Abstract
Soluble extracts prepared from Xenopus eggs have been used extensively to study various aspects of cellular and developmental biology. During early egg development, transcription of the zygotic genome is suppressed. As a result, traditional extracts derived from unfertilized and early stage eggs possess little or no intrinsic transcriptional activity. In this study, we show that Xenopus nucleoplasmic extract (NPE) supports robust transcription of a chromatinized plasmid substrate. Although prepared from eggs in a transcriptionally inactive state, the process of making NPE resembles some aspects of egg fertilization and early embryo development that lead to transcriptional activation. With this system, we observed that promoter-dependent recruitment of transcription factors and RNA polymerase II leads to conventional patterns of divergent transcription and pre-mRNA processing, including intron splicing and 3′ cleavage and polyadenylation. We also show that histone density controls transcription factor binding and RNA polymerase II activity, validating a mechanism proposed to regulate genome activation during development. Together, these results establish a new cell-free system to study the regulation, initiation, and processing of mRNA transcripts.
Keywords: plasmid, Xenopus, transcription, histone, RNA processing, cell-free system, gene regulation, genome activation, in vitro assay, nucleoplasmic extract
Introduction
The eggs of Xenopus laevis frogs contain a high concentration of maternal factors that support early embryo development after fertilization (1, 2). Soluble extracts prepared from Xenopus eggs have been used extensively to study various aspects of cellular and developmental biology, including nuclear formation (3–5), DNA replication and repair (6–9), cellular and checkpoint signaling (10–13), mitosis (14–16), and apoptosis (17). However, these extracts have been found to possess little or no intrinsic transcriptional activity (18), limiting study of a fundamental biological process with this model system.
The primary characteristics of Xenopus egg extracts are determined by the developmental stage of the eggs from which they are derived (described in Fig. S1) (19). Newly laid eggs are arrested in metaphase II of meiosis. After fertilization, eggs progress to an interphase state that is transcriptionally inactive. Chromatin then undergoes decondensation and is enveloped by membranes to form a nucleus. Although limited transcription of the nuclear genome can occur, further development depends on maternal proteins and mRNA provided by the egg cytoplasm (1, 2). The single-cell embryo then undergoes multiple rounds of rapid DNA synthesis and cellular division to form a fluid-filled sphere of cells called a blastula. At this point in embryo development, the genome transitions to a transcriptionally active state through a process referred to as the mid-blastula transition (MBT)2 (20).
Recent studies have identified histones as an important regulator of the MBT, suggesting they act as a sensor for the number of cellular divisions (21–23). DNA is bound by histones to form chromatin, which supports DNA compaction and acts as a scaffold for regulating various aspects of transcription (24, 25). During early embryo development, the concentration of maternal histones remains constant. However, each round of DNA synthesis increases the ratio of DNA to histones. As histones become limiting, promoter elements throughout the genome are thought to become more accessible to transcription factors that trigger a wave of transcriptional activity. The MBT is characterized by several cellular changes that promote differentiation and further embryo development, including slower cell cycles with extended S phase, asynchronous cellular divisions, and cellular motility (20).
Previously, a nucleoplasmic extract (NPE) was developed that contains a highly concentrated fraction of nuclear proteins (26). NPE supports highly efficient chromatinization and synthesis of plasmid DNA substrates and has led to seminal discoveries in DNA replication and repair (27–31). However, the transcriptional activity of NPE has not been determined. Although prepared from eggs in a transcriptionally inactive state, the process of making NPE recapitulates several events during egg fertilization and early embryo development that lead to transcriptional activation (Fig. S1). When eggs are crushed by centrifugation, calcium release drives the extract into interphase, mimicking the events following egg fertilization. Addition of sperm chromatin then leads to nuclear formation and chromatin condensation, followed by progression into S phase and DNA synthesis.
In this study, we demonstrate that NPE readily supports transcription from endogenous gene elements on a naturally chromatinized plasmid substrate. Promoter-dependent recruitment of transcription factors and RNA polymerase II (RNAPII) leads to conventional patterns of divergent transcription and pre-mRNA processing, including intron splicing and 3′ cleavage and polyadenylation. We also show that histone density regulates transcription in NPE by limiting the recruitment of transcription factors to DNA, validating a mechanism proposed to control genome activation during early development (21). Together, these results establish a new cell-free system that supports multiple mechanisms involved in the regulation, initiation, and processing of mRNA transcripts.
Results
Nucleoplasmic extract supports robust transcription of plasmid DNA
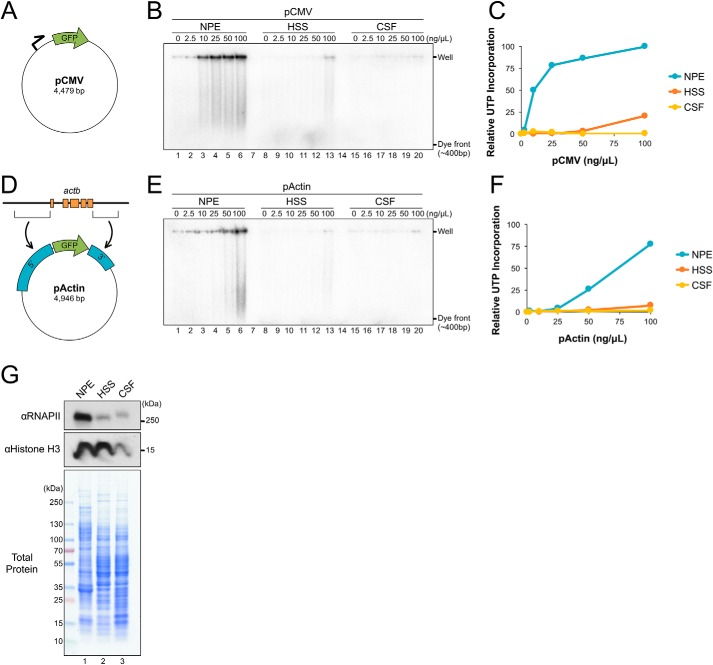
To determine the relative efficiency of transcription in NPE, we compared its activity with other Xenopus egg extracts shown to have limited transcriptional activity, including HSS (a high-speed supernatant of interphase-arrested eggs) and CSF (a mitotic extract from eggs arrested in metaphase II by a cytostatic factor). Each extract was incubated with increasing concentrations of a GFP reporter plasmid that contains a cytomegalovirus (CMV) promoter (Fig. 1A; pCMV). The CMV promoter was previously shown to support transcription in Xenopus oocytes (18) and cultured somatic cell lysate (32), indicating that it is recognized by Xenopus transcription machinery. Extracts were supplemented with [α-32P]UTP, and its incorporation into RNA transcripts was visualized by agarose gel electrophoresis and autoradiography (Fig. 1B). When pCMV was incubated in HSS or CSF, there was little or no UTP incorporation, respectively (Fig. 1B, lanes 8–13 and 15–20). In contrast, incubation in NPE led to a large accumulation of radiolabeled product (Fig. 1B, lanes 1–6), indicating that transcription of plasmid DNA readily occurs in NPE.
Figure 1.
NPE supports robust transcription of plasmid substrates. A, pCMV schematic. Relative location of promoter and GFP are indicated. B, different concentrations of pCMV were incubated in NPE, HSS, or CSF extract in the presence of [α-32P]UTP. Samples were withdrawn at 180 min, resolved by agarose gel electrophoresis, and visualized by autoradiography. C, total UTP incorporation from (B) was quantified and graphed. D, pActin schematic showing the 5′ and 3′ regions cloned from Xenopus actb. E, pActin was incubated in NPE, HSS, or CSF and UTP incorporation was analyzed in parallel to (B) to allow a direct comparison. F, total UTP incorporation from (E) was quantified and graphed relative to peak intensity in (B). G, total protein from each extract was resolved by SDS-PAGE and visualized with Coomassie Brilliant Blue stain or by Western blotting using the indicated antibodies.
To investigate transcription of a promoter native to the Xenopus genome, we replaced the 5′ and 3′ regions of pCMV with those from the Xenopus laevis actb gene to form pActin (Fig. 1D). actb encodes β-actin, one of three major actin isoforms found in vertebrates, and is known to be transcriptionally activated during egg development (33). pActin was incubated in NPE, HSS, and CSF extracts and UTP incorporation was visualized as described above. As seen with pCMV, pActin was readily transcribed in NPE, but showed little or no UTP incorporation in HSS or CSF (Fig. 1E). At the highest DNA concentration tested (100 ng/μl), pCMV and pActin had similar levels of UTP incorporation (compare blue traces in Fig. 1, C and F). However, at lower DNA concentrations, pActin produced relatively fewer products. These results suggest that transcription from the actb promoter is suppressed in NPE and that the effect can be alleviated with excess DNA.
To compare the relative levels of transcription machinery in each extract, equal volumes of NPE, HSS, and CSF were analyzed by Western blotting and Coomassie Brilliant Blue stain. Although total protein levels were relatively similar in each extract, RNAPII was highly enriched in NPE compared with HSS and CSF (Fig. 1G). We also saw that the level of histone H3 was enriched in NPE and HSS, compared with CSF. Thus, compared with other Xenopus egg extracts, NPE is enriched for both RNAPII and histones.
Transcription is driven by regulated recruitment of RNAPII to the promoter
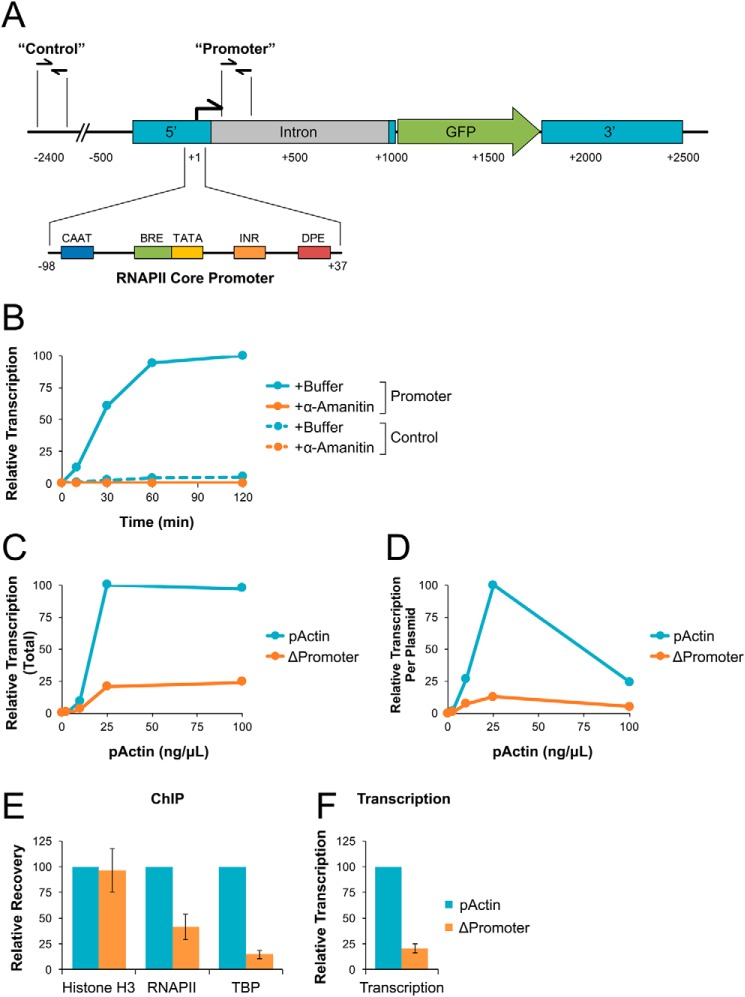
To quantify transcription originating from the actb promoter, RNA products were isolated from NPE and analyzed by reverse transcription quantitative PCR (RT-qPCR). RNA levels were measured using primers that amplify a region ∼150 bp downstream of the actb promoter region (Promoter) or ∼2400 bp upstream (Control) (Fig. 2A). Primers were also used to amplify endogenous Xenopus 18S rRNA that is retained during the preparation of NPE to serve as an internal control for RNA recovery between different samples.
Figure 2.
NPE supports regulated and promoter-dependent transcription. A, pActin schematic. Sequence elements are shown relative to the transcription start site (+1). “Control” and “Promoter” primer pair locations are indicated. B, pActin was incubated at 10 ng/μl in NPE supplemented with buffer or α-amanitin. RNA was isolated at the indicated time points and quantified by RT-qPCR. C, different concentrations of pActin or ΔPromoter plasmid were incubated in NPE for 120 min. RNA was isolated and quantified by RT-qPCR using the Promoter primers. D, transcription from (C) was normalized based on starting plasmid concentration. E, pActin or ΔPromoter plasmid were incubated in NPE at 25 ng/μl. At 30 min, DNA-bound protein was analyzed by ChIP with the indicated antibodies. F, at 120 min, RNA was isolated from the reactions in (E) and quantified by RT-qPCR using the Promoter primers. Error bars represent ± 1 S.D. See Fig. S2 for experimental replicates.
When pActin was incubated in NPE, transcription from the actb promoter increased over time, peaking at ∼60 min (Fig. 2B, solid cyan trace). In comparison, transcription of the control region was relatively low, reaching only ∼5% of that detected at the promoter (Fig. 2B, dashed cyan trace). We then supplemented NPE with α-amanitin, a highly selective inhibitor of RNAPII (34). In the presence of α-amanitin, transcription from both the promoter and control regions was reduced to <1% of that detected from the promoter in buffer-treated samples (Fig. 2B, solid and dashed orange traces), indicating that transcription at both sites is RNAPII-dependent.
We showed that total UTP incorporation was sensitive to the concentration of pActin incubated in NPE (Fig. 1, E and F). To directly test how DNA concentration affected transcription from the actb promoter, we incubated NPE with increasing concentrations of pActin and then analyzed the accumulation of RNA products by RT-qPCR. For comparison, we also analyzed transcription from a pActin control plasmid that contained a deletion of the RNAPII core promoter elements (ΔPromoter). Total transcription from the actb promoter increased with pActin concentration up to 25 ng/μl and then plateaued (Fig. 2C, cyan trace). At all concentrations tested, transcription from the actb promoter region was severely reduced with the ΔPromoter plasmid compared with pActin (Fig. 2C, orange trace), showing that NPE supports promoter-driven transcription. By calculating the amount of transcription per plasmid, we saw that transcription efficiency peaked at 25 ng/μl and was severely reduced at both lower and higher DNA concentrations (Fig. 2D, cyan trace). Similar results were also seen when a fixed amount of pActin was incubated with increasing amounts of a “carrier” plasmid that has no sequence homology (Fig. S2, C and D), indicating that transcription efficiency was determined by total DNA concentration and not the number of actb promoters present in the reaction. Thus, at low DNA concentrations, transcription from the actb promoter is suppressed in NPE. At high DNA concentrations, the transcriptional machinery likely becomes limiting, reducing overall efficiency but not total product produced.
To verify that the actb promoter supports regulated transcription in NPE, we analyzed recruitment of histone H3, RNAPII, and the transcription factor TATA-binding protein (TBP) to the 5′ region of both the pActin and ΔPromoter plasmids by chromatin immunoprecipitation (ChIP). Compared with pActin, binding of RNAPII and TBP to the ΔPromoter plasmid was severely reduced (Fig. 2E), consistent with the decrease in transcription at the promoter region (Fig. 2F). In contrast, histone H3 levels remained similar for both plasmids. These results indicate that the actb promoter supports sequence-specific recruitment of bona fide transcription factors to initiate transcription in NPE.
Regulation of transcriptional activity by histone density
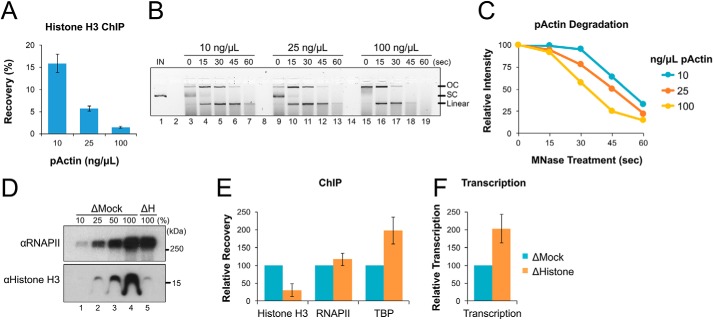
During early development, relative histone levels play an important role in regulating the onset of transcription during the MBT. When plasmid DNA is incubated in NPE, it becomes spontaneously chromatinized within ∼30 min (Fig. S3). To investigate whether the level of histone binding in NPE was responsible for decreased transcription at low DNA concentrations, we first incubated different amounts of pActin in NPE for 30 min and then analyzed DNA-bound histone H3 by ChIP. With increasing concentrations of pActin, the percentage of histone-bound DNA recovered was reduced by more than 10-fold (Fig. 3A), indicating a dramatic decrease in the number of histones bound to each plasmid.
Figure 3.
Histone occupancy regulates transcriptional activity in NPE. A, pActin was incubated in NPE at the indicated concentrations for 30 min. DNA-bound protein was then analyzed by ChIP using histone H3 antibodies. B, pActin was incubated in NPE at the indicated concentrations for 60 min. Next, reaction samples were diluted in MNase buffer and treated with 100 units MNase at 37°C for the indicated time. DNA was then isolated and resolved by agarose gel electrophoresis. Input DNA (IN) and topological isoforms of the resolved plasmids are indicated: open circular (OC), supercoiled (SC), and linear. C, the total intensity of all three full-length plasmid molecules identified in (B) was quantified and graphed. D, mock-depleted (ΔMock) and histone-depleted (ΔH) NPE were analyzed by Western blotting with the indicated antibodies. E, pActin was incubated in ΔMock or ΔH extract at 10 ng/μl. At 30 min, DNA-bound protein was analyzed by ChIP using the indicated antibodies. F, RNA was isolated from the reactions in (E) at 120 min and quantified by RT-qPCR using the Promoter primers. Error bars represent ± 1 S.D.
We then tested whether plasmid concentration also affected DNA accessibility. pActin was again incubated in NPE at various concentrations. After 60 min, reactions were supplemented with micrococcal nuclease (MNase), which exhibits both exonuclease and endonuclease activity against exposed dsDNA. Reaction samples containing equal amounts of DNA were withdrawn at different times after MNase addition, separated by agarose gel electrophoresis, and then visualized with SYBR Gold stain. As the concentration of pActin incubated in NPE increased, we saw that its sensitivity to MNase digestion also increased (Fig. 3, B and C). Together, these results suggest that changes in DNA concentration affect DNA accessibility by altering histone density.
To determine whether histone availability controls the access of transcription machinery to DNA, we immunodepleted NPE using pre-immune (Mock) or anti-histone H4K12ac (ΔH) antibodies, which co-depleted more than 75% of total histone H3 from extract (Fig. 3D) (35). pActin was then incubated in each extract, and samples were withdrawn after 30 min to measure protein binding by ChIP. Compared with mock-depleted reactions, histone depletion reduced the level of DNA-bound histones by ∼3-fold (Fig. 3E). In contrast, histone depletion caused TBP binding to increase ∼2-fold, consistent with greater access to nucleosome-free DNA. Although RNAPII levels were not significantly changed, transcription also increased ∼2-fold in histone-depleted reactions (Fig. 3F), suggesting that a greater fraction of DNA-associated RNAPII complexes were activated by TBP and able to transcribe downstream from the promoter. Together, these results suggest that reduced histone abundance allows increased transcription factor binding and stimulates RNAPII activity, consistent with models developed for Xenopus development(21–23).
RNA sequencing reveals characteristics of actb promoter regulation
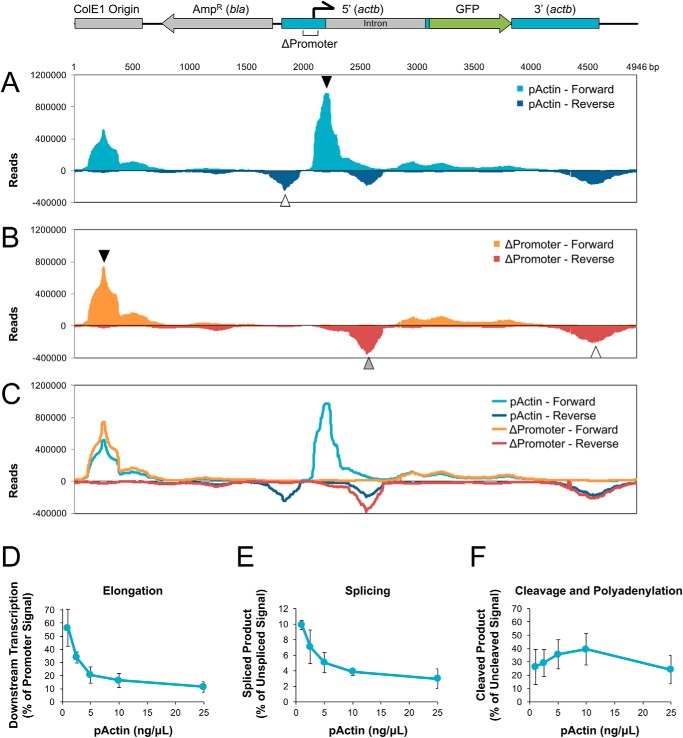
To further investigate how NPE supports transcription of the actb promoter, RNA products were analyzed by RNA sequencing (RNA-Seq). pActin or the ΔPromoter plasmid were each incubated in NPE at 25 ng/μl for 120 min. Total RNA was then isolated and analyzed using paired-end RNA-Seq. RNA reads were aligned to the pActin sequence and both forward (+) and reverse (−) reads were graphed. Consistent with RT-qPCR results (Fig. 2C), pActin showed a large accumulation of RNA immediately downstream of the actb promoter in the forward orientation (Fig. 4A, closed arrowhead). A smaller RNA peak was also present upstream of the actb promoter in the reverse orientation (Fig. 4A, open arrowhead). This pattern of divergent transcription from a promoter is thought to be important for maintaining a nucleosome-free region for initiation and has been observed in organisms ranging from yeast to mammals (36). Both divergent peaks were completely absent in the ΔPromoter reads (Fig. 4B), indicating that formation of both RNA products depended on the actb promoter.
Figure 4.
Analysis of whole-plasmid transcription and pre-mRNA processing in NPE. A, pActin was incubated at 25 ng/μl in NPE for 120 min. RNA was then purified and analyzed by paired-end RNA-Seq. The total of all mapped reads were graphed for both the forward (+) and reverse (−) orientations. B, the ΔPromoter plasmid was incubated in NPE and analyzed by RNA-Seq as described in (A). C, reads from (A) and (B) were overlaid onto the same graph for direct comparison. A diagram of pActin showing the relative position of major sequence elements is shown above. See text for description of arrowheads. D–F, pActin was incubated in NPE at the indicated concentrations for 120 min. RNA was then isolated and analyzed by RT-qPCR to determine the efficiency of (D) elongation, (E) splicing, and (F) cleavage and polyadenylation. Elongation was graphed as a percentage of amplification with Elongation primers versus Promoter primers. Splicing and cleavage and polyadenylation were graphed as a percentage of amplification with Unspliced and Uncleaved primers, respectively. Error bars represent ± 1 S.D.
There were three major regions of actb-independent transcription found on pActin. The largest peak was localized to the ColE1 origin (Fig. 4B, closed arrowhead) and was adjacent to a smaller peak in the opposite orientation (Fig. 4B, open arrowhead). These reads were likely produced by divergent transcription originating from an A/T-rich region within the origin sequence (37). The third peak originated within the 5′ actb intron (Fig. 4B, gray arrowhead) and faced toward the major actb promoter peak. In a previous study analyzing actb expression, deletion analysis identified a negative transcriptional element in this region (38). Together with our RNA-Seq data, these results suggest that the intron promoter may interfere with expression of actb(39). Interestingly, transcription from both the ColE1 origin and the actb intron regions increased in the ΔPromoter plasmid relative to pActin (Fig. 4C, compare cyan with orange and blue with red traces), suggesting that the actb promoter competes with nearby promoters.
We noted that forward transcription from the actb promoter was limited in length. Transcripts showed highly efficient initiation and extension to ∼250 nucleotides, well beyond the short transcripts associated with abortive transcription (up to ∼15 nucleotides) (40, 41). Roughly 16% of established transcripts escaped the promoter region, extending further to ∼310 nucleotides. Extension beyond this point failed rapidly, with the vast majority of transcripts terminating by ∼500 nucleotides. This phenomenon was not specific to the actb promoter, as forward transcription from the ColE1 origin (Fig. 4B, closed arrowhead) and reverse transcription from the intron region (Fig. 4B, gray arrowhead) also showed similar lengths of elongation.
Transcription elongation and pre-mRNA processing in NPE
We reasoned that some factors involved in transcription elongation might be limiting in extract. To test this hypothesis, pActin was incubated in NPE at different concentrations for 120 min, and then RT-qPCR was used to measure transcription at the promoter and another site ∼600 bp downstream. Because total transcription levels vary with plasmid concentration (as seen in Fig. 2C), we graphed “elongation” as the percentage of transcripts measured downstream versus at the promoter (Fig. 4D). At 25 ng/μl, elongation efficiency was ∼11%. However, with decreasing pActin concentration, the efficiency of elongation increased dramatically, reaching ∼56% at 1 ng/μl. Thus, transcription elongation improved at lower DNA concentrations in NPE, despite higher levels of histone binding (Fig. 3A).
The 5′ region of actb contains an intron that is typically spliced during expression of the actb gene. However, analysis of the pActin RNA-Seq identified only a trace amount of reads corresponding to spliced products (data not shown). To test whether splicing improved with increased transcription elongation, RNA produced at different pActin concentrations was analyzed by RT-qPCR using primers that amplify either unspliced or spliced transcripts (Fig. S4, B and C). As with elongation, splicing efficiency increased when the concentration of pActin was reduced from 25 to 1 ng/μl (Fig. 4E), suggesting that the two processes are linked during transcription in NPE (42).
To examine 3′ cleavage and polyadenylation, we first used an unbiased approach to identify potential cleavage sites. pActin was incubated in NPE at 5 ng/μl for 120 min and RNA was isolated to generate cDNA. Transcripts were then amplified using a forward primer that hybridizes upstream of the consensus polyadenylation sequence (43) and an anchored oligo-dT reverse primer. The major PCR product was gel purified and sequenced. New primers were then designed that amplify either uncleaved transcripts or those that had been cleaved and polyadenylated (Fig. S4, D and E). RNA produced at different pActin concentrations was then analyzed to measure the efficiency of cleavage and polyadenylation at this site. Unlike elongation and splicing, the efficiency of cleavage and polyadenylation remained relatively constant at each pActin concentration tested (Fig. 4F). Taken together, these results indicate that NPE supports regulated transcription from endogenous promoters and subsequent pre-mRNA processing required to generate mature mRNA transcripts.
Discussion
Xenopus egg extracts have been used extensively to study numerous biological processes in a highly tractable system. A major advantage of extracts over cell-based models is the ability to study direct effects in the absence of global gene expression and cell cycle changes. However, traditional Xenopus egg extracts have been found to support only limited transcription without the addition of exogenous transcription machinery or the removal of endogenous histones to prevent DNA chromatinization (18, 21–23). In this study, we establish a new cell-free system using NPE that supports robust transcription of chromatinized substrates by endogenous factors.
Transcription activity and efficiency in NPE
NPE contains a highly concentrated fraction of soluble, nuclear proteins. Relative to other Xenopus egg extracts, NPE is enriched for both transcription machinery and histones (Fig. 1G). This combination of factors supports robust transcription (Fig. 1B and E) in the context of chromatinized DNA (Figs. S3 and 3A). NPE is prepared from nuclei undergoing active DNA synthesis. As such, transcription in NPE likely resembles that of cells within S phase. Although the primary mechanics of transcription are similar throughout the cell cycle, the regulation of specific genes and transcription factors may be influenced by these characteristics of NPE.
Xenopus eggs contain an abundance of histone dimers that are coupled to molecular chaperones (44, 45). In NPE, these complexes promote spontaneous loading of histones onto DNA within ∼30 min (Fig. S3). Although transcription of pActin begins prior to complete chromatinization, robust transcription continues up to ∼60 min before leveling off (Fig. 2B). The decline in transcription activity over time is not because of limited availability of ribonucleotides or ATP (data not shown). This limited window of transcription suggests that factors required for initiating transcription become inactivated or suppressed over time, possibly because of changes in chromatin signaling (25).
Based on the final quantity of RNA detected by qPCR and the efficiencies of RNA isolation and cDNA amplification (determined using samples of known concentration), we estimate that the actb promoter produced ∼2.5 extended transcripts per molecule of pActin when incubated in NPE. Transcriptional efficiency of the actb promoter peaked at 25 ng/μl (Fig. 2D and S1B), suggesting a balance between fully chromatinized DNA that suppresses transcription and underchromatinized DNA that supports increased transcription factor binding (Fig. 3E). These conditions are likely analogous to the cellular states of chromatin referred to as heterochromatin and euchromatin, respectively (46).
Histone density and developmental regulation
During early embryo development, histones act as a sensor for the number of cellular divisions. As histones become limiting, it triggers the MBT, which marks the onset of transcription and developmental progression (20–22). In NPE, the ratio of DNA to extract plays a similar role in regulating transcription, which can be modulated by altering the concentration of plasmid present within the reaction (Fig. 2C). Previous studies have shown that the MBT can be altered by adding or depleting histones (21, 22), arguing that histone levels control the onset of transcription. Our results indicate that transcription in NPE is regulated by the same mechanism. Increasing plasmid DNA concentration reduced histone binding (Fig. 3A) and increased DNA accessibility (Fig. 3, B and C). We also showed that histone binding limited recruitment of TBP to the actb promoter (Fig. 3E), adding support to the model that histone occupancy suppresses transcription by obscuring genes during early embryo development.
Promoter activity and pre-RNA processing
Using RNA-Seq, we performed a detailed analysis of transcripts produced by pActin and the ΔPromoter plasmids. We saw traditional patterns of divergent transcription originating from the actb promoter and plasmid origin sequence (Fig. 4A). Promoters that support divergent transcription initiate bidirectional transcripts read from opposite DNA strands. Although some antisense transcripts are unstable and quickly degraded, others have important regulatory functions (47). Despite the prevalence of divergent transcription throughout higher order species, the mechanism of its regulation and biological purpose remain poorly understood.
Within the 5′ intron of actb, we identified a putative promoter (Fig. 4B, gray arrowhead) oriented toward the actb promoter (Fig. 4A, closed arrowhead). Transcription from the intron promoter was weaker than that of actb, but increased when the core promoter elements of actb were deleted (Fig. 4C). Although this region of actb was proposed to contain a negative regulatory element (38), its potential role in regulating gene expression has not been explored. Notably, the intrinsic activity of weak or dormant promoters may also be elevated in this system because of the absence of other genomic elements that normally compete for access to limited transcription factors and machinery.
Interestingly, we found that pre-mRNA processing events responded differently to changes in plasmid concentration. The efficiency of cleavage and polyadenylation remained relatively constant at different plasmid concentrations (Fig. 4F), suggesting that the required factors are present in excess or able to function independently of other transcriptional events. In contrast, the efficiency of transcription elongation and splicing both improved in a nonlinear fashion with decreasing pActin concentration (Fig. 4D and E), arguing that a threshold must be achieved for full stimulation. We propose that highly chromatinized DNA is required for efficient elongation and splicing to occur in NPE. Although histones generally play a negative role in transcription initiation (Fig. 3F), they are also critical for chromatin signaling that regulates many downstream events (48–51).
Concluding remarks
Together, the results described in this study add another fundamental process to NPE's repertoire. As such, it provides a unique tool to examine the interplay between different cellular processes and the various pathways that regulate them. Decades of research have revealed an array of dynamic regulatory networks that control each phase of gene expression (48, 52, 53). When these mechanisms fail, it can result in the development of numerous diseases, including cancer (51, 54–56). Understanding how different signaling events directly impact the initial phases of gene expression will provide new insight into the mechanisms of disease and identify new strategies for treatment.
Experimental procedures
Plasmid substrates
The parent pCMV vector was purchased from Addgene (no. 11153). The 5′ and 3′ regions of actb were amplified from Xenopus laevis sperm chromatin (prepared as described in Ref. 57) using the following primer pairs: 5′ region, CAGGAACTAGTAGAACAGGGAAGCAATGGAT and TAGACCATGGTGGCCTGAAAAGAGAATTAGATT; 3′ region, ATAGCGGCCGCAGGACAGACCCTTTCAACATG and GCGCTGCCTAGGTTTGTTTGAGTGCACCACC.
The resulting fragments were then cloned into pCMV using SpeI and NcoI (5′ region) or NotI and AvrII (3′ region). The carrier plasmid (pCarrier) utilized in Fig. S2, C and D was a pFastBac1 vector (Thermo Fisher) carrying the Xenopus BARD1 gene (58). To generate the ΔPromoter plasmid, actb core promoter elements were deleted by site-directed mutagenesis (Agilent Technologies) using the following primers: forward, CTTCGTCCGCAGTTCCTACGTCCAACCCTCAGGC and reverse, GCCTGAGGGTTGGACGTAGGAACTGCGGACGAAG.
Incubation in Xenopus egg extract
HSS, CSF, and NPE were produced as described previously (26, 57). The care and use of Xenopus laevis followed established protocols approved by IACUC with AAALAC accreditation. In all reactions, extracts were supplemented with ATP regenerating mix (6.5 mm phosphocreatine, 0.65 mm ATP, and 1.6 μg/ml creatine phosphokinase). NPE was also supplemented with 1 mm DTT, and CSF was supplemented with 0.3 mm Ca2+ to promote entry into interphase. Reactions were incubated at 21 °C for 10 min prior to the addition of plasmid DNA, which represents 0 min. Where indicated, extracts were supplemented with [α-32P]UTP to label nascent RNA, or 10 μm α-amanitin to inhibit RNAPII. All experiments were performed at least two times with representative or averaged data shown.
UTP incorporation gels
Three h after addition of plasmid DNA to each extract, samples were withdrawn from the reaction and added to Stop Buffer (3.6% SDS, 18 mm EDTA, 90 mm Tris-HCl, 9% Ficoll). Samples were then mixed with RNA Gel Loading Dye (Thermo Fisher), incubated at 65 °C for 20 min, and resolved by agarose gel electrophoresis. Radiolabeled transcripts were visualized and quantified using a phosphorimager.
RT-qPCR
RNA was isolated from extract using the EZNA RNA Purification kit (Omega Bio-tek) and cDNA was generated using the QuantiTect Reverse Transcription kit (Qiagen). Samples were then analyzed by quantitative real-time PCR with the following primer pairs: control, CCCAACCAGTGTTACACCACTTCC and ATGCCTGGGAGCGGCCTTAT; promoter, TATGGGCTGCATGAAATGG and AATTGCGCGACCTACAACTC; elongation, GCGCTTTACGTTAGCAATCC and AGGCTTTCAGTGAGCCAGTC; unspliced, GCGGGTCCTCACCTTCAATCTAATTCTC and TCGCCGGACACGCTGAACTT; spliced, TCCAACCCTCAGGCCACCAT and TCGCCGGACACGCTGAACTT; uncleaved, CACATCTGTTTCTTGCTATGAGGTG and CAAAACCCATTCATTTTGCCA; cleaved, CACATCTGTTTCTTGCTATGAGGTG and TTTTTTTTTTTTTTTTTTTTGCCA; 18S rRNA, GACCGGCGCAAGACGAACCA and TGCTCGGCGGGTCATGGGAA.
To quantify RNA processing, the level of nonspecific amplification of spliced and cleaved primers was determined using a pActin standard curve, which only contains unspliced and uncleaved sequences. The background amplification was then subtracted from each sample based on the level of total unprocessed RNA present in that sample. PCR fragments containing spliced or cleaved sequences were also generated using the spliced and cleaved primer pairs to create a standard curve for processed samples.
To identify cleavage and polyadenylation sites downstream of the actb promoter, pActin was incubated in NPE for 120 min at 5 ng/μl and cDNA was generated as described above. Cleaved and polyadenylated transcripts were amplified by PCR with the following primers: forward primer, GGGCTCATTCTCTTTAACATCTGGAAG; anchored oligo(dT)20, TTTTTTTTTTTTTTTTTTTTVN (Integrated DNA Technologies). PCR reaction products were then resolved by agarose gel electrophoresis, purified using a gel extraction kit (Qiagen), and sequenced (Genewiz).
Chromatin immunoprecipitation (ChIP)
ChIP was performed as described previously (59). Briefly, reaction samples were crosslinked in Egg Lysis Buffer (10 mm HEPES-KOH, pH 7.7, 2.5 mm MgCl2, 50 mm KCl, and 250 mm sucrose) containing 1% formaldehyde. Crosslinking was stopped by the addition of 125 mm glycine, and formaldehyde was removed using a Micro Bio-Spin 6 chromatography column (Bio-Rad). Samples were then sonicated (Diagenode Bioruptor UCD-600 TS) and immunoprecipitated with the indicated antibody. Following immunoprecipitation, crosslinks were reversed and DNA was isolated by phenol/chloroform extraction and ethanol precipitation. Total (INPUT) and recovered DNA were then analyzed by qPCR to determine percent recovery using the following primer pairs: pActin, CCTCCTTCGTCCGCAGTTCC and GCTGGCGAACCGCTACTTGC and Δpromoter, GAAAATACGGGGCGTGGAAGATT and GCTGGCGAACCGCTACTTGC.
Micrococcal nuclease digestion
pActin was incubated in NPE at 10, 25, or 100 ng/μl for 60 min at 21 °C. Equal amounts of pActin were withdrawn from each reaction and mixed with 1× Micrococcal Nuclease Reaction Buffer (New England Biolabs). Additional NPE was also added to the 25 and 100 ng/μl mixes so that all three treatments contained equal amounts of DNA and extract. 100 units of micrococcal nuclease (New England Biolabs) was then added and reactions were incubated at 37 °C. At the indicated time, samples were withdrawn, mixed with an equal volume of STOP Buffer, and then treated with proteinase K (Thermo Fisher) for 120 min at 37 °C. Undigested DNA was then resolved by agarose gel electrophoresis and visualized with SYBR Gold stain.
Plasmid pulldown
Plasmids were isolated from NPE as described previously(59). Briefly, reaction samples were withdrawn at the indicated time and added to LacI-coupled magnetic beads (Dynabeads M-280; Invitrogen) suspended in LacI pulldown buffer (10 mm HEPES, pH 7.7, 2.5 mm MgCl2, 50 mm KCl, 250 mm sucrose, 0.25 mg/ml BSA, and 0.02% Tween 20). Samples were incubated for 20 min, rotating at 4 °C. Beads were then washed three times with LacI wash buffer (10 mm HEPES, pH 7.7, 2.5 mm MgCl2, 50 mm KCl, 0.25 mg/ml BSA, and 0.02% Tween 20), dried, and suspended in 2× SDS sample buffer (100 mm Tris-HCl, pH 6.8, 4% SDS, 0.2% bromphenol blue, 20% glycerol, and 200 mm β-mercaptoethanol). DNA-bound proteins were then resolved by SDS-PAGE and visualized by Western blotting with the indicated antibodies.
Antibodies and immunodepletion
RNA polymerase II antibodies were purchased from Bethyl Laboratories (A300-653A for Western blotting, A304-405A for ChIP). TBP antibodies were purchased from Boster Biological Technology (PA1534). Histone H3 antibodies were purchased from Thermo Fisher (PA5-16183). To deplete histones from NPE, two rounds of depletion were performed by incubating 10 μl of extract with Protein-A Sepharose beads (GE Healthcare) bound to 50 μg of purified histone H4 K12Ac antibodies(35) for 1 h at 4 °C.
RNA sequencing analysis
RNA was isolated from extract using the E.Z.N.A. RNA Purification kit (Omega Bio-tek). Total RNA samples were then analyzed by Novogene after rRNA removal using paired-end RNA-Seq. A total of ∼10,000,000 clean reads were obtained for both the pActin and ΔPromoter plasmids. Output FASTQ files were aligned to the pActin sequence using bowtie version 2.3.5(60). One nucleotide was removed from the 3′ and 5′ end of reads and the subseeding length was 20. Bam files were sorted by SAMtools (61) and output alignments were analyzed using Integrative Genomics Viewer.
Author contributions
J. K. B. and D. T. L. conceptualization; J. K. B. and D. T. L. data curation; J. K. B. investigation; J. K. B. and D. T. L. methodology; J. K. B. writing-original draft; J. K. B. and D. T. L. writing-review and editing; D. T. L. formal analysis; D. T. L. supervision; D. T. L. funding acquisition.
Supplementary Material
Acknowledgments
We thank Hiroshi Kimura for providing the histone H4K12 acetylation antibody. We thank James Dewar and members of the laboratory for critical reading of the manuscript.
This work was supported by National Institutes of Health Grant R35 GM119512 (to D. T. L.) and National Center for Advancing Translational Sciences (NCATS), National Institutes of Health Grants TL1 TR001451 and UL1 TR001450 (to J. K. B.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S4.
- MBT
- mid-blastula transition
- NPE
- nucleoplasmic extract
- RNAPII
- RNA polymerase II
- HSS
- high-speed supernatant
- CMV
- cytomegalovirus
- qPCR
- quantitative PCR
- TBP
- TATA-binding protein
- MNase
- micrococcal nuclease.
References
- 1. Heasman J. (2006) Patterning the early Xenopus embryo. Development (Camb.) 133, 1205–1217 10.1242/dev.02304 [DOI] [PubMed] [Google Scholar]
- 2. Jones C. M., and Smith J. C. (2008) An overview of Xenopus development. Methods Mol. Biol. 461, 385–394 10.1007/978-1-60327-483-8_28 [DOI] [PubMed] [Google Scholar]
- 3. Blow J. J., and Sleeman A. M. (1990) Replication of purified DNA in Xenopus egg extract is dependent on nuclear assembly. J. Cell Sci. 95, 383–391 [DOI] [PubMed] [Google Scholar]
- 4. Macaulay C., and Forbes D. J. (1996) Assembly of the nuclear pore: Biochemically distinct steps revealed with NEM, GTP gamma S, and BAPTA. J. Cell Biol. 132, 5–20 10.1083/jcb.132.1.5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Newport J. (1987) Nuclear reconstitution in vitro: Stages of assembly around protein-free DNA. Cell 48, 205–217 10.1016/0092-8674(87)90424-7 [DOI] [PubMed] [Google Scholar]
- 6. Hashimoto Y., and Costanzo V. (2011) Studying DNA replication fork stability in Xenopus egg extract. Methods Mol. Biol. 745, 437–445 10.1007/978-1-61779-129-1_25 [DOI] [PubMed] [Google Scholar]
- 7. You Z., Bailis J. M., Johnson S. A., Dilworth S. M., and Hunter T. (2007) Rapid activation of ATM on DNA flanking double-strand breaks. Nature Cell Biol. 9, 1311–1318 10.1038/ncb1651 [DOI] [PubMed] [Google Scholar]
- 8. Leno G. H., and Laskey R. A. (1991) DNA replication in cell-free extracts from Xenopus laevis. Methods Cell Biol. 36, 561–579 10.1016/S0091-679X(08)60297-6 [DOI] [PubMed] [Google Scholar]
- 9. Blow J. J. (2001) Control of chromosomal DNA replication in the early Xenopus embryo. EMBO J. 20, 3293–3297 10.1093/emboj/20.13.3293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Félix M. A., Labbé J. C., Dorée M., Hunt T., and Karsenti E. (1990) Triggering of cyclin degradation in interphase extracts of amphibian eggs by cdc2 kinase. Nature 346, 379–382 10.1038/346379a0 [DOI] [PubMed] [Google Scholar]
- 11. Hyde A. S., Hang B. I., and Lee E. (2016) Reconstitution of the cytoplasmic regulation of the Wnt signaling pathway using Xenopus egg extracts. Methods Mol. Biol. 1481, 101–109 10.1007/978-1-4939-6393-5_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Minshull J., Golsteyn R., Hill C. S., and Hunt T. (1990) The A- and B-type cyclin associated cdc2 kinases in Xenopus turn on and off at different times in the cell cycle. EMBO J. 9, 2865–2875 10.1002/j.1460-2075.1990.tb07476.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Srinivasan S. V., and Gautier J. (2011) Study of cell cycle checkpoints using Xenopus cell-free extracts. Methods Mol. Biol. 782, 119–158 10.1007/978-1-61779-273-1_10 [DOI] [PubMed] [Google Scholar]
- 14. Desai A., Murray A., Mitchison T. J., and Walczak C. E. (1999) The use of Xenopus egg extracts to study mitotic spindle assembly and function in vitro. Methods Cell Biol. 61, 385–412 10.1016/s0091-679x(08)61991-3 [DOI] [PubMed] [Google Scholar]
- 15. Cross M. K., and Powers M. A. (2009) Learning about cancer from frogs: Analysis of mitotic spindles in Xenopus egg extracts. Dis. Models Mech. 2, 541–547 10.1242/dmm.002022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Maresca T. J., and Heald R. (2006) Methods for studying spindle assembly and chromosome condensation in Xenopus egg extracts. Methods Mol. Biol. 322, 459–474 10.1007/978-1-59745-000-3_33 [DOI] [PubMed] [Google Scholar]
- 17. Newmeyer D. D., Farschon D. M., and Reed J. C. (1994) Cell-free apoptosis in Xenopus egg extracts: Inhibition by Bcl-2 and requirement for an organelle fraction enriched in mitochondria. Cell 79, 353–364 10.1016/0092-8674(94)90203-8 [DOI] [PubMed] [Google Scholar]
- 18. Wang W. L., and Shechter D. (2016) Chromatin assembly and transcriptional cross-talk in Xenopus laevis oocyte and egg extracts. Int. J. Dev. Biol. 60, 315–320 10.1387/ijdb.160161ds [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hutchison C. J., Brill D., Cox R., Gilbert J., Kill I., and Ford C. C. (1989) DNA replication and cell cycle control in Xenopus egg extracts. J. Cell Sci. Suppl. 12, 197–212 10.1242/jcs.1989.supplement_12.17 [DOI] [PubMed] [Google Scholar]
- 20. Newport J., and Kirschner M. (1982) A major developmental transition in early Xenopus embryos: I. characterization and timing of cellular changes at the midblastula stage. Cell 30, 675–686 10.1016/0092-8674(82)90272-0 [DOI] [PubMed] [Google Scholar]
- 21. Amodeo A. A., Jukam D., Straight A. F., and Skotheim J. M. (2015) Histone titration against the genome sets the DNA-to-cytoplasm threshold for the Xenopus midblastula transition. Proc. Natl. Acad. Sci. U.S.A. 112, E1086–E1095 10.1073/pnas.1413990112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Joseph S. R., Palfy M., Hilbert L., Kumar M., Karschau J., Zaburdaev V., Shevchenko A., and Vastenhouw N. L. (2017) Competition between histone and transcription factor binding regulates the onset of transcription in zebrafish embryos. Elife 6, e23326 10.7554/eLife.23326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Prioleau M. N., Huet J., Sentenac A., and Méchali M. (1994) Competition between chromatin and transcription complex assembly regulates gene expression during early development. Cell 77, 439–449 10.1016/0092-8674(94)90158-9 [DOI] [PubMed] [Google Scholar]
- 24. Kornberg R. D., and Thomas J. O. (1974) Chromatin structure; oligomers of the histones. Science 184, 865–868 10.1126/science.184.4139.865 [DOI] [PubMed] [Google Scholar]
- 25. Li B., Carey M., and Workman J. L. (2007) The role of chromatin during transcription. Cell 128, 707–719 10.1016/j.cell.2007.01.015 [DOI] [PubMed] [Google Scholar]
- 26. Walter J., Sun L., and Newport J. (1998) Regulated chromosomal DNA replication in the absence of a nucleus. Mol. Cell 1, 519–529 10.1016/S1097-2765(00)80052-0 [DOI] [PubMed] [Google Scholar]
- 27. Dewar J. M., Budzowska M., and Walter J. C. (2015) The mechanism of DNA replication termination in vertebrates. Nature 525, 345–350 10.1038/nature14887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Long D. T., Raschle M., Joukov V., and Walter J. C. (2011) Mechanism of RAD51-dependent DNA interstrand cross-link repair. Science 333, 84–87 10.1126/science.1204258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Räschle M., Knipscheer P., Enoiu M., Angelov T., Sun J., Griffith J. D., Ellenberger T. E., Schärer O. D., and Walter J. C. (2008) Mechanism of replication-coupled DNA interstrand crosslink repair. Cell 134, 969–980 10.1016/j.cell.2008.08.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fu Y. V., Yardimci H., Long D. T., Ho T. V., Guainazzi A., Bermudez V. P., Hurwitz J., van Oijen A., Schärer O. D., and Walter J. C. (2011) Selective bypass of a lagging strand roadblock by the eukaryotic replicative DNA helicase. Cell 146, 931–941 10.1016/j.cell.2011.07.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu R. A., Semlow D. R., Kamimae-Lanning A. N., Kochenova O. V., Chistol G., Hodskinson M. R., Amunugama R., Sparks J. L., Wang M., Deng L., Mimoso C. A., Low E., Patel K. J., and Walter J. C. (2019) TRAIP is a master regulator of DNA interstrand crosslink repair. Nature 567, 267–272 10.1038/s41586-019-1002-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Toyoda T., and Wolffe A. P. (1992) Characterization of RNA polymerase II-dependent transcription in Xenopus extracts. Dev. Biol. 153, 150–157 10.1016/0012-1606(92)90099-3 [DOI] [PubMed] [Google Scholar]
- 33. Session A. M., Uno Y., Kwon T., Chapman J. A., Toyoda A., Takahashi S., Fukui A., Hikosaka A., Suzuki A., Kondo M., van Heeringen S. J., Quigley I., Heinz S., Ogino H., Ochi H., et al. (2016) Genome evolution in the allotetraploid frog Xenopus laevis. Nature 538, 336–343 10.1038/nature19840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lindell T. J., Weinberg F., Morris P. W., Roeder R. G., and Rutter W. J. (1970) Specific inhibition of nuclear RNA polymerase II by α-amanitin. Science 170, 447–449 10.1126/science.170.3956.447 [DOI] [PubMed] [Google Scholar]
- 35. Zierhut C., Jenness C., Kimura H., and Funabiki H. (2014) Nucleosomal regulation of chromatin composition and nuclear assembly revealed by histone depletion. Nat. Struct. Mol. Biol. 21, 617–625 10.1038/nsmb.2845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Seila A. C., Core L. J., Lis J. T., and Sharp P. A. (2009) Divergent transcription: A new feature of active promoters. Cell Cycle 8, 2557–2564 10.4161/cc.8.16.9305 [DOI] [PubMed] [Google Scholar]
- 37. Inoue N., and Uchida H. (1991) Transcription and initiation of ColE1 DNA replication in Escherichia coli K-12. J. Bacteriol. 173, 1208–1214 10.1128/jb.173.3.1208-1214.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Liu Z. J., Zhu Z. Y., Roberg K., Faras A., Guise K., Kapuscinski A. R., and Hackett P. B. (1990) Isolation and characterization of β-actin gene of carp (Cyprinus carpio). DNA Seq. 1, 125–136 10.3109/10425179009016040 [DOI] [PubMed] [Google Scholar]
- 39. Shearwin K. E., Callen B. P., and Egan J. B. (2005) Transcriptional interference—a crash course. Trends Genet. 21, 339–345 10.1016/j.tig.2005.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carpousis A. J., and Gralla J. D. (1980) Cycling of ribonucleic acid polymerase to produce oligonucleotides during initiation in vitro at the lac UV5 promoter. Biochemistry 19, 3245–3253 10.1021/bi00555a023 [DOI] [PubMed] [Google Scholar]
- 41. Munson L. M., and Reznikoff W. S. (1981) Abortive initiation and long ribonucleic acid synthesis. Biochemistry 20, 2081–2085 10.1021/bi00511a003 [DOI] [PubMed] [Google Scholar]
- 42. Bentley D. L. (2014) Coupling mRNA processing with transcription in time and space. Nat. Rev. Genet. 15, 163–175 10.1038/nrg3662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tian B., and Graber J. H. (2012) Signals for pre-mRNA cleavage and polyadenylation. Wiley Interdiscip. Rev. RNA 3, 385–396 10.1002/wrna.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Earnshaw W. C., Honda B. M., Laskey R. A., and Thomas J. O. (1980) Assembly of nucleosomes: The reaction involving X. laevis nucleoplasmin. Cell 21, 373–383 10.1016/0092-8674(80)90474-2 [DOI] [PubMed] [Google Scholar]
- 45. Kleinschmidt J. A., Fortkamp E., Krohne G., Zentgraf H., and Franke W. W. (1985) Co-existence of two different types of soluble histone complexes in nuclei of Xenopus laevis oocytes. J. Biol. Chem. 260, 1166–1176 [PubMed] [Google Scholar]
- 46. Woodcock C. L., and Ghosh R. P. (2010) Chromatin higher-order structure and dynamics. Cold Spring Harb. Perspect. Biol. 2, a000596 10.1101/cshperspect.a000596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wu X., and Sharp P. A. (2013) Divergent transcription: A driving force for new gene origination? Cell 155, 990–996 10.1016/j.cell.2013.10.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jimeno-González S., and Reyes J. C. (2016) Chromatin structure and pre-mRNA processing work together. Transcription 7, 63–68 10.1080/21541264.2016.1168507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Tanny J. C. (2014) Chromatin modification by the RNA polymerase II elongation complex. Transcription 5, e988093 10.4161/21541264.2014.988093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Nagai S., Davis R. E., Mattei P. J., Eagen K. P., and Kornberg R. D. (2017) Chromatin potentiates transcription. Proc. Natl. Acad. Sci. U.S.A. 114, 1536–1541 10.1073/pnas.1620312114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morgan M. A., and Shilatifard A. (2015) Chromatin signatures of cancer. Genes Dev. 29, 238–249 10.1101/gad.255182.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Lelli K. M., Slattery M., and Mann R. S. (2012) Disentangling the many layers of eukaryotic transcriptional regulation. Annu. Rev. Genet. 46, 43–68 10.1146/annurev-genet-110711-155437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Klemm S. L., Shipony Z., and Greenleaf W. J. (2019) Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 10.1038/s41576-018-0089-8 [DOI] [PubMed] [Google Scholar]
- 54. Bradner J. E., Hnisz D., and Young R. A. (2017) Transcriptional addiction in cancer. Cell 168, 629–643 10.1016/j.cell.2016.12.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Read A., and Natrajan R. (2018) Splicing dysregulation as a driver of breast cancer. Endocr. Relat. Cancer 25, R467–R478 10.1530/ERC-18-0068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lee T. I., and Young R. A. (2013) Transcriptional regulation and its misregulation in disease. Cell 152, 1237–1251 10.1016/j.cell.2013.02.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gillespie P. J., Gambus A., and Blow J. J. (2012) Preparation and use of Xenopus egg extracts to study DNA replication and chromatin associated proteins. Methods 57, 203–213 10.1016/j.ymeth.2012.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Joukov V., Groen A. C., Prokhorova T., Gerson R., White E., Rodriguez A., Walter J. C., and Livingston D. M. (2006) The BRCA1/BARD1 heterodimer modulates Ran-dependent mitotic spindle assembly. Cell 127, 539–552 10.1016/j.cell.2006.08.053 [DOI] [PubMed] [Google Scholar]
- 59. Fullbright G., Rycenga H. B., Gruber J. D., and Long D. T. (2016) p97 promotes a conserved mechanism of helicase unloading during DNA cross-link repair. Mol. Cell. Biol. 36, 2983–2994 10.1128/MCB.00434-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Langmead B., Trapnell C., Pop M., and Salzberg S. L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10, R25 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., and Durbin R. (2009) The sequence alignment/map format and SAMtools. Bioinformatics (Oxf.) 25, 2078–2079 10.1093/bioinformatics/btp352 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.