Key Points
The tetraspanin CD53 is required for normal B cell development.
CD53 promotes B cell development by maintaining IL-7R signaling.
CD53 functions cell autonomously during B lymphopoiesis.
Abstract
The tetraspanin CD53 has been implicated in B cell development and function. CD53 is a transcriptional target of EBF1, a critical transcription factor for early B cell development. Further, human deficiency of CD53 results in recurrent infections and reduced serum Igs. Although prior studies have indicated a role for CD53 in regulating mature B cells, its role in early B cell development is not well understood. In this study, we show that CD53 expression, which is minimal on hematopoietic stem and progenitor cells, increases throughout bone marrow B cell maturation, and mice lacking CD53 have significantly decreased bone marrow, splenic, lymphatic, and peripheral B cells. Mixed bone marrow chimeras show that CD53 functions cell autonomously to promote B lymphopoiesis. Cd53−/− mice have reduced surface expression of IL-7Rα and diminished phosphatidylinositol 3 kinase and JAK/STAT signaling in prepro- and pro-B cells. Signaling through these pathways via IL-7R is essential for early B cell survival and transition from the pro-B to pre-B cell developmental stage. Indeed, we find increased apoptosis in developing B cells and an associated reduction in pre-B and immature B cell populations in the absence of CD53. Coimmunoprecipitation and proximity ligation studies demonstrate physical interaction between CD53 and IL-7R. Together, these data, to our knowledge, suggest a novel role for CD53 during IL-7 signaling to promote early B cell differentiation.
Introduction
B lymphopoiesis follows a series of well-defined, highly regulated processes to confer broad immunity to foreign pathogens and simultaneously prevent self-recognition (1, 2). Developing B cells depend on extracellular cues to facilitate maturation from the common lymphoid progenitor (CLP) to a mature plasma cell (3, 4). B cell development begins in the bone marrow, but emigration from the marrow to the spleen is required for complete differentiation. Commitment to B cell lineage during transition from CLP to prepro–B cell requires IL-7 to induce expression of early B cell factor 1 (EBF1) (5). EBF1, along with E2A and PU.1, directs the expression of necessary B cell transcription factors, including Pax5, to specify and commit prepro–B cells to the B lineage (6, 7). The next developmental hurdle is the formation of a functional pre-BCR through the pro–B and pre–B cell stages (8). Signaling through the pre-BCR on pre–B cells initiates formation of the mature BCR, which drives the transition to IgM-expressing immature B cells. Immature B cells then egress from the bone marrow, enter circulation, and migrate to the spleen, where they form transitional B cells. Transitional B cells continue to mature and differentiate into follicular (FO) or marginal zone (MZ) B cells, which are both capable of forming immunocompetent B cells (9).
In mice, IL-7 signaling is necessary for B cell development, specifically to promote progression from a pro–B cell to the pre–B cell developmental stage. IL-7−/− and IL-7R−/− mice have a significant impairment of early lymphocyte expansion and progression past the pro–B cell stage (10, 11). Ligation of IL-7 to its receptor activates phosphatidylinositol 3 kinase (PI3K) and Janus-associated kinase (JAK)–STAT signaling (12). Homeostatic IL-7 signaling promotes cell survival, proliferation, and differentiation through transcription of Ccnd1/3, Rag1/2, Pax5, and Il-7r, while also repressing the expression of proapoptotic genes, Bax, Bad, and Bcl2l11 (BIM) (13–15). Lymphocytes are primed for IL-7 signaling, as p-STAT5 translocation to the nucleus after IL-7R ligation occurs within minutes. This primed state is achieved by the formation of microdomains in the plasma membrane surrounding the IL-7R, which organizes interacting proteins for near immediate signal transduction (16).
Tetraspanins are a family of transmembrane proteins important for organization of the plasma membrane and regulation of cellular migration, adhesion, and activation (17, 18). These small hydrophobic proteins, bearing four transmembrane domains, two short cytoplasmic tails, and two extracellular domains, are known to associate with other proteins in the membrane and cytosol, as well as other tetraspanins, to form specialized tetraspanin-enriched microdomains (TEMs). Of the 33 identified tetraspanins, CD53 is one of four to be exclusively expressed on hematopoietic cells and is highly expressed on mature B cells (19, 20). A case of familial CD53 deficiency was reported, with patients suffering recurrent bacterial, viral, and fungal infections, as well as reduced serum Ig levels, suggesting a role for CD53 in immune system function (21). CD53 is capable of organizing MHC class II on B cells through TEMs into functional clusters on the cell surface, suggesting that CD53 may interact with other surface proteins to modulate B cell activity (22). Stimulation of CD53 influences calcium influx, apoptosis, and proliferation in various lymphocytes. A recent study showed that CD53 recruits protein kinase C (PKC) to the plasma membrane to facilitate BCR-dependent PKC signaling (19, 23–25). Thus, multiple prior studies have suggested a role for CD53 in regulating B cells. However, the natural ligands for CD53 in B cells and the mechanisms by which it influences B cell development and function are largely unknown.
In this study, we present the requirement of CD53 for normal bone marrow B cell development. Although the enhancer for Cd53 has been observed to be a direct transcriptional target of EBF1, a key regulator of early B cell development, a specific role for CD53 in this process has not been described (26). In this study, we show that Cd53−/− mice have significantly reduced bone marrow, splenic, lymphatic, and peripheral B cells compared with wild-type (WT) littermate controls. In addition, hematopoietic stem cells (HSCs) isolated from Cd53−/− mice give rise to fewer B cells compared with controls in vitro, yet there is no difference in NK or myeloid cell production. Mixed bone marrow chimeras reveal that CD53 functions cell autonomously during early B cell development. Analysis of bone marrow B cell development demonstrates that this loss of B cells originates with early B cell progenitors, which display reduced IL-7Rα expression and signaling. Specifically, we observe reduced PI3K and STAT5 activation in prepro– and pro–B cells in the absence of CD53, with a consequent increase in apoptosis in these populations. Finally, coimmunoprecipitation and proximity ligation studies demonstrate a physical interaction between CD53 and IL-7Rα, suggesting that these proteins associate at the cell surface to maintain the homeostatic early B cell signaling necessary for normal B cell development.
Materials and Methods
Mice
Cd53−/− mice were generated using CRISPR/Cas9 reagents designed and validated by the Genome Engineering Center at Washington University School of Medicine (St. Louis, MO), as previously described (27). Briefly, Cas9 mRNA and a guide RNA targeting the third exon of Cd53 were injected into C57BL/6J embryos. Cas9 cleavage generated offspring with a 4-bp deletion in the targeted exon of Cd53, and the generation of an SmlI restriction site that was used for genotyping. Newly generated Cd53−/− mice were back-crossed to WT mice (C57BL/6J; The Jackson Laboratory, Bar Harbor, ME) prior to experimental use, and WT littermates were used throughout the study as controls. Il-7r−/− mice (stock no. 002295) were purchased from The Jackson Laboratory and maintained as homozygous knockouts or bred to C57BL/6J mice (10). Sex- and age-matched mice were used for each experiment in accordance with the guidelines of the Washington University Animal Studies Committee.
Flow cytometry
Peripheral blood was obtained by cardiac puncture. Bone marrow hematopoietic cells were isolated by centrifugation of femurs and tibiae at 6000 rpm for 3 min. Spleen, thymus, and lymphoid cells were harvested by gentle crushing through a 100-μM strainer. Cells were processed for staining, as previously described (28), and stained using the Abs listed in Supplemental Table I. Cell counts were determined using a Hemavet HV950 (Drew Scientific, Erba Diagnostics, Miami Lake, FL). Stained cells were analyzed on a BD flow cytometer (BD Biosciences, San Jose, CA), a Gallios flow cytometer (Beckman Coulter, Indianapolis, IN), an Attune NxT flow cytometer (Thermo Fisher, Santa Clara, CA), or a NovoCyte flow cytometer (Acea Bioscience, San Diego, CA). Data were analyzed with FlowJo software (version 10.5.3; TreeStar, Ashland, OR).
Cell sorting
Bone marrow cells were obtained by crushing long bones (femurs, tibiae, and humerii), pelvic bones, and vertebrae with a mortar and pestle in FACS buffer supplemented with 2% BSA. Single-cell suspensions were filtered using a 40-μm filter prior to staining with primary conjugated Abs (Supplemental Table I) on ice for 40 min. B220+ cells were enriched prior to sorting by selection with B220-conjugated paramagnetic beads using the autoMACS Pro Separator (Miltenyi Biotec, Auburn, CA). Nonviable cells were excluded from analyses by DAPI staining.
IgM and IgG serum measurements
Blood obtained by cardiac puncture was allowed to clot at room temperature for 1 h, then centrifuged for 10 min at 6000 rpm. Collected serum was assessed for IgG and IgM levels using the Mouse Total IgG Uncoated ELISA Kit and the Mouse IgM Uncoated ELISA Kit (Invitrogen, Thermo Fisher Scientific, Santa Clara, CA) following the manufacturer’s instructions.
In vitro differentiation
OP9-GFP stromal cells were cultured in α-MEM (Life Technologies) with 20% FBS and penicillin/streptomycin. Two hundred fifty HSCs from WT or Cd53−/− mice were sorted directly into 24-well plates seeded with 5 × 104 OP9-GFP cells. Cells were cultured for 12–14 d in cytokine-supplemented media [B cells: FMS-like tyrosine kinase three ligand (Flt3-L; 5 ng ml−1; Miltenyi Biotec) and IL-7 (5 ng ml−1; Invitrogen); NK cells: Flt3-L (5 ng ml−1), stem cell factor (SCF; 10 ng ml−1; PeproTech, Rocky Hill, NJ), CXCL12 (20 ng ml−1; PeproTech), IL-2 (10 ng ml−1; PeproTech), and IL-15 (10 ng ml−1); myeloid cells: Flt3-L (5 ng ml−1); IL-7 (5 ng ml−1); SCF (10 ng ml−1); IL-3 (10 ng ml−1; PeproTech), IL-6 (10 ng ml−1; PeproTech), M-CSF (10 ng ml−1; PeproTech), G-CSF (10 ng ml−1; PeproTech), and GM-CSF (10 ng ml−1; PeproTech)]. Half-media changes were performed every fourth day.
Chimera generation
Chimeric mice were generated by transplanting a 1:1 mixture of whole bone marrow cells from Cd53+/+ (CD45.2) and WT (CD45.1) mice into WT (CD45.1/CD45.2) recipients or Cd53−/− (CD45.2) and WT (CD45.1) mice into WT (CD45.1/CD45.2) recipients. A total of 2 × 106 cells were injected retro–orbitally into lethally irradiated recipients. Cells were allowed to engraft for 4–6 wk prior to analysis.
Cell cycle analysis
For Ki-67 staining, bone marrow cells were stained for surface markers (Supplemental Table I), fixed, using the BD Cytofix/Cytoperm kit (BD Biosciences), blocked with 5% goat serum, stained with mouse anti-human Ki-67 (clone B56; BD Pharmingen) as per the manufacturer’s instructions, washed, and resuspended in DAPI-containing FACS buffer.
Cell death analysis
For annexin V detection, cells were stained for surface markers (Supplemental Table I). Cells were then washed, and 2 × 106 cells were stained according to the APC Annexin V Apoptosis Detection Kit (Affymetrix, Thermo Fisher Scientific, Santa Clara, CA) prior to flow cytometry analysis.
Proximity ligation assay
Protein interaction of CD53 and IL-7Rα was measured by Duolink proximity ligation assay (PLA; Sigma-Aldrich, St. Louis, MO). Purified anti-CD53 and anti-CD127 Abs were conjugated with Duolink In Situ Probemaker PLUS and MINUS kits, respectively, following the manufacturer’s instructions (Sigma-Aldrich). Duolink flow cytometry protocol was followed with a few deviations. Briefly, bone marrow cells were stained for flow cytometry surface markers (Supplemental Table I), fixed using the BD Cytofix/Cytoperm Kit (BD Biosciences), then blocked with 5% goat serum and mouse Fc receptor block (BioLegend, San Diego, CA). Samples were stained with CD53-PLUS and IL-7Rα-MINUS Abs sequentially, each for 30 min at room temperature. Ligation, amplification, and detection steps followed the manufacturer’s instructions. Samples were then analyzed by flow cytometry.
Mass cytometry
For all samples, EQ Four Element Calibration Beads were used during collection, according to the manufacturer’s instructions (Fluidigm, San Francisco, CA). Data were normalized using Fluidigm CyTOF2 Bead Normalization Tool. Metal-tagged Abs were purchased from Fluidigm or custom conjugated using the Maxpar X8 Antibody Labeling Kit, according to the manufacturer’s instructions (Fluidigm). All custom-conjugated Abs were titrated. Staining was comparable to flow cytometry (Kaluza 2.0, Beckman Coulter, Gallios). Whole bone marrow cells from WT or Cd53−/− mice were stained for viability with 2.5 μM cisplatin (Sigma-Aldrich), washed in serum-supplemented media, and fixed in 1.6% paraformaldehyde. For staining, 3 × 106 bone marrow cells were stained with surface Abs (Supplemental Table I) for 1 h at 4°C in CyFACS buffer (0.1% BSA, 0.02% NaN2, 2 mM EDTA in CyPBS; Rockland Immunochemicals, Gilbertsville, PA). Surface Abs were washed away using CyPBS. The cells were then permeabilized in ice-cold methanol. Intracellular staining was performed in CyFACS buffer for 1 h at 4°C. Cells were washed and stained with a Cell-ID Intercalator, according to the manufacturer’s instructions (Fluidigm). Samples were analyzed on a CyTOF2 mass cytometer (Fluidigm), and data were analyzed with the Cytobank online platform (Cytobank, Santa Clara, CA).
Quantitative RT-PCR analysis
RNA from sorted prepro–B and pro–B cells from individual WT or CD53−/− mice (age 5 wk) was prepared using the NucleoSpin RNA XS Kit (MACHERY-NAGEL, Bethlehem, PA) and quantitative RT-PCR (qRT-PCR) was performed with a TaqMan RNA-to-Ct 1-Step Kit (Thermo Fisher Scientific). Gene probes are listed in Supplemental Table I.
Western blotting
Pro–B cells from 5-wk-old WT or Cd53−/− mice were sorted into 10% trichloroacetic acid, pelleted at 14,000 rpm for 10 min at 4°C, and washed twice with acetone. Dried pellets were resuspended in solubilization buffer (9 M urea, 2% Triton X-100, and 1% DTT), and incubated with loading dye at 70°C for 10 min. Samples were run in 4–12% Bolt Bis-Tris Plus Gels (Invitrogen), following the manufacturer’s instructions and transferred to PVDF membranes. Membranes were probed with p-STAT5 (1:1000; stock no. 9314; Cell Signaling Technology, Danvers, MA), STAT5 (1:1000; stock no. 94205; Cell Signaling Technology), tubulin (1:1000; stock no. 3873; Cell Signaling Technology), and rabbit anti-mouse HRP-conjugated secondary Ab (1:1000; stock no. 7074; Cell Signaling Technology). Proteins were detected using Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) on chemiluminescence film. Densitometry was performed using ImageJ (National Institutes of Health, Bethesda, MD).
Coimmunoprecipitation and immunoblot
Human CD53 and human IL-7Rα, with V5-tag or FLAG-tag at the C terminus, were subcloned into pCMV6-Entry vector and transfected into HEK-293T cells. Cells were grown in DMEM with 10% FBS and 1% penicillin/streptomycin (Life Technologies). After 48 h of transfection, cells were lysed for 30 min in lysis buffer (20 mM Tris [pH 7.5], 150 mM NaCl, 2% DDM, and a protease-inhibitor mixture [Roche]). Lysates were centrifuged at 20,000 × g for 10 min at 4°C. The supernatant was collected and incubated with anti-M2 FLAG beads for 5 h. Beads were recovered, washed three times (20 mM Tris [pH 7.5], 150 mM NaCl, 0.05% DDM), and boiled with SDS loading buffer. Samples were run through polyacrylamide gel and transferred to PVDF membranes. After blocking with 5% nonfat milk, the membranes were incubated with primary Abs (Sigma-Aldrich) overnight at 4°C. Proteins were detected by HRP-conjugated secondary Abs (DakoCytomation) and ECL reagents (GE Healthcare).
Statistical analysis
Data are presented as mean ± SEM, unless otherwise stated. Statistical significance was assessed using an unpaired, two-tailed Student t test or two-way ANOVA. GraphPad Prism (Version 8.2.1) was used for all statistical analyses (GraphPad Software, La Jolla, CA). The p values in all instances indicate: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Results
CD53 expression increases with B cell development
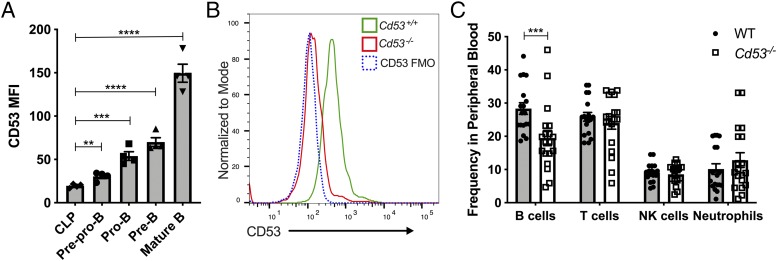
Consistent with prior reports showing an increase in CD53 transcript levels throughout B cell development, we find that CD53 surface expression, although relatively low on hematopoietic stem and progenitor cells, increases throughout maturation in the bone marrow (26, 29). Specifically, we find that B cell progenitors, defined as CLP (lineage− [Lin−; B220−, CD3e−, Ter-119−, Gr-1−], CD27+, Flk2+, CD127+, Ly-6D−); prepro-B (CD11b−, NK1.1−, B220+, IgM−, CD19−, CD43+); pro-B (CD11b−, NK1.1−, B220+, IgM−, CD19+, CD43+); pre-B (CD11b−, NK1.1−, B220+, IgM−, CD19+, CD43low); and mature B cells (CD11b−, NK1.1−, B220high, IgMhigh), express increasing amounts of CD53 as determined by surface flow cytometry (Fig. 1A).
FIGURE 1.
CD53 surface expression increases throughout bone marrow B cell development. (A) CD53 expression is negligible on CLPs, but after commitment to a B cell fate, CD53 is significantly upregulated through maturation. Shown is surface expression of CD53 on the indicated cell types (see Supplemental Fig. 2 for gating strategy) as determined by flow cytometry (n = 4 mice, age 8 wk). (B) Representative flow plot of CD53 expression on mature B cells in the bone marrow of WT and Cd53−/− mice. A CD53 FMO control was included as a negative control. (C) The frequency of B220+ lymphocytes in the peripheral blood is significantly reduced in Cd53−/− mice compared with WT (n = 18–19 mice per group, age 6–8 wk) over six independent experiments. Error bars represent mean ± SEM. **p < 0.01, ***p < 0.001, ****p < 0.0001 by an unpaired Student t test. FMO, fluorescence minus one; MFI, median fluorescent intensity; WBC, white blood cell.
To determine the role of CD53 in B cell development, we used CRISPR–Cas9 technology to generate mice deficient for CD53. These mice have a 4-bp deletion in the third exon, predicted to abolish CD53 production. Indeed, we do not detect CD53 expression in the leukocytes of our Cd53−/− mice by flow cytometry or qRT-PCR (Fig. 1B, Supplemental Fig. 1). Cd53−/− mice are born at Mendelian ratios, are viable without any gross abnormalities, and have complete blood counts within the standard range across various ages (data not shown). Investigation into the hematopoietic effects of CD53 loss, however, revealed significantly reduced B cells (B220+) in the peripheral blood of these mice (Fig. 1C). There were no overt differences in other leukocyte populations, including T cells (CD3e+), NK cells (NK1.1+), and neutrophils (Gr-1+), suggesting that the loss of CD53 specifically affects B cell production.
Dependence on CD53 for normal B cell development
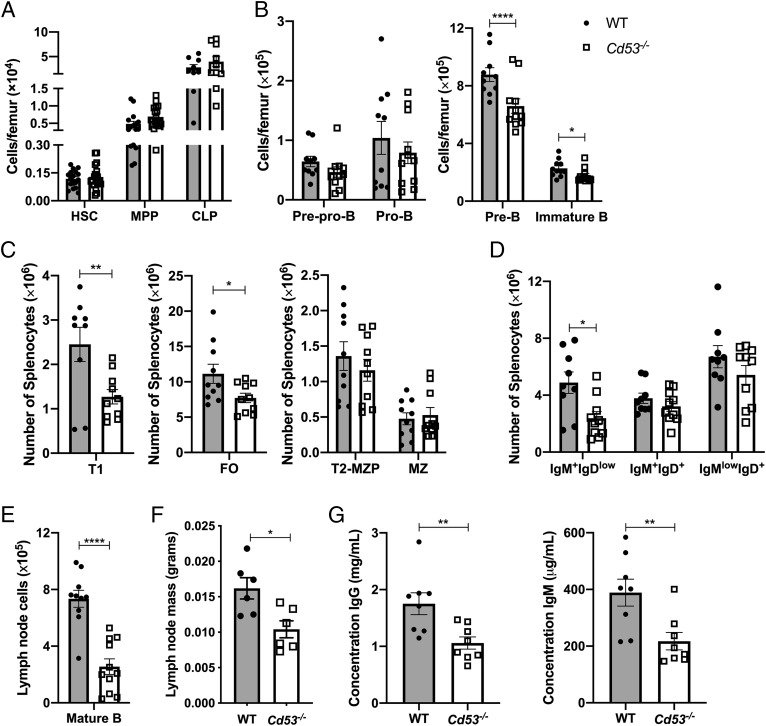
B cell development begins in the bone marrow with the differentiation of HSCs into multipotent progenitors (MPPs) and CLPs before specification of the B cell lineage, although there is evidence that lymphoid-primed MPPs are capable of directly differentiating into prolymphocytes without a CLP intermediate (30). Consistent with the fact that CD53 expression is low prior to B cell commitment, Cd53−/− mice have similar numbers of HSCs (Lin−, c-kit+, Sca-1+, CD150+, CD48−), MPPs (c-kit+, Sca-1+, Lin−, Flk2+, CD34+) and CLPs to their WT littermates (Fig. 2A). However, once the B cell lineage was established, Cd53−/− mice demonstrated a significant reduction in pre-B and immature B cells in the bone marrow (Fig. 2B).
FIGURE 2.
Loss of CD53 significantly impairs B cell development. Shown are absolute numbers of hematopoietic stem and progenitor cells in the bone marrow (A) and B cells in the bone marrow (B), spleen (C and D), and lymph nodes (E) (n = 6–19 mice per group, age 5–8 wk) over two to six independent experiments. Gating strategies for defined populations are outlined in Supplemental Fig. 2. (F) Inguinal, axial, and cervical lymph nodes dissected from Cd53−/− mice are significantly smaller than age-matched WT mice (n = 6 mice per group, age 5 wk) over four independent experiments. (G) Serum concentrations of IgG and IgM are significantly reduced in Cd53−/− mice, as measured by ELISA (n = 8 mice per group, age 8 wk) over three independent experiments. Error bars represent mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001 by an unpaired Student t test. T1, transitional 1 B cells; T2-MZP, transitional 2 MZ progenitor B cells.
This reduction was continued during splenic development of B cells, as immature B cells egress from the bone marrow and home to the spleen, where they mature into transitional 1 B cells (Fig. 2C). The number of more immature IgM+IgDlow cells was still significantly decreased in Cd53−/− mice, but as this population continued maturation through IgM+IgD+ to IgMlowIgD+ cells, the reduction in Cd53−/− mice became less pronounced (Fig. 2D). Further dissection of maturing splenic B cells revealed that, whereas MZ progenitors and MZ cells were equivalent between WT and Cd53−/− mice, FO B cells were significantly reduced (Fig. 2C). Upon emigration from the spleen, mature B cells may traffic to the lymph nodes and enter the lymphatic system. Analysis of B cells in the lymph nodes of Cd53−/− mice again revealed decreased mature B cells as well as a reduction in overall lymph node size (Fig. 2E, 2F). Parallel to the reduction in tissue-resident mature B cells in secondary lymphoid organs, serum concentrations of IgG and IgM were both reduced in Cd53−/− mice, suggesting a decrease in mature B cell function (Fig. 2G). Thus, with loss of CD53 expression, B cell development is altered beginning at the pre–B cell stage in the bone marrow and continuing through splenic maturation.
CD53 is necessary for normal B cell development in vitro
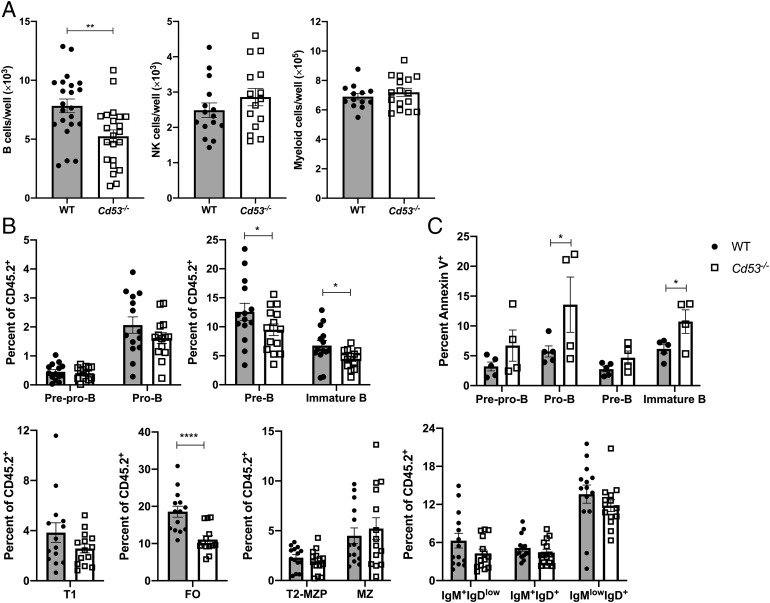
Tetraspanins are known to regulate and interact with surface proteins important for plasma membrane organization and cellular migration and adhesion and, as a family, have been highly studied for their importance during cancer metastasis (17, 31). Via TEMs, tetraspanins are capable of recruiting and stabilizing protein and receptor interactions; furthermore, CD53 has been shown to coordinate protein localization and interactions on the surface of B cells (22, 32). Based on these data, B cell in vitro differentiation from sorted HSCs was performed to discern whether CD53 was necessary only for proper cellular localization within the bone marrow microenvironment or if differentiation required proper organization of proteins within the membrane. When cultured with IL-7 and Flt3-L, Cd53−/− HSCs gave rise to ∼30% fewer B cells (B220+CD19+ cells) in vitro than WT HSCs (Fig. 3A, Supplemental Fig. 2F). Therefore, even when given the same external stimuli, Cd53−/− cells are impaired in their ability to differentiate along the B cell lineage. This differentiation deficit was specific to B cells, as NK (NK1.1+CD3−) and myeloid (Gr-1+CD11b+) development were unaffected between WT and Cd53−/− HSCs (Fig. 3A).
FIGURE 3.
CD53 functions cell autonomously to promote B cell differentiation. (A) HSCs from WT or Cd53−/− mice (age 5 wk) were sorted directly onto seeded OP-9 cells in cytokine-supplemented media and assessed by flow cytometry after 14 d in culture. Shown are the numbers of B cells (B220+ CD19+), NK cells (NK1.1+ CD3−), and myeloid cells (Gr-1+ CD11b+) generated per well of 250 sorted HSCs (n = 15–23 wells per group) over three independent experiments. (B) Cd53 chimeras were generated, and the contribution of Cd53+/+ and Cd53−/− developing B cells to overall CD45.2 chimerism was assessed. Cd53−/− bone marrow gave rise to significantly fewer pre-B, immature B, and FO B cells compared with WT populations (n = 14 mice per group) over four independent experiments and three independent transplants. (C) Chimeric Cd53 mice have increased annexin V staining on Cd53−/− cells compared with Cd53+/+ cells (n = 4–5 mice per group) over two independent experiments. Error bars represent mean ± SEM. *p < 0.05, **p < 0.01, ****p < 0.0001. by an unpaired Student t test.
Chimeric B cell development supports a cell-autonomous function of CD53
To further assess whether CD53 functions through a cell-autonomous or cell-nonautonomous mechanism, Cd53 chimeric mice were generated. To mitigate any potential effect because of allelic variants, CD45.2+ CD53−/− bone marrow was competed against CD45.1+ WT bone marrow, and independently, CD45.2+ WT bone marrow was competed against CD45.1+ WT bone marrow. Thus, the ability of a transplanted CD45.2+ cell to give rise to developing B cells was investigated. Matching the observed phenotype in naive mice, Cd53−/− cells gave rise to significantly fewer pre-B and immature B cells in the bone marrow of transplant recipients (Fig. 3B). Upon maturation to the spleen, the reduction in B cells from Cd53−/− marrow became less pronounced, yet there were still significantly fewer Cd53−/− FO cells compared with WT (Fig. 3B). In addition to B cell production, a subset of chimeric mice was assayed for apoptosis of developing B cells. Indeed, Cd53−/− cells had increased cell death in the early developing bone marrow B cells, compared with WT transplants (Fig. 3C). Although the phenotype does not exactly mimic that of naive mice, it is evident that cell-autonomous Cd53 expression is necessary for normal production of B cells.
Loss of CD53 reduces IL-7R expression and induces B cell progenitor apoptosis
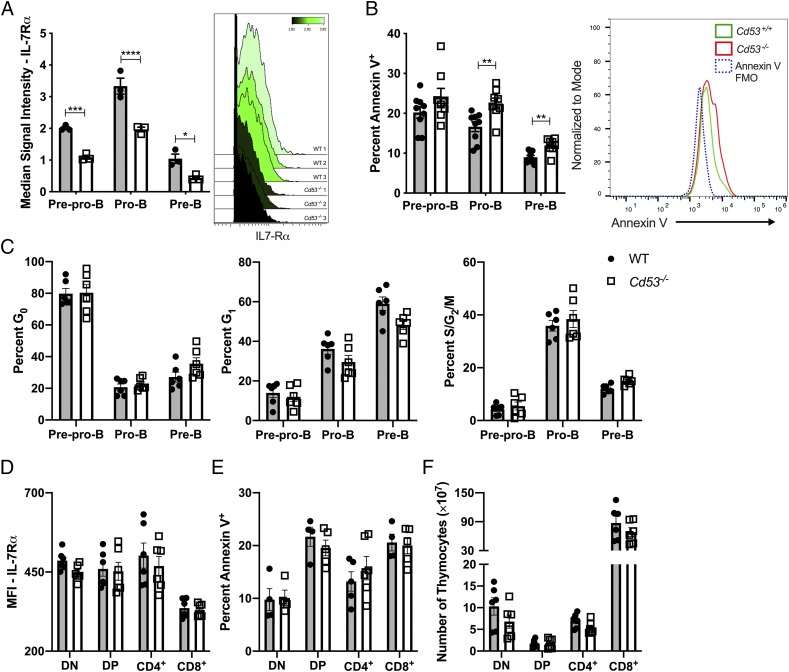
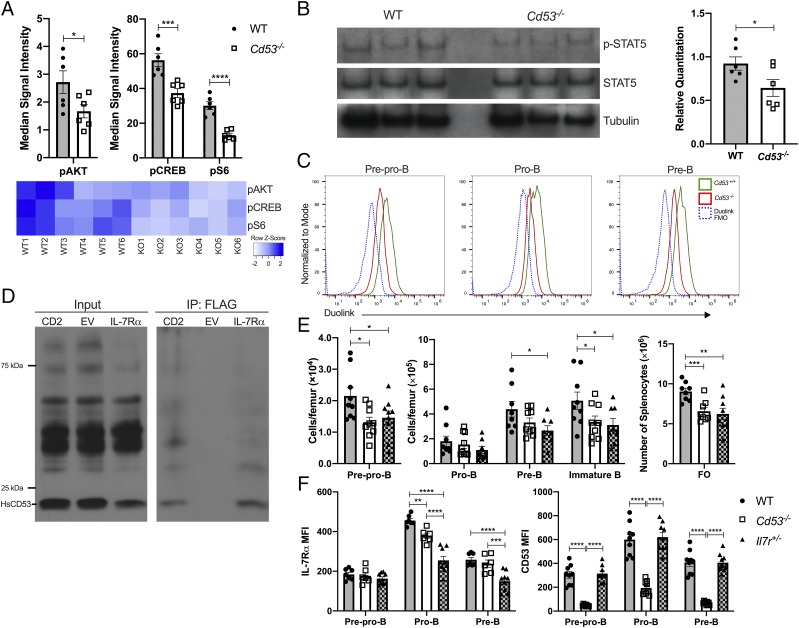
With the observation that loss of CD53 expression leads to reduced B cell numbers at the pre–B cell stage, cellular signaling prior to the loss of pre–B cells was investigated. We specifically interrogated IL-7 signaling, as this is crucial for early B cell development, and the loss of IL-7 signaling blocks further B cell maturation and induces cell death (10). Analysis of bone marrow B cell progenitors by mass cytometry (CyTOF) revealed significantly reduced IL-7Rα expression on the surface of prepro-B, pro-B, and pre–B cells of Cd53−/− mice (Fig. 4A). Additionally, conventional flow cytometry showed significantly reduced IL-7Rα surface expression on pro–B cells (Fig. 5F). Cell death, as measured by annexin V staining, was increased in pro– and pre–B cells in the absence of CD53 (Fig. 4B), but cell cycling was not altered in the Cd53−/− cells (Fig. 4C, Supplemental Fig. 2G). Given that IL-7 signaling is also crucial for cell survival in normal T cell development (33), we assessed thymocyte numbers, IL-7Rα surface expression, and apoptosis in Cd53−/− mice. In contrast to our findings regarding developing B cells, we find that IL-7Rα surface expression is not significantly altered on T cells in the absence of CD53 (Fig. 4D). Furthermore, thymocyte numbers and survival are grossly normal (Fig. 4E, 4F, Supplemental Fig. 2E). These data suggest that CD53 maintains normal IL-7Rα expression and preserves cell survival, specifically in early B lymphopoiesis.
FIGURE 4.
Surface IL-7Rα expression is reduced in the absence of CD53. (A) Surface expression of IL-7Rα is significantly downregulated on prepro-B, pro-B, and pre–B cells of Cd53−/− mice compared with littermate controls. Shown on the right is a representative plot of IL-7Rα MSI on pro–B cells (n = 3 mice per group, age 5 wk). (B) Cd53−/− B cell progenitors have increased annexin V staining by flow cytometry, compared with WT controls (n = 8–9 mice per group, age 5 wk) over three independent experiments. Example flow plot is shown on the right. (C) The frequency of bone marrow precursor B cells in G0, G1, and S/G2/M phase of the cell cycle as determined by Ki-67 and DAPI staining revealed no difference in cell cycling between WT and Cd53−/− mice (n = 6 mice per group, age 5 wk) over three independent experiments. Surface expression of IL-7Rα on developing T cells (D) shows no difference between WT and Cd53−/− mice by flow cytometry, with no change in apoptosis (E) or total cell number (F) between WT and Cd53−/− thymocytes (n = 6 mice per group) over two independent experiments. Gating strategy for T cell populations are outlined in Supplemental Fig. 2E. Error bars represent mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 by an unpaired Student t test. DN, double negative (CD4− CD8−); DP, double positive (CD4+ CD8+); IL-7Rα, IL-7R α subunit; MSI, median signal intensity.
FIGURE 5.
CD53−/− B cell progenitors have diminished IL-7 signal transduction. (A) CyTOF was performed to assess signaling in bone marrow B cell progenitors. Shown are the reduction in PI3K signaling in prepro–B cells as measured by decreased levels of phosphorylation of Akt, CREB, and S6. See Supplemental Fig. 3 for gating strategy and further results (n = 6 mice per group, age 5 wk) over two independent experiments. (B) Western blot analysis revealed reduced phosphorylation of STAT5 in sorted Cd53−/− pro–B cells, yet revealed no loss of total STAT5 expression. Relative quantitation of p-STAT5 compared with total STAT5 levels is shown below (n = 6 mice per group, age 5 wk). Data represent two independent experiments. (C) Representative flow plot of PLA indicating that CD53 and IL-7Rα interact on the membrane of early B cell progenitors (over two independent experiments). Duolink FMO contained neither CD53-PLUS nor IL-7Rα–MINUS Abs. Cd53−/− mice were used as a biological negative control. (D) Immunoblot of whole cell lysate from 293T cells transfected with human Cd53-V5 and Il-7r-FLAG shows an interaction of the proteins. CD2, a known binding partner of CD53, was used as a positive control. (E) Analysis of Il-7r+/− mice reveals a similar phenotype to Cd53−/− mice of bone marrow B cell progenitors with significantly fewer pre-B, immature B, and FO B cells compared with WT controls (n = 7–9 mice per group, age 5 wk) over two independent experiments. (F) Surface expression of IL-7Rα and CD53 on early B cells indicates that heterozygous loss of IL-7Rα does not affect surface levels of CD53 compared with WT cells but is sufficient to affect B lymphopoiesis (n = 7–9 mice per group, age 5wk). Data represent two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. by an unpaired Student t test or two-way ANOVA. Error bars represent mean ± SEM. CREB, cAMP response element-binding protein; EV, empty vector; FMO, fluorescence minus one.
Cd53−/− B cells have impaired PI3K and STAT5 signaling
The reduction in B cell number across multiple compartments, coupled with decreased IL-7Rα expression, suggested differential downstream signaling between WT and Cd53−/− B cells. CyTOF was used to query canonical B cell signaling pathways, including PI3K, MAPK, JAK-STAT, and NF-κB signaling. IL-7 predominantly signals through two pathways, PI3K and JAK-STAT, and the activation of each is regulated temporally, dependent on developmental stage (12). PI3K signaling coordinates survival signals downstream of IL-7R. PI3K signaling at the prepro–B cell stage was consistently and significantly impaired in Cd53−/− mice across experimental replicates (Fig. 5A, Supplemental Fig. 3A). STAT5 signaling is critical for progression from the prepro-B to pro–B cell, with STAT5-deficient mice halting B lymphopoiesis at the prepro–B cell stage (34). Similar to reduced PI3K signaling, Cd53−/− pro–B cells have reduced phosphorylation of STAT5 compared with WT pro–B cells, with no changes in total STAT5 levels (Fig. 5B).
CD53 interacts with IL-7Rα on the plasma membrane
Reduced activation of PI3K and STAT5 in Cd53−/− B cell progenitors suggests that IL-7R signaling is impaired; however, an interaction between CD53 and the IL-7R has not been previously reported. To investigate whether there is a physical interaction, PLA was performed and shows that in WT prepro–, pro–, and pre–B cells, CD53 and IL-7Rα reside within 40 nm of each other (theoretical maximum, Fig. 5C). Cd53−/− mice were used as a biological negative control. The interaction of CD53 and IL-7Rα was also measured for human homologs. Human Cd53-V5 and Il-7r-FLAG were coexpressed in 293T cells. Immunoprecipitation of IL-7Rα and probing for CD53 revealed that CD53 and IL-7Rα are associated in the plasma membrane (Fig. 5D). Thus, CD53 interacts with IL-7Rα on the surface of developing murine B cells, and human CD53 has the capability to interact with IL-7Rα.
Heterozygous loss of IL-7Rα reduces early B cell development
Although loss of CD53 results in reduced IL-7Rα expression on developing B cells, B lymphopoiesis in Il-7r+/− mice has not been reported in the literature. Upon investigation of early B cell progenitors in the bone marrow, Il-7r+/− mice produce significantly fewer pre-B, immature B, and FO B cells compared with WT, yet they produce similar numbers as Cd53−/− mice (Fig. 5E). Although Il-7r+/− and Cd53−/− mice have a similar B lymphopoiesis phenotype, the expression of IL-7R, a critical signaling receptor, is significantly lower in Il-7r+/− mice than Cd53−/− mice (Fig. 5F), suggesting that CD53 exerts its function through maintenance of signaling and not just IL-7R expression. Of note, although IL-7R is differentially expressed, there is no reduction of CD53 surface expression in Il-7r+/− mice (Fig. 5F).
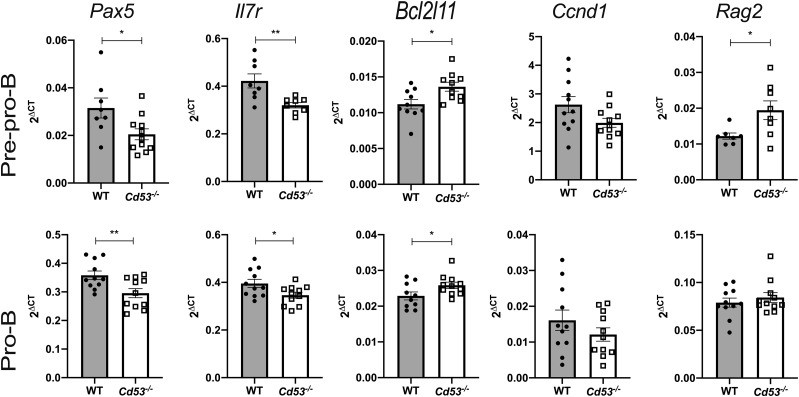
PI3K target gene expression is disrupted upon loss of CD53
Consistent with the reduction in PI3K signaling in early B cells, qRT-PCR of total RNA isolated from sorted prepro-B and pro–B cells from WT and Cd53−/− mice reveals differential target gene expression (Fig. 6). Pax5, which is induced by IL-7/PI3K signaling, restricts cells to the B cell lineage and promotes activation of B cell–specific genes through chromatin modifications (35). Reduction in Pax5 signaling impairs further maturation of developing B cells, congruent with observed developmental deficits in Cd53−/− mice. As discussed, IL-7 signaling is vital for B cell development, and Cd53−/− mice have reduced surface expression and mRNA levels for IL-7Rα (Fig. 4A). This likely occurs through a self-enforced feedback loop, whereby IL-7Rα signals to induce transcription of Ebf, and EBF then promotes Il-7r transcription; however, we cannot rule out that there may be an alternative mechanism by which Il-7r transcription is abrogated, resulting in reduced protein expression (12, 36–38). Cd53−/− mice have increased expression of BIM, which is proapoptotic. BIM is typically repressed by IL-7/PI3K signaling to promote cell survival. The increase in BIM transcripts is consistent with decreased IL-7/PI3K signaling and increased apoptosis in Cd53−/− B cell progenitors. IL-7 signaling also negatively regulates Rag1/2 expression, and Rag2 expression is increased in prepro–B cells of Cd53−/− mice, which have reduced IL-7 signaling (39). Consistent with the lack of observed changes in cell cycling, there was no difference in cyclin D1 expression between WT and Cd53−/− B cells. Together, these differences in gene expression of key mediators of B cell development and survival reflect the reduction of IL-7/PI3K signaling in early B cells in the absence of CD53 (Fig. 7).
FIGURE 6.
Loss of CD53 alters downstream gene expression. Gene expression of canonical PI3K target genes. RNA was isolated from sorted B cell populations and qRT-PCR was performed for Pax5, Il-7r, BIM, Ccnd1, and Rag2, relative to Actb (n = 11 mice per group, age 5 wk) over three independent experiments. Error bars represent mean ± SEM. *p < 0.05, **p < 0.01 by an unpaired Student t test.
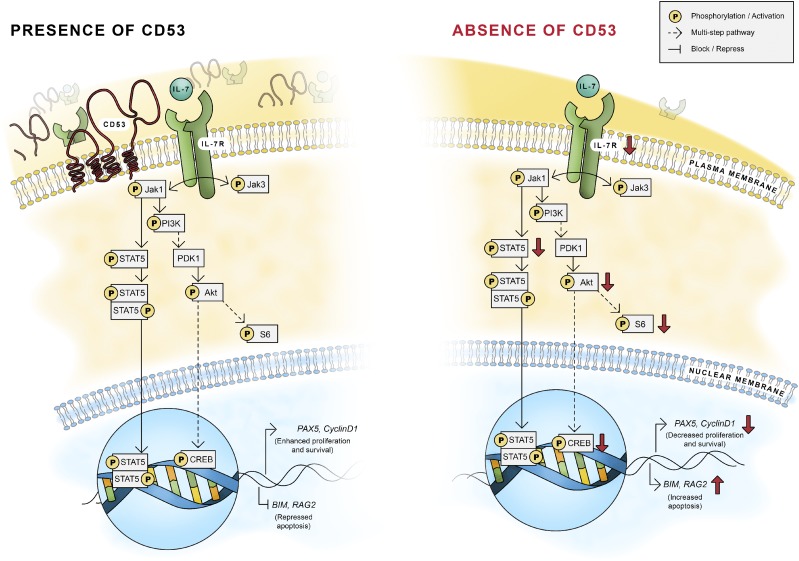
FIGURE 7.
CD53 maintains early B cell development through IL-7R signaling. Working model of the necessity of CD53 for normal B cell development. Without CD53, IL-7R on the cellular membrane is reduced, leading to decreased IL-7Rα expression, reduced PI3K signaling and STAT5 phosphorylation, and increased apoptosis of B cell progenitors.
Discussion
In this study, the necessity of the tetraspanin CD53 for normal IL-7 signaling during B cell development is demonstrated. During normal B lymphopoiesis, CD53 expression increases through B cell maturation, consistent with activation of EBF1, which has been shown to directly target and enhance CD53 expression (26). Through association with IL-7Rα, CD53 maintains signaling through the PI3K and STAT5 pathways to support early B cell growth, promote B cell differentiation, and reduce B cell death. Additionally, CD53 functions, at least in part, cell autonomously, as chimeric mice showed a specific reduction of B cell progenitors in Cd53−/− cells. Our data support that CD53 has B cell–specific functions, as no changes in T cells or NK cells are observed in Cd53−/− mice and we observed no difference between in vitro NK cell production. CD53 is expressed on T and NK cells, and prior studies have implicated a potential role for CD53 in regulating the function of these cells; however, further studies will be needed to determine the interacting partners of CD53 in these cell types and elucidate the specific consequences of these interactions (40, 41). Of particular note, IL-7 signaling is crucial for normal T cell development, but given the grossly normal numbers of T cells in the Cd53−/− mice and the normal IL-7Rα expression on these cells, we postulate that other factors besides CD53 promote IL-7R stability in these cells.
In the bone marrow of Cd53−/− mice, there is a clear deficit in B lymphopoiesis, yet these differences in B cell development become less prominent during splenic maturation (Fig. 2B, 2C). Our data show that FO cell development is impacted, but MZ development appears unchanged upon constitutional or chimeric loss of CD53. Further investigation into MZ versus FO cell fate would need to be performed to delineate the observed differences. Although a clear reduction in bone marrow B cell development is outlined, additional signaling differences in more mature populations cannot be ruled out, and perhaps, there are continued impediments in splenic or lymph node B cell development not captured by these analyses. Future study into the immune competence of Cd53−/− mice could suggest additional disparities in B cell function, as serum Igs are significantly reduced in Cd53−/− mice.
B lymphopoiesis occurs within distinct niches of the bone marrow, and CD53 has been shown to interact with integrins important for cellular localization (42). Our in vitro B cell differentiation data, however, suggests that the observed reduction in B cell development is location independent, as even when WT and Cd53−/− cultures were supplemented with equivalent IL-7 and Flt3-L cytokines, Cd53−/− HSCs had impaired B cell development. Homing or localization defects could be present in the absence of CD53 during immature B cell immigration to the spleen, but as detectable immunophenotypic splenic B cells are present, it does not seem that Cd53−/− mice have any gross homing defects.
Together, our data support a model whereby CD53 associates with IL-7Rα on the surface of developing bone marrow B cells to help maintain its presence and signaling. Immunoprecipitation and PLA show that CD53 and IL-7Rα associate on the surface of prepro–, pro–, and pre–B cells in a TEM complex (Fig. 5C, 5D). Without CD53, IL-7Rα surface levels are reduced, and there is a significant reduction in downstream IL-7 signaling through both PI3K and p-STAT5 pathways, resulting in altered gene expression and increased apoptosis of developing B cells. Our data suggest that, whereas loss of CD53 moderately impairs IL-7Rα expression, the reduction in IL-7 signaling is significantly diminished (Figs. 4A, 5A, 5F). The change in IL-7Rα expression between WT and Cd53−/− B cells is modest, suggesting that the effects observed are related to the influence of CD53 on IL-7R signaling itself.
During IL-7R signaling, CD53 supports IL-7R–dependent phosphorylation of PI3K proteins and STAT5 in prepro– and pro–B cells. Thus, IL-7R transcriptionally coordinates enhanced Pax5 and Il-7r expression and repressed BIM expression, resulting in progression through B cell maturation and prolonged survival (Fig. 7). Beyond the pre–B cell stage, formation of the pre-BCR and BCR preserves those B cells that mature past IL-7 dependency. Interestingly, whereas Cd53−/− and Il-7r+/− mice have similarly reduced developing B cell populations, surface IL-7Rα expression is significantly lower in Il-7r+/− mice (Fig. 5E, 5F). These data then suggest that IL-7 signaling, more so than expression, is maintained by CD53 to promote B cell development.
The association of CD53 and IL-7Rα suggests that the TEM formation is required for homeostatic maturation of B cells. Current studies are underway to identify the essential amino acid residues of CD53 for proper interaction with the IL-7R and to determine other interacting proteins. Finally, with these observed changes in B cell development in the absence of CD53, the ability to modify CD53 stimulation or inhibition could provide a framework for clinical investigation. CD53 overexpression has been observed in several B cell malignancies, including B cell–precursor leukemias, and thus, inhibition of CD53 signaling or interactions may represent innovative therapies for B lineage cancers (43). To our knowledge, we report a novel function of CD53 during B cell development that will promote further study into the potential clinical roles of CD53.
Supplementary Material
Acknowledgments
We thank the Alvin J. Siteman Cancer Center at Washington University School of Medicine and Barnes-Jewish Hospital for the use of the Siteman Flow Cytometry Core and Immunomonitoring Laboratory, which provided assistance with CyTOF experiments. We thank Jillian Dunbar for graphical design assistance, Jackie Tucker-Davis and Al Sorensen for animal care, Kathryn Leonard for technical assistance, and Dr. S. Celeste Morley (Washington University) for valuable discussions. We thank the Genome Engineering and iPSC Center at Washington University for generation of the Cd53−/− mouse.
This work was supported by funding from the Children’s Discovery Institute of Washington University and St. Louis Children’s Hospital (to L.G.S.), and the Training Program in Cellular and Molecular Biology (T32 GM007067-44) (to Z.J.G.).
The online version of this article contains supplemental material.
- BIM
- Bcl2l11
- CLP
- common lymphoid progenitor
- CyTOF
- mass cytometry
- EBF1
- early B cell factor 1
- Flt3-L
- FMS-like tyrosine kinase three ligand
- FO
- follicular
- HSC
- hematopoietic stem cell
- JAK
- Janus-associated kinase
- Lin−
- lineage−
- MPP
- multipotent progenitor
- MZ
- marginal zone
- PI3K
- phosphatidylinositol 3 kinase
- PLA
- proximity ligation assay
- qRT-PCR
- quantitative RT-PCR
- TEM
- tetraspanin-enriched microdomain
- WT
- wild-type.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.LeBien T. W., Tedder T. F. 2008. B lymphocytes: how they develop and function. Blood 112: 1570–1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eibel H. 2015. Early B cell development. In Agammaglobulinemia. Plebani A., Lougaris V., eds. Springer International Publishing, Switzerland, p. 1–17. [Google Scholar]
- 3.Wang L. D., Clark M. R. 2003. B-cell antigen-receptor signalling in lymphocyte development. Immunology 110: 411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sudo T., Nishikawa S., Ohno N., Akiyama N., Tamakoshi M., Yoshida H., Nishikawa S. 1993. Expression and function of the interleukin 7 receptor in murine lymphocytes. Proc. Natl. Acad. Sci. USA 90: 9125–9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kikuchi K., Kasai H., Watanabe A., Lai A. Y., Kondo M. 2008. IL-7 specifies B cell fate at the common lymphoid progenitor to pre-proB transition stage by maintaining early B cell factor expression. J. Immunol. 181: 383–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh H., Medina K. L., Pongubala J. M. 2005. Contingent gene regulatory networks and B cell fate specification. Proc. Natl. Acad. Sci. USA 102: 4949–4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O’Riordan M., Grosschedl R. 1999. Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11: 21–31. [DOI] [PubMed] [Google Scholar]
- 8.Mårtensson I. L., Almqvist N., Grimsholm O., Bernardi A. I. 2010. The pre-B cell receptor checkpoint. FEBS Lett. 584: 2572–2579. [DOI] [PubMed] [Google Scholar]
- 9.Pieper K., Grimbacher B., Eibel H. 2013. B-cell biology and development. J. Allergy Clin. Immunol. 131: 959–971. [DOI] [PubMed] [Google Scholar]
- 10.Peschon J. J., Morrissey P. J., Grabstein K. H., Ramsdell F. J., Maraskovsky E., Gliniak B. C., Park L. S., Ziegler S. F., Williams D. E., Ware C. B., et al. 1994. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J. Exp. Med. 180: 1955–1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.von Freeden-Jeffry U., Vieira P., Lucian L. A., McNeil T., Burdach S. E., Murray R. 1995. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J. Exp. Med. 181: 1519–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corfe S. A., Paige C. J. 2012. The many roles of IL-7 in B cell development; mediator of survival, proliferation and differentiation. Semin. Immunol. 24: 198–208. [DOI] [PubMed] [Google Scholar]
- 13.Heltemes-Harris L. M., Willette M. J., Vang K. B., Farrar M. A. 2011. The role of STAT5 in the development, function, and transformation of B and T lymphocytes. Ann. N. Y. Acad. Sci. 1217: 18–31. [DOI] [PubMed] [Google Scholar]
- 14.Mandal M., Powers S. E., Ochiai K., Georgopoulos K., Kee B. L., Singh H., Clark M. R. 2009. Ras orchestrates exit from the cell cycle and light-chain recombination during early B cell development. Nat. Immunol. 10: 1110–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oliver P. M., Wang M., Zhu Y., White J., Kappler J., Marrack P. 2004. Loss of Bim allows precursor B cell survival but not precursor B cell differentiation in the absence of interleukin 7. J. Exp. Med. 200: 1179–1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tamarit B., Bugault F., Pillet A. H., Lavergne V., Bochet P., Garin N., Schwarz U., Thèze J., Rose T. 2013. Membrane microdomains and cytoskeleton organization shape and regulate the IL-7 receptor signalosome in human CD4 T-cells. [Published erratum appears in 2019 J. Biol. Chem. 294: 5212–5213.] J. Biol. Chem. 288: 8691–8701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hemler M. E. 2005. Tetraspanin functions and associated microdomains. Nat. Rev. Mol. Cell Biol. 6: 801–811. [DOI] [PubMed] [Google Scholar]
- 18.Boucheix C., Rubinstein E. 2001. Tetraspanins. Cell. Mol. Life Sci. 58: 1189–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rasmussen A. M., Blomhoff H. K., Stokke T., Horejsi V., Smeland E. B. 1994. Cross-linking of CD53 promotes activation of resting human B lymphocytes. J. Immunol. 153: 4997–5007. [PubMed] [Google Scholar]
- 20.Tarrant J. M., Robb L., van Spriel A. B., Wright M. D. 2003. Tetraspanins: molecular organisers of the leukocyte surface. Trends Immunol. 24: 610–617. [DOI] [PubMed] [Google Scholar]
- 21.Mollinedo F., Fontán G., Barasoain I., Lazo P. A. 1997. Recurrent infectious diseases in human CD53 deficiency. Clin. Diagn. Lab. Immunol. 4: 229–231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuidscherwoude M., Göttfert F., Dunlock V. M., Figdor C. G., van den Bogaart G., van Spriel A. B. 2015. The tetraspanin web revisited by super-resolution microscopy. Sci. Rep. 5: 12201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zuidscherwoude M., Dunlock V. E., van den Bogaart G., van Deventer S. J., van der Schaaf A., van Oostrum J., Goedhart J., In ’t Hout J., Hämmerling G. J., Tanaka S., et al. 2017. Tetraspanin microdomains control localized protein kinase C signaling in B cells. Sci. Signal. 10: eaag2755. [DOI] [PubMed] [Google Scholar]
- 24.Olweus J., Lund-Johansen F., Horejsi V. 1993. CD53, a protein with four membrane-spanning domains, mediates signal transduction in human monocytes and B cells. J. Immunol. 151: 707–716. [PubMed] [Google Scholar]
- 25.Yunta M., Lazo P. A. 2003. Apoptosis protection and survival signal by the CD53 tetraspanin antigen. Oncogene 22: 1219–1224. [DOI] [PubMed] [Google Scholar]
- 26.Månsson R., Lagergren A., Hansson F., Smith E., Sigvardsson M. 2007. The CD53 and CEACAM-1 genes are genetic targets for early B cell factor. Eur. J. Immunol. 37: 1365–1376. [DOI] [PubMed] [Google Scholar]
- 27.Sentmanat M. F., Peters S. T., Florian C. P., Connelly J. P., Pruett-Miller S. M. 2018. A survey of validation strategies for CRISPR-Cas9 editing. Sci. Rep. 8: 888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuettpelz L. G., Gopalan P. K., Giuste F. O., Romine M. P., van Os R., Link D. C. 2012. Kruppel-like factor 7 overexpression suppresses hematopoietic stem and progenitor cell function. Blood 120: 2981–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagger F. O., Sasivarevic D., Sohi S. H., Laursen L. G., Pundhir S., Sønderby C. K., Winther O., Rapin N., Porse B. T. 2016. BloodSpot: a database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 44: D917–D924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dias S., Månsson R., Gurbuxani S., Sigvardsson M., Kee B. L. 2008. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity 29: 217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zöller M. 2009. Tetraspanins: push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 9: 40–55. [DOI] [PubMed] [Google Scholar]
- 32.Mattila P. K., Feest C., Depoil D., Treanor B., Montaner B., Otipoby K. L., Carter R., Justement L. B., Bruckbauer A., Batista F. D. 2013. The actin and tetraspanin networks organize receptor nanoclusters to regulate B cell receptor-mediated signaling. Immunity 38: 461–474. [DOI] [PubMed] [Google Scholar]
- 33.Fry T. J., Mackall C. L. 2005. The many faces of IL-7: from lymphopoiesis to peripheral T cell maintenance. J. Immunol. 174: 6571–6576. [DOI] [PubMed] [Google Scholar]
- 34.Yao Z., Cui Y., Watford W. T., Bream J. H., Yamaoka K., Hissong B. D., Li D., Durum S. K., Jiang Q., Bhandoola A., et al. 2006. Stat5a/b are essential for normal lymphoid development and differentiation. Proc. Natl. Acad. Sci. USA 103: 1000–1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Medvedovic J., Ebert A., Tagoh H., Busslinger M. 2011. Pax5: a master regulator of B cell development and leukemogenesis. Adv. Immunol. 111: 179–206. [DOI] [PubMed] [Google Scholar]
- 36.Medina K. L., Pongubala J. M., Reddy K. L., Lancki D. W., Dekoter R., Kieslinger M., Grosschedl R., Singh H. 2004. Assembling a gene regulatory network for specification of the B cell fate. Dev. Cell 7: 607–617. [DOI] [PubMed] [Google Scholar]
- 37.Kikuchi K., Lai A. Y., Hsu C. L., Kondo M. 2005. IL-7 receptor signaling is necessary for stage transition in adult B cell development through up-regulation of EBF. J. Exp. Med. 201: 1197–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pongubala J. M., Northrup D. L., Lancki D. W., Medina K. L., Treiber T., Bertolino E., Thomas M., Grosschedl R., Allman D., Singh H. 2008. Transcription factor EBF restricts alternative lineage options and promotes B cell fate commitment independently of Pax5. Nat. Immunol. 9: 203–215. [DOI] [PubMed] [Google Scholar]
- 39.Johnson K., Chaumeil J., Micsinai M., Wang J. M., Ramsey L. B., Baracho G. V., Rickert R. C., Strino F., Kluger Y., Farrar M. A., Skok J. A. 2012. IL-7 functionally segregates the pro-B cell stage by regulating transcription of recombination mediators across cell cycle. J. Immunol. 188: 6084–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Todros-Dawda I., Kveberg L., Vaage J. T., Inngjerdingen M. 2014. The tetraspanin CD53 modulates responses from activating NK cell receptors, promoting LFA-1 activation and dampening NK cell effector functions. PLoS One 9: e97844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tomlinson M. G., Hanke T., Hughes D. A., Barclay A. N., Scholl E., Hünig T., Wright M. D. 1995. Characterization of mouse CD53: epitope mapping, cellular distribution and induction by T cell receptor engagement during repertoire selection. Eur. J. Immunol. 25: 2201–2205. [DOI] [PubMed] [Google Scholar]
- 42.Mannion B. A., Berditchevski F., Kraeft S. K., Chen L. B., Hemler M. E. 1996. Transmembrane-4 superfamily proteins CD81 (TAPA-1), CD82, CD63, and CD53 specifically associated with integrin alpha 4 beta 1 (CD49d/CD29). J. Immunol. 157: 2039–2047. [PubMed] [Google Scholar]
- 43.Barrena S., Almeida J., Yunta M., López A., Fernández-Mosteirín N., Giralt M., Romero M., Perdiguer L., Delgado M., Orfao A., Lazo P. A. 2005. Aberrant expression of tetraspanin molecules in B-cell chronic lymphoproliferative disorders and its correlation with normal B-cell maturation. Leukemia 19: 1376–1383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.