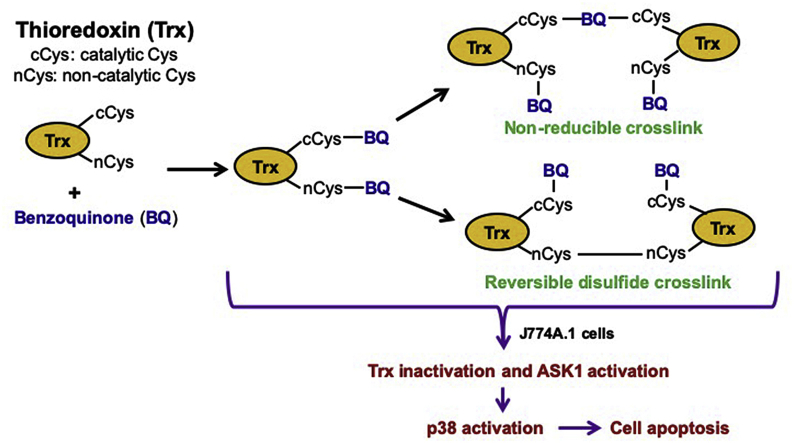
Abstract
Quinones can modify biological molecules through both redox-cycling reactions that yield radicals (semiquinone, superoxide and hydroxyl) and via covalent adduction to nucleophiles (e.g. thiols and amines). Kinetic data indicate that Cys residues in GSH and proteins are major targets. In the studies reported here, the interactions of a prototypic quinone compound, p-benzoquinone (BQ), with the key redox protein, thioredoxin-1 (Trx1) were examined. BQ binds covalently with isolated Trx1 forming quinoprotein adducts, resulting in a concentration-dependent loss of enzyme activity and crosslink formation. Mass spectrometry peptide mass mapping data indicate that BQ forms adducts with all of the Trx1 Cys residues. Glutathione (GSH) reacts competitively with BQ, and thereby modulates the loss of activity and crosslink formation. Exposure of macrophage-like (J774A.1) cells to BQ results in a dose-dependent loss of Trx and thioredoxin reductase (TrxR) activities, quinoprotein formation, and a decrease in GSH levels without a concomitant increase in oxidized glutathione. GSH depletion aggravates the loss of Trx and TrxR activity. These data are consistent with adduction of GSH to BQ being a primary protective pathway. Reaction of BQ with Trx in cells resulted in the activation of apoptosis signal-regulating kinase 1 (ASK1), and p38 mitogen-activated protein kinase (MAPK) leading to apoptotic cell death. These data suggest that BQ reacts covalently with Cys residues in Trx, including at the active site, leading to enzyme inactivation and protein cross-linking. Modification of the Cys residues in Trx also results in activation of the ASK1/p38-MAPK signalling pathway and promotion of apoptotic cell death.
Keywords: Quinone, Thioredoxin, Michael addition, Thioredoxin reductase, Apoptosis, Quinoprotein
Graphical abstract
Highlights
-
•
Quinone (e.g. p-benzoquinone, BQ) toxicity is linked to Michael adduction reactions.
-
•
Adduction of BQ to Cys residues in proteins are rapid (≤105 M−1 s−1) and selective.
-
•
BQ reaction with Cys inactivates thioredoxin (Trx) and yields quinone- and disulfide-linked dimers.
-
•
GSH reacts competitively with BQ and modulates damage, without GSSG formation.
-
•
BQ activates ASK1 and p38 pathways and induced apoptosis in cells via Trx damage.
Abbreviations
- ASK1
apoptosis signal-regulating kinase 1
- BEI
biotin ethylenediamine iodoacetamide
- BQ
p-benzoquinone
- BSO
buthionine sulfoximine
- CAA
2-chloroacetamide
- DMEM
Dulbecco's modified Eagle's medium
- DTNB
5,5′-dithiobis(2-nitrobenzoic acid)
- EDTA
ethylenediaminetetraacetic acid
- GSH
reduced glutathione
- GSSG
glutathione disulfide
- JNK
c-Jun N-terminal kinase
- MAPK
mitogen-activated protein kinase
- NBT
nitrobluetetrazolium chloride
- Trx
thioredoxin
- TrxR
thioredoxin reductase
1. Introduction
Quinones are a common structural motif in many biological molecules [1]. Human exposure can arise from the oxidation of environmental pollutants, such as benzene [2], cigarette smoke [3], and polychlorinated biphenyls [4], as well as through enzymatic metabolism of endogenous compounds (e.g. estrogens, dopamine [5,6]) and commercial drugs (e.g. acetaminophen, diclofenac [7]). Quinones are commonplace in foods and can also be formed from metabolism of phenols and polyphenols [8]. Synthetic and natural product quinones are also widely employed in the clinic, or are under investigation as therapeutic drugs (e.g. doxorubicin, daunorubicin, epirubicin, idarubicin, mitoxantrone, rhein, atovoquone, emodin, juglone, tanoshinones) [1].
Quinones can react as electrophiles to modify nucleophiles such as the cysteine (Cys) and lysine (Lys) residues in peptides (e.g. GSH) and proteins, or the amine-containing nucleobases of DNA/RNA via Michael addition reactions [1]. Alternatively, they can undergo redox cycling in the presence of O2 and biological reductants to generate semiquinone and superoxide radical anions, and downstream species such as hydrogen peroxide (H2O2) and hydroxyl radicals (HO.) [1]. Our previous kinetic data indicate that quinones react with GSH and other thiols with higher rate constants than some widely studied oxidants (e.g. H2O2, ONOOH, HOSCN), suggesting that these compounds are likely to be significant thiol modifying agents in biological systems [9,10]. Reaction with Cys residues on proteins results in the formation of protein-hydroquinone adducts that can undergo ready oxidation to give quinone-protein (quinoprotein) species [11]; the latter can undergo further reactions resulting in protein cross-linking [9,10]. Thus, reaction of quinones with proteins can result in significant changes to protein structure and function [9,10]. The active site thiol residue of calcium-ATPase has been shown to be modified by quinones leading to a loss of enzyme activity [12]. Similar reactions of quinone methide metabolites of 2,6-di-tert-butyl-4-methylphenol (BHT) can modify Cys residues in structural and antioxidant proteins, resulting in an enhancement of intracellular oxidative stress and inflammation; such reactions have been reported to underlie the lung tumorigenesis property of BHT [13]. A strong correlation has been reported between the covalent binding reactivity of substituted p-benzoquinones (BQ) and their hepatotoxicity, and the rate of soft electrophilic reactivity with GSH has been used to predict the potential cytotoxicity of p-benzoquinones in rat hepatocytes [14].
The thioredoxin system is the major disulfide reductase system in mammals, with this composed of NADPH, thioredoxin (Trx), and thioredoxin reductase (TrxR). Trx is a small (12 kDa) reductase, present in both the cytosol (Trx1) and mitochondria (Trx2). It contains a conserved two Cys-containing sequence (-CGPC-) at its catalytic centre. The substrates for this system include peroxiredoxins (Prx), methionine sulfoxide reductases (MSR), and some redox-sensitive transcription factors [15]. Trx reduces disulfide bonds in client proteins by transferring electrons from the two reactive Cys residues in its active site, with concomitant formation of a disulfide bond in the catalytic centre. TrxR uses NADPH as the electron donor to reduce the oxidized Trx, thus maintaining it in a reduced form and completing its catalytic cycle [16]. We have recently reported that TrxR is susceptible to modification by benzoquinone via modification of both its selenocysteine (Sec) and Cys residues [10]. The GSH/GSSG-glutaredoxin-glutathione reductase-NAPDH reduction system, another critical antioxidant system, undergoes cross-talk, and has some overlap, with the Trx system, as GSH can reduce oxidized Trx in the presence of glutaredoxin (Grx) [17], and the thioredoxin system can also reduce oxidized GSH [18].
In addition to acting as a direct antioxidant system, Trx also acts as a central redox regulator mediating the activation of intracellular transcription factors including apoptosis signal regulating kinase 1 (ASK1). ASK1 belongs to the mitogen-activated protein kinase kinase kinase (MAPKKK) family. Activation of MAPKKK ultimately leads to the activation of MAPK including c-Jun N-terminal kinase (JNK) and the p38 MAPK family, therefore modulating both the stress response and apoptosis [19,20]. Trx can negatively regulate ASK1 function by binding to the N-terminal noncatalytic region with formation of an inactive Trx-ASK1 complex. Oxidation of Cys32 and Cys35 of Trx, generates a disulfide bond that releases ASK1, thus activating ASK1 and the downstream cell apoptosis pathway [21]. Several compounds have been reported to induce cell apoptosis via the activation of Trx-ASK1-MAPK pathway. Thus, mercury compounds can oxidize Trx, resulting in the phosphorylation of ASK1, activation of caspase-3, and neuronal cell death via apoptosis [22]. Paraquat can induce oxidative stress, leading to the oxidation of Trx, thereby activating ASK1 and downstream JNK and p38-MAPK pathways, resulting in the induction of apoptosis [23].
Previous studies have predominantly attributed Trx-ASK1 mediated cell death to Trx oxidation and subsequent release of ASK1. However, as quinones can modify both Cys and Sec residues (in the case of TrxR) by acting as electrophiles [9,10], we hypothesized that BQ might modify the Cys residues of Trx by this pathway, and thereby affect the function and oligomerization state of Trx, with this resulting in downstream cellular apoptosis via activation of the Trx-ASK1-MAPK pathway.
2. Materials and methods
2.1. Reagents and cell culture
Human recombinant thioredoxin-1, and thioredoxin reductase-1 (from rat) were purchased from IMCO (Stockholm, Sweden). Bovine serum albumin (BSA), GSH, GSSG, NADPH, glutathione reductase (GR) from yeast, recombinant human insulin, 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), 5-sulfosalicylic acid dehydrate, Triton X-100, 2-vinylpyridine, nitroblue tetrazolium (NBT), 2-chloroacetamide (CAA) and p-benzoquinone (BQ) were purchased from Sigma (St. Louis, MO, USA). BQ solutions were prepared fresh for each experiment by dissolving the compound in dimethyl sulfoxide (DMSO) to give a stock solution, with this then diluted into the relevant reaction systems. Lys-C was obtained from Promega (Madison, WI, USA). ThioGlo-1 (MMBC, methyl maleimidobenzochromenecarboxylate) was obtained from Berry and Associates (Dexter, MI, USA). Antibodies against ASK1 (702278) and phospho-ASK1 (Thr838, PA5-64541) were obtained from Thermo Fisher (Waltham, MA, USA). Antibodies against JNK (9252s), phospho-JNK (4668s), p38 MAPK (8690s) and phospho-p38 MAPK (Thr180/Tyr182, 4511s) were purchased from Cell Signalling Technology (Danvers, MA, USA). All other reagents were of the highest available quality. High purity (MQ) water was obtained from a Milli-Q System (Millipore, Bedford, MA).
Murine macrophage-like J774A.1 cells (American Type Culture Collection; No. 915051511) were cultured in Dulbecco's modified Eagle's medium (DMEM, Sigma) with 10% (v/v) fetal bovine serum (Invitrogen), and 2 mM l-glutamine (Thermo, Waltham, MA, USA), in a humidified incubator at 37 °C and 5% CO2.
2.2. Enzyme activity assay
Trx activity was measured according to a previously reported method [24] with some modifications. Purified Trx1 protein or cell samples were incubated with the reaction mixture buffer (790 μM insulin, 2 mM EDTA, 2 mM NAPDH, 0.5 μM TrxR in 210 mM HEPES, pH 7.6) at 37 °C for 30 min, then the reaction was terminated by adding the stopping buffer (10 mM DTNB, 8 M guanidine hydrochloride in 0.2 M Tris-HCl, pH 8.0). Absorbance changes at 412 nm were recorded using a SpectraMax i3 plate reader (Molecular Devices, CA, USA).
TrxR activity was determined using a previously established method [25] with some modifications. Equal volumes of sample and reaction buffer (10 mM EDTA, 0.2 mg mL−1 BSA, 5 mM DTNB, and 240 μM NADPH in 50 mM potassium phosphate, pH 7.4) were mixed at 22 °C. The conversion of DTNB to TNB was recorded at 412 nm every 30 s over a 20 min period using a plate reader (SpectraMax i3, Molecular Devices, CA, USA). Data were corrected for changes detected in the presence of the TrxR inhibitor auranofin (500 nM) added into the reaction system.
2.3. Inactivation of Trx and TrxR by BQ
Recombinant human Trx1 (100 μM) was incubated with DTT (10 mM) in reaction buffer (50 mM Tris-HCl containing 2 mM EDTA, pH 7.6) at 22 °C in the dark for 40 min. Excess DTT was removed by elution of the protein through a NAP5 column (GE healthcare, Chicago, IL, USA). Fully reduced Trx1 (2.5 μM) was treated with different concentrations of BQ (0.5–10 μM) for 5 min, before measuring the remaining enzyme activity using the method described above.
For the experiment involving GSH as the protective reagent, GSH was co-incubated with reduced Trx1 and BQ for 5 min, or Trx1 was pre-incubated with BQ for 5 min before adding different concentrations of GSH. After this, excess GSH and BQ were removed using Zeba desalting columns (Thermo, Waltham, MA, USA), and the residual enzyme activity in the protein fraction measured as described above.
J774A.1 cells were seeded in 12-well plate at a density of 1 × 105 cells well−1 and cultured overnight in media. After removing the media and washing with HBSS, the cells were then incubated with different concentrations of BQ (2–80 μM) in HBSS for 1 h at 37 °C. The cells were then washed twice with cold PBS, and lysed in lysis buffer (0.2 mg mL−1 BSA, 10 mM EDTA in 50 mM Tris-HCl, pH 7.6). The Trx and TrxR activities were measured using the methods described above. The protein concentration was determined by the bicinchoninic acid (BCA) assay.
2.4. Assessment of Trx1 oligomerization
Trx (human or E. coli) was reduced using DTT as described above, then 5 μM protein was incubated with different concentrations of BQ (2–80 μM) for 5 min. Aliquots of samples were then removed and diluted 4-fold with loading buffer (LDS Sample Buffer (4X) from Thermo Fisher, containing lithium dodecyl sulfate at pH 8.5 with SERVA Blue G250 and phenol red) with or without DTT. Samples were loaded on to 12-well SDS-PAGE gels (4–12% Bis-Tris Gel, Invitrogen) and subjected to electrophoresis using NuPAGE MES SDS running buffer (Invitrogen) at 200 V for 40 min. The proteins were subsequently developed using silver staining [26] and scanned using a flatbed scanner.
2.5. Detection of quinoprotein formation using glycinate redox-cycling staining
Quinone-adducted proteins were detected by nitrobluetetrazolium (NBT)-glycinate redox staining using a previously reported method [27] with some modifications. After reduction with DTT, Trx1 (10 μM) was incubated with different concentrations of BQ (0–40 μM) for 5 min, then protein samples were prepared by diluting with loading buffer (as above). After separation on SDS-PAGE gels, the proteins were transferred onto the PVDF membranes using an iBlot 2 dry blotting system (Thermo, Waltham, MA) at 20 V for 7 min, and then incubated with NBT (0.73 μM in 2 M potassium glycinate buffer, pH 10.0) for 40 min in the dark. The membranes were subsequently washed twice with sodium borate solution (0.16 M, pH 10.0) and scanned using a flatbed scanner.
2.6. Identification of BQ-modified sites in Trx using biotin ethylenediamine iodoacetamide (BEI)
Fully reduced Trx1 (5 μM, see above) was incubated with BQ (0–80 μM) for 5 min at 22 °C. After this, samples were mixed with BEI (100 μM) at pH 7.6 followed by incubation at 37 °C in the dark for another 30 min to alkylate residual –SH groups. Samples were then diluted 4-fold with loading buffer, and separated on 12-well SDS-PAGE gels (4–12% Bis-Tris Gel, Invitrogen) using NuPAGE MES SDS running buffer (Invitrogen) at 200 V for 40 min. The separated proteins were transferred to PVDF membranes using an iBlot 2 dry blotting system (Thermo, Waltham, MA) at 20 V for 7 min. The BEI-labelled protein was detected using horseradish peroxidase-conjugated streptavidin and enhanced chemiluminescence detection.
2.7. Protein digestion and identification by LC-MS/MS
Fully reduced Trx1 (10 μM, see above) was incubated with BQ (50 μM) for 5 min at 22 °C. The samples were then incubated with 20 mM 2-chloroacetamide (CAA) to alkylate residual thiols in 6 M urea solution containing 50 mM Tris-HCl (pH 8.0) for 30 min at 22 °C in the dark, then digested using Lys-C (Lys-C: protein ratio of 1:50) for 16 h at 37 °C. Samples were desalted using C-18 disk StageTips (Empore C18, 47 mm Solid Phase Extraction Disks #2215, 3 M Purification, Eagan, MN, USA), then separated using a nanoUPLC system (Thermo, Waltham, MA) equipped with a C18 column (75 μm × 15 cm, 2 μm particle size, Thermo), held at 35 °C, using a gradient elution system consisting of mobile phase A (0.1% formic acid in high purity water) and B (80% acetonitrile containing 0.1% formic acid in high purity water). The gradient employed was: 4% B for 5 min, linearly increased to 60% B over 30 min, then linearly increased to 99% B over 10 min, maintained at 99% B for 5 min, then returned to 4% B over 10 min, maintained at 4% B for another 10 min, with a flow rate of 0.3 μL min−1 and a total run time of 65 min. The eluted peptides were transferred to a Captivespray source of an Impact II Q-TOF mass spectrometer (Bruker, Billerica, MA). The capillary voltage was 1400 V, the dry gas flow was 3.0 L min−1 and the temperature 180 °C. MaxQuant (version 1.6.3.4) was used for peptide sequence identification, and database searching. BQ-induced alkylation and S-carbamidomethylation at Cys were set as variable modifications to identify spectra of adducted peptides.
2.8. Cell metabolic activity assay
J774A.1 cells were seeded in 96-well plates at a density of 8 × 103 cells well−1 and cultured overnight in media. The cells were then treated with different concentrations of BQ (2–80 μM) for 24 h at 37 °C. MTS solution (0.3 mg mL−1) was then added to every well and incubated for 4 h, after which the absorbance at 490 nm was quantified using a plate reader (SpectraMax i3, Molecular Devices, CA, USA).
2.9. Determination of total cellular thiols
J774A.1 cells were seeded in 12-well plates at a density of 1 × 105 cells well−1 and cultured overnight as described above. Cells were then washed with HBSS and incubated with different concentration of BQ (2–80 μM) for 1 h in HBSS at 37 °C. After this, cells were washed twice with cold PBS and lysed in Milli-Q water on ice. The cell lysates were transferred to a 96-well plate and incubated with ThioGlo-1 reagent (26 μM) for 5 min in dark at 22 °C. The resulting fluorescence was recorded using λex 384 nm, λem 513 nm using a plate reader (SpectraMax i3, Molecular Devices, CA, USA). The resulting thiol concentration was normalized to the protein concentration and then expressed as a % of the control.
2.10. Determination of total GSH and GSSG
The total intracellular GSH and GSSG levels were measured using the enzymatic recycling method described in Ref. [28] with some modifications. Cells were seeded in 12-well plate at density of 1 × 105 cells well−1 and cultured overnight. Then cells were treated with different concentrations of BQ (0–40 μM) for 30 min as described above. After this, cells were washed twice with cold Ca2+- and Mg2+-free PBS, and harvested by centrifugation at 1000g for 5 min at 4 °C. The cell pellets were then resuspended in cold extraction buffer (0.1% Triton-X and 0.6% sulfosalicylic acid in 0.1 M potassium phosphate buffer with 5 mM EDTA disodium salt, pH 7.5) and sonicated for 2 min with vortexing every 30 s. The homogenate was then centrifuged at 3000 g for 10 min at 4 °C, and the supernatant taken for analysis. Total GSH levels were quantified by mixing the samples with assay solution (0.3 mg mL−1 DTNB, 0.3 mg mL−1 NADPH, and 2 U mL−1 GR), with the absorbance at 412 nm recorded every 30 s for 4 min. For quantification of GSSG, the samples were incubated with 10% 2-vinylpyridine at 22 °C in the dark for 1 h. The derivatized samples were then analysed by the same procedure as described for the total GSH measurements. Standard GSH, GSSG and blank samples were run in parallel.
2.11. Preparation of cell lysates and immunoblotting analysis
J774A.1 cells were plated in 6-well plates at the density of 1 × 106 cells well−1 and cultured overnight in media. After removal of the media and washing with HBSS, the cells were treated with different concentrations of BQ (0–10 μM) or H2O2 (0.3 mM) in HBSS for 15 or 30 min, and then washed with cold PBS, lysed in RIPA buffer containing a protease and phosphatase inhibitor cocktail. After centrifugation (16000 g for 10 min at 4 °C) to obtain the soluble fraction, samples (40 μg protein) were separated by SDS-PAGE under non-reducing conditions, followed by transfer to PVDF membranes using an iBlot 2 dry blotting system (Thermo, Waltham, MA) at 20 V for 7 min. The membranes were blocked with 5% BSA prior to probing with specific antibodies (see section 2.1). Phosphorylated protein levels were normalized to the total concentration of phosphorylated and unphosphorylated material. Levels of a β-actin, used as loading control were also quantified to confirm equal protein loading on the gels.
2.12. Apoptosis identification
J774A.1 cells were cultured overnight in eight-well chamber glass culture slides at a density of 8 × 103 cells per well as described above. Cells were then treated with BQ (20 μM) for 4 or 24 h, and cell apoptosis was detected using an APC Annexin V Apoptosis Kit (Nordic BioSite, Sweden). In brief, after the treatment, cells were washed twice with the binding buffer and double-stained with Annexin V and propidium iodide (PI) for 20 min in the dark. The samples were then rinsed twice with binding buffer and cover slips added. Cell images were captured from multiple fields using a fluorescence microscope (Olympus, Japan) equipped with cellSense Entry v1.5 software.
2.13. Statistics
Results were analysed using one-way analysis of variance (ANOVA) followed by Tukey's post hoc test using SPSS 25 (IBM, Armonk, NY, USA). Data are presented as mean ± standard deviations (SD) from at least triplicate independent experiments unless otherwise noted. Significance was set at p < 0.05.
3. Results
3.1. BQ-induced inhibition of Trx1 activity
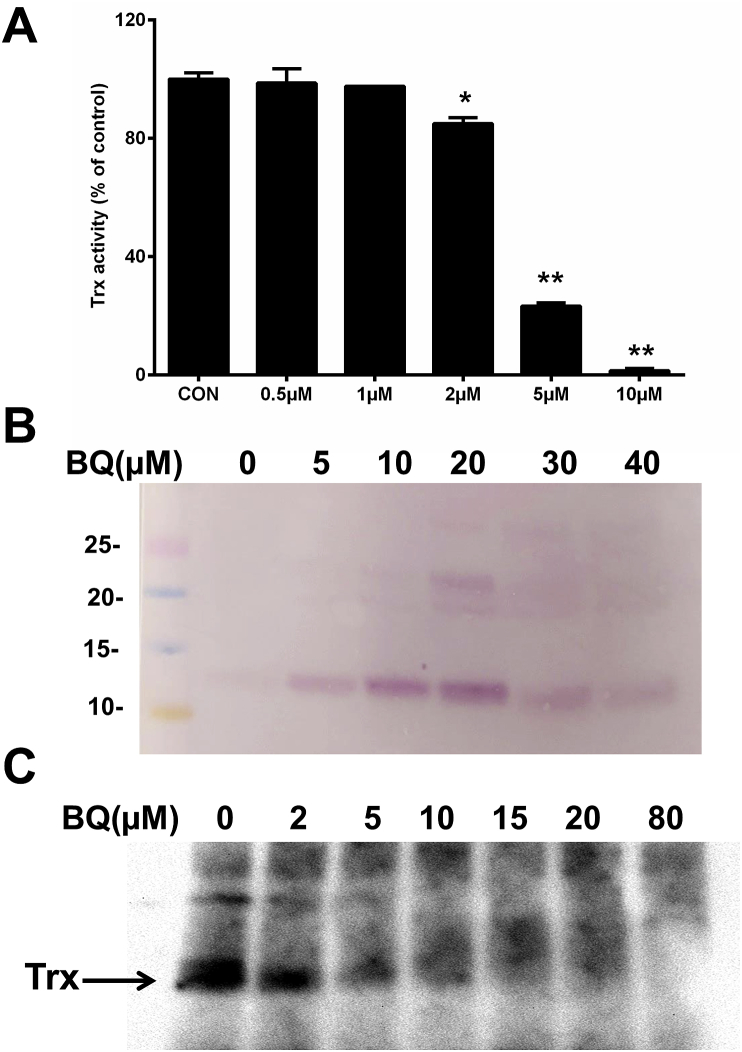
Incubation of the Trx1 with BQ for 5 min resulted in a rapid and concentration-dependent decrease in Trx1 activity (Fig. 1A). A significant loss of Trx1 activity was detected when the BQ concentration was two-fold or greater compared to the Trx1 concentration.
Fig. 1.
BQ-induced enzyme inactivation and covalent modification of purified Trx1 protein. (A) Concentration-dependent inhibition of Trx1 activity by BQ. DTT-reduced Trx1 (2.5 μM) was incubated with BQ (0.5–10 μM) in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6), with Trx1 activity determined after 5 min of treatment. (B) BQ-induced quinoprotein detection. DTT-reduced Trx1 (10 μM) was incubated with BQ (5–40 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6). Samples were analysed by SDS-PAGE followed by NBT redox staining. (C) Free thiol detection using BEI labelling. DTT-reduced Trx1 (5 μM) was incubated with BQ (2–80 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6). Samples were incubated with BEI (250 μM) in dark for 30 min at 37 °C, then analysed by SDS-PAGE followed by incubation with horseradish peroxidase-conjugated streptavidin and enhanced chemiluminescence detection. *p < 0.05, **p < 0.01 vs. control (CON) samples with no added BQ. Representative images from 3 independent experiments are shown in panels (B) and (C).
3.2. Detection of BQ adduction to Trx1 and identification of sites of modification
NBT-redox staining revealed that BQ rapidly formed adducts with Trx1, with significant bands from quinoprotein species detected within 5 min of the initiation of the reaction (Fig. 1B). When the BQ concentration was 3-fold or greater compared to the Trx1 concentration, a decrease in the quinoprotein band intensity was detected, possibly as a result of changes in Trx1 oligomerization (see below).
As Cys residues have been identified as kinetically important sites for quinone reaction [9,11], further studies were carried out to examine the role of Trx Cys residues in the formation of the quinoprotein species, with the Cys residues on Trx labelled using BEI. As shown in Fig. 1C, treatment of Trx1 with BQ prior to incubation with BEI, resulted in a decreased intensity of the bands arising from BEI labelling. This decrease occurred in a concentration-dependent manner with increasing BQ levels, with 80 μM BQ resulting in a complete blocking of available –SH groups.
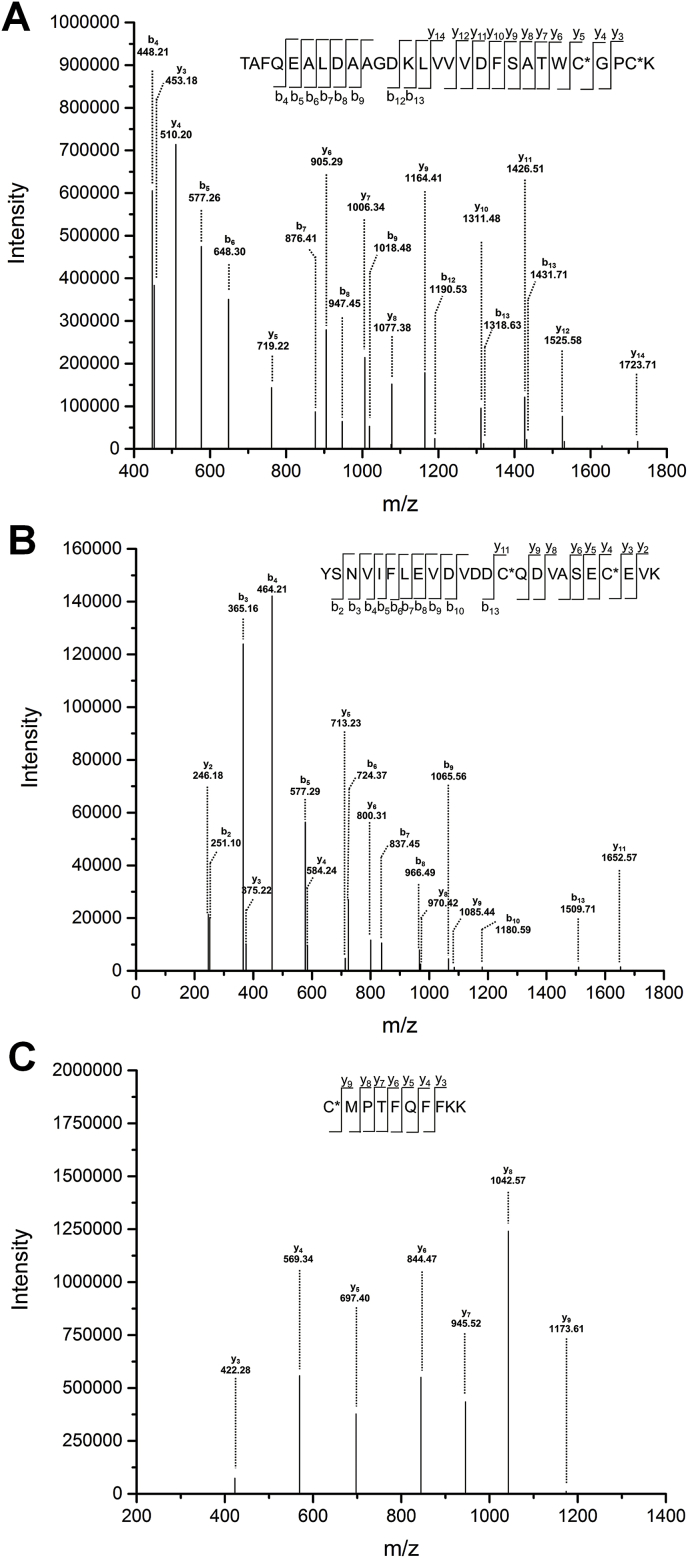
The sites of modification were determined using mass spectrometry (MS) peptide mass mapping. BQ-modified Trx1 (generated as described above), was digested with Lys-C and subjected to LC-MS analysis. Peptide coverage for both the native and modified protein was >90%. All peptides yielded abundant and characteristic fragmentation ions with good signal intensities in MS2 experiments that could be readily analysed. Three peptides containing all of the Trx Cys residues were identified for both native and modified protein, and every residue was detected as a BQ-modified species (Table 1, Fig. 2), supporting the hypothesis that modification of the catalytic Cys residues is, at least partly, responsible for the observed loss of activity. These data are consistent with the BEI labelling experiments.
Table 1.
Sites of p-benzoquinone modification in Trx1.a.
| Peptide number | Position | Peptide Sequence | Theoretical MS | Observed MS | Cys |
|---|---|---|---|---|---|
| 1 | 9–36 | TAFQEALDAAGDKLVVVDFSATWC*GPC*K | 3154.4099 | 3154.4359 | Cys32 Cys35 |
| 2 | 49–72 | YSNVIFLEVDVDDC*QDVASEC*EVK | 2931.2150 | 2931.2319 | Cys62 Cys69 |
| 3 | 73–82 | C*MPTFQFFKK | 1383.6355 | 1383.6464 | Cys73 |
Average sequence coverage of the control samples was 93.3%, for the BQ treated samples 90.3%.
Fig. 2.
Identification of BQ-modified Cys residues in Trx1. DTT-reduced Trx1 (10 μM) was incubated with BQ (50 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6). The BQ-treated Trx1 was then digested using Lys-C at 37 °C overnight and analysed by LC-MS/MS (seen Materials and methods for further details). (A) Peptide 1, (B) Peptide 2, (C) Peptide 3 (cf. Table 1).
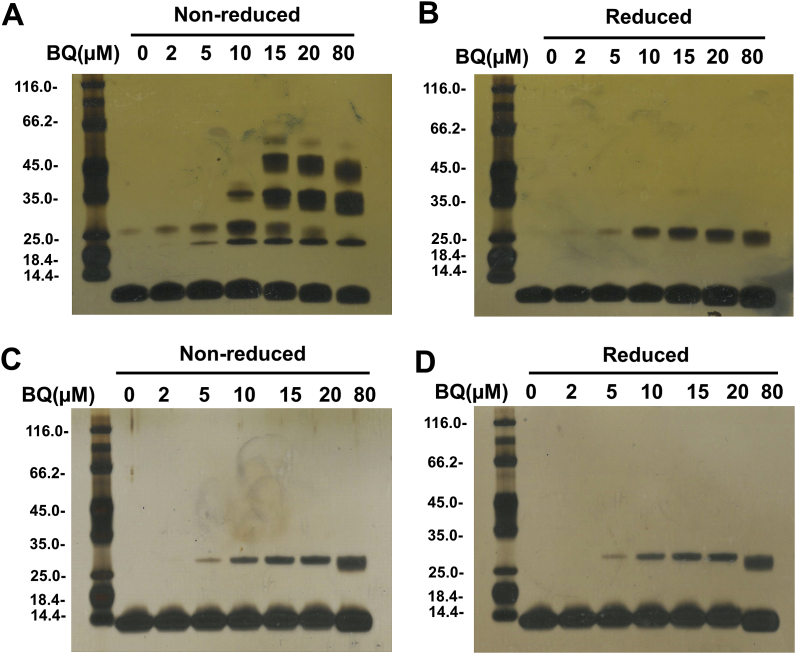
3.3. Effect of BQ treatment on Trx1 oligomer formation and cross-linking
To characterize modifications to Trx1 induced by BQ, fully reduced Trx1 was treated with different concentrations of BQ at 22 °C in the dark for 5 min, with samples subsequently subjected to SDS-PAGE, under non-reducing and reducing conditions, and silver staining. The gels run under non-reducing conditions (Fig. 3A), indicate that BQ induces formation of multiple protein oligomers and loss of the parent protein band, particularly with a 3-fold, or greater, molar excess of BQ compared to the Trx1 concentration.
Fig. 3.
BQ-induced protein cross-linking. Human Trx1 (5 μM, A and B) and E. coli Trx (5 μM, C and D), pre-treated with DTT, were incubated with BQ (2–80 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6). Samples were analysed under non-reducing (A and C) and reducing (B and D) conditions, and then analysed by SDS-PAGE with silver staining. Representative images from 3 independent experiments are shown in each panel.
Inclusion of DTT as a reducing agent, significantly decreased the intensity of most of the oligomer bands, although a band with a mass consistent with the presence of a dimer species showed a BQ concentration-dependent increase (Fig. 3B). These data suggest that inter-protein disulfide bonds are a major contributor to the protein cross-linking, but that at least one dimer species contains non-reducible cross-links.
Trx from E. coli contains only two of the Cys residues present in the human protein, which are present in the catalytic centre [29]. Consequently, limited studies were carried out to examine the effect of BQ treatment on this isoform. As shown in Fig. 3C and D, BQ induced only non-reducible dimers with E. coli Trx, suggesting that the disulfide-bonded oligomers detected with the human protein arise from the non-catalytic Cys residues.
3.4. Effect of GSH on BQ-induced loss of enzyme activity, oligomer formation and cross-linking
Previous data indicate that BQ-induced modification of Cys residues on GAPDH can be prevented by competitive reaction of GSH with BQ, and (to a more limited extent) reversed by S-transarylation reactions [9,10]. Consequently, we hypothesized that BQ modification of Trx1 may also be prevented or reversed by GSH. This was investigated by incubating Trx1 (2.5 μM) and BQ (10 μM) for 5 min in the absence or presence of different concentrations of GSH (0–40 μM) before determination of residual enzyme activity. Co-incubation with GSH showed a concentration-dependent protection against BQ-induced loss of Trx1 activity (Fig. 4A), with near complete enzyme activity retained with a 16-fold molar ratio of GSH (i.e. 40 μM) compared to Trx1 concentration; the significance of this ratio is discussed further below.
Fig. 4.
Effect of GSH on BQ-induced enzyme activity loss, protein cross-linking and modification. Panels A, C, E: DTT-reduced Trx1 (2.5 μM) was incubated with BQ (20 μM) in the presence of GSH (5–1000 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6). The samples were split into three portions. The remaining Trx activity was measured (panel A), samples were analysed by SDS-PAGE followed by silver staining (panel C), or incubated with BEI (250 μM) in dark for 30 min at 37 °C, then analysed by SDS-PAGE followed by treatment with horseradish peroxidase-conjugated streptavidin and enhanced chemiluminescence detection (panel E). Panels B, D, F: DTT-reduced Trx1 (2.5 μM) was incubated with BQ (20 μM) for 5 min in 50 mM Tris-HCl containing 2 mM EDTA (pH 7.6), then GSH (5–1000 μM) was added and incubated for another 5 min. The samples were then split into three portions and analysed, as described above, for remaining Trx activity (panel B), by SDS-PAGE followed by silver staining (panel D), or incubated with BEI (250 μM) in dark for 30 min at 37 °C, then analysed by SDS-PAGE followed by treatment with horseradish peroxidase-conjugated streptavidin and enhanced chemiluminescence detection (panel F). Data (mean ± standard deviations) from 3 independent experiments are presented in panels A and B. Representative images from 3 independent experiments are shown in panels (C - F). *p < 0.05, **p < 0.01 vs. control (CON) samples with no added BQ; #p < 0.05, ##p < 0.01 vs. samples with no added GSH.
To confirm this competitive effect of GSH, Trx1 (5 μM) was co-incubated with BQ (20 μM) in the absence or presence of GSH (0–1000 μM) with the samples analysed by SDS-PAGE with silver staining under non-reducing conditions. GSH was observed to decrease BQ-induced cross-linking of Trx1 in a concentration-dependent manner (Fig. 4C), indicating that GSH can preserve both Trx activity and prevent changes in functional integrity.
S-transarylation, which can reverse initial adduct formation, was investigated by pre-incubation of Trx1 (2.5 μM) with BQ (10 μM) for 5 min, to allow adduct formation, then different concentrations of GSH (0–40 μM) were added to examine potential recovery of enzyme activity. Over the concentration range tested, GSH could not reactivate Trx1 (Fig. 4B). Analysis by SDS-PAGE of similar samples (Fig. 4D), indicated that adding GSH to BQ-adducted Trx1 marginally decreased the intensity of the BQ-induced protein oligomer bands, with a new cross-linked species generated.
To examine whether the prevention of loss of activity and protein cross-linking was associated with sparing of Trx Cys residues, Trx1 (5 μM) was incubated with BQ (20 μM) for 5 min in the absence or presence of GSH (0–40 μM). The samples were subsequently incubated with BEI (250 μM) and the BEI-labelled proteins were subjected to SDS-PAGE and immunoblotting. The data obtained indicate that co-incubation with GSH could alleviate BQ-induced band intensity loss in a concentration-dependent manner (Fig. 4E). This confirms that GSH reacts competitively with BQ, resulting in a decrease in the loss of Cys residues on Trx1, retention of enzyme activity and a decrease in oligomer formation/cross-linking. Addition of GSH after pre-exposure Trx1 to BQ did not reverse the observed loss of BEI-labelling, consistent with the activity data (Fig. 4F).
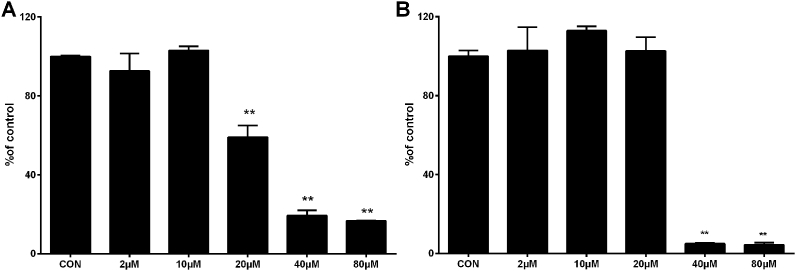
3.5. BQ-induced loss of metabolic activity in J774A.1 cells
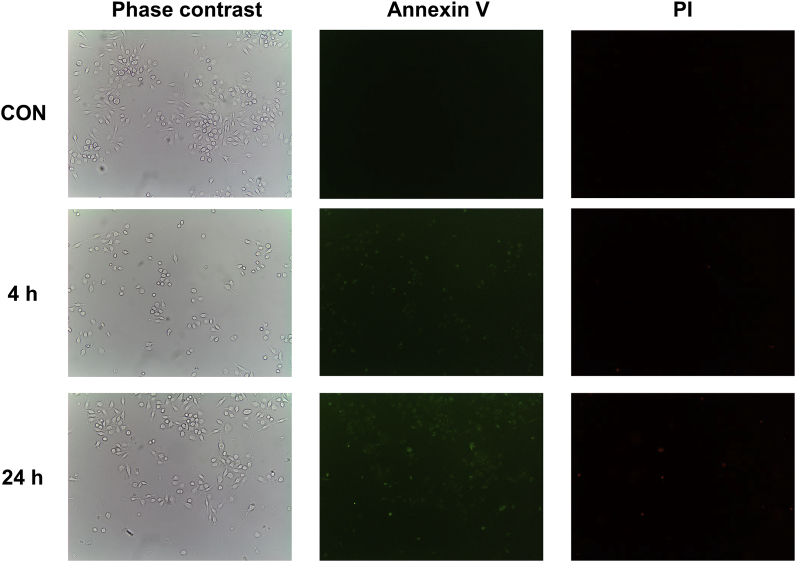
BQ-Trx interactions were studied further with J774A.1 cells. Different concentrations of BQ (2–80 μM) were used to treat cells (8 × 103 cells well−1) for 24 h, before assessment of cellular metabolic activity using the MTS assay. As shown in Fig. 5A, BQ induced a dose-dependent decrease in cellular metabolic activity, with a significant loss detected for the 20, 40 and 80 μM treatment groups. To determine whether this was associated with the induction of apoptosis, cells were dual stained with Annexin V and PI and imaged by fluorescence microscopy. The data obtained indicate that after 4 h incubation, BQ induced significant cellular apoptosis, as detected by fluorescence in FITC channel, with longer incubation times (up to 24 h) increasing the fluorescence intensity and hence extent of apoptosis (Fig. 6).
Fig. 5.
Cell metabolic activity assay and intracellular total thiol measurement. J774A.1 cells (8 × 103 cells well−1) were exposed for 24 h to different concentrations of BQ (2–80 μM). Cell viability was determined by MTS assay (A). J774A.1 cells (1 × 105 cells well−1) were exposed for 1 h to different concentrations of BQ (2–80 μM). Total intracellular thiol level was measured using the ThioGlo-1 assay (B), with levels normalized to the total protein concentration. *p < 0.05, **p < 0.01 vs. control (CON) samples with no added BQ.
Fig. 6.
BQ-induced apoptosis identification. J774A.1 cells (8 × 103 cells well−1) were incubated with BQ (20 μM) for 4 h and 24 h, then the cells were washed with binding buffer and stained with Annexin-V and propidium iodide (PI) for 20 min. Images were captured by a phase contrast inverted microscope. Representative images from 3 independent experiments are shown in the panels.
To investigate possible pathways associated with this induction of apoptosis, cells were incubated with BQ for 1 h, and total cellular thiol concentrations were quantified. BQ treatment significantly decreased intracellular thiol levels, consistent with a rapid perturbation of cellular redox balance after BQ treatment (Fig. 5B). This imbalance may act as the trigger for apoptosis.
3.6. Effect of BQ on antioxidant systems in J774A.1 cells
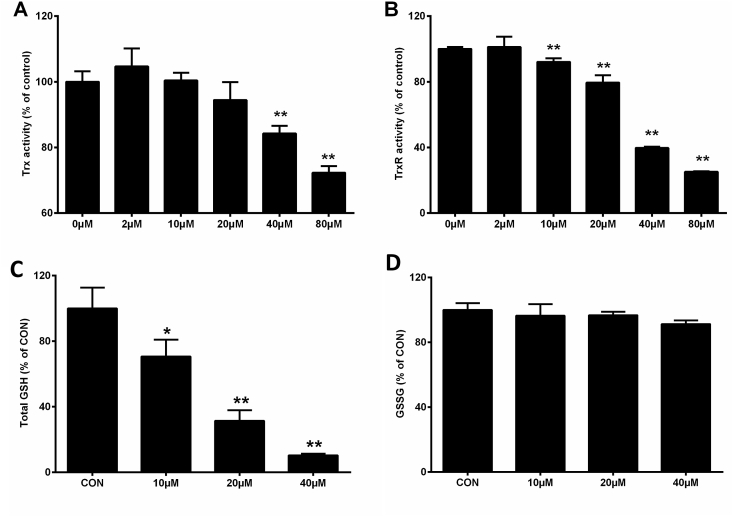
The effect of BQ on the activity of Trx and TrxR in J774A.1 cells was investigated. The data indicate that BQ reduced the activity of both Trx (Fig. 7A) and TrxR (Fig. 7B) after 1 h treatment. Concomitant studies examined the intracellular concentrations of GSH and GSSG by an enzymatic recycling method. As shown in Fig. 7C, BQ treatment for 30 min significantly decreased the total GSH level, without a corresponding increase in GSSG concentration (Fig. 7D). These data suggest that the major pathway to loss of GSH is via formation of GSH-BQ adducts, which cannot be reduced by the GR, rather than via redox cycling and oxidant formation. This conclusion is supported by data obtained from experiments which compared the rate of TNB formation from DTNB by untreated GSH, and GSH pretreated with BQ, with the latter showing much slower reaction kinetics (Supplementary Fig. 1). The GSH system also appears to be more vulnerable to BQ than the Trx system, as 20 μM BQ dramatically decreased the concentration of GSH while having little effect on Trx activity.
Fig. 7.
Effect of BQ on antioxidant systems in J774A.1 cells. Cells (1 × 105 cells well−1) were exposed to different concentrations of BQ (2–80 μM) for 1 h, then harvested and lysed. Trx activity was measured by insulin reduction assay (A) and TrxR activity was measured by the DTNB assay (B). Cells were incubated with different concentrations of BQ (2–80 μM) for 30 min, the intracellular total GSH (C) and GSSG (D) level were measured by an enzymatic recycling method. *p < 0.05, **p < 0.01 vs. control (CON) samples with no added BQ.
3.7. BQ-induced quinoprotein formation in J774A.1 cells
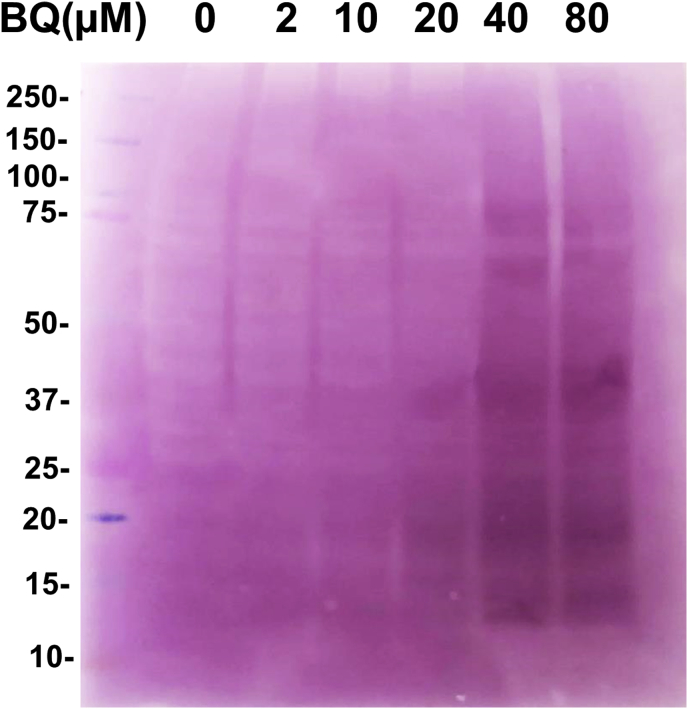
BQ binds to isolated proteins to produce quinoprotein adducts as detected by NBT redox staining (Fig. 1). To test whether this also occurs in cells, and especially in the presence of GSH, J774A.1 cells were exposed to varying concentrations of BQ (2–80 μM) for 1 h followed by analysis for the presence of quinoproteins (Fig. 8). The data obtained indicate that BQ binds to multiple intracellular proteins, with at least five bands detected with 40 and 80 μM BQ. The identity of these proteins has not been determined due to the technical complexity of proteomic analysis of whole cell lysates separated on 1-dimensional gels.
Fig. 8.
Quinoprotein detection in J774A.1 cells. Cells (1 × 106 cells well−1) were incubated with different concentrations of BQ (2–80 μM) for 1 h. Cells were harvested and lysed in RIPA buffer containing protease and phosphatase inhibitor cocktail. Then samples were analysed by SDS-PAGE followed by NBT redox staining. A representative image from 3 independent experiments is shown.
3.8. Effect of GSH-depletion on BQ-induced cell viability loss and Trx system impairment
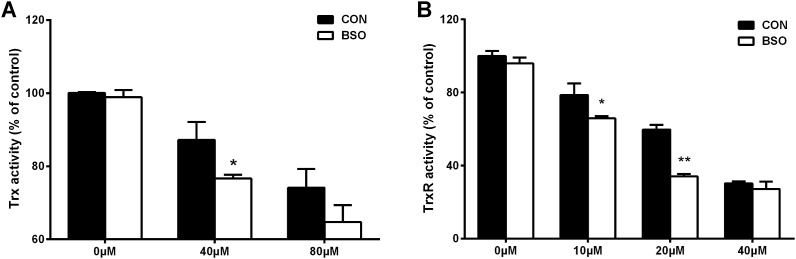
Previous studies have provided evidence that the GSH system can substitute for TrxR, when the latter is eliminated, to reduce Trx thus preventing cell death [17]. The cross-talk of these two systems was therefore studied here with regard to protection against BQ-induced damage. Intracellular GSH was depleted by treating cells with 25 μM l-buthionine-sulfoximine (BSO) for 24 h, and then cells were exposed to different concentrations of BQ for 1 h. Following exposure to BQ, BSO pre-treated cells had lower Trx and TrxR activity (Fig. 9), suggesting that the GSH system can supplement the role of TrxR in protecting Trx from BQ-induced damage.
Fig. 9.
Effect of GSH depletion on the activity of the Trx system in J774A.1 cells. Cells (1 × 105 cells well−1) were treated with 25 μM BSO for 24 h, and then incubated with different concentrations of BQ (10–80 μM) for another 1 h. Trx activity was measured using the insulin reduction assay (A), TrxR activity was measured by the DTNB assay (B). *p < 0.05, **p < 0.01 vs. control (CON) samples with no BSO pretreatment. The relative activities for all groups are referenced to the control group without BSO and BQ treatment (indicated as CON; 0 μM, black bars), with this value taken as 100%.
3.9. BQ-mediated activation of ASK1/p38 signalling pathway
Trx can serve as a negative regulator of ASK1 by directly binding with it via an inter-protein disulfide bond [30]. We therefore postulated that BQ might impair this function of Trx system, by modifying the critical Trx Cys residues, and hence that BQ might activate ASK1 and the downstream MAPK signalling pathway to promote cell apoptosis. Immunoblotting studies clearly indicate the phosphorylation of ASK1 at Thr838 upon treatment of cells with BQ (5 μM) for 15 min (Fig. 10A and B), consistent with the decreased Trx activity reported above. This activation of ASK1 also resulted in increased phosphorylation of the downstream MAPK protein, with a significant increase in the phosphorylation of p38-MAPK detected. In contrast, no obvious activation of JNK was detected (Fig. 10C–E), though positive controls with H2O2 showed marked increases. This last observation also suggests that the concentrations of BQ employed do not give rise to marked intracellular concentrations of H2O2 via redox cycling.
Fig. 10.
Activation of ASK1/p38-MAPK signalling pathway in J774A.1 cells. Cells (1 × 106 cells well−1) were incubated with different concentrations of BQ (2 and 5 μM) for 15 min, and then cells were harvested and lysed in RIPA buffer containing protease and phosphatase inhibitor cocktail. Samples were analysed by immunoblotting to quantify activation of ASK1 by the phosphorylation of Thr838 (A and B). Cells were incubated with different concentrations of BQ (5 and 10 μM) for 30 min, and then cells were harvested and lysed in RIPA buffer containing a protease and phosphatase inhibitor cocktail. Samples were analysed by immunoblotting (representative images from 3 independent experiments are shown in (A) and (C) and activation of JNK and p38-MAPK measured by the phosphorylation of Thr183/Tyr185 and Thr180/Tyr182, respectively, quantified in panels (B), (D) and (E). The phosphorylated protein levels were normalized to the total of the phosphorylated and unphosphorylated) forms to give the ratio shown in the panels. Levels of β-actin were also determined as a loading controls to confirm equal protein loading (data not shown). H2O2 (300 μM) was used as a positive control. *p < 0.05, **p < 0.01 vs. control (CON) samples with no added BQ.
4. Discussion
The strong electrophilic property of BQ results in rapid reactions with proteins to form Michael adducts. Previous studies have reported, for example, covalent adduction to serum albumin [3,9], haemoglobin [31], α-crystallin [32], GAPDH [9], creatine kinase [9], papain [9] and TrxR [10] with downstream effects on protein integrity and function. Evidence has been presented for a concomitant loss of enzymatic activity for GAPDH and creatine kinase, as a result of modifications at the catalytic Cys residues, and for TrxR as a result of adduction at the Sec or Cys residues [9,10]. Trx has a conserved active site -Cys-Gly-Pro-Cys-motif, which mediates electron transfer to substrates. These Cys residues bind ASK1, with this resulting in a negative regulation of its function. Previous studies have shown that oxidation of the active site Cys residues in Trx results in the release and activation of ASK1, increased MAPK activity and the induction of apoptosis [21]. Based on these data, we hypothesized that BQ might covalently bind to these Cys residues, and thereby change the nature of the protein, modulate the activity of Trx, and induce apoptosis.
BQ induced a rapid and concentration-dependent inhibition of purified Trx1 activity, with a 4-fold excess of BQ treatment inducing >90% loss of activity (Fig. 1A). This is consistent with the Cys labelling data obtained with BEI, where the same excess of BQ fully blocked the Cys residue (Fig. 1C), and the presence of 5 Cys residues in human Trx (ExPASy entry P10599). The MS analyses also yielded data consistent with BQ adduction (rather than oxidation) at both the catalytic Cys residues, and the other three Cys residues in the sequence (Fig. 2 and Table 1), and the NBT staining indicated the presence of quinoproteins after BQ treatment (Fig. 1B). These data suggest that BQ binds covalently to Trx1 at each of the Cys residues, including those in catalytic centre, resulting in enzyme inactivation. Modification of Trx Cys residues, and associated inactivation, has been reported for other electrophiles. Thus, (−)-epigallocatechin-3-gallate (EGCG) has been reported to selectively modify Cys32 of Trx1 after oxidation [33], and a covalent adduct has been reported between 4-hydroxy-2-nonenal (HNE) and Trx, with the catalytic Cys residues (Cys32 and Cys35) being the primary targets [34]. The cytotoxicity of HNE has therefore been proposed to arise from Trx inhibition [34].
BQ modification of Trx results in oligomer formation and cross-linking, as observed with other proteins [9,10], with cross-links detected between Trx1 molecules (Fig. 3). The active site Cys32 and Cys35 residues of Trx are partly exposed on the protein surface (Fig. 11), allowing reduction of client proteins. This accessibility, which is a required for enzymatic activity, also makes these residues susceptible to modification. Of these two residues, Cys32 has a lower pKa value [35], and has been reported to be more vulnerable to alteration; whether this is also true with BQ cannot be determined from the current data as both Cys residues appear in the same peptide after Lys-C digestion.
Fig. 11.
Location of Cys residues in human Trx1 (PDB structure: 1ert). Crystal structure data rendered using the software program Jmol, using 100% van der Waals spheres, with the two views of the protein structure rotated by 180° around the vertical axis. Cys residues numbered from the protein sequence with Cys32 and Cys35 being the catalytic site pair present in both mammals and Escherichia coli. The latter form lacks the Cys residues present at positions analogous to 62, 69 and 73.
Treatment of Trx with equimolar or a two-fold excess of BQ, generated predominantly non-reducible dimers (Fig. 3). This is inconsistent with the formation of inter-molecular disulfides as detected with reactive oxidants [35]. However, previous studies have reported BQ-induced dimerization of annexin V via a quinone bridge [36]. This can arise from initial Michael addition to form a catechol adduct, oxidation of this adduct to a quinoprotein, and then a second Michael addition with another protein molecule. We therefore postulate that Cys32 and Cys35 are major targets for BQ, with modification of these two Cys residues giving Trx1 homodimers involving a quinone bridge. Whether this involves two Cys32 residues, two Cys35 residues, or a mixed inter-protein Cys32-Cys35 species is unknown and warrants further investigation. The involvement of Cys32 and/or Cys35 is supported by the detection of cross-links with the E. coli protein that only contains these Cys residues, and not the three additional Cys residues present in the human protein [29]. The non-reducible cross-links detected for the E. coli protein (Fig. 3C vs. D) are consistent with similar quinone-bridged dimers.
With higher excesses of BQ (>2-fold, Fig. 3A and B) formation of reducible dimers was also detected for human Trx. These may be disulfide linked species, or quinone-linked species that are susceptible to S-transarylation [37] repair reactions. Whether these reversible adducts involve Cys32 and/or Cys35 cannot be determined for the human protein, but the data obtained with the E. coli protein suggest that disulfide cross-links that involve only these residues (i.e. Cys32-Cys32, Cys35-Cys35 or Cys32-Cys35) are not formed. However, we cannot exclude disulfide cross-links involving one of these residues and another Cys residue in the human protein.
Of the non-catalytic Cys residues in human Trx1, Cys73 is closest to the catalytic centre and surface exposed (Fig. 11) [38]. It has been reported that homodimers formed between two Cys73 residues make the catalytic centre inaccessible to TrxR, thereby inhibiting Trx activity [39]. Previous studies have also proposed that Cys73 is a primary target for modification [[39], [40], [41]]. However, the detection of higher oligomers (trimers, tetramers) suggest that Cys73 cannot be the only site through which inter-protein disulfides are generated. The formation of these higher oligomers also appears to be associated with lower levels of quinoprotein formation (and hence potential further reaction) as detected using NBT redox staining (Fig. 1B).
GSH modulates BQ adduction to Trx1 and its consequences. This protection appears to occur mainly via competitive conjugation to give GSH-BQ adducts. Co-incubation of GSH and Trx1 with BQ resulted in lower levels of thiol depletion on Trx, decreased cross-link formation, and diminished loss of Trx1 activity (Fig. 4). This competition between GSH and Trx1 for BQ will depend, at least in part, on the concentrations of these compounds and their relative rates of reaction with BQ. Whilst the GSH concentration in cells is typically 1–5 mM [42], the cellular concentration of Trx appears to be more variable and cell-type dependent [43,44]. Recent (minimally-invasive) NMR analyses indicate that normal physiological levels of Trx are 10–70 μM, with this dependent on external factors/stimuli [43], and much lower than for GSH. We have reported kinetic data for the reaction of GSH with BQ, with k2 ~ 6 × 105 M−1 s−1 at 10 °C and pH 7.0 [9]. Corresponding data for Trx are not yet available, but the data obtained here indicate that slightly lower than stoichiometric, or equal concentrations of GSH relative to Trx1, provide significant protection (as measured by loss of activity, Cys depletion and aggregate formation), suggesting that the rate constant for reaction of BQ with Trx1 is likely to be of a similar magnitude to that for GSH (i.e. close to 1–5 × 105 M−1 s−1 at 10 °C, pH 7.0). This value is significantly (~10-fold) higher than for the other proteins reported to date [9]. These data suggest that within cells reaction of BQ with GSH is a major reaction. This conclusion is supported by the cell data, where BQ treatment induced a more extensive loss of GSH, than Trx and TrxR activity, though reaction with both Trx and TrxR can also occur to a significant extent, and have biological consequences (Fig. 7 and [10]). Depletion of GSH (using BSO) resulted in an increased loss of Trx and TrxR activity (Fig. 9), indicating that reaction of BQ with Trx and TxrR is of particular significance in cells where GSH is depleted. Whilst GSH is often viewed as a key defence against intracellular oxidants [45], with resulting formation of GSSG, the latter was not detected in the current studies, despite marked GSH loss. These data indicate that GSH protects Trx and TrxR by direct conjugation with BQ, rather than via prevention of redox cycling of BQ, and consequent formation of semiquinone and superoxide radicals, and H2O2. In the light of these data, we propose that BQ-induced cytotoxicity may arise primarily via direct adduction to intracellular proteins, and subsequent modulation of protein function, rather than via redox cycling and radical/peroxide formation.
GSH has been reported to reverse quinone-induced enzyme inactivation via S-transarylation [46], and we have reported GSH-mediated reversal of BQ-induced damage to some (e.g. GAPDH and papain) but not all proteins [9,10]. In the current study, addition of GSH to Trx1 pre-treated with BQ did not reverse either protein Cys consumption or activity loss, suggesting the S-transarylation may be selective to certain proteins and is not a ubiquitous process. This potential selectivity warrants further study, and may be due to steric or electronic factors.
Trx regulates ASK1 in a negative manner via binding at Cys32 and/or Cys35 [47]. It was therefore of interest to examine whether BQ modulated this process in cells, and whether this resulted in phosphorylation of ASK1 at Thr838 (Fig. 7A). Thr838 is present in the catalytic domain of ASK1, with phosphorylation resulting in full activation [48], and subsequent induction of the mitogen-activated protein kinase kinase (MKK) 4/7-JNK and MKK3/6-p38 pathways [49]. BQ induced activation of the p38-MAPK, but not the JNK, pathway while H2O2 (used as a positive control) activated both (Fig. 10); this effect of H2O2 arises from Trx oxidation and dissociation of ASK1 from the ASK1-Trx complex [50]. These differences between BQ and H2O2, confirm that BQ has a differential mode of action, independent of H2O2 generation, with this ascribed to direct modifications at the Trx catalytic centre, and activation of specific downstream signalling pathways.
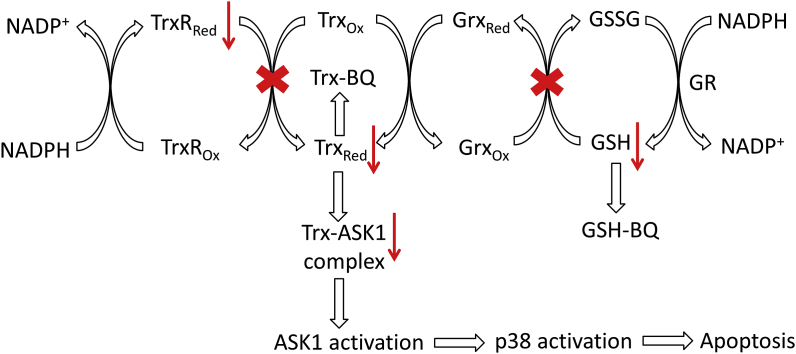
The findings reported here are summarized in Fig. 12. BQ treatment results in inhibition of Trx and TrxR activity as well as depleting GSH levels. The toxic effects of BQ on cells are attributed to the electrophilic property of BQ, which can covalently bind with GSH and Trx. The modification of Cys residues in Trx results in the release of ASK1 from the ASK1-Trx complex with subsequent activation of ASK1, the p38-MAPK pathway and cell apoptosis. BQ is a relatively non-specific modifying agent which generates adducts at all Trx1 Cys residues, with this resulting in protein cross-linking and enzyme inactivation. In macrophages, this ultimately results in apoptosis. This work suggested a new mechanism of ASK1 activation, and indicates that the ASK1-p38-MAPK pathway is important in quinone-induced cytotoxicity.
Fig. 12.
Proposed mechanism of interaction between BQ and Trx1 leading to ASK1/p38-MAPK signalling pathway activation. The red-cross indicates the inhibition effect, and the down arrow indicates the level decrease. Abbreviations: Trx, thioredoxin; TrxR, thioredoxin reductase, Grx, glutaredoxin; GSH, glutathione; GSSG, glutathione disulfide; GR, glutathione reductase; Ox, oxidized; Red, reduced; ASK1, apoptosis signal-regulating kinase 1. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
Declaration of competing interest
The authors declare no conflicts of interest with regard to the data presented.
Acknowledgements
The authors are grateful for financial support from the Novo Nordisk Foundation (Laureate Research Grant NNF13OC0004294 to MJD) and the China Scholarship Council (PhD scholarships to NS and HC).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101400.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Bolton J.L., Trush M.A., Penning T.M., Dryhurst G., Monks T.J. Role of quinones in toxicology. Chem. Res. Toxicol. 2000;13(3):135–160. doi: 10.1021/tx9902082. [DOI] [PubMed] [Google Scholar]
- 2.Monks T.J., Hanzlik R.P., Cohen G.M., Ross D., Graham D.G. Quinone chemistry and toxicity. Toxicol. Appl. Pharmacol. 1992;112(1):2–16. doi: 10.1016/0041-008x(92)90273-u. [DOI] [PubMed] [Google Scholar]
- 3.Ghosh A., Choudhury A., Das A., Chatterjee N.S., Das T., Chowdhury R., Panda K., Banerjee R., Chatterjee I.B. Cigarette smoke induces p-benzoquinone–albumin adduct in blood serum: implications on structure and ligand binding properties. Toxicology. 2012;292(2–3):78–89. doi: 10.1016/j.tox.2011.11.014. [DOI] [PubMed] [Google Scholar]
- 4.Srinivasan A., Robertson L., Ludewig G. Sulfhydryl binding and topoisomerase inhibition by PCB metabolites. Chem. Res. Toxicol. 2002;15(4):497–505. doi: 10.1021/tx010128+. [DOI] [PubMed] [Google Scholar]
- 5.O'brien P. Molecular mechanisms of quinone cytotoxicity. Chem. Biol. Interact. 1991;80(1):1–41. doi: 10.1016/0009-2797(91)90029-7. [DOI] [PubMed] [Google Scholar]
- 6.Stokes A.H., Hastings T.G., Vrana K.E. Cytotoxic and genotoxic potential of dopamine. J. Neurosci. Res. 1999;55(6):659–665. doi: 10.1002/(SICI)1097-4547(19990315)55:6<659::AID-JNR1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 7.Klopčič I., Dolenc M.S. Chemicals and drugs forming reactive quinone and quinone imine metabolites. Chem. Res. Toxicol. 2018;32(1):1–34. doi: 10.1021/acs.chemrestox.8b00213. [DOI] [PubMed] [Google Scholar]
- 8.Le Bourvellec C., Renard C. Interactions between polyphenols and macromolecules: quantification methods and mechanisms. Crit. Rev. Food Sci. Nutr. 2012;52(3):213–248. doi: 10.1080/10408398.2010.499808. [DOI] [PubMed] [Google Scholar]
- 9.Shu N., Lorentzen L.G., Davies M.J. Reaction of quinones with proteins: kinetics of adduct formation, effects on enzymatic activity and protein structure, and potential reversibility of modifications. Free Radic. Biol. Med. 2019;137:169–180. doi: 10.1016/j.freeradbiomed.2019.04.026. [DOI] [PubMed] [Google Scholar]
- 10.Shu N., Cheng Q., Arner E.S.J., Davies M.J. Inhibition and crosslinking of the selenoprotein thioredoxin reductase-1 by p-benzoquinone. Redox Biol. 2020;28 doi: 10.1016/j.redox.2019.101335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li Y., Jongberg S., Andersen M.L., Davies M.J., Lund M.N. Quinone-induced protein modifications: kinetic preference for reaction of 1, 2-benzoquinones with thiol groups in proteins. Free Radic. Biol. Med. 2016;97:148–157. doi: 10.1016/j.freeradbiomed.2016.05.019. [DOI] [PubMed] [Google Scholar]
- 12.Boots A.W., Haenen G.R., den Hartog G.J., Bast A. Oxidative damage shifts from lipid peroxidation to thiol arylation by catechol-containing antioxidants. Biochim. Biophys. Acta Mol. Cell Biol. Lipids. 2002;1583(3):279–284. doi: 10.1016/s1388-1981(02)00247-0. [DOI] [PubMed] [Google Scholar]
- 13.Meier B.W., Gomez J.D., Kirichenko O.V., Thompson J.A. Mechanistic basis for inflammation and tumor promotion in lungs of 2, 6-di-tert-butyl-4-methylphenol-treated mice: electrophilic metabolites alkylate and inactivate antioxidant enzymes. Chem. Res. Toxicol. 2007;20(2):199–207. doi: 10.1021/tx060214f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chan K., Jensen N., O'Brien P.J. Structure–activity relationships for thiol reactivity and rat or human hepatocyte toxicity induced by substituted p‐benzoquinone compounds. J. Appl. Toxicol. 2008;28(5):608–620. doi: 10.1002/jat.1312. [DOI] [PubMed] [Google Scholar]
- 15.Lu J., Holmgren A. The thioredoxin antioxidant system. Free Radic. Biol. Med. 2014;66:75–87. doi: 10.1016/j.freeradbiomed.2013.07.036. [DOI] [PubMed] [Google Scholar]
- 16.Matsuzawa A. Thioredoxin and redox signaling: roles of the thioredoxin system in control of cell fate. Arch. Biochem. Biophys. 2017;617:101–105. doi: 10.1016/j.abb.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 17.Du Y., Zhang H., Lu J., Holmgren A. Glutathione and glutaredoxin act as a backup of human thioredoxin reductase 1 to reduce thioredoxin 1 preventing cell death by aurothioglucose. J. Biol. Chem. 2012;287(45):38210–38219. doi: 10.1074/jbc.M112.392225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tan S.-X., Greetham D., Raeth S., Grant C.M., Dawes I.W., Perrone G.G. The thioredoxin-thioredoxin reductase system can function in vivo as an alternative system to reduce oxidized glutathione in Saccharomyces cerevisiae. J. Biol. Chem. 2010;285(9):6118–6126. doi: 10.1074/jbc.M109.062844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ichijo H., Nishida E., Irie K., ten Dijke P., Saitoh M., Moriguchi T., Takagi M., Matsumoto K., Miyazono K., Gotoh Y. Induction of apoptosis by ASK1, a mammalian MAPKKK that activates SAPK/JNK and p38 signaling pathways. Science. 1997;275(5296):90–94. doi: 10.1126/science.275.5296.90. [DOI] [PubMed] [Google Scholar]
- 20.Ichijo H. From receptors to stress-activated MAP kinases. Oncogene. 1999;18(45):6087. doi: 10.1038/sj.onc.1203129. [DOI] [PubMed] [Google Scholar]
- 21.Lu J., Holmgren A. Thioredoxin system in cell death progression. Antioxidants Redox Signal. 2012;17(12):1738–1747. doi: 10.1089/ars.2012.4650. [DOI] [PubMed] [Google Scholar]
- 22.Branco V., Coppo L., Solá S., Lu J., Rodrigues C.M., Holmgren A., Carvalho C. Impaired cross-talk between the thioredoxin and glutathione systems is related to ASK-1 mediated apoptosis in neuronal cells exposed to mercury. Redox biology. 2017;13:278–287. doi: 10.1016/j.redox.2017.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Niso-Santano M., González-Polo R.A., Bravo-San Pedro J.M., Gómez-Sánchez R., Lastres-Becker I., Ortiz-Ortiz M.A., Soler G., Morán J.M., Cuadrado A., Fuentes J.M. Activation of apoptosis signal-regulating kinase 1 is a key factor in paraquat-induced cell death: modulation by the Nrf2/Trx axis. Free Radic. Biol. Med. 2010;48(10):1370–1381. doi: 10.1016/j.freeradbiomed.2010.02.024. [DOI] [PubMed] [Google Scholar]
- 24.Arnér E.S., Holmgren A. Measurement of thioredoxin and thioredoxin reductase. Curr. Protoc. Toksikol. 2005;24(1) doi: 10.1002/0471140856.tx0704s05. 7.4. 1-7.4. 14. [DOI] [PubMed] [Google Scholar]
- 25.Rahmanto A.S., Pattison D.I., Davies M.J. Photo-oxidation-induced inactivation of the selenium-containing protective enzymes thioredoxin reductase and glutathione peroxidase. Free Radic. Biol. Med. 2012;53(6):1308–1316. doi: 10.1016/j.freeradbiomed.2012.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Shevchenko A., Wilm M., Vorm O., Mann M. Mass spectrometric sequencing of proteins from silver-stained polyacrylamide gels. Anal. Chem. 1996;68(5):850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- 27.Paz M., Flückiger R., Boak A., Kagan H., Gallop P.M. Specific detection of quinoproteins by redox-cycling staining. J. Biol. Chem. 1991;266(2):689–692. [PubMed] [Google Scholar]
- 28.Rahman I., Kode A., Biswas S.K. Assay for quantitative determination of glutathione and glutathione disulfide levels using enzymatic recycling method. Nat. Protoc. 2006;1(6):3159. doi: 10.1038/nprot.2006.378. [DOI] [PubMed] [Google Scholar]
- 29.Jeng M.-F., Campbell A.P., Begley T., Holmgren A., Case D.A., Wright P.E., Dyson H.J. High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin. Structure. 1994;2(9):853–868. doi: 10.1016/s0969-2126(94)00086-7. [DOI] [PubMed] [Google Scholar]
- 30.Saitoh M., Nishitoh H., Fujii M., Takeda K., Tobiume K., Sawada Y., Kawabata M., Miyazono K., Ichijo H. Mammalian thioredoxin is a direct inhibitor of apoptosis signal‐regulating kinase (ASK) 1. EMBO J. 1998;17(9):2596–2606. doi: 10.1093/emboj/17.9.2596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mitra A., Mandal A.K. Conjugation of para-benzoquinone of cigarette smoke with human hemoglobin leads to unstable tetramer and reduced cooperative oxygen binding. J. Am. Soc. Mass Spectrom. 2018;29(10):2048–2058. doi: 10.1007/s13361-018-2011-1. [DOI] [PubMed] [Google Scholar]
- 32.Chowdhury A., Choudhury A., Chakraborty S., Ghosh A., Banerjee V., Ganguly S., Bhaduri G., Banerjee R., Das K., Chatterjee I.B. p-Benzoquinone-induced aggregation and perturbation of structure and chaperone function of α-crystallin is a causative factor of cigarette smoke-related cataractogenesis. Toxicology. 2018;394:11–18. doi: 10.1016/j.tox.2017.11.018. [DOI] [PubMed] [Google Scholar]
- 33.Zhang H., Cao D., Cui W., Ji M., Qian X., Zhong L. Molecular bases of thioredoxin and thioredoxin reductase-mediated prooxidant actions of (−)-epigallocatechin-3-gallate. Free Radic. Biol. Med. 2010;49(12) doi: 10.1016/j.freeradbiomed.2010.09.031. [DOI] [PubMed] [Google Scholar]
- 34.Fang J., Holmgren A. Inhibition of thioredoxin and thioredoxin reductase by 4-hydroxy-2-nonenal in vitro and in vivo. J. Am. Chem. Soc. 2006;128(6):1879–1885. doi: 10.1021/ja057358l. [DOI] [PubMed] [Google Scholar]
- 35.Holmgren A. Thioredoxin structure and mechanism: conformational changes on oxidation of the active-site sulfhydryls to a disulfide. Structure. 1995;3(3):239–243. doi: 10.1016/s0969-2126(01)00153-8. [DOI] [PubMed] [Google Scholar]
- 36.Yu P., Strug I., Cafarella T.R., Seaton B.A., Krantz A. Site-specific crosslinking of annexin proteins by 1, 4-benzoquinone: a novel crosslinker for the formation of protein dimers and diverse protein conjugates. Org. Biomol. Chem. 2012;10(23):4500–4504. doi: 10.1039/c2ob25460c. [DOI] [PubMed] [Google Scholar]
- 37.Toyama T., Shinkai Y., Yazawa A., Kakehashi H., Kaji T., Kumagai Y. Glutathione-mediated reversibility of covalent modification of ubiquitin carboxyl-terminal hydrolase L1 by 1, 2-naphthoquinone through Cys152, but not Lys4. Chem. Biol. Interact. 2014;214:41–48. doi: 10.1016/j.cbi.2014.02.008. [DOI] [PubMed] [Google Scholar]
- 38.Forman-Kay J.D., Clore G.M., Wingfield P.T., Gronenborn A.M. High-resolution three-dimensional structure of reduced recombinant human thioredoxin in solution. Biochemistry. 1991;30(10):2685–2698. doi: 10.1021/bi00224a017. [DOI] [PubMed] [Google Scholar]
- 39.Weichsel A., Gasdaska J.R., Powis G., Montfort W.R. Crystal structures of reduced, oxidized, and mutated human thioredoxins: evidence for a regulatory homodimer. Structure. 1996;4(6):735–751. doi: 10.1016/s0969-2126(96)00079-2. [DOI] [PubMed] [Google Scholar]
- 40.Gasdaska J.R., Kirkpatrick D.L., Montfort W., Kuperus M., Hill S.R., Berggren M., Powis G. Oxidative inactivation of thioredoxin as a cellular growth factor and protection by a Cys73→ Ser mutation. Biochem. Pharmacol. 1996;52(11):1741–1747. doi: 10.1016/s0006-2952(96)00595-3. [DOI] [PubMed] [Google Scholar]
- 41.Shinkai Y., Iwamoto N., Miura T., Ishii T., Cho A.K., Kumagai Y. Redox cycling of 1, 2-naphthoquinone by thioredoxin1 through Cys32 and Cys35 causes inhibition of its catalytic activity and activation of ASK1/p38 signaling. Chem. Res. Toxicol. 2012;25(6):1222–1230. doi: 10.1021/tx300069r. [DOI] [PubMed] [Google Scholar]
- 42.Meister A., Anderson M.E. Glutathione. Annu. Rev. Biochem. 1983;52:711–760. doi: 10.1146/annurev.bi.52.070183.003431. [DOI] [PubMed] [Google Scholar]
- 43.Mochizuki A., Saso A., Zhao Q., Kubo S., Nishida N., Shimada I. Balanced regulation of redox status of intracellular thioredoxin revealed by in-cell NMR. J. Am. Chem. Soc. 2018;140(10):3784–3790. doi: 10.1021/jacs.8b00426. [DOI] [PubMed] [Google Scholar]
- 44.Spector A., Yan G.Z., Huang R.R., McDermott M.J., Gascoyne P.R., Pigiet V. The effect of H2O2 upon thioredoxin-enriched lens epithelial cells. J. Biol. Chem. 1988;263(10):4984–4990. [PubMed] [Google Scholar]
- 45.Forman H.J., Zhang H., Rinna A. Glutathione: overview of its protective roles, measurement, and biosynthesis. Mol. Asp. Med. 2009;30(1–2):1–12. doi: 10.1016/j.mam.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Miura T., Kakehashi H., Shinkai Y., Egara Y., Hirose R., Cho A.K., Kumagai Y. GSH-mediated S-transarylation of a quinone glyceraldehyde-3-phosphate dehydrogenase conjugate. Chem. Res. Toxicol. 2011;24(11):1836–1844. doi: 10.1021/tx200025y. [DOI] [PubMed] [Google Scholar]
- 47.Liu Y., Min W. Thioredoxin promotes ASK1 ubiquitination and degradation to inhibit ASK1-mediated apoptosis in a redox activity-independent manner. Circ. Res. 2002;90(12):1259–1266. doi: 10.1161/01.res.0000022160.64355.62. [DOI] [PubMed] [Google Scholar]
- 48.Liu H., Nishitoh H., Ichijo H., Kyriakis J.M. Activation of apoptosis signal-regulating kinase 1 (ASK1) by tumor necrosis factor receptor-associated factor 2 requires prior dissociation of the ASK1 inhibitor thioredoxin. Mol. Cell. Biol. 2000;20(6):2198–2208. doi: 10.1128/mcb.20.6.2198-2208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sui X., Kong N., Ye L., Han W., Zhou J., Zhang Q., He C., Pan H. p38 and JNK MAPK pathways control the balance of apoptosis and autophagy in response to chemotherapeutic agents. Cancer Lett. 2014;344(2):174–179. doi: 10.1016/j.canlet.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 50.Katagiri K., Matsuzawa A., Ichijo H. Regulation of apoptosis signal-regulating kinase 1 in redox signaling. Methods Enzymol. 2010;474:277–288. doi: 10.1016/S0076-6879(10)74016-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.