG accumulation and massive DNA hypomethylation [165]. On top of that, methylated dC flanking 8-oxo-dG shows a reduced affinity for methyl-CpG binding proteins thus reducing their transcription suppression efficiency [166], which suggests that 8-oxo-dG formation may exert a long-lasting effect on gene expression.
Notably, oxidative damage also occurs directly at RNA and it has been shown to be mediated by the same oxidative agents that act on DNA [167]. It is estimated that up to half mRNAs bear at least one 8-oxo-G adduct; thus, 8-oxo-G content is ten-fold higher than 8-oxo-dG [168]. RNA oxidative damage affects protein synthesis process, promoting both ribosome stalling, premature translation termination, and accumulation of short polypeptides [169,170]. The oxidation of rRNA and tRNA is also documented and leads to decrease in protein production rate [171] tRNA degradation [172] respectively.
Therefore, even though DNA oxidation may exert an activating effect on gene expression, oxidized RNAs are only associated to decreased protein synthesis, thus suggesting multiple control layers of the gene expression process upon oxidative stress.
9. Genome-independent functions of OGG1 and 8-oxo-dG
OGG1 and MTH1 expression appear stable during the cell cycle [173]. In addition to promoting DNA repair, OGG1 has been shown to possess other roles involved in multiple aspects of cellular physiology. Phosphorylated OGG1 enzyme sticks to mitotic chromatin [93] and associates with microtubules and with the mitotic spindle during interphase and mitosis respectively. The interaction with the mitotic machinery allows a faithful distribution of the repair enzyme pool to daughter cells [174]. Moreover, time-lapse microscopy upon OGG1 siRNA-mediated silencing revealed a critical role of OGG1 in chromosome alignment and segregation during mitosis [175]. Moreover, cytosolic OGG1 shows the capacity to form a complex with free 8-Oxo-7,8-dihydroguanine. This complex interacts with Rho GTPase leading to the phosphorylation of the mitogen-activated kinases and to the activation of the related signaling cascade [176]. In addition, the same complex induces α-smooth muscle actin polymerization resulting in cytoskeletal modulation [177].
Notably, the pool of 8-oxo-GTP,deriving from the hydroxyl radical attack to the cytosolic 2-dG 5′- triphosphate (dGTP) [178], inhibits the activity of soluble guanylyl cyclases and thus alters the cellular signaling upon oxidative stress [179]. 8-oxo-dG in mitochondrial genome has been shown to promote profound changes in liver metabolism. Indeed, recent works showed that upon OGG1 knockout, the increased mitochondrial 8-oxo-dG levels impacts on the electron transport chain (ETC) efficiency in an age-, tissue-, and strain-dependent manner [180,181]. However, this situation should not be ascribed to mutations in the ETC genes. Rather, the authors suggested a specific role for the interaction between 8-oxo-dG and OGG1 in promoting efficient mitochondrial metabolism.
10. Lessons from 8-oxo-dG repair defective mice
OGG1 knock-out mice (OGG1−/−) have been generated [70,182]. Despite the lack of any evident pathological phenotype, they show 3- to 7-fold increase in 8-oxo-dG levels at 2–4 months of age. Interestingly, while heterozygous mice (OGG1+/−) show two-fold reduction of OGG1 activity, they do not display abnormal accumulation of 8-oxo-dG, thus suggesting that the wild-type (OGG1+/+) 8-oxo-dG repair rate exceeds the rate of 8-oxo-dG formation. In addition, OGG1−/− mice do not display neither increased mutation burden nor enhanced tumorigenic potential [70,183,184]. Notably, OGG1−/− mice display improved resistance to gastrointestinal infection [185] and to systemic LPS-induced inflammation [186]. However, double mutant OGG1−/− and MTH1−/− (mice are short living due to severe incidence of tumor [187].
OGG1 deletion has been also shown to exert profound effects on the mitochondrial genome. Liver mitochondrial DNA of OGG1−/− mice bears 20-fold more 8-oxo-dG than the wild-type counterpart [188]: interestingly, this phenotype is completely abolished in fasting OGG1−/− mice, which display the same mitochondrial 8-oxo-dG levels as compared to wild-type ones [180].
Another interesting phenotype of OGG1−/− mice is the downregulation of fatty acids oxidation genes and the consequent switch towards lipogenesis in the liver. Consistently, OGG1−/− mice accumulate more fat upon high fat-enriched diet and they are more susceptible to develop glucose intolerance suggesting 8-oxo-dG processing may affect metabolism at systemic level [101]. In this scenario, a recent study has suggested that OGG−/- mice skeletal muscles are much more sensitive to palmitate induced mtDNA damage [189].
In addition, OGG1−/− mice display increased macrophage apoptosis, cytokine release and subsequent inflammatory response. This situation is particularly severe in the atherogenesis process, since impaired macrophage physiology leads both to atherosclerosis and thrombotic events [190].
Eventually, recent studies on MTH1/OGG1 double knock-out mice have suggested a role for these two proteins in neuronal physiology. In this model, neurons exhibit mitochondrial dysfunction and poor neurite outgrowth [191]. Given the prominent role of mitochondrial homeostasis in Alzheimer disease onset, OGG1 and MTH1 alterations have been also suggested to be key players in the development of this pathology [192].
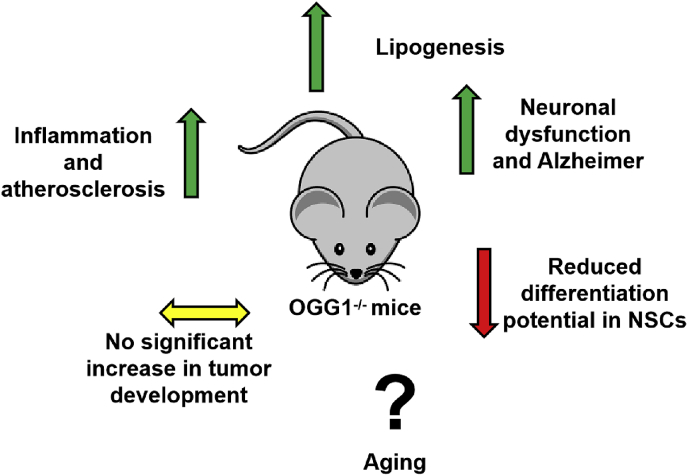
A summary of main dysfunction associated with OGG1 deletion in mice is depicted in Fig. 7.
Fig. 7.
Phenotypical alteration identified in OGG1 defective mice. Even though OGG1 absence has not been associated to enhanced tumor onset, it may significantly impair several aspects of mouse physiology, with the induction of inflammation, lipogenesis, and neuron and neural stem cell alteration. Intriguingly, OGG1 deletion has not been demonstrated to have a functional role in aging promotion.
11. Oxidative signaling in the mitochondrial genome
The effective rate of 8-oxo-dG formation in mtDNA was discussed long time ago [193]. Some laboratories estimated in mtDNA up to 1-3 8-oxo-dG per 105 dG, a frequency much higher than in the nuclear DNA. It would account approximately for one adduct every 20 mitochondrial genomes [194] and, when considering the number of mitochondrial genomes per cell [195], at least 8-oxo-dG might be present in each cell. This phenotype might be due to mtDNA proximity to the ROS produced by ETC leakage and to the lack of histone shields in mtDNA. However, in other studies, oxidative damage to mtDNA was indicated to be much less frequent, even lower than in nuclear DNA [196]. This might be explained by the efficient 8-oxo-dG repair mitochondrial machinery [197].
The response of mtDNA to oxidative stress is also controversial. Pro-oxidant treatment of human fibroblasts induces mtDNA damage and a significant increase in mtDNA copy number [198], though the mtDNA-specific Polymerase-γ is known to be inhibited by 8-oxo-dG [199]. In addition, hypoxia-mediated oxidative stress leads to mtDNA copy number increase in many tissues. Indeed, oxidative damage induces OGG1 accumulation in the D-loop region of mtDNA, eventually leading to the recruitment of TFAM, the main factor in mtDNA transcription and replication [200].
On the contrary, oxidative stress or BER inhibition result in degradation of damaged mtDNA molecules [201] in tumoral cell lines. Consistently, 8-oxo-dG adducts localize preferentially on fragmented mtDNA upon oxidative stress [202]. High susceptibility of mtDNA to strand breaks and Endonuclease G-mediated degradation was observed upon oxidative stress and proposed as a mechanism for protecting from deleterious accumulation of mutated mitochondrial genomes [203]. Intriguingly, recent works have shown that oxidative stress may also inhibit Endonuclease G activity, thus preventing mtDNA degradation and affecting cell death process [204].
These results suggest that both mtDNA copy number increase and degradation represent major responses to oxidative stress conditioning. The specific role of the two different processes in mitochondrial response to oxidative stress has still to be elucidated. Moreover, regardless the peculiarity of mtDNA with respect to oxidative stress, a regulatory function of 8-oxo-dG within mitochondria still needs to be validated.
12. 8-Oxo-dG during aging and tissue remodeling
The implications of oxidative damage in aging are well known and 8-oxo-dG has been shown to be the most common and abundant oxidative DNA base lesion in all aged cell types [205]. Several-fold increase of 8-oxo-dG in nuclear DNA of old compared to young animals has been observed in major organs [206,207]. Intriguingly, studies conducted both on mice and on rats suggest that nervous tissue displays the highest accumulation of 8-oxo-dG during the aging process [208,209]. Furthermore, measures of urinary secreted 8-oxo-G confirmed in human the aging-associated increase of this adduct [210]. Aging-related 8-oxo-dG accumulation was attributed to both higher levels of ROS and decreased activity of OGG1 in the elderly [211]. A dynamic role of 8-oxo-dG repair throughout life span is supported by several observations. In particular, levels of total OGG1 have been shown to be unaltered over time across individuals [212], while the acetylated active form displays critical decrease in old individuals [102]. In addition, mitochondrial OGG1 displays a pattern of decreased expression in old mice [213]. However, even though 8-oxo-dG accumulation represents a clear hallmark of aging, its functional role in the promotion or in the progression of this process has still to be elucidated.
In parallel, 8-oxo-dG accumulation has also been shown to take part into the differentiation process. Indeed, interfering with OGG1 expression resulted in the impairment of embryonic stem cell neuronal differentiation [214] suggesting that 8-oxo-dG repair might have a role in the control of stem cell and tissue homeostasis. To this purpose, the contribution of Sirt3 on OGG1 gene expression and on the maintenance of low 8-oxo-dG levels was found to be critical for the suppression of fibroblast differentiation [215]. Moreover, the commitment of embryonic stem cells toward myocardial differentiation has been also associated with accumulation of 8-oxo-dG and G∙C to C∙G transversion, particularly in the promoter region of transcription factors involved in cardiac specification such as Tbx5, Mef2C, Nkx2-5, and Gata4 [74]. Notably, this mutation-mediated effect of DNA oxidation on the expression pattern is transmissible and may represent a mechanism of epigenetic adaptation to oxidative stress conditions. Eventually, accumulation of 8-oxo-dG in mtDNA of OGG1−/− neural stem cells (NSCs) leads to a reduced mitochondrial activity which is in turn associated to a limited differentiation potential of NSCs [216].
13. Evolutionary perspective and conclusions
Epigenetics is defined as the study of heritable changes in genome function that are not due to alterations in the DNA sequence. 5 mC is a typical epigenetic mark that extends the 4 bases code and provides a new information level. The oxidized form of guanosine represents a stable change in the DNA structure that, similarly to 5 mC, may encode additional information to the primary DNA sequence dictated by the redox environment surrounding the genome.
8-oxo-dG is produced by nonspecific fluxes of H2O2 from the cytosol to the nucleus, thus reflecting overall intracellular redox balance. 8-oxo-dG is also produced by local reactions taking place at the chromatin level, such as LSD1-mediated demethylation, and is involved in the control of expression of specific genes. Whether these two pathways of 8-oxo-dG formation cooperate is unknown. Given the implication of DNA oxidation in mutagenesis and aging, it is likely that marking dG by oxy group represents a tightly regulated process.
The importance of this regulation is also suggested by the conservation of critical residues in the glycosylases aimed at removing 8-oxo-dG from bacteria to eukaryotes [217,218]. In parallel, phylogeny studies have shown a consistent difference in the number of G in the DNA across species. The positive correlation between G content and environmental temperatures observed in some sea organisms or in birds versus reptiles is interpreted as a selective advantage of the more stable GC than AT pair, though the importance of the 3 versus 2 hydrogen bonds for helix stability is debated [219,220]. In addition, a possible benefit of G enrichment is the reduced risk of thymine dimer formation upon sun exposure.
Generally, external temperature, thermogenesis, and aerobic metabolism involve higher ROS levels that in turn lead to 8-oxo-dG formation. Counterintitively, the genomes of aerobic organisms show high levels of G compared to anaerobes [221]. In this scenario, higher G content could act as a ROS buffer and therefore reflect an adaptation aimed at improving the genomic response to potentially dangerous peaks of oxidant generated within cells by metabolism. Even the intron-exon architecture of different genomes has been suggested to be selected in order to make genes resistant to oxidative stress [222]. In this view, DNA oxidation is an accidental by-product of aerobic metabolism that triggers undesirable genotoxic damage. However, “physiological” DNA oxidation may have important consequences for the fitness as revealed by the regulation of pro-angiogenic factor in hypoxic conditions. Available information supports the view that cells use controlled production of 8-oxo-dG to regulate gene expression and therefore DNA oxidation should not be regarded solely as an accidental by-product. However, further studies are still required to understand whether guanosine oxidation codes for a transmissible information capable to durably and specifically alter cellular behavior.
Funding
This work was supported by the Italian Association for Cancer Research (AIRC) and by the National Institute on Aging - NIH U.S.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
We thank Stefania Averaimo for assistance in writing the manuscript and Rani Pallavi for grammar check.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101398.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Domej W., Oettl K., Renner W. Oxidative stress and free radicals in COPD--implications and relevance for treatment. Int. J. Chronic Obstr. Pulm. Dis. 2014;9:1207–1224. doi: 10.2147/COPD.S51226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Steenken S., Jovanovic S.V. How easily oxidizable is DNA? One-electron reduction potentials of adenosine and guanosine radicals in aqueous solution. J. Am. Chem. Soc. 1997;119:617–618. [Google Scholar]
- 3.Baik M.H., Silverman J.S., Yang I.V., Ropp P.A., Szalai V.A., Yang W., Thorp W.H.J. Using density functional theory to design DNA base analogues with low oxidation potentials. J. Phys. Chem. B. 2001;105(27):6437–6444. [Google Scholar]
- 4.Burak M.J., Guja K.E., Garcia-Diaz M. Nucleotide binding interactions modulate dNTP selectivity and facilitate 8-oxo-dGTP incorporation by DNA polymerase lambda. Nucleic Acids Res. 2015;43(16):8089–8099. doi: 10.1093/nar/gkv760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hall D.B., Holmlin R.E., Barton J.K. Oxidative DNA damage through long-range electron transfer. Nature. 1996;382:731–735. doi: 10.1038/382731a0. [DOI] [PubMed] [Google Scholar]
- 6.Margolin Y., Cloutier J.F., Shafirovich V., Geacintov N.E., Dedon P.C. Paradoxical hotspots for guanine oxidation by a chemical mediator of inflammation. Nat. Chem. Biol. 2006;2:365–366. doi: 10.1038/nchembio796. [DOI] [PubMed] [Google Scholar]
- 7.Gedik C.M., Collins A. Establishing the background level of base oxidation in human lymphocyte DNA: results of an inter laboratory validation study. FASEB J. 2005;19:82–84. doi: 10.1096/fj.04-1767fje. [DOI] [PubMed] [Google Scholar]
- 8.Yamaguchi R., Hirano T., Asami S., Chung M.H., Sugita A., Kasai H. Increased 8-hydroxyguanine levels in DNA and its repair activity in rat kidney after administration of a renal carcinogen, ferric nitrilotriacetate. Carcinogenesis. 1996;17:2419–2422. doi: 10.1093/carcin/17.11.2419. [DOI] [PubMed] [Google Scholar]
- 9.van Loon B., Markkanen E., Hubscher U. Oxygen as a friend and enemy: how to combat the mutational potential of 8-oxo-guanine. DNA Repair. 2010;9:604–616. doi: 10.1016/j.dnarep.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 10.Jena N.R., Mishra P.C. Mechanisms of formation of 8-oxoguanine due to reactions of one and two OH• radicals and the H2O2 molecule with guanine: a quantum computational study. J. Phys. Chem. B. 2005;109:14205–14218. doi: 10.1021/jp050646j. [DOI] [PubMed] [Google Scholar]
- 11.Jena N.R., Mishra P.C. Formation of 8-nitroguanine and 8-oxoguanine due to reactions of peroxynitrite with guanine. J. Comput. Chem. 2007;28:1321–1335. doi: 10.1002/jcc.20607. [DOI] [PubMed] [Google Scholar]
- 12.Rokhlenko Y., Geacintov N.E., Vladimir Shafirovich V. Lifetimes and reaction pathways of guanine radical cations and neutral guanine radicals in an oligonucleotide in aqueous solutions. J. Am. Chem. Soc. 2012;134:4955–4962. doi: 10.1021/ja212186w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fenton H.J.H. Oxidation of tartaric acid in presence of iron. J. Chem. Soc. 1894;65:899–910. [Google Scholar]
- 14.Yin R., Zhang D., Song Y., Zhu B.Z., Wang H. Potent DNA damage by polyhalogenated quinones and H2O2 via a metal-independent and Intercalation-enhanced oxidation mechanism. Sci. Rep. 2013;3:1269. doi: 10.1038/srep01269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rae T.D., Schmidt P.J., Pufahl R.A., Culotta V.C., O'Halloran T.V. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- 16.Banci L., Bertini I., Ciofi-Baffoni S., Kozyreva T., Zovo K., Palumaa P. Affinity gradients drive copper to cellular destinations. Nature. 2010;465:645–648. doi: 10.1038/nature09018. [DOI] [PubMed] [Google Scholar]
- 17.Carballal S., Bartesaghi S., Radi R. Kinetic and mechanistic considerations to assess the biological fate of peroxynitrite. Biochim. Biophys. Acta. 2014;1840(2):768–780. doi: 10.1016/j.bbagen.2013.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lymar S.V., Khairutdinov R.F., Hurst J.K. Hydroxyl radical formation by O-O bond homolysis in peroxynitrous acid. Inorg. Chem. 2003;42:5259–5266. doi: 10.1021/ic030104l. [DOI] [PubMed] [Google Scholar]
- 19.Aust A.E., Eveleigh J.F. Mechanisms of DNA oxidation. Proc. Soc. Exp. Biol. Med. 1999;222:246–252. doi: 10.1046/j.1525-1373.1999.d01-141.x. [DOI] [PubMed] [Google Scholar]
- 20.Dizdaroglu M., Jaruga P. Mechanisms of free radical-induced damage to DNA. Free Radic. Res. 2012;46(4):382–419. doi: 10.3109/10715762.2011.653969. [DOI] [PubMed] [Google Scholar]
- 21.Denicola A., Souza J.M., Radi R. Diffusion of peroxynitrite across erythrocyte membranes. Proc. Natl. Acad. Sci. U. S. A. 1998;95:3566–3571. doi: 10.1073/pnas.95.7.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Giorgio M., Trinei M., Migliaccio E., Pelicci P.G. Hydrogen peroxide: a metabolic by-product or a common mediator of ageing signals? Nat. Rev. Mol. Cell Biol. 2007;8:722–728. doi: 10.1038/nrm2240. [DOI] [PubMed] [Google Scholar]
- 23.Malinouski M., Zhou Y., Belousov V.V., Hatfield D.L., Gladyshev V.N. Hydrogen peroxide probes directed to different cellular compartments. PLoS One. 2011;6 doi: 10.1371/journal.pone.0014564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cejková J., Stípek S., Crkovská J., Ardan T., Midelfart A. Reactive oxygen species (ROS)-generating oxidases in the normal rabbit cornea and their involvement in the corneal damage evoked by UVB rays. Histol. Histopathol. 2001;16:523–533. doi: 10.14670/HH-16.523. [DOI] [PubMed] [Google Scholar]
- 25.Fukai T., Ushio-Fukai M. Superoxide dismutases: role in redox signaling, vascular function, and diseases. Antioxidants Redox Signal. 2011;15:1583–1606. doi: 10.1089/ars.2011.3999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda J., Nakagawa K., Yamasaki T., Nakamura K., Takeya R., Kuribayashi F., Imajoh-Ohmi S., Igarashi K., Shibata Y., Sueishi K., Sumimoto H. The superoxide-producing NAD(P)H oxidase Nox4 in the nucleus of human vascular endothelial cells. Genes Cells. 2005;10:1139–1151. doi: 10.1111/j.1365-2443.2005.00907.x. [DOI] [PubMed] [Google Scholar]
- 27.Matsushima S., Kuroda J., Ago T., Zhai P., Park J.Y., Xie L.H., Tian B., Sadoshima J. Increased oxidative stress in the nucleus caused by Nox4 mediates oxidation of HDAC4 and cardiac hypertrophy. Circ. Res. 2013;112:651–663. doi: 10.1161/CIRCRESAHA.112.279760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gordillo G., Fang H., Park H., Roy S. Nox-4-dependent nuclear H2O2 drives DNA oxidation resulting in 8-OHdG as urinary biomarker and hemangioendothelioma formation. Antioxidants Redox Signal. 2010;12:933–943. doi: 10.1089/ars.2009.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guida M., Maraldi T., Beretti F., Follo M.Y., Manzoli L., De Pol A. Aberrant expression on Nox4 in the nucleus was reported to be associated with high levels of 8-oxo-dG critical for tumor progression. BioMed Res. Int. 2014;2014:456937. doi: 10.1155/2014/456937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsang C.K., Liu Y., Thomas J., Zhang Y., Zheng X.F. Superoxide dismutase 1 acts as a nuclear transcription factor to regulate oxidative stress resistance. Nat. Commun. 2014;5:3446. doi: 10.1038/ncomms4446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shi Y., Lan F., Matson C., Mulligan P., Whetstine J.R., Cole P.A., Casero R.A., Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941–953. doi: 10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 32.Fagan R., Palfey B. vol. 7. Elsevier Ltd; 2010. Flavin-dependent Enzymes. Comprehensive Natural Products II: Chemistry and Biology; pp. 37–113. [Google Scholar]
- 33.Forneris F., Binda C., Battaglioli E., Mattevi A. LSD1: oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem. Sci. 2008;33:181–189. doi: 10.1016/j.tibs.2008.01.003. [DOI] [PubMed] [Google Scholar]
- 34.Luch A., Frey F.C., Meier R., Fei J., Naegeli H. Low-dose formaldehyde delays DNA damage. recognition and DNA excision repair in human cells. PLoS One. 2014;9 doi: 10.1371/journal.pone.0094149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okkelman I.A., Sukaeva A.Z., Kirukhina E.V., Korneenko T.V., Pestov N.B. Nuclear translocation of lysyl oxidase is promoted by interaction with transcription repressor p66β. Cell Tissue Res. 2014;358(2):481–489. doi: 10.1007/s00441-014-1972-z. [DOI] [PubMed] [Google Scholar]
- 36.Cervelli M., Amendola R., Polticelli F., Mariottini P. Spermine oxidase: ten years after. Amino Acids. 2012;42:441–450. doi: 10.1007/s00726-011-1014-z. [DOI] [PubMed] [Google Scholar]
- 37.Roschzttardtz H., Grillet L., Isaure M.P., Conéjéro G., Ortega R., Curie C., Mari S. Plant cell nucleolus as a hot spot for iron. J. Biol. Chem. 2011;286:27863–27866. doi: 10.1074/jbc.C111.269720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robinson I., Yang Y., Zhang F., Lynch C., Yusuf M., Cloetens P. Nuclear incorporation of iron during the eukaryotic cell cycle. J. Synchrotron Radiat. 2016;23:1490–1497. doi: 10.1107/S1600577516012807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Faa G., Terlizzo M., Gerosa C., Congiu T., Angelucci E. Patterns of iron distribution in liver cells in beta-thalassemia studied by X-ray microanalysis. Haematologica. 2002;87:479–484. PMID:12010660. [PubMed] [Google Scholar]
- 40.Fabrini R., Bocedi A., Pallottini V., Canuti L., De Canio M., Urbani A., Marzano V., Cornetta T., Stano P., Giovanetti A., Stella L., Canini A., Federici G., Ricci G. Nuclear shield: a multi-enzyme task-force for nucleus protection. PLoS One. 2010;5 doi: 10.1371/journal.pone.0014125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hatem E., Berthonaud V., Dardalhon M., Lagniel G., Baudouin-Cornu P., Huang M.E., Labarre J., Chédin S. Glutathione is essential to preserve nuclear function and cell survival under oxidative stress. Free Radic. Biol. Med. 2014;67:103–114. doi: 10.1016/j.freeradbiomed.2013.10.807. [DOI] [PubMed] [Google Scholar]
- 42.Wu Y., Brosh R.M., Jr. DNA helicase and helicase-nuclease enzymes with a conserved iron-sulfur cluster. Nucleic Acids Res. 2012;40:4247–4260. doi: 10.1093/nar/gks039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thompson K.J., Fried M.G., Ye Z., Boyer P., Connor J.R. Regulation, mechanisms and proposed function of ferritin translocation to cell nuclei. J. Cell Sci. 2002;115:2165–2177. doi: 10.1242/jcs.115.10.2165. PMID: 11973357. [DOI] [PubMed] [Google Scholar]
- 44.Kohlgrüber S., Upadhye A., Dyballa-Rukes N., McNamara C.A., Altschmied J. Regulation of transcription factors by reactive oxygen species and nitric oxide in vascular physiology and pathology. Antioxidants Redox Signal. 2017;26:679–699. doi: 10.1089/ars.2016.6946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Go Y.M., Jones D.P. Redox control systems in the nucleus: mechanisms and functions. Antioxidants Redox Signal. 2010;13:489–509. doi: 10.1089/ars.2009.3021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barascu A., Le Chalony C., Pennarun G., Genet D., Zaarour N., Bertrand P. Oxydative stress alters nuclear shape through lamins dysregulation: a route to senescence. Nucleus. 2012;3:411–417. doi: 10.4161/nucl.21674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koga Y., Taniguchi Y., Sasaki S. Synthesis of the oligoribonucleotides incorporating 8-oxo-guanosine and evaluation of their base pairing properties. Nucleosides Nucleotides Nucleic Acids. 2013;32:124–136. doi: 10.1080/15257770.2013.767461. [DOI] [PubMed] [Google Scholar]
- 48.Boiteux S., Coste F., Castaing B. Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: properties and biological roles of the Fpg and OGG1 DNA N-glycosylases. Free Radic. Biol. Med. 2017;107 doi: 10.1016/j.freeradbiomed.2016.11.042. 179-20. [DOI] [PubMed] [Google Scholar]
- 49.Batra V.K., Beard W.A., Hou E.W., Pedersen L.C., Prasad R., Wilson S.H. Mutagenic conformation of 8-oxo-7,8-dihydro-2'-dGTP in the confines of a DNA polymerase active site. Nat. Struct. Mol. Biol. 2010;17:889–890. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markkanen E., Castrec B., Villani G., Hübscher U. A switch between DNA polymerases δ and λ promotes error-free bypass of 8-oxo-G lesions. Proc. Natl. Acad. Sci. U. S. A. 2012;109:20401–20406. doi: 10.1073/pnas.1211532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.David S.S., O'Shea V.L., Kundu S. Base-excision repair of oxidative DNA damage. Nature. 2007;447:941–950. doi: 10.1038/nature05978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Oda H., Taketomi A., Maruyama R., Itoh R., Nishioka K., Yakushiji H., Suzuki T., Sekiguchi M., Nakabeppu Y. Multi-forms of human MTH1 polypeptides produced by alternative translation initiation and single nucleotide polymorphism. Nucleic Acids Res. 1999;27:4335–4343. doi: 10.1093/nar/27.22.4335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Radicella J.P., Dherin C., Desmaze C., Fox M.S., Boiteux S. Cloning and characterization of hOGG1, a human homolog of the OGG1 gene of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 1997;94:8010–8015. doi: 10.1073/pnas.94.15.8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Abbotts R., Madhusudan S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat Rev. 2010;36:425–435. doi: 10.1016/j.ctrv.2009.12.006. [DOI] [PubMed] [Google Scholar]
- 55.Iyama T., Wilson D.M. DNA repair mechanisms in dividing and non-dividing cells. DNA Repair. 2013;12:620–636. doi: 10.1016/j.dnarep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Loeb L.A., Monnat R.J. DNA polymerases and human disease. Nat. Rev. Genet. 2008;9:594–604. doi: 10.1038/nrg2345. [DOI] [PubMed] [Google Scholar]
- 57.Robertson A.B., Klungland A., Rognes T., Leiros I. DNA repair in mammalian cells: base excision repair: the long and short of it. Cell. Mol. Life Sci. 2009;66:981–993. doi: 10.1007/s00018-009-8736-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Horton J.K., Seddon H.J., Zhao M.L., Gassman N.R., Janoshazi A.K., Stefanick D.F., Wilson S.H. Role of the oxidized form of XRCC1 in protection against extreme oxidative stress. Free Radic. Biol. Med. 2017;107:292–300. doi: 10.1016/j.freeradbiomed.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Quesnelle K.M., Bystrom P.V., Toledo-Pereyra L.H. Molecular responses to ischemia and reperfusion in the liver. Arch. Toxicol. 2015;89:651–657. doi: 10.1007/s00204-014-1437-x. [DOI] [PubMed] [Google Scholar]
- 60.Ferrari R.S., Andrade C.F. Oxidative stress and lung ischemia-reperfusion injury. Oxid. Med. Cell Longev. 2015:590987. doi: 10.1155/2015/590987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Curi R., de Siqueira Mendes R., de Campos Crispin L.A., Norata G.D., Sampaio S.C., Newsholme P. A past and present overview of macrophage metabolism and functional outcomes. Clin. Sci. 2017;131:1329–1342. doi: 10.1042/CS20170220. [DOI] [PubMed] [Google Scholar]
- 62.Sena L.A., Chandel N.S. Physiological roles of mitochondrial reactive oxygen species. Mol. Cell. 2012;48:158–167. doi: 10.1016/j.molcel.2012.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boveris A., Chance B. The mitochondrial generation of hydrogen peroxide: general properties and effect of hyperbaric oxygen. Biochem. J. 1973;134:707–716. doi: 10.1042/bj1340707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Brand M.D. The efficiency and plasticity of mitochondrial energy transduction. Biochem. Soc. Trans. 2005;33:897–904. doi: 10.1042/BST0330897. [DOI] [PubMed] [Google Scholar]
- 65.Calabrese V., Cornelius C., Mancuso C., Lentile R., Stella A.M., Butterfield D.A. Redox homeostasis and cellular stress response in aging and neurodegeneration. Methods Mol. Biol. 2010;610:285–308. doi: 10.1007/978-1-60327-029-8_17. [DOI] [PubMed] [Google Scholar]
- 66.Circu M.L., Aw T.Y. Reactive oxygen species, cellular redox systems, and apoptosis. Free Radic. Biol. Med. 2010;48:749–762. doi: 10.1016/j.freeradbiomed.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jang S., Imlay J.A. Micromolar intracellular hydrogen peroxide disrupts metabolism by damaging iron-sulfur enzymes. J. Biol. Chem. 2007;282:929–937. doi: 10.1074/jbc.M607646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lamberts T., Cuppen H.M., Ioppolo S., Linnartz H. Water formation at low temperatures by surface O2 hydrogenation III: Monte Carlo simulation. Phys. Chem. Chem. Phys. 2013;15:8287–8302. doi: 10.1039/c3cp00106g. [DOI] [PubMed] [Google Scholar]
- 69.Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D.E. Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. U. S. A. 1999;96:13300–13305. doi: 10.1073/pnas.96.23.13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Blainey P.C., van Oijen A.M., Banerjee A., Verdine G.L., Xie X.S. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ding Y., Fleming A.M., Burrows C.J. Sequencing the mouse genome for the oxidatively modified base 8-Oxo-7,8-dihydroguanine by OG-seq. J. Am. Chem. Soc. 2017;139(7):2569–2572. doi: 10.1021/jacs.6b12604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shichiri M. The role of lipid peroxides in neurological disorders. J. Clin. Biochem. Nutr. 2014;54:151–160. doi: 10.3164/jcbn.14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Selvaratnam J., Robaire B. Overexpression of catalase in mice reduces age-related oxidative stress and maintains sperm production. Exp. Gerontol. 2016;84:12–20. doi: 10.1016/j.exger.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 74.Park J., Park J.W., Oh H., Maria F.S., Kang J., Tian X. Gene-specific assessment of guanine oxidation as an epigenetic modulator for cardiac specification of mouse embryonic stem cells. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Will O., Mahler H.C., Arrigo A.P., Epe B. Influence of glutathione levels and heat-shock on the steady-state levels of oxidative DNA base modifications in mammalian cells. Carcinogenesis. 1999;20:333–337. doi: 10.1093/carcin/20.2.333. [DOI] [PubMed] [Google Scholar]
- 76.Reliene R., Fischer E., Schiestl R.H. Effect of N-acetyl cysteine on oxidative DNA damage and the frequency of DNA deletions in atm-deficient mice. Cancer Res. 2004;64:5148–5153. doi: 10.1158/0008-5472.CAN-04-0442. [DOI] [PubMed] [Google Scholar]
- 77.Jiang L., Zhong Y., Akatsuka S., Liu Y.T., Dutta K.K., Lee W.H., Onuki J., Masumura K., Nohmi T., Toyokuni S. Deletion and single nucleotide substitution at G:C in the kidney of gpt delta transgenic mice after ferric nitrilotriacetate treatment. Cancer Sci. 2006;97:1159–1167. doi: 10.1111/j.1349-7006.2006.00301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Beddowes E.J., Faux S.P., Chipman J.K. Chloroform, carbon tetrachloride and glutathione depletion induce secondary genotoxicity in liver cells via oxidative stress. Toxicology. 2003;187:101–115. doi: 10.1016/s0300-483x(03)00058-1. [DOI] [PubMed] [Google Scholar]
- 79.Szmidt M., Sawosz E., Urbańska K., Jaworski S., Kutwin M., Hotowy A., Wierzbicki M., Grodzik M., Lipińska L., Chwalibog A. Toxicity of different forms of graphene in a chicken embryo model. Environ. Sci. Pollut. Res. Int. 2016;23:19940–19948. doi: 10.1007/s11356-016-7178-z. [DOI] [PubMed] [Google Scholar]
- 80.Sonmez E., Cacciatore I., Bakan F., Turkez H., Mohtar Y.I., Togar B., Stefano A.D. Toxicity assessment of hydroxyapatite nanoparticles in rat liver cell model in vitro. Hum. Exp. Toxicol. 2016;35:1073–1083. doi: 10.1177/0960327115619770. [DOI] [PubMed] [Google Scholar]
- 81.Wang R., Hao W., Pan L., Boldogh I., Ba X. The roles of base excision repair enzyme OGG1 in gene expression. Cell. Mol. Life Sci. 2018;75:3741–3750. doi: 10.1007/s00018-018-2887-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jaiswal M., LaRusso N.F., Nishioka N., Nakabeppu Y., Gores G.J. Human OGG1, a protein involved in the repair of 8-oxoguanine, is inhibited by nitric oxide. Cancer Res. 2001;61:6389–6393. PMID: 11522631. [PubMed] [Google Scholar]
- 83.Bravard A., Vacher M., Gouget B., Coutant A., de Boisferon F.H., Marsin S., Chevillard S., Radicella J.P. Redox regulation of human OGG1 activity in response to cellular oxidative stress. Mol. Cell. Biol. 2006;26:7430–7436. doi: 10.1128/MCB.00624-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Bravard A., Campalans A., Vacher M., Gouget B., Levalois C., Chevillard S., Radicella J.P. Inactivation by oxidation and recruitment into stress granules of hOGG1 but not APE1 in human cells exposed to sub-lethal concentrations of cadmium. Mutat. Res. 2010;685:61–69. doi: 10.1016/j.mrfmmm.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 85.Saitoh T., Shinmura K., Yamaguchi S., Tani M., Seki S., Murakami H., Nojima Y., Yokota J. Enhancement of OGG1 protein AP lyase activity by increase of APEX protein. Mutat. Res. 2001;486:31–40. doi: 10.1016/s0921-8777(01)00078-7. [DOI] [PubMed] [Google Scholar]
- 86.Yang J.L., Tadokoro T., Keijzers G., Mattson M.P., Bohr V.A. Neurons efficiently repair glutamate-induced oxidative DNA damage by a process involving CREB-mediated up-regulation of apurinic endonuclease 1. J. Biol. Chem. 2010;285:28191–28199. doi: 10.1074/jbc.M109.082883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bhakat K.K., Mokkapati S.K., Boldogh I., Hazra T.K., Mitra S. Acetylation of human8-oxoguanine-DNA glycosylase by p300 and its role in 8-oxoguanine repair in vivo. Mol. Cell. Biol. 2006;26:1654–1665. doi: 10.1128/MCB.26.5.1654-1665.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jain S., Wei J., Mitrani L.R., Bishopric N.H. Auto-acetylation stabilizes p300 in cardiac myocytes during acute oxidative stress, promoting STAT3 accumulation and cell survival. Breast Canc. Res. Treat. 2012;135:103–114. doi: 10.1007/s10549-012-2069-6. [DOI] [PubMed] [Google Scholar]
- 89.Tini M., Benecke A., Um S.J., Torchia J., Evans R.M., Chambon P. Association of CBP/p300 acetylase and thymine DNA glycosylase links DNA repair and transcription. Mol. Cell. 2002;9:265–277. doi: 10.1016/s1097-2765(02)00453-7. [DOI] [PubMed] [Google Scholar]
- 90.Sarga L., Hart N., Koch L.G., Britton S.L., Hajas G., Boldogh I., Ba X., Radak Z. Aerobic endurance capacity affects spatial memory and SIRT1 is a potent modulator of 8-oxoguanine repair. Neuroscience. 2013;252:326–336. doi: 10.1016/j.neuroscience.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yamamori T., DeRicco J., Naqvi A., Hoffman T.A., Mattagajasingh I., Kasuno K., Jung S.B., Kim C.S., Irani K. SIRT1 deacetylates APE1 and regulates cellular base excision repair. Nucleic Acids Res. 2010;38:832–845. doi: 10.1093/nar/gkp1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Cheng Y., Ren X., Gowda A.S., Shan Y., Zhang L., Yuan Y.S., Patel R., Wu H., Huber-Keener K., Yang J.W., Liu D., Spratt T.E., Yang J.M. Interaction of Sirt3 with OGG1 contributes to repair of mitochondrial DNA and protects from apoptotic cell death under oxidative stress. Cell Death Dis. 2013;4 doi: 10.1038/cddis.2013.254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Dantzer F., Luna L., Bjørås M., Seeberg E. Human OGG1 undergoes serine phosphorylation and associates with the nuclear matrix and mitotic chromatin in vivo. Nucleic Acids Res. 2002;30:2349–2357. doi: 10.1093/nar/30.11.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cosentino-Gomes D., Rocco-Machado N., Meyer-Fernandes J.R. Cell signaling through protein kinase C oxidation and activation. Int. J. Mol. Sci. 2012;13:10697–10721. doi: 10.3390/ijms130910697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rojas F., Gonzalez D., Cortes N., Ampuero E., Hernández D.E., Fritz E., Abarzua S., Martinez A., Elorza A.A., Alvarez A., Court F., van Zundert B. Reactive oxygen species trigger motoneuron death in non-cell-autonomous models of ALS through activation of c-Abl signaling. Front. Cell. Neurosci. 2015;9:203. doi: 10.3389/fncel.2015.00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kim H.S., Kim B.H., Jung J.E., Lee C.S., Lee H.G., Lee J.W., Lee K.H., You H.J., Chung M.H., Ye S.K. Potential role of 8-oxoguanine DNA glycosylase 1 as a STAT1 coactivator in endotoxin-induced inflammatory response. Free Radic. Biol. Med. 2016;93:12–22. doi: 10.1016/j.freeradbiomed.2015.10.415. [DOI] [PubMed] [Google Scholar]
- 97.de Prati A.C., Ciampa A.R., Cavalieri E., Zaffini R., Darra E., Menegazzi M., Suzuki H., Mariotto S. STAT1 as a new molecular target of anti-inflammatory treatment. Curr. Med. Chem. 2005;12:1819–1828. doi: 10.2174/0929867054546645. [DOI] [PubMed] [Google Scholar]
- 98.Aguilera-Aguirre L., Hao W., Pan L., Li X., Saavedra-Molina A., Bacsi A., Radak Z., Sur S., Brasier A.R., Ba X., Boldogh I. Pollen-induced oxidative DNA damage response regulates miRNAs controlling allergic inflammation. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;313:1058–1068. doi: 10.1152/ajplung.00141.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Morreall J., Limpose K., Sheppard C., Kow Y.W., Werner E., Doetsch P.W. Inactivation of a common OGG1 variant by TNF-alpha in mammalian cells. DNA Repair (Amst) 2015;26:15–22. doi: 10.1016/j.dnarep.2014.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Chen Z., Wang D., Liu X., Pei W., Li J., Cao Y., Zhang J., An Y., Nie J., Tong J. Oxidative DNA damage is involved in cigarette smoke-induced lung injury in rats. Environ. Health Prev. Med. 2015;20:318–324. doi: 10.1007/s12199-015-0469-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sampath H., Vartanian V., Rollins M.R., Sakumi K., Nakabeppu Y., Lloyd R.S. 8-Oxoguanine DNA glycosylase (OGG1) deficiency increases susceptibility to obesity and metabolic dysfunction. PLoS One. 2012;7 doi: 10.1371/journal.pone.0051697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Radak Z., Bori Z., Koltai E., Fatouros I.G., Jamurtas A.Z., Douroudos, Terzis G., Nikolaidis M.G., Chatzinikolaou A., Sovatzidis A., Kumagai S., Naito H., Boldogh I. Age-dependent changes in 8-oxoguanine-DNA glycosylase activity are modulated by adaptive responses to physical exercise in human skeletal muscle. Free Radic. Biol. Med. 2011;51:417–423. doi: 10.1016/j.freeradbiomed.2011.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Siggens L., Figg N., Bennett M., Foo R. Nutrient deprivation regulates DNA damage repair in cardiomyocytes via loss of the base-excision repair enzyme OGG1. FASEB J. 2012;26:2117–2124. doi: 10.1096/fj.11-197525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fantini D., Moritz E., Auvré F., Amouroux R., Campalans A., Epe B., Bravard A., Radicella J.P. Rapid inactivation and proteasome-mediated degradation of OGG1 contribute to the synergistic effect of hyperthermia on genotoxic treatments. DNA Repair. 2013;12:227–237. doi: 10.1016/j.dnarep.2012.12.006. [DOI] [PubMed] [Google Scholar]
- 105.Dalen M.L., Alme T.N., Bjørås M., Munkeby B.H., Rootwelt T., Saugstad O.D. Reduced expression of DNA glycosylases in post-hypoxic newborn pigs undergoing therapeutic hypothermia. Brain Res. 2010;1363:198–205. doi: 10.1016/j.brainres.2010.09.080. [DOI] [PubMed] [Google Scholar]
- 106.Ohno M., Miura T., Furuichi M., Tominaga Y., Tsuchimoto D., Sakumi K., Nakabeppu Y. A genome-wide distribution of 8-oxoguanine correlates with the preferred regions for recombination and single nucleotide polymorphism in the human genome. Genome Res. 2006;16:567–575. doi: 10.1101/gr.4769606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yoshihara M., Jiang L., Akatsuka S., Suyama M., Toyokuni S. Genome-wide profiling of 8-oxoguanine reveals its association with spatial positioning in nucleus. DNA Res. 2014;6:603–612. doi: 10.1093/dnares/dsu023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Amente S., Di Palo G., Scala G., Castrignanò T., Gorini F., Cocozza S., Moresano A., Pucci P., Ma B., Stepanov I., Lania L., Pelicci P.G., Dellino G.I., Majello B. Genome-wide mapping of 8-oxo-7,8-dihydro-2'-deoxyguanosine reveals accumulation of oxidatively-generated damage at DNA replication origins within transcribed long genes of mammalian cells. Nucleic Acids Res. 2019;47:221–236. doi: 10.1093/nar/gky1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sagelsdorff P., Lutz W.K. Sensitivity of DNA and nucleotides to oxidation by permanganate and hydrogen peroxide. Arch. Toxicol Suppl. 1987;11:84–88. doi: 10.1007/978-3-642-72558-6_11. [DOI] [PubMed] [Google Scholar]
- 110.Emerit I., Cerutti P.A. Tumour promoter phorbol-12-myristate-13-acetate induces chromosomal damage via indirect action. Nature. 1981;293:144–146. doi: 10.1038/293144a0. [DOI] [PubMed] [Google Scholar]
- 111.Schiavone D., Jozwiakowski S.K., Romanello M., Guilbaud G., Guilliam T.A., Bailey L.J., Sale J.E., Doherty A.J. PrimPol is required for replicative tolerance of G quadruplexes in vertebrate cells. Mol. Cell. 2016;61:161–169. doi: 10.1016/j.molcel.2015.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Amouroux R., Campalans A., Epe B., Radicella J.P. Oxidative stress triggers the preferential assembly of base excision repair complexes on open chromatin regions. Nucleic Acids Res. 2010;38:2878–2890. doi: 10.1093/nar/gkp1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Menoni H., Wienholz F., Theil A.F., Janssens R.C., Lans H., Campalans A., Radicella J.P., Marteijn J.A., Vermeulen W. The transcription-coupled DNA repair-initiating protein CSB promotes XRCC1 recruitment to oxidative DNA damage. Nucleic Acids Res. 2018;46:7747–7756. doi: 10.1093/nar/gky579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rhee D.B., Ghosh A., Lu J., Bohr V.A., Liu Y. Factors that influence telomeric oxidative base damage and repair by DNA glycosylase OGG1. DNA Repair (Amst) 2011;10(1):34–44. doi: 10.1016/j.dnarep.2010.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.O'Callaghan N., Baack N., Sharif R., Fenech M. A qPCR-based assay to quantify oxidized guanine and other FPG-sensitive base lesions within telomeric DNA. Biotechniques. 2011;51(6):403–411. doi: 10.2144/000113788. [DOI] [PubMed] [Google Scholar]
- 116.von Zglinicki T. Oxidative stress shortens telomeres. Trends Biochem. Sci. 2002;27(7):339–344. doi: 10.1016/s0968-0004(02)02110-2. [DOI] [PubMed] [Google Scholar]
- 117.Opresko P.L., Fan J., Danzy S., Wilson D.M., 3rd, Bohr V.A. Oxidative damage in telomeric DNA disrupts recognition by TRF1 and TRF2. Nucleic Acids Res. 2005;33(4):1230–1239. doi: 10.1093/nar/gki273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang Z., Rhee D.B., Lu J., Bohr C.T., Zhou F., Vallabhaneni H., de Souza-Pinto N.C., Liu Y. Characterization of oxidative guanine damage and repair in mammalian telomeres. PLoS Genet. 2010;6(5) doi: 10.1371/journal.pgen.1000951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Fouquerel E., Barnes R.P., Uttam S., Watkins S.C., Bruchez M.P., Opresko P.L. Targeted and persistent 8-oxoguanine base damage at telomeres promotes telomere loss and crisis. Mol. Cell. 2019;75(1):117–130. doi: 10.1016/j.molcel.2019.04.024. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Fouquerel E., Lormand J., Bose A., Lee H.T., Kim G.S., Li J., Sobol R.W., Freudenthal B.D., Myong S., Opresko P.L. Oxidative guanine base damage regulates human telomerase activity. Nat. Struct. Mol. Biol. 2016;23(12):1092–1100. doi: 10.1038/nsmb.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Allgayer J., Kitsera N., von der Lippen C., Epe B., Khobta A. Modulation of base excision repair of 8-oxoguanine by the nucleotide sequence. Nucleic Acids Res. 2013;41:8559–8571. doi: 10.1093/nar/gkt620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.David-Cordonnier M.H., Boiteux S., O'Neill P. Efficiency of excision of 8-Oxo-guanine within DNA clustered damage by XRS5 nuclear extracts and purified human OGG1 protein. Biochemistry. 2001;40:11811–11818. doi: 10.1021/bi0112356. [DOI] [PubMed] [Google Scholar]
- 123.Enright H., Miller W.J., Hays R., Floyd R.A., Hebbel R.P. Preferential targeting of oxidative base damage to internucleosomal DNA. Carcinogenesis. 1996;17:1175–1177. doi: 10.1093/carcin/17.5.1175. [DOI] [PubMed] [Google Scholar]
- 124.Menoni H., Shukla M.S., Gerson V., Dimitrov S., Angelov D. Base excision repair of 8-oxoG in dinucleosomes. Nucleic Acids Res. 2012;40:692–700. doi: 10.1093/nar/gkr761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuraoka I., Endou M., Yamaguchi Y., Wada T., Handa H., Tanaka K. Effects of endogenous DNA base lesions on transcription elongation by mammalian RNA polymerase II. Implications for transcription-coupled DNA repair and transcriptional mutagenesis. J. Biol. Chem. 2003;278:7294–7299. doi: 10.1074/jbc.M208102200. [DOI] [PubMed] [Google Scholar]
- 126.Charlet-Berguerand N., Feuerhahn S., Kong S.E., Ziserman H., Conaway J.W., Conaway R., Egly J.M. RNA polymerase II bypass of oxidative DNA damage is regulated by transcription elongation factors. EMBO J. 2006;25:5481–5491. doi: 10.1038/sj.emboj.7601403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pastoriza-Gallego M., Sarasin A. Transcription-coupled repair of 8-oxoguanine in human cells and its deficiency in some DNA repair diseases. Biochimie. 2003;85:1073–1082. doi: 10.1016/j.biochi.2003.11.005. [DOI] [PubMed] [Google Scholar]
- 128.Kuraoka I., Suzuki K., Ito S., Hayashida M., Kwei J.S., Ikegami T., Handa H., Nakabeppu Y., Tanaka K. RNA polymerase II bypasses 8-oxoguanine in the presence of transcription elongation factor TFIIS. DNA Repair (Amst) 2007;6:841–851. doi: 10.1016/j.dnarep.2007.01.014. [DOI] [PubMed] [Google Scholar]
- 129.Kathe S.D., Shen G.P., Wallace S.S. Single-stranded breaks in DNA but not oxidative DNA base damages block transcriptional elongation by RNA polymerase II in HeLa cell nuclear extracts. J. Biol. Chem. 2004;279:18511–18520. doi: 10.1074/jbc.M313598200. [DOI] [PubMed] [Google Scholar]
- 130.Kitsera N., Stathis D., Lühnsdorf B., Müller H., Carell T., Epe B., Khobta A. 8-Oxo-7,8-dihydroguanine in DNA does not constitute a barrier to transcription, but is converted into transcription-blocking damage by OGG1. Nucleic Acids Res. 2011;39:5926–5934. doi: 10.1093/nar/gkr163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Larsen E., Kwon K., Coin F., Egly J.M., Klungland A. Transcription activities at 8-oxoG lesions in DNA. DNA Repair. 2004;3:1457–1468. doi: 10.1016/j.dnarep.2004.06.008. [DOI] [PubMed] [Google Scholar]
- 132.Tornaletti S., Maeda L.S., Kolodner R.D., Hanawalt P.C. Effect of 8-oxoguanine on transcription elongation by T7 RNA polymerase and mammalian RNA polymerase II. DNA Repair (Amst) 2004;3:483–494. doi: 10.1016/j.dnarep.2004.01.003. [DOI] [PubMed] [Google Scholar]
- 133.Pastoriza-Gallego M., Armier J., Sarasin A. Transcription through 8-oxoguanine in DNA repair-proficient and Csb(-)/Ogg1(-) DNA repair-deficient mouse embryonic fibroblasts is dependent upon promoter strength and sequence context. Mutagenesis. 2007;22:343–351. doi: 10.1093/mutage/gem024. [DOI] [PubMed] [Google Scholar]
- 134.Fleming A.M., Burrows C.J. 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair. 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Hailer-Morrison M.K., Kotler J.M., Martin B.D., Sugden K.D. Oxidized guanine lesions as modulators of gene transcription. Altered p50 binding affinity and repair shielding by 7,8-dihydro-8-oxo-2'-deoxyguanosine lesions in the NF-kappaB promoter element. Biochemistry. 2003;42:9761–9770. doi: 10.1021/bi034546k. [DOI] [PubMed] [Google Scholar]
- 137.Fedeles B.I. G-quadruplex-forming promoter sequences enable transcriptional activation in response to oxidative stress. Proc. Natl. Acad. Sci. U. S. A. 2017;114:2788–2790. doi: 10.1073/pnas.1701244114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Hänsel-Hertsch R., Beraldi D., Lensing S.V., Marsico G., Zyner K., Parry A., Di Antonio M., Pike J., Kimura H., Narita M., Tannahill D., Balasubramanian S. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016;48:1267–1272. doi: 10.1038/ng.3662. [DOI] [PubMed] [Google Scholar]
- 139.Armas P., David A., Calcaterra N.B. Transcriptional control by G-quadruplexes: in vivo roles and perspectives for specific intervention. Transcription. 2017;8:21–25. doi: 10.1080/21541264.2016.1243505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Burra S., Marasco D., Malfatti M.C., Antoniali G., Virgilio A., Esposito V., Demple B., Galeone A., Tell G. Human AP-endonuclease (Ape1) activity on telomeric G4 structures is modulated by acetylatable lysine residues in the N-terminal sequence. DNA Repair (Amst) 2019;73:129–143. doi: 10.1016/j.dnarep.2018.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Fleming A.M., Ding Y., Burrows C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Natl. Acad. Sci. U. S. A. 2017;114(10):2604–2609. doi: 10.1073/pnas.1619809114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Fleming A.M., Zhu J., Howpay Manage S.A., Burrows C.J. Human NEIL3 gene expression regulated by epigenetic-like oxidative DNA modification. J. Am. Chem. Soc. 2019;141(28):11036–11049. doi: 10.1021/jacs.9b01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Fleming A.M., Burrows C.J. 8-Oxo-7,8-dihydroguanine, friend and foe: epigenetic-like regulator versus initiator of mutagenesis. DNA Repair (Amst) 2017;56:75–83. doi: 10.1016/j.dnarep.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Pastukh V., Ruchko M., Gorodnya O., Wilson G.L., Gillespie M.N. Sequence-specific oxidative base modifications in hypoxia-inducible genes. Free Radic. Biol. Med. 2007;43:1616–1626. doi: 10.1016/j.freeradbiomed.2007.08.027. [DOI] [PubMed] [Google Scholar]
- 145.Redstone S.C.J., Fleming A.M., Burrows C.J. Oxidative modification of the potential G-quadruplex sequence in the PCNA gene promoter can turn on transcription. Chem. Res. Toxicol. 2019;32(3):437–446. doi: 10.1021/acs.chemrestox.8b00332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Fleming A.M., Zhu J., Howpay Manage S.A., Burrows C.J. Human NEIL3 gene expression regulated by epigenetic-like oxidative DNA modification. J. Am. Chem. Soc. 2019;141(28):11036–11049. doi: 10.1021/jacs.9b01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Zhu J., Fleming A.M., Burrows C.J. The RAD17 promoter sequence contains a potential tail-dependent G-quadruplex that downregulates gene expression upon oxidative modification. ACS Chem. Biol. 2018;13(9):2577–2584. doi: 10.1021/acschembio.8b00522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cogoi S., Xodo L.E. G4 DNA in ras genes and its potential in cancer therapy. Biochim. Biophys. Acta. 2016;1859(4):663–674. doi: 10.1016/j.bbagrm.2016.02.002. [DOI] [PubMed] [Google Scholar]
- 149.Cogoi S., Ferino A., Miglietta G., Pedersen E.B., Xodo L.E. The regulatory G4 motif of the Kirsten ras (KRAS) gene is sensitive to guanine oxidation: implications on transcription. Nucleic Acids Res. 2018;46(2):661–676. doi: 10.1093/nar/gkx1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Membrino A., Cogoi S., Pedersen E.B., Xodo L.E. G4-DNA formation in the HRAS promoter and rational design of decoy oligonucleotides for cancer therapy. PLoS One. 2011;6(9) doi: 10.1371/journal.pone.0024421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Perillo B., Ombra M.N., Bertoni A., Cuozzo C., Sacchetti S., Sasso A., Chiari-otti L., Malorni A., Abbondanza C., Avvedimento E.V. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 152.Yang S., Zhang J., Zhang Y., Wan X., Zhang C., Huang X., Huang W., Pu H., Pei C., Wu H., Huang Y., Huang S., Li Y. KDM1A triggers androgen-induced miRNA transcription via H3K4me2 demethylation and DNA oxidation. The Prostate. 2015;75:936–946. doi: 10.1002/pros.22977. [DOI] [PubMed] [Google Scholar]
- 153.Amente, Bertoni A., Morano A., Lania L., Avvedimento E.V., Majello B. LSD1-mediated demethylation of histone H3 lysine 4 triggers Myc-induced transcription. Oncogene. 2010;29:3691–3702. doi: 10.1038/onc.2010.120. [DOI] [PubMed] [Google Scholar]
- 154.Pan L., Zhu B., Hao W., Zeng X., Vlahopoulos S.A., Hazra T.K., Hegde M.L., Radak Z., Bacsi A., Brasier A.R., Ba X., Boldogh I. Oxidized guanine base lesions function in 8-oxoguanine DNA glycosylase-1-mediated epigenetic regulation of nuclear factor κB-driven gene expression. J. Biol. Chem. 2016;291:25553–25566. doi: 10.1074/jbc.M116.751453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Pan L., Hao W., Zheng X., Zeng X., Ahmed Abbasi A., Boldogh I., Ba X. OGG1-DNA interactions facilitate NF-κB binding to DNA targets. Sci. Rep. 2017;7:43297. doi: 10.1038/srep43297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Ba X., Boldogh I. 8-Oxoguanine DNA glycosylase 1: beyond repair of the oxidatively modified base lesions. Redox Biol. 2018;14:669–678. doi: 10.1016/j.redox.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ba X., Bacsi A., Luo J., Aguilera-Aguirre L., Zeng X., Radak Z., Brasier A.R., Boldogh I. 8-oxoguanine DNA glycosylase-1 augments proinflammatory gene expression by facilitating the recruitment of site-specific transcription factors. J. Immunol. 2014;192:2384–2394. doi: 10.4049/jimmunol.1302472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Pastukh V., Roberts J.T., Clark D.W., Bardwell G.C., Patel M., Al-Mehdi A.B., Borchert G.M., Gillespie M.N. An oxidative DNA "damage" and repair mechanism localized in the VEGF promoter is important for hypoxia-induced VEGF mRNA expression. Am. J. Physiol. Lung Cell Mol. Physiol. 2015;309:L1367–L1375. doi: 10.1152/ajplung.00236.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Holliday R. DNA methylation and epigenetic defects in carcinogenesis. Mutat. Res. 1987;181:215–217. doi: 10.1016/0027-5107(87)90098-4. [DOI] [PubMed] [Google Scholar]
- 160.Weitzman S.A., Turk P.W., Milkowski D.H., Kozlowski K. Free radical adducts induce alterations in DNA cytosine methylation. Proc. Natl. Acad. Sci. U. S. A. 1994;91:1261–1264. doi: 10.1073/pnas.91.4.1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Maltseva D.V., Baykov A.A., Jeltsch A., Gromova E.S. Impact of 7,8-dihydro-8-oxoguanine on methylation of the CpG site by Dnmt3a. Biochemistry. 2009;48(6):1361–1368. doi: 10.1021/bi801947f. [DOI] [PubMed] [Google Scholar]
- 162.Traoré A., Ruiz S., Baudrimont I., Sanni A., Dano S.D., Guarigues P., Narbonne J.F., Creppy E.E. Combined effects of okadaic acid and cadmium on lipid peroxidation and DNA bases modifications (m5dC and 8-(OH)-dG) in Caco-2 cells. Arch. Toxicol. 2000;74:79–84. doi: 10.1007/s002040050656. [DOI] [PubMed] [Google Scholar]
- 163.Franco R., Schoneveld O., Georgakilas A., Panayiotidis M. Oxidative stress, DNA methylation and carcinogenesis. Cancer Lett. 2008;266:6–11. doi: 10.1016/j.canlet.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 164.Guz J., Foksinski M., Siomek A., Gackowski D., Rozalski R., Dziaman T., Szpila A., Olinski R. The relationship between 8-oxo-7,8-dihydro-2'-deoxyguanosine level and extent of cytosine methylation in leukocytes DNA of healthy subjects and in patients with colon adenomas and carcinomas. Mutat. Res. 2008;640:170–173. doi: 10.1016/j.mrfmmm.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 165.Bhusari S.S., Dobosy J.R., Fu V., Almassi N., Oberley T., Jarrard D.F. Superoxide dismutase 1 knockdown induces oxidative stress and DNA methylation loss in the prostate. Epigenetics. 2010;5:402–409. doi: 10.4161/epi.5.5.11853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Valinluck V., Tsai H.H., Rogstad D.K., Burdzy A., Bird A., Sowers L.C. Oxidative damage to methyl-CpG sequences inhibits the binding of the methyl-CpG binding domain (MBD) of methyl-CpG binding protein 2 (MeCP2) Nucleic Acids Res. 2004;32:4100–4108. doi: 10.1093/nar/gkh739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nunomura A., Lee H.G., Zhu X., Perry G. Consequences of RNA oxidation on protein synthesis rate and fidelity: implications for the pathophysiology of neuropsychiatric disorders. Biochem. Soc. Trans. 2017;45:1053–1066. doi: 10.1042/BST20160433. [DOI] [PubMed] [Google Scholar]
- 168.Hofer T., Badouard C., Bajak E., Ravanat J.L., Mattsson A., Cotgreave I.A. Hydrogen peroxide causes greater oxidation in cellular RNA than in DNA. Biol. Chem. 2005;386:333–337. doi: 10.1515/BC.2005.040. [DOI] [PubMed] [Google Scholar]
- 169.Shan X., Chang Y., Lin C.L. Messenger RNA oxidation is an early event preceding cell death and causes reduced protein expression. FASEB J. 2007;21:2753–2764. doi: 10.1096/fj.07-8200com. [DOI] [PubMed] [Google Scholar]
- 170.Tanaka M., Chock P.B., Stadtman E.R. Oxidized messenger RNA induces translation errors. Proc. Natl. Acad. Sci. U. S. A. 2007;104:66–71. doi: 10.1073/pnas.0609737104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Honda K., Smith M.A., Zhu X., Baus D., Merrick W.C., Tartakoff A.M., Hattier T., Harris P.L., Siedlak S.L., Fujioka H., Liu Q., Moreira P.I., Miller F.P., Nunomura A., Shimohama S., Perry G. Ribosomal RNA in Alzheimer disease is oxidized by bound redox-active iron. J. Biol. Chem. 2005;280:20978–20986. doi: 10.1074/jbc.M500526200. [DOI] [PubMed] [Google Scholar]
- 172.Mishima E., Inoue C., Saigusa D., Inoue R., Ito K., Suzuki Y., Jinno D., Tsukui Y., Akamatsu Y., Araki M., Araki K., Shimizu R., Shinke H., Suzuki T., Takeuchi Y., Shima H., Akiyama Y., Toyohara T., Suzuki C., Saiki Y., Tominaga T., Miyagi S., Kawagisihi N., Soga T., Ohkubo T., Yamamura K., Imai Y., Masuda S., Sabbisetti V., Ichimura T., Mount D.B., Bonventre J.V., Ito S., Tomioka Y., Itoh K., Abe T. Conformational change in transfer RNA is an early indicator of acute cellular damage. J. Am. Soc. Nephrol. 2014;25:2316–2326. doi: 10.1681/ASN.2013091001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Mjelle R., Hegre S.A., Aas P.A., Slupphaug G., Drabløs F., Saetrom P., Krokan H.E. Cell cycle regulation of human DNA repair and chromatin remodeling genes. DNA Repair. 2015;30:53–67. doi: 10.1016/j.dnarep.2015.03.007. [DOI] [PubMed] [Google Scholar]
- 174.Conlon K.A., Zharkov D.O., Berrios M. Cell cycle regulation of the murine 8-oxoguanine DNA glycosylase (mOGG1): mOGG1 associates with microtubules during interphase and mitosis. DNA Repair. 2004;3:1601–1615. doi: 10.1016/j.dnarep.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 175.Neumann B., Walter T., Hériché J.K., Bulkescher J., Erfle H., Conrad C., Rogers P., Poser I., Held M., Liebel U., Cetin C., Sieckmann F., Pau G., Kabbe R., Wünsche A., Satagopam V., Schmitz M.H., Chapuis C., Gerlich D.W., Schneider R., Eils R., Huber W., Peters J.M., Hyman A.A., Durbin R., Pepperkok R., Ellenberg J. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature. 2010;464:721–727. doi: 10.1038/nature08869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Boldogh I., Hajas G., Aguilera-Aguirre L., Hegde M.L., Radak Z., Bacsi A., Sur S., Hazra T.K., Mitra S. Activation of ras signaling pathway by 8-oxoguanine DNA glycosylase bound to its excision product, 8-oxoguanine. J. Biol. Chem. 2012;287:20769–20773. doi: 10.1074/jbc.C112.364620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Luo J., Hosoki K., Bacsi A., Radak Z., Hegde M.L., Sur S., Hazra T.K., Brasier A.R., Ba X., Boldogh I. 8-Oxoguanine DNA glycosylase-1-mediated DNA repair is associated with Rho GTPase activation and α-smooth muscle actin polymerization. Free Radic. Biol. Med. 2014;73:430–438. doi: 10.1016/j.freeradbiomed.2014.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Hayakawa H., Taketomi A., Sakumi K., Kuwano M., Sekiguchi M. Generation and elimination of 8-oxo-7,8-dihydro-2'-deoxyguanosine 5'-triphosphate, a mutagenic substrate for DNA synthesis, in human cells. Biochemistry. 1995;34:89–95. doi: 10.1021/bi00001a011. [DOI] [PubMed] [Google Scholar]
- 179.Bolin C., Cardozo-Pelaez F. Characterization of oxidized guanosine 5'-triphosphate as a viable inhibitor of soluble guanylyl cyclase. Free Radic. Biol. Med. 2009;46:828–835. doi: 10.1016/j.freeradbiomed.2008.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Scheffler K., Rachek L., You P., Rowe A.D., Wang W., Kuśnierczyk A., Kittelsen L., Bjørås M., Eide L. 8-oxoguanine DNA glycosylase (Ogg1) controls hepatic gluconeogenesis. DNA Repair. 2018;61:56–62. doi: 10.1016/j.dnarep.2017.11.008. [DOI] [PubMed] [Google Scholar]
- 181.Stuart J.A., Bourque B.M., de Souza-Pinto N.C., Bohr V.A. No evidence of mitochondrial respiratory dysfunction in OGG1-null mice deficient in removal of 8-oxodeoxyguanine from mitochondrial DNA. Free Radic. Biol. Med. 2005;38:737–745. doi: 10.1016/j.freeradbiomed.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 182.Minowa O., Arai T., Hirano M., Monden Y., Nakai S., Fukuda M., Itoh M., Takano H., Hippou Y., Aburatani H., Masumura K., Nohmi T., Nishimura S., Noda T. Mmh/Ogg1 gene inactivation results in accumulation of 8-hydroxyguanine in mice. Proc. Natl. Acad. Sci. U. S. A. 2000;97:4156–4161. doi: 10.1073/pnas.050404497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Arai T., Kelly V.P., Minowa O., Noda T., Nishimura S. High accumulation of oxidative DNA damage, 8-hydroxyguanine, in Mmh/Ogg1 deficient mice by chronic oxidative stress. Carcinogenesis. 2002;23:2005–2010. doi: 10.1093/carcin/23.12.2005. [DOI] [PubMed] [Google Scholar]
- 184.Larsen E., Reite K., Nesse G., Gran C., Seeberg E., Klungland A. Repair and mutagenesis at oxidized DNA lesions in the developing brain of wild-type and Ogg1-/- mice. Oncogene. 2006;25:2425–2432. doi: 10.1038/sj.onc.1209284. [DOI] [PubMed] [Google Scholar]
- 185.Touati E., Michel V., Thiberge J.M., Ave P., Huerre M., Bourgade F., Klungland A., Labigne A. Deficiency in OGG1 protects against inflammation and mutagenic effects associated with H. pylori infection in mouse. Helicobacter. 2006;11:494–505. doi: 10.1111/j.1523-5378.2006.00442.x. [DOI] [PubMed] [Google Scholar]
- 186.Mabley J.G., Pacher P., Deb A., Wallace R., Elder R.H., Szabo C. Potential role for 8-oxoguanine DNA glycosylase in regulating inflammation. FASEB J. 2005;19:290–292. doi: 10.1096/fj.04-2278fje. [DOI] [PubMed] [Google Scholar]
- 187.Xie Y., Yang H., Cunanan C., Okamoto K., Shibata D., Pan J., Barnes D.E., Lindahl T., McIlhatton M., Fishel R., Miller J.H. Deficiencies in mouse Myh and Ogg1 result in tumor predisposition and G to T mutations in codon 12 of the K-ras oncogene in lung tumors. Cancer Res. 2004;64:3096–3102. doi: 10.1158/0008-5472.can-03-3834. [DOI] [PubMed] [Google Scholar]
- 188.de Souza-Pinto N.C., Eide L., Hogue B.A., Thybo T., Stevnsner T., Seeberg E., Klungland A., Bohr V.A. Repair of 8-oxodeoxyguanosine lesions in mitochondrial dna depends on the oxoguanine dna glycosylase (OGG1) gene and 8-oxoguanine accumulates in the mitochondrial dna of OGG1-defective mice. Cancer Res. 2001;61:5378–5381. PMID:11454679. [PubMed] [Google Scholar]
- 189.Yuzefovych L.V., Schuler A.M., Chen J., Alvarez D.F., Eide L., Ledoux S.P., Wilson G.L., Rachek L.I. Alteration of mitochondrial function and insulin sensitivity in primary mouse skeletal muscle cells isolated from transgenic and knockout mice: role of ogg1. Endocrinology. 2013;154:2640–2649. doi: 10.1210/en.2013-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Tumurkhuu G., Shimada K., Dagvadorj J., Crother T.R., Zhang W., Luthringer D., Gottlieb R.A., Chen S., Arditi M. Ogg1-Dependent DNA repair regulates NLRP3 inflammasome and prevents atherosclerosis. Circ. Res. 2016;119:76–90. doi: 10.1161/CIRCRESAHA.116.308362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Leon J., Sakumi K., Castillo E., Sheng Z., Oka S., Nakabeppu Y. 8-Oxoguanine accumulation in mitochondrial DNA causes mitochondrial dysfunction and impairs neuritogenesis in cultured adult mouse cortical neurons under oxidative conditions. Sci. Rep. 2016;6:22086. doi: 10.1038/srep22086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Abolhassani N., Leon J., Sheng Z., Oka S., Hamasaki H., Iwaki T., Nakabeppu Y. Molecular pathophysiology of impaired glucose metabolism, mitochondrial dysfunction, and oxidative DNA damage in Alzheimer's disease brain. Mech. Ageing Dev. 2017;161:95–104. doi: 10.1016/j.mad.2016.05.005. [DOI] [PubMed] [Google Scholar]
- 193.Beckman K.B., Ames B.N. Endogenous oxidative damage of mtDNA. Mutat. Res. 1999;424:51–58. doi: 10.1016/s0027-5107(99)00007-x. [DOI] [PubMed] [Google Scholar]
- 194.Nakamoto H., Kaneko T., Tahara S., Hayashi E., Naito H., Radak Z., Goto S. Regular exercise reduces 8-oxodG in the nuclear and mitochondrial DNA and modulates the DNA repair activity in the liver of old rats. Exp. Gerontol. 2007;42:287–295. doi: 10.1016/j.exger.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 195.Wiesner R.J., Rüegg J.C., Morano I. Counting target molecules by exponential polymerase chain reaction: copy number of mitochondrial DNA in rat tissues. Biochem. Biophys. Res. Commun. 1992;183:553–559. doi: 10.1016/0006-291x(92)90517-o. [DOI] [PubMed] [Google Scholar]
- 196.Lim K.S., Jeyaseelan K., Whiteman M., Jenner A., Halliwell B. Oxidative damage in mitochondrial DNA is not extensive. Ann. N. Y. Acad. Sci. 2005;1042:210–220. doi: 10.1196/annals.1338.023. [DOI] [PubMed] [Google Scholar]
- 197.Akhmedov A.T., Marín-García J. Mitochondrial DNA maintenance: an appraisal. Mol. Cell. Biochem. 2015;409:283–305. doi: 10.1007/s11010-015-2532-x. [DOI] [PubMed] [Google Scholar]
- 198.Lee H.C., Yin P.H., Lu C.Y., Chi C.W., Wei Y.H. Increase of mitochondria and mitochondrial DNA in response to oxidative stress in human cells. Biochem. J. 2000;348:425–432. [PMC free article] [PubMed] [Google Scholar]
- 199.Pinz K.G., Shibutani S., Bogenhagen D.F. Action of mitochondrial DNA polymerase gamma at sites of base loss or oxidative damage. J. Biol. Chem. 1995;270:9202–9206. doi: 10.1074/jbc.270.16.9202. [DOI] [PubMed] [Google Scholar]
- 200.Pastukh V.M., Gorodnya O.M., Gillespie M.N., Ruchko M.V. Regulation of mitochondrial genome replication by hypoxia: the role of DNA oxidation in D-loop region. Free Radic. Biol. Med. 2016;96:78–88. doi: 10.1016/j.freeradbiomed.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Shokolenko I., Venediktova N., Bochkareva A., Wilson G.L., Alexeyev M.F. Mutations in mtDNA are an underlying factor in many mitochondrial diseases and aging. Nucleic Acids Res. 2009;37:2539–2548. doi: 10.1093/nar/gkp100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Suter M., Richter C. Fragmented mitochondrial DNA is the predominant carrier of oxidized DNA bases. Biochemistry. 1999;38:459–464. doi: 10.1021/bi9811922. [DOI] [PubMed] [Google Scholar]
- 203.Wiehe R.S., Gole B., Chatre L., Walther P., Calzia E., Ricchetti M., Wiesmüller L. Endonuclease G promotes mitochondrial genome cleavage and replication. Oncotarget. 2018;9:18309–18326. doi: 10.18632/oncotarget.24822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Lin J.L.J., Nakagawa A., Skeen-Gaar R., Yang W.Z., Zhao P., Zhang Z., Ge X., Mitani S., Xue D., Yuan H.S. Oxidative stress impairs cell death by repressing the nuclease activity of mitochondrial endonuclease. G. Cell Rep. 2016;16:279–287. doi: 10.1016/j.celrep.2016.05.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Dianov G.L., Souza-Pinto N., Nyaga S.G., Thybo T., Stevnsner T., Bohr V.A. Base excision repair in nuclear and mitochondrial DNA. Prog. Nucleic Acid Res. Mol. Biol. 2001;68:285–297. doi: 10.1016/s0079-6603(01)68107-8. [DOI] [PubMed] [Google Scholar]
- 206.Kaneko T., Tahara S., Matsuo M. Non-linear accumulation of 8-hydroxy-2'-deoxyguanosine, a marker of oxidized DNA damage, during aging. Mutat. Res. 1996;316:277–285. doi: 10.1016/s0921-8734(96)90010-7. [DOI] [PubMed] [Google Scholar]
- 207.Izzotti A., Cartiglia C., Taningher M., De Flora S., Balansky R. Age-related increases of 8-hydroxy-2'-deoxyguanosine and DNA-protein crosslinks in mouse organs. Mutat. Res. 1999;446:215–223. doi: 10.1016/s1383-5718(99)00189-8. [DOI] [PubMed] [Google Scholar]
- 208.Sattarova E.A., Sinitsyna O.I., Vasyunina E.A., Duzhak A.B., Kolosova N.G., Zharkov D.O., Nevinsky G.A. Age-dependent guanine oxidation in DNA of different brain regions of Wistar rats and prematurely aging OXYS rats. Biochim. Biophys. Acta. 2013;1830:3542–3552. doi: 10.1016/j.bbagen.2013.01.027. [DOI] [PubMed] [Google Scholar]
- 209.Gan W., Nie B., Shi F., Xu X.M., Qian J.C., Takagi Y., Hayakawa H., Sekiguchi M., Cai J.P. Age-dependent increases in the oxidative damage of DNA, RNA, and their metabolites in normal and senescence-accelerated mice analyzed by LC-MS/MS: urinary 8-oxoguanosine as a novel biomarker of aging. Free Radic. Biol. Med. 2012;52:1700–1707. doi: 10.1016/j.freeradbiomed.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 210.Ichiba M., Yamada S., Ishii K., Gonda K., Murai R., Shimomura T., Saeki T., Kanbe T., Tanabe Y., Yoshida Y., Tsuchiya H., Hoshikawa Y., Kurimasa A., Kishimoto Y., Kawasaki H., Shiota G. Significance of urinary excretion of 8-hydroxy-2'-deoxyguanosine in healthy subjects and liver disease patients. Hepato-Gastroenterology. 2007;54:1736–1740. PMID:18019707. [PubMed] [Google Scholar]
- 211.Chen S.K., Hsieh W.A., Tsai M.H., Chen C.C., Hong A.I., Wei Y.H., Chang W.P. Age-associated decrease of oxidative repair enzymes, human 8-oxoguanine DNA glycosylases (hOgg1), in human aging. J. Radiat. Res. 2003;44:31–35. doi: 10.1269/jrr.44.31. [DOI] [PubMed] [Google Scholar]
- 212.Mikkelsen L., Bialkowski K., Risom L., Løhr M., Loft S., Møller P. Aging and defense against generation of 8-oxo-7,8-dihydro-2'-deoxyguanosine in DNA. Free Radic. Biol. Med. 2009;47:608–615. doi: 10.1016/j.freeradbiomed.2009.05.030. [DOI] [PubMed] [Google Scholar]
- 213.Tian F., Tong T.J., Zhang Z.Y., McNutt M.A., Liu X.W. Age-dependent down-regulation of mitochondrial 8-oxoguanine DNA glycosylase in SAM-P/8 mouse brain and its effect on brain aging. Rejuvenation Res. 2009;12:209–215. doi: 10.1089/rej.2009.0849. [DOI] [PubMed] [Google Scholar]
- 214.Reis A., Hermanson O. The DNA glycosylases OGG1 and NEIL3 influence differentiation potential, proliferation, and senescence-associated signs in neural stem cells. Biochem. Biophys. Res. Commun. 2012;423:621–626. doi: 10.1016/j.bbrc.2012.04.125. [DOI] [PubMed] [Google Scholar]
- 215.Bindu S., Pillai V.B., Kanwal A., Samant S., Mutlu G.M., Verdin E., Dulin N., Gupta M.P. SIRT3 blocks myofibroblast differentiation and pulmonary fibrosis by preventing mitochondrial DNA damage. Am. J. Physiol. Lung Cell Mol. Physiol. 2017;312:L68–L78. doi: 10.1152/ajplung.00188.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Wang W., Esbensen Y., Kunke D., Suganthan R., Rachek L., Bjørås M., Eide L. Mitochondrial DNA damage level determines neural stem cell differentiation fate. J. Neurosci. 2011;31:9746–9751. doi: 10.1523/JNEUROSCI.0852-11.2011. https://doi:10.1523/JNEUROSCI.0852-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Lavrukhin O.V., Lloyd R.S. Involvement of phylogenetically conserved acidic amino acid residues in catalysis by an oxidative DNA damage enzyme formamidopyrimidine glycosylase. Biochemistry. 2000;39:15266–15271. doi: 10.1021/bi001587x. [DOI] [PubMed] [Google Scholar]
- 218.Kim B.M., Rhee J.S., Seo J.S., Kim I.C., Lee Y.M., Lee J.S. 8-Oxoguanine DNA glycosylase 1 (OGG1) from the copepod Tigriopus japonicus: molecular characterization and its expression in response to UV-B and heavy metals. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012;155:290–299. doi: 10.1016/j.cbpc.2011.09.010. [DOI] [PubMed] [Google Scholar]
- 219.Wada A., Suyama A. Local stability of DNA and RNA secondary structure and its relation to biological functions. Prog. Biophys. Mol. Biol. 1986;47:113–115. doi: 10.1016/0079-6107(86)90012-x. [DOI] [PubMed] [Google Scholar]
- 220.Galtier N., Lobry Relationships between genomic G+C content, RNA secondary structures, and optimal growth temperature in prokaryotes. J. Mol. Evol. 1997;44:632–636. doi: 10.1007/pl00006186. [DOI] [PubMed] [Google Scholar]
- 221.Naya H., Romero H., Zavala A., Alvarez B., Musto H. Aerobiosis increases the genomic guanine plus cytosine content (GC%) in prokaryotes. J. Mol. Evol. 2002;55:260–264. doi: 10.1007/s00239-002-2323-3. [DOI] [PubMed] [Google Scholar]
- 222.Friedman K.A., Heller A. Guanosine distribution and oxidation resistance in eight eukaryotic genomes. J. Am. Chem. Soc. 2004;126:2368–2371. doi: 10.1021/ja038217r. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.