Abstract
Proteins of nuclear receptor subfamily 4 group A (NR4A), including NR4A1/NGFI-B, NR4A2/Nurr1, and NR4A3/NOR-1, are nuclear transcription factors that play important roles in metabolism, apoptosis, and proliferation. NR4A proteins recognize DNA response elements as monomers or dimers to regulate the transcription of a variety of genes involved in multiple biological processes. In this study, we determined two crystal structures of the NR4A2 DNA-binding domain (NR4A2-DBD) bound to two Nur-responsive elements: an inverted repeat and an everted repeat at 2.6–2.8 Å resolution. The structures revealed that two NR4A2-DBD molecules bind independently to the everted repeat, whereas two other NR4A2-DBD molecules form a novel dimer interface on the inverted repeat. Moreover, substitution of the interfacial residue valine 298 to lysine as well as mutation of DNA bases involved in the interactions abolished the dimerization. Overall, our structural, biochemical, and bioinformatics analyses provide a molecular basis for the binding of the NR4A2 protein dimers to NurREs and advance our understanding of the dimerization specificity of nuclear receptors.
Keywords: nuclear receptor, protein crystallization, protein–DNA interaction, protein–protein interaction, dimerization, crystal structure, gene regulation, nuclear receptor subfamily 4 group A member 2, Nur-responsive elements, transcription factor
Introduction
The NR4A subfamily, including NR4A1 (NGFI-B), NR4A2 (Nurr1), and NR4A3 (NOR-1), belongs to the nuclear receptor superfamily (1, 2). Most nuclear receptor members are regulated by small lipophilic ligands such as retinoids and steroids, whereas NR4A proteins are orphan members for which no ligand has been identified (3). NR4A proteins function as transcription factors (TFs)3 that regulate the expression of many key genes involved in proliferation (Wnt/β-catenin) (4), metabolism (peroxisome proliferator-activated receptor γ coactivator 1a (PGC1a)), apoptosis (Fas-ligand, TNF-related apoptosis-inducing ligand), inflammation (interleukin-8), DNA repair (arginase-1) (5), and angiogenesis (vascular endothelial growth factor) (2). Altered expression of NR4A receptors has been implicated in a wide variety of cancers, such as melanoma, colon cancer, and oral squamous cell carcinoma (6–8). Increasing evidence has shown roles of the NR4A receptors in cancer immunity. NR4A receptors are expressed at high levels in CD8+ T cells from humans with cancer or chronic viral infection and are linked to induction of T cell dysfunction (9–11). NR4A receptors repress effector gene expression by inhibiting the function of the TF AP-1 and activate tolerance-related genes by promoting acetylation of histone 3 at lysine 27 (11). Treatment of tumor-bearing mice with T cells expressing chimeric antigen receptors lacking all three NR4A receptors in vivo results in tumor regression and prolonged survival (12).
Nuclear receptors engage the hormone response elements of target genes to regulate transcription. Most nuclear receptors dimerize on target DNA, which contains two repeats of the hexamer AGAACA or AGGTCA (13). The two half-sites can be organized as direct, inverted, or everted repeats with spacers of varying lengths (14) (Fig. S1). For example, steroid receptors bind as homodimers to inverted palindromic repeats separated by a three-nucleotide spacer (IR3). Nonsteroid nuclear receptors, such as retinoic acid receptors, retinoid X receptors (RXRs), and vitamin D3 receptors (VDRs), form homodimers or heterodimers with RXR on response elements consisting of direct repeats separated by a one-to-five nucleotide spacer (DR1–DR5).
NR4A receptors were initially found to bind as monomers to a NGFI-B–responsive element (NBRE, AAAGGTCA), an octanucleotide consisting of the canonical nuclear receptor binding motif AGGTCA preceded by two adenines (15). In addition, NR4A1 and NR4A2 (but not NR4A3) can bind as homodimers or heterodimers to the Nur-responsive element (NurRE), which consists of two repeats of the NBRE-related octanucleotides (16). NR4A1 and NR4A2 can also form heterodimers with the RXR (17). A large sample analysis of most human TFs by high-throughput Systematic Evolution of Ligands by Exponential Enrichment (SELEX) and ChIP sequencing revealed that NR4A2 can bind to two different NurRE motifs. One motif, named ER0, consists of two everted, palindromic, 8-nt half-sites with no spacer. The other motif is IR5, which consists of two inverted 8-nt repeats spaced by five nucleotides (Fig. 1A) (18). NR4A proteins also bind to the promoter regions of the pituitary proopiomelanocortin gene to mediate the physiological response of the proopiomelanocortin gene to corticotropin-releasing hormone (19). Corticotropin-releasing hormone signals rapidly activate the nuclear DNA binding activity of NR4A dimers but not monomers (20). In turn, NR4A dimers synergistically enhance the transcription of NurRE reporters (16).
Figure 1.
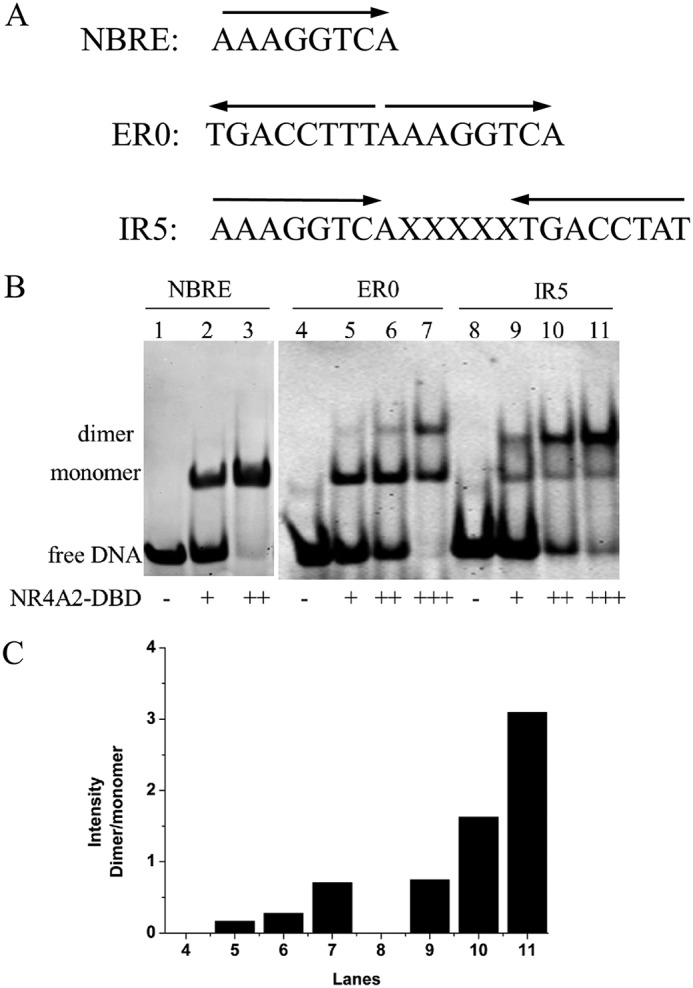
Binding ability of NR4A2-DBD with different DNAs. A, DNA sequences of the response elements for NR4A2. B, binding features of NR4A2-DBD with different DNAs were determined by EMSA. Lanes 1, 4, and 8 show free DNA without protein. The primary concentrations of DNA and protein used were both 45 μm. + indicates that the molar ratio of protein to DNA is 1:1, and ++ indicates that the molar ratio is 2:1. C, quantification of the intensity of dimer/monomer bands. The graph shows the relative density of the dimer/monomer bands detected by EMSA.
The three NR4A receptors all share a common structural organization with other nuclear receptors. These receptors share a high degree of sequence homology in their ligand-binding domains (LBDs) and DNA-binding domains (DBDs) but exhibit divergent N-terminal regions containing the activation function 1 (AF1) domain (15). A previously reported three-dimensional structure of a rat NR4A1-DBD protein shows the DNA-binding mode of the NR4A1 monomer (15), whereas the DNA-binding mode of NR4A receptors as dimers remains unclear.
In this study, we report two crystal structures of NR4A2-DBD bound to ER0 and IR5 at 2.5 Å and 2.7 Å, respectively. These structures reveal that NR4A2-DBD can bind the two DNAs in different manners. We further analyzed the roles of the protein–protein interactions between two NR4A2 molecules and protein–DNA interactions in the promotion of formation of the NR4A2-DBD–IR5 complex. Then a bioinformatic analysis of the endogenous NR4A-binding motif identified the existence and ratio of the ER0- and IR5-binding motifs in vivo. Overall, our structural, biochemical, and bioinformatics analyses will help elucidate the molecular basis of the DNA binding specificity of NR4A dimers.
Results
Ability of NR4A2 to bind to different DNAs
As mentioned above, NR4A2 can bind to different response elements as a monomer or dimer (15, 18). Here we carried out an Electrophoretic Mobility Shift Assay (EMSA) to analyze the ability of NR4A2-DBD to bind to three different DNAs in vitro (Fig. 1A). Purified recombinant NR4A2-DBD protein was incubated with NBRE, ER0, and IR5. Then samples were detected on a native polyacrylamide gel. As shown in Fig. 1B, NR4A2-DBD formed a complex band with NBRE (Fig. 1B, lanes 2 and 3). In contrast, NR4A2-DBD formed two mobility complex bands when incubated with ER0 or IR5 (Fig. 1B, lanes 6 and 10). The lower band migrated to the same position as the NR4A2-DBD monomer bound to NBRE. The upper, slowly migrating band presumably represented the dimeric complex. With the decrease in free DNA levels upon protein binding, NR4A2-DBD formed an enhanced dimeric complex with ER0 and IR5 when the amount of NR4A2 protein increased (Fig. 1B, lanes 7 and 11). The dimer/monomer ratio was quantified using ImageJ (Fig. 1C). For site ER0, the dimer/monomer ratio was 0.17, 0.28, and 0.70 with a 1-, 2- and 3-fold increase in protein concentration, respectively. For site IR5, the ratio increased to 0.74, 1.62, and 3.09 for the same protein concentrations. These results indicated that NR4A-DBD could bind as a dimer to the ER0 and IR5 sequences in a cooperative manner and that the IR5 site was more cooperative and stronger than site ER0.
Overall structures of NR4A2-DBD bound to two NurREs
To better characterize the mechanism by which NR4A2 proteins recognize NurRE dimers, we determined the crystal structures of human NR4A2-DBD bound to ER0 and IR5, respectively. The methods of crystallization and structure determination are described under “Experimental procedures.” The final refinement statistics are summarized in Table 1. The NR4A2-DBD–IR5 structure was solved at 2.8 Å and crystallized in the P 21 space group with one complex per asymmetric unit. The NR4A2-DBD–ER0 structure was solved at 2.6 Å and crystallized in the P 43 space group with one complex per asymmetric unit. The initially screened NR4A2-DBD–ER0 crystals were twinned. After multiple rounds of optimization, the twinning problem was not solved. The best set of diffraction data for the NR4A2-DBD–ER0 crystals was collected, which was estimated to be 47.8% twinned by phenix.xtriage (21). Although we applied detwinning during refinement, the R-factors of the final model remained higher than that expected for nontwinned crystals of a similar resolution range (Rwork/Rfree, 0.25/0.29; resolution, 2.60 Å).
Table 1.
Data collection and refinement statistics
| NR4A2-DBD–ER0 complex | NR4A2-DBD–IR5 complex | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.97915 | 0.97915 |
| Space group | P 43 | P 21 |
| a, b, c (Å) | 35.696, 35.696, 259.273 | 42.405, 64.315, 60.776 |
| α, β, γ (°) | 90, 90, 90 | 90, 96.84, 90 |
| Resolution (Å) | 50–2.601 (2.693–2.601) | 50–2.781 (2.88–2.781) |
| Unique reflections | 9915 (990) | 8163 (737) |
| Redundancy | 13.8 (13.3) | 3.3 (3.0) |
| Completeness (%) | 99.95 (100.00) | 98.67 (89.99) |
| Mean I/σI | 16.81 (4.42) | 6.69 (2.01) |
| Refinement | ||
| Resolution (Å) | 32.41–2.601 | 35.23–2.781 |
| Wilson B-factor | 47.51 | 37.19 |
| Rwork/Rfree | 0.25/0.29 | 0.22/0.25 |
| Number of non-hydrogen atoms | 2172 | 2205 |
| Macromolecules | 2147 | 2177 |
| Protein residues | 210 | 211 |
| RMS (bonds) | 0.008 | 0.008 |
| RMS (angles) | 1.17 | 1.12 |
| Ramachandran favored (%) | 95 | 96 |
| Ramachandran outliers (%) | 0 | 0 |
| Clash score | 15.47 | 11.25 |
| Average B-factor | 53.10 | 41.70 |
| Macromolecules | 53.20 | 41.90 |
| Ligands | 42.70 | 31.10 |
| Solvent | 39.30 | 20.50 |
The global conformation of NR4A2-DBD had few structural variations in these two complex structures, which were composed of two highly conserved α-helical zinc modules (H1 and H2), an N-terminal loop, a loop linking H1 and H2, and a C-terminal extension (CTE) (Fig. 2A). The core DBD comprises two highly conserved four-cysteine/zinc–nucleated modules. The CTE includes a T-box and an A-box (Fig. 2C). The structure of NR4A2-DBD also showed an analogous tertiary structure with the previously determined rat NR4A1-DBD structure (15), and the root mean square deviation for the superimposition of Cα atoms was ∼0.334 Å.
Figure 2.
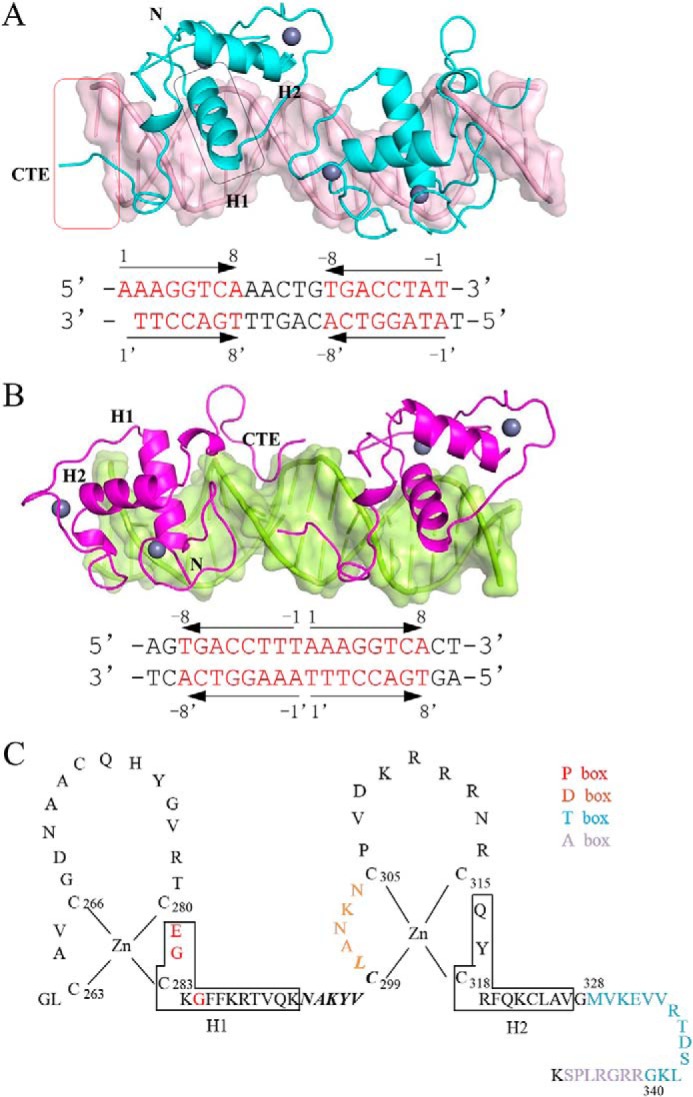
Overall structures of NR4A2-DBD–DNA complexes. A, overall structure of the NR4A2-DBD–IR5 complex. NR4A2-DBD is colored cyan, and DNA targets are colored light pink. B, overall structure of the NR4A2-DBD–ER0 complex. NR4A2-DBD is colored magenta, and ER0 is colored lemon. The sequences of the DNA duplex used in the structures are listed below, and the core octanucleotides are colored red. The arrows show the orientation of each core half-site. Secondary structure elements (H1 and H2) are labeled, and the region in the box is involved in the protein–DNA interactions. C, amino acid sequences of NR4A2-DBD. Arrangement of residues 261–348 of NR4A2 shows the classic nuclear receptor-type zinc-finger motif and a CTE. The red and orange letters correspond to the P-box and D-box, respectively. The blue and purple letters correspond to the T-box and A-box, respectively. Solid-line boxes indicate α-helical segments. The residues in bold and italic are involved in dimerization.
In the structure of the NR4A2-DBD–ER0 complex, two NR4A2-DBD molecules were bound to the same face of the dsDNA in a tail-to-tail orientation (Fig. 2B). The two molecules were bound independently to the half-sites. In the structure of the NR4A2-DBD–IR5 complex, two NR4A2-DBD molecules were arranged in a head-to-head orientation. Moreover, the two NR4A2-DBD molecules formed an interface through the reversely arrayed loop from residues 294–300. The protein–protein interactions might help NR4A2-DBD homodimers cooperatively bind IR5, and we will analyze this aspect later. Overall, NR4A2 bound in different manners to the two NurRE sequences. The difference might result from the relative orientation (inverted or everted repeats) and number of spacer nucleotides of the half-sites.
DNA recognition by NR4A2
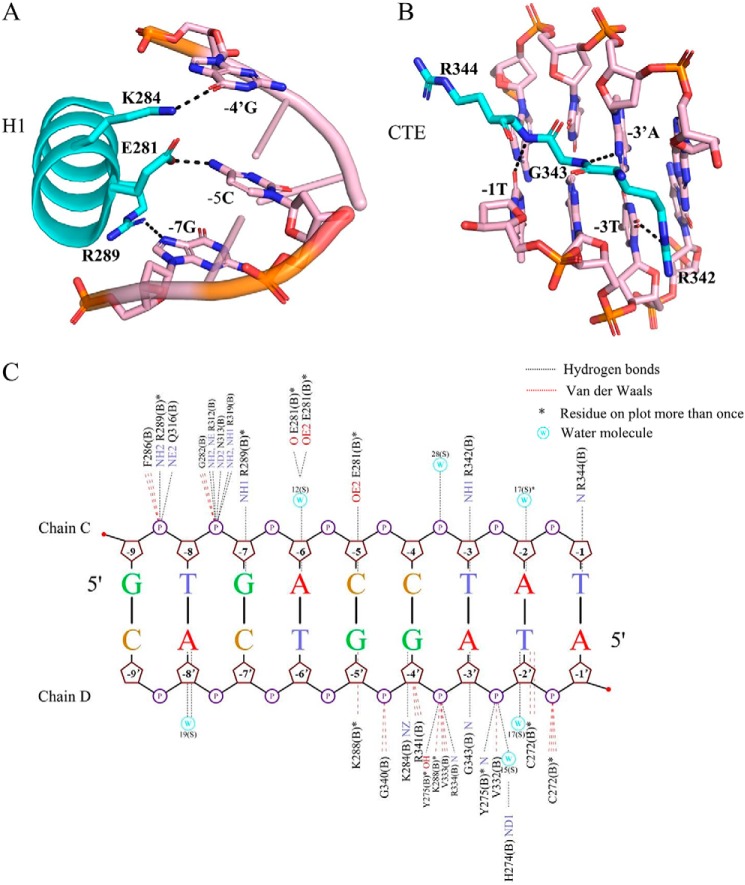
We analyzed the detailed information of protein-DNA interactions by NUCPLOT (22), as shown in Fig. 3. The interactions were basically conserved for half-site recognition, and we used the NR4A2-DBD bound downstream of IR5 as a representative example. The N-terminal helix H1 of NR4A2-DBD docked into and predominantly interacted with the major groove. Three conserved residues, Glu-281, Lys-284, and Arg-289, formed hydrogen bonds with -5Cyd, -4′Gua, and -7Gua, respectively (Fig. 3A). The C terminus of NR4A2-DBD formed a second independent DNA-binding surface to interact with the minor groove. The conserved Arg-Gly-Arg motif (Arg-342, Gly-343, and Arg-344) made hydrogen bonds contacts with the flanking extension bases -3Thy, -3′Ade, and -1Thy, respectively (Fig. 3B). In addition to specific base contacts, the NR4A2-DBD protein also formed numerous hydrogen bonds and van der Waals interactions with the phosphate backbone to further stabilize DNA binding (Fig. 3C).
Figure 3.
Detailed protein–base contacts in the NR4A2-DBD–IR5 structure. A, stereo diagram of major groove recognition by helix H1 of NR4A2-DBD. B, stereo diagram of minor groove recognition by the C-terminal extension of NR4A2-DBD. The color is as shown in Fig. 2. All hydrogen bonds are shown as black dotted lines. C, summary of protein–DNA interactions between the NR4A2-DBD and the upstream half-site DNA generated by NUCPLOT. Hydrogen bonds are shown as black dotted lines. van der Waals interactions are shown as red dotted lines. Asterisks indicate residues that are on the plot more than once. W, water molecule.
Protein–protein interactions in the NR4A2-DBD–IR5 complex
In the structure of the NR4A2-DBD–IR5 complex, a dimerization interface between two NR4A2-DBD molecules was identified. This interface involves the same region of the two NR4A2 molecules (amino acids 294–300) in an antiparallel manner (Fig. 4A). From an analysis of PBDePISA (23), the dimer interface buried a solvent-accessible area of ∼419 Å2. The dimerization contacts involved residues Asn-294, Val-298, and Leu-300 of the upstream subunit and residues Asn-294, Lys-296, Val-298, and Leu-300 of the downstream subunit (Fig. 4B).
Figure 4.
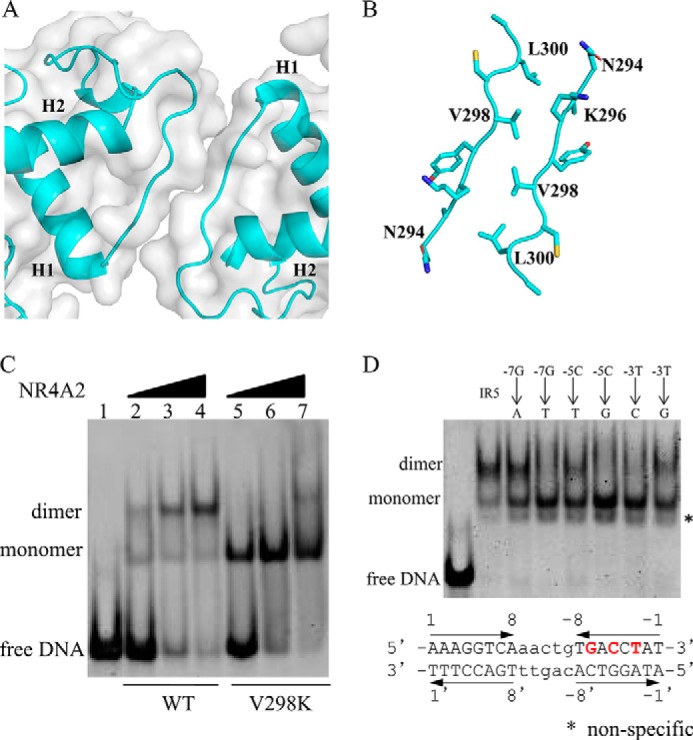
Importance of protein–protein interactions and protein–DNA interactions in the NR4A2-DBD–IR5 structure. A, surface representation of the dimerization interface in two NR4A2-DBD molecules. B, detailed stereo diagram of the residues (sticks) involved in the dimer interface. C, the DNA binding ability of the WT and V298K variant of NR4A2-DBD was measured by EMSA. The concentration of the DNA and proteins used here was 45 μm. D, binding features of NR4A2-DBD with different IR5 DNA variants. The core sequences of the IR5 DNA duplex used are listed. The arrows show the orientation of each core half-site. The bases in red and bold are involved in the base–protein interaction and are mutated in the EMSA. The concentration of DNA used here were 45 μm, and the proteins were used at 90 μm.
To identify the role of the dimer interface, we mutated the key hydrophobic residue Val-298 to a charged lysine. Then, the effect of mutant V298K on DNA binding ability was measured using EMSA. Compared with WT NR4A2-DBD, the dimeric complex was hardly detected when the mutant V298K was incubated with IR5 (Fig. 4C). The results indicate that Val-298 is crucial for NR4A2 dimerization on IR5 and that cooperative assembly is inhibited without the dimer interaction.
We also investigated the effect of the IR5 sequence on the DNA binding properties of NR4A2. The three bases (-7Gua, -5Cyd, and -3Thy) in the downstream half-site involved in the specific protein–base interactions were mutated to other bases. As shown in Fig. 4D, the dimeric complexes were abolished when WT NR4A2-DBD was incubated with these IR5 variants, apart from a -7Gua-to-Ade mutant. The results indicate the crucial roles of these bases for NR4A2 dimerization. Taken together, these results suggest that the protein–protein interactions between two NR4A2 molecules as well as the specific protein–base interactions, play similar important roles in favoring cooperative formation of the NR4A2-DBD–IR5 complex.
Analysis of the NR4A2 and RXR heterodimer
To investigate whether NR4A2 can heterodimerize with RXR on the IR5 response element, we modeled the NR4A2–RXR heterodimer bound to IR5 by superposition of one NR4A2 molecule with an RXR structure (PDB code 4CN2) (Fig. 5A). Given the sequence similarities and high homology, the RXR exhibited a good fit and showed no obvious clashes. Then we carried out an EMSA to analyze the heterodimeric abilities of NR4A2 and RXR bound to IR5 response elements (Fig. 5B). NR4A2 or RXR alone could form a monomer band with IR5 (Fig. 5B, lanes 2 and 3). When IR5 was incubated with NR4A2 and RXR at the same time, a higher complex band was observed (Fig. 5B, lane 4). The results suggested that NR4A2 could form a heterodimer with RXR on the IR5 response element in vitro.
Figure 5.
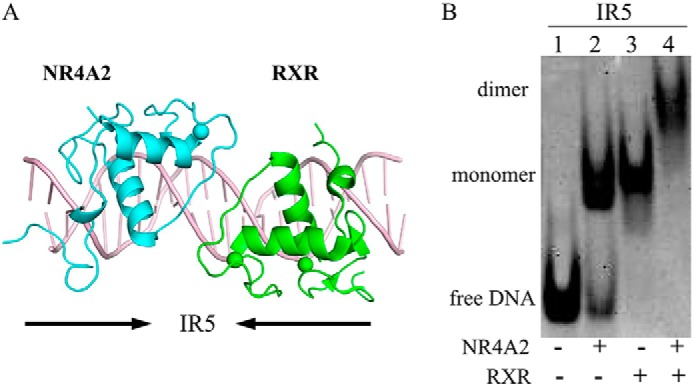
Analysis of the NR4A2–RXR heterodimer. A, model of the NR4A2–RXR heterodimer on IR5. The RXR structure was obtained from a previously reported RXR–DR1 structure (PDB code 4CN2) and superimposed onto our NR4A2–IR5 complex structure. The RXR and NR4A2 structures were aligned to create the NR4A2–RXR–DNA structural model. C, DNA binding ability of the NR4A2–RXR heterodimer bound to DNAs.
Identification of the endogenous NR4A-binding motifs ER0 and IR5
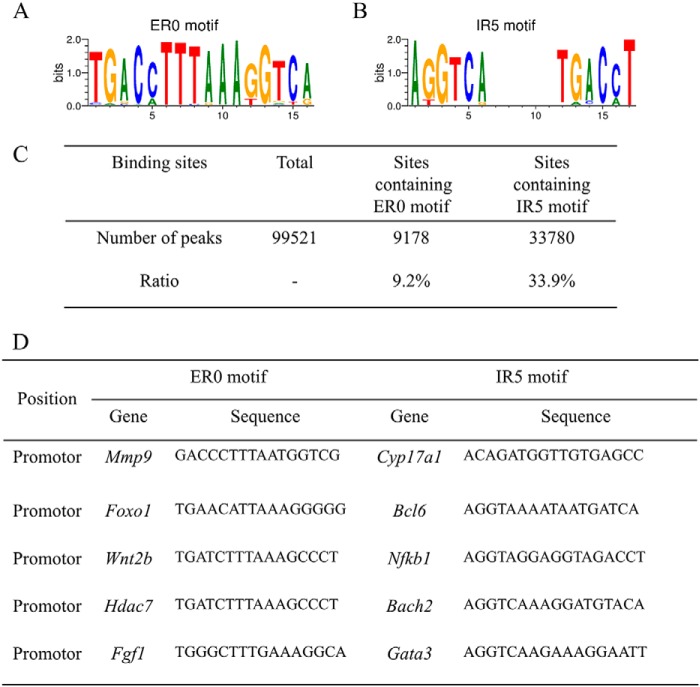
To identify whether the NR4A protein binds ER0 and IR5 in vivo, we analyzed the occurrence of these motifs in endogenous NR4A-binding sites. We first obtained the ER0 and IR5 matrixes from the footprintDB database (Fig. 6, A and B) (46). Then we searched for the motif IR5 or ER0 in the NR4A ChIP-seq database (GSE123629) using these two matrixes. For motif ER0, 9178 peaks were found, accounting for 9.2% of the total NR4A-binding sites. For motif IR5, 33,780 peaks were found, accounting for 33.9% of the total NR4A-binding sites (Fig. 6C). The ER0 motif was identified in the promoters of genes such as Mmp9, Foxo1, Wnt2b, Hdac7, and Fgf1 (Fig. 6D). The IR5 motif was identified in the promoters of genes such as Cyp17a1, Bcl6, Nfkb1, Bach2, and Gata3 (Fig. 6D). These data suggest that NR4A proteins can indeed bind ER0 or IR5 motifs in vivo.
Figure 6.
Occurrence of the IR5 and ER0 binding motifs in NR4A-binding sites in human cells. A, analysis of the ratio of motifs IR5 and ER0 in the NR4A ChIP-seq data. B, representative gene promoters near NR4A-binding sites containing motif IR5.
Discussion
In this paper, we first report the cocrystal structures of NR4A2-DBD bound to two different NurREs. Both structures demonstrate that NR4A2 makes similar base interactions with half-sites as those seen in the structure of rat NR4A1 bound to NBRE (15). The highly conserved helix H1 forms specific contacts with the identity elements of the major groove (Fig. 3). The C-terminal residues form a unique substructure to interact extensively with the minor groove, in particular with the characteristic 5′-flanking extended A-T base pair of the canonical nuclear receptor binding motif AGGTCA (Fig. 3).
In addition to exhibiting similar base recognition, NR4A2 subunits were bound to these two NurREs in distinct modes: the everted repeats mode, in which two molecules bind independently to each half-site of DNA in a tail-to-tail orientation (Fig. 2B), and the inverted repeats mode, in which the two molecules form a dimerization interface and synergistically bind to each half-site of DNA in a head-to-head orientation (Fig. 2A). The primary association across the interface is via van der Waals contacts. Val-298 contributes to the hydrophobic character of the interface, and substitution of the hydrophobic valine with a charged lysine is likely to strongly inhibit homodimer formation (Fig. 4). In addition to the dimer interface, the presence of two inverted DNA-binding sites also plays important roles in driving formation of the dimer on DNA (Fig. 4). Moreover, bioinformatics analysis of ChIP-seq data revealed that 33.9% and 9.2% of the total NR4A-binding sites contain IR5 and ER0 sites, respectively (Fig. 6). These results suggest that NR4A proteins can bind these sites in vivo.
Previous study reported that NR4A2 can heterodimerize with RXRα or RXRγ in midbrain dopaminergic neurons (24). The important role of the NR4A2–RXR heterodimer in vivo has been well studied, and several synthetic ligands that bind to the RXR-binding pocket are thought to be potential therapeutic agents that act by activating NR4A2–RXR heterodimers (25). Based on our structure of the NR4A2 homodimer, we modeled the structures of the NR4A2–RXR heterodimer bound to the IR5 motif (Fig. 5). It will help explain the molecular mechanism by which the NR4A2–RXR heterodimer binds to DNA. Moreover, our EMSA results verified that NR4A2 could heterodimerize with RXR on the IR5 response element (Fig. 5).
Nuclear receptors form dimers on their target DNAs via highly cooperative assembly of their DBDs. With highly conserved core DBDs and similar response elements, nuclear receptors can recognize and differentiate between specific DNAs. This specificity can be partly attributed to the ability of nuclear receptors to make distinct protein–protein contacts to reinforce their protein–DNA contacts, as observed for the glucocorticoid receptor (GR), RXR, and VDR (Fig. S2). GR forms the largest dimerization interface on IR3 (PDB code 3G6P) (26). Salt bridges, numerous hydrogen bonds, and van der Waals forces contribute to the dimer interactions (Fig. S2A). In the structure of the RXR–DR1 complex (PDB code 4CN2) (27), Glu-207 of the downstream subunit makes hydrogen bond interactions with Arg-182 of the upstream subunit, and Gln-210 of the downstream subunit makes contacts with residues Arg-172 and Arg-186 of the upstream subunit (Fig. S2B). VDR binds to DR3 with a dimerization interface involving the side chains of Pro-61, Phe-62, and His-75 of the upstream subunit and residues Asn-37, Glu-92, and Phe-93 of the downstream subunit (Fig. S2C) (28). In the NR4A2–IR5 structure, the hydrophobic residue Val-298 plays a key role in the dimerization interaction, as well as numerous surrounding van der Waals contacts (Fig. S2D). Compared with these previously published nuclear receptor dimer structures, the NR4A2-DBD homodimer utilizes a different region to form a compact and nonpolar dimer interface.
The polarity and strength of dimers are modulated by the spacing and relative orientation of the half-sites. DNAs with direct repeats, inverted repeats, and everted repeats exhibit nuclear receptor binding as head-to-tail, head-to-head, and tail-to-tail dimers, respectively. For example, RXR (27), Rev-Erb (29), and VDR (28) engage with DR1, DR2, and DR3, respectively, as head-to-tail homodimers (Fig. S3, A–C). The structure was similar in the case of DR4 sequences recognized by a RXR–Thyroid hormone receptor heterodimer (30) (Fig. S3D). A dimer interface was formed between the CTE sequence of the downstream subunit and the second zinc finger of the upstream subunit in all of these structures. The 1- to 4-bp spacers between two half-sites diversified the relative displacement of the two protein subunits on the DNAs. For inverted repeats, steroid receptors such as GR (26) and NR4A2 bind to IR3 and IR5 sites as head-to-head homodimers, respectively. The different spacers drove the participation of varying residues in the dimer interaction. Two GR subunits form an extensive dimer interface through residues along with the zinc ions in the second zinc-binding motif (Fig. S3E), whereas two NR4A2 subunits engaged with the IR5 sequence form a narrow interface involving N-terminal residues close to the second zinc-binding motif (Fig. 2A). For everted repeats, the structure of NR4A2–ER0 determined here demonstrated that two NR4A2 molecules formed a tail-to-tail orientation (Fig. 2B). Overall, the relative orientation and diverse spacers of the two half-sites dictated the different displacements and dimer interfaces of nuclear receptor homodimers.
In addition to DNA recognition by DBDs of nuclear receptors, LBDs can also affect the DNA binding properties of these receptors (31). Classic LBD-mediated dimerization interactions have been observed in the structure of the full-length PPARγ–RXRα heterodimer bound to DR1 (PDB code 3DZY) (31). The LBD of PPARγ not only forms contacts with the LBD of RXRα through helices 7, 9, and 10 of each receptor but also interacts with the DBD CTE region of RXRα. These dimerization interfaces contribute to stabilizing DR1 binding. Although the structure of full-length NR4A receptors has not been determined, the sequences that share homology with other nuclear receptors indicate that the LBDs might also help in defining the preferred dimerization to bring the full-length proteins in close proximity to DNA.
In summary, we determined two crystal structures of NR4A2-DBD–DNA complexes and provided the molecular basis for DNA recognition by NR4A dimers. We revealed a new mode of nuclear receptor binding as a dimer to IR5. The two DBDs formed a novel dimer interface, primarily via van der Waals contacts between residues formed by the loop preceding the second zinc-finger motif of each subunit. The dimer interface and protein–DNA interactions both play important roles in favoring cooperative dimer formation. Our structural, biochemical, and bioinformatics analyses may provide a better understanding of the dimerization specificity of nuclear receptors.
Experimental procedures
Expression and purification
Human NR4A2-DBD (residues 259–348) was cloned into a modified pMAL-C5X vector (32). The NR4A2 plasmid was transformed into Escherichia coli Rosetta (DE3) cells. After purification by an amylose resin column (BioLabs), the N-terminal maltose-binding protein tag was removed by PreScission protease at 4 °C overnight. The cleaved protein was further purified by a Mono S cation exchange column and a Superdex 75 column (33). The final protein was concentrated to ∼20 mg/ml in 20 mm HEPES (pH 7.5), 200 mm NaCl, and 0.5 mm tris(2-carboxyethyl)phosphine (34). DNA was synthesized by Genewiz (Nanjing, China) and purified as described previously (35). Human RXR-DBD was cloned into the pGEX-6P1 vector and purified as described previously (27).
Crystallization and data collection
Protein and DNA complexes were prepared by mixing protein and DNA at a 5:3 molar ratio. Crystals of NR4A2-DBD–ER0 were grown at 18 °C by the hanging drop method with a reservoir buffer containing 150 mm NaCl, 50 mm MES (pH 5.93), 10 mm MgCl2, 5 mm CaCl2, and 10%–12% PEG4K (w/v). Crystals of NR4A2-DBD–IR5 were grown at 18 °C with a reservoir buffer containing 50 mm Na acetate (pH 4.7), 200 mm NaCl, 10 mm MgCl2, 5 mm Li2SO4, 5 mm DTT, and 15%–19% PEG4K (w/v). Then crystals were transferred into a well solution containing an additional 20% glycerol (v/v) and flash-frozen in liquid nitrogen. Data were collected at the BL17U1 beamline and BL19U1 beamline of the Shanghai Synchrotron Radiation Facility.
Structure determination
Data were reduced using HKL2000 (36). The structures were solved by molecular replacement using Phaser from the PHENIX package (37). A previously solved rat NR4A1 structure (PDB code 1CIT) (15) was used as an initial search model. Further refinements were performed using Phenix and CCP4 (38, 39). Graphical representations of structures were generated using PyMOL (40). The statistics of the crystallographic analysis are presented in Table 1. Schematics of protein–DNA interactions were generated by NUCPLOT (22). The figures were generated using PyMOL (41).
Site-directed mutagenesis
Site-directed mutagenesis of NR4A2-DBD was performed according to the manufacturer's instructions for the ClonExpress II One Step Cloning Kit (Vazyme) using the pMAL-C5X-NR4A2 plasmid as the template (42). These mutants were verified by DNA sequencing (Tsingke, Changsha, China). Mutant NR4A2 proteins were expressed and purified as WT protein.
EMSA
Protein and DNA samples were prepared at a concentration of 45 μm. DNA was incubated with protein in a total volume of 6 μl in a buffer containing 20 mm HEPES (pH 7.5), 10 mm MgCl2, 200 mm NaCl, and 0.01% Triton X-100 for 20 min at room temperature. A native 8% (w/v) PAGE in 0.5× Tris borate-EDTA buffer was used to separate the free DNA from the protein–DNA complex (43). The gel was visualized after staining with GoldView.
Bioinformatics analysis
The bed files of the ChIP-seq data (GSE123629) were downloaded (12). The peaks were annotated, and the sequences were fetched using the R package ChIPpeakAnno (44). The core sequences ER0 and IR5 were searched against the ChIP-seq peak sequences using the R package TFBSTools (45).
Author contributions
L. J. and H. W. writing-original draft; L. J. writing-review and editing; S. D., J. L., X. L., X. C., and M. G. software; L. Q. and Y. C. supervision; Z. C. and Y. C. resources; L. C., H. W., and Y. C. validation; Y. C. project administration.
Supplementary Material
Acknowledgments
We thank the staff of the BL17U1 and BL19U1 beamlines of the Shanghai Synchrotron Radiation facility for help with data collection. We thank Dr. Michael R. Stallcup for proofreading.
This work was supported by National Natural Science Foundation of China Grants 81372904 and 81570537 (to Y. C.) and 81272971 (to Z. C.) and the Science and Technology Department of Hunan Province Grant 2015SK2037 (to Y. C.). The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
The atomic coordinates and structure factors (codes 6L6Q and 6L6L) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- TF
- transcription factor
- RXR
- retinoid X receptor
- VDR
- vitamin D3 receptor
- NBRE
- NGFI-B–responsive element
- NurRE
- Nur-responsive element
- LBD
- ligand-binding domain
- DBD
- DNA-binding domain
- CTE
- C-terminal extension
- ChIP-seq
- ChIP sequencing
- GR
- glucocorticoid receptor.
References
- 1. Wang J., Yang J., Zou Y., Huang G. L., and He Z. W. (2013) Orphan nuclear receptor nurr1 as a potential novel marker for progression in human prostate cancer. Asian Pac. J. Cancer Prev. 14, 2023–2028 10.7314/APJCP.2013.14.3.2023 [DOI] [PubMed] [Google Scholar]
- 2. Mohan H. M., Aherne C. M., Rogers A. C., Baird A. W., Winter D. C., and Murphy E. P. (2012) Molecular pathways: the role of NR4A orphan nuclear receptors in cancer. Clin. Cancer Res. 18, 3223–3228 10.1158/1078-0432.CCR-11-2953 [DOI] [PubMed] [Google Scholar]
- 3. Zhan Y. Y., Chen Y., Zhang Q., Zhuang J. J., Tian M., Chen H. Z., Zhang L. R., Zhang H. K., He J. P., Wang W. J., Wu R., Wang Y., Shi C., Yang K., Li A. Z., et al. (2012) The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK. Nat. Chem. Biol. 8, 897–904 10.1038/nchembio.1069 [DOI] [PubMed] [Google Scholar]
- 4. Berwick D. C., and Harvey K. (2014) The regulation and deregulation of Wnt signaling by PARK genes in health and disease. J. Mol. Cell Biol. 6, 3–12 10.1093/jmcb/mjt037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Yin K., Chhabra Y., Tropée R., Lim Y. C., Fane M., Dray E., Sturm R. A., and Smith A. G. (2017) NR4A2 promotes DNA double-strand break repair upon exposure to UVR. Mol. Cancer Res. 15, 1184–1196 10.1158/1541-7786.MCR-17-0002 [DOI] [PubMed] [Google Scholar]
- 6. Zhang L., Wang Q., Liu W., Liu F., Ji A., and Li Y. (2018) The orphan nuclear receptor 4A1: a potential new therapeutic target for metabolic diseases. J. Diabetes Res. 2018, 9363461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Holla V. R., Mann J. R., Shi Q., and DuBois R. N. (2006) Prostaglandin E2 regulates the nuclear receptor NR4A2 in colorectal cancer. J. Biol. Chem. 281, 2676–2682 10.1074/jbc.M507752200 [DOI] [PubMed] [Google Scholar]
- 8. Shigeishi H., Higashikawa K., Hatano H., Okui G., Tanaka F., Tran T. T., Rizqiawan A., Ono S., Tobiume K., and Kamata N. (2011) PGE 2 targets squamous cell carcinoma cell with the activated epidermal growth factor receptor family for survival against 5-fluorouracil through NR4A2 induction. Cancer Lett. 307, 227–236 10.1016/j.canlet.2011.04.008 [DOI] [PubMed] [Google Scholar]
- 9. Philip M., Fairchild L., Sun L., Horste E. L., Camara S., Shakiba M., Scott A. C., Viale A., Lauer P., Merghoub T., Hellmann MD, Wolchok J. D., Leslie C. S., and Schietinger A. (2017) Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Scott-Browne J. P., López-Moyado I. F., Trifari S., Wong V., Chavez L., Rao A., and Pereira R. M. (2016) Dynamic changes in chromatin accessibility occur in CD8+ T cells responding to viral infection. Immunity 45, 1327–1340 10.1016/j.immuni.2016.10.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Liu X., Wang Y., Lu H., Li J., Yan X., Xiao M., Hao J., Alekseev A., Khong H., Chen T., Huang R., Wu J., Zhao Q., Wu Q., Xu S., et al. (2019) Genome-wide analysis identifies NR4A1 as a key mediator of T cell dysfunction. Nature 567, 525–529 10.1038/s41586-019-0979-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J., López-Moyado I. F., Seo H., Lio C. J., Hempleman L. J., Sekiya T., Yoshimura A., Scott-Browne J. P., and Rao A. (2019) NR4A transcription factors limit CAR T cell function in solid tumours. Nature 567, 530–534 10.1038/s41586-019-0985-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Khorasanizadeh S., and Rastinejad F. (2001) Nuclear-receptor interactions on DNA-response elements. Trends Biochem. Sci. 26, 384–390 10.1016/S0968-0004(01)01800-X [DOI] [PubMed] [Google Scholar]
- 14. Helsen C., Kerkhofs S., Clinckemalie L., Spans L., Laurent M., Boonen S., Vanderschueren D., and Claessens F. (2012) Structural basis for nuclear hormone receptor DNA binding. Mol. Cell. Endocrinol. 348, 411–417 10.1016/j.mce.2011.07.025 [DOI] [PubMed] [Google Scholar]
- 15. Meinke G., and Sigler P. B. (1999) DNA-binding mechanism of the monomeric orphan nuclear receptor NGFI-B. Nat. Struct. Biol. 6, 471–477 10.1038/8276 [DOI] [PubMed] [Google Scholar]
- 16. Maira M., Martens C., A, Philips A., and Drouin J. (1999) Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell. Biol. 19, 7549–7557 10.1128/MCB.19.11.7549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Aarnisalo P., Kim C. H., Lee J. W., and Perlmann T. (2002) Defining requirements for heterodimerization between the retinoid X receptor and the orphan nuclear receptor Nurr1. J. Biol. Chem. 277, 35118–35123 10.1074/jbc.M201707200 [DOI] [PubMed] [Google Scholar]
- 18. Jolma A., Yan J., Whitington T., Toivonen J., Nitta K. R., Rastas P., Morgunova E., Enge M., Taipale M., Wei G., Palin K., Vaquerizas J. M., Vincentelli R., Luscombe N. M., Hughes T. R., et al. (2013) DNA-binding specificities of human transcription factors. Cell 152, 327–339 10.1016/j.cell.2012.12.009 [DOI] [PubMed] [Google Scholar]
- 19. Murphy E. P., and Conneely O. M. (1997) Neuroendocrine regulation of the hypothalamic pituitary adrenal axis by the nurr1/nur77 subfamily of nuclear receptors. Mol. Endocrinol. 11, 39–47 10.1210/mend.11.1.9874 [DOI] [PubMed] [Google Scholar]
- 20. Maira M., Martens C., Batsché E., Gauthier Y., and Drouin J. (2003) Dimer-specific potentiation of NGFI-B (Nur77) transcriptional activity by the protein kinase A pathway and AF-1-dependent coactivator recruitment. Mol. Cell Biol. 23, 763–776 10.1128/MCB.23.3.763-776.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zwart P. H., Grosse-Kunstleve R. W., Lebedev A. A., Murshudov G. N., and Adams P. D. (2008) Surprises and pitfalls arising from (pseudo)symmetry. Acta Crystallogr. D Biol. Crystallogr. 64, 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Luscombe N. M., Laskowski R. A., and Thornton J. M. (1997) NUCPLOT: a program to generate schematic diagrams of protein-nucleic acid interactions. Nucleic Acids Res. 25, 4940–4945 10.1093/nar/25.24.4940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Krissinel E., and Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 10.1016/j.jmb.2007.05.022 [DOI] [PubMed] [Google Scholar]
- 24. Wallen-Mackenzie A., Mata de Urquiza A., Petersson S., Rodriguez F. J., Friling S., Wagner J., Ordentlich P., Lengqvist J., Heyman R. A., Arenas E. and Perlmann T. (2003) Nurr1-RXR heterodimers mediate RXR ligand-induced signaling in neuronal cells. Genes Dev. 17, 3036–3047 10.1101/gad.276003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Scheepstra M., Andrei S. A., de Vries R. M. J. M., Meijer F. A., Ma J.-N., Burstein E. S., Olsson R., Ottmann C., Milroy L.-G., and Brunsveld L. (2017) Ligand dependent switch from RXR homo- to RXR-NURR1 heterodimerization. ACS Chem. Neurosci. 8, 2065–2077 10.1021/acschemneuro.7b00216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Meijsing S. H., Pufall M. A., So A. Y., Bates D. L., Chen L., and Yamamoto K. R. (2009) DNA binding site sequence directs glucocorticoid receptor structure and activity. Science 324, 407–410 10.1126/science.1164265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Osz J., McEwen A. G., Poussin-Courmontagne P., Moutier E., Birck C., Davidson I., Moras D., and Rochel N. (2015) Structural basis of natural promoter recognition by the retinoid X nuclear receptor. Sci. Rep. 5, 8216 10.1038/srep08216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Shaffer P. L., and Gewirth D. T. (2002) Structural basis of VDR–DNA interactions on direct repeat response elements. EMBO J. 21, 2242–2252 10.1093/emboj/21.9.2242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhao Q., Khorasanizadeh S., Miyoshi Y., Lazar M. A., and Rastinejad F. (1998) Structural elements of an orphan nuclear receptor-DNA complex. Mol. Cell 1, 849–861 10.1016/S1097-2765(00)80084-2 [DOI] [PubMed] [Google Scholar]
- 30. Rastinejad F., Perlmann T., Evans R. M., and Sigler P. B. (1995) Structural determinants of nuclear receptor assembly on DNA direct repeats. Nature 375, 203–211 10.1038/375203a0 [DOI] [PubMed] [Google Scholar]
- 31. Chandra V., Huang P., Hamuro Y., Raghuram S., Wang Y., Burris T. P., and Rastinejad F. (2008) Structure of the intact PPAR-γ-RXR-nuclear receptor complex on DNA. Nature 456, 350–356 10.1038/nature07413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Jiang L., Liang X., Qu L., Wu D., Chen X., Guo M., Chen Z., Chen L., and Chen Y. (2017) DNA-binding properties of FOXP3 transcription factor. Acta Biochim. Biophys. Sin. 49, 792–799 10.1093/abbs/gmx079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen Y., Chen C., Zhang Z., Liu C. C., Johnson M. E., Espinoza C. A., Edsall L. E., Ren B., Zhou X. J., Grant S. F., Wells A. D., and Chen L. (2015) DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res. 43, 1268–1282 10.1093/nar/gku1373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Yan N., Li J., Chen X., Chen Y., Chen L., and Chen Z. (2015) Expression and purification of DNA binding domain of NR4A1. Zhong Nan Da Xue Bao Yi Xue Ban 40, 345–350 [DOI] [PubMed] [Google Scholar]
- 35. Li J., Dantas Machado A. C., Guo M., Sagendorf J. M., Zhou Z., Jiang L., Chen X., Wu D., Qu L., Chen Z., Chen L., Rohs R., and Chen Y. (2017) Structure of the forkhead domain of FOXA2 bound to a complete DNA consensus site. Biochemistry 56, 3745–3753 10.1021/acs.biochem.7b00211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Otwinowski Z., and Minor W. (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- 37. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Echols N., Headd J. J., Hung L. W., Jain S., Kapral G. J., Grosse Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R. D., Read R. J., Richardson D. C., et al. (2011) The Phenix software for automated determination of macromolecular structures. Methods 55, 94–106 10.1016/j.ymeth.2011.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 39. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., and Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 40. DeLano W. L. (2012) The PyMOL Molecular Graphics System, version 1.5.0.1, Schroedinger, LLC, New York [Google Scholar]
- 41. Bramucci E., Paiardini A., Bossa F., and Pascarella S. (2012) PyMod: sequence similarity searches, multiple sequence-structure alignments, and homology modeling within PyMOL. BMC Bioinformatics 13, S2 10.1186/1471-2105-13-S4-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wu D., Guo M., Min X., Dai S., Li M., Tan S., Li G. Q., Chen X., Ma Y., and Li J. (2018) LY2874455 potently inhibits FGFR gatekeeper mutant and overcomes mutation-based resistance. Chem. Commun. 54, 12089–12092 10.1039/C8CC07546H [DOI] [PubMed] [Google Scholar]
- 43. Chen X., Wei H., Li J., Liang X., Dai S., Jiang L., Guo M., Qu L., Chen Z., Chen L., and Chen Y. (2019) Structural basis for DNA recognition by FOXC2. Nucleic Acids Res. 47, 3752–3764 10.1093/nar/gkz077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhu L. J., Gazin C., Lawson N. D., Pagès H., Lin S. M., Lapointe D. S., and Green M. R. (2010) ChIPpeakAnno: a Bioconductor package to annotate ChIP-seq and ChIP-chip data. BMC Bioinformatics 11, 237 10.1186/1471-2105-11-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tan G., and Lenhard B. (2016) TFBSTools: an R/bioconductor package for transcription factor binding site analysis. Bioinformatics 32, 1555–1556 10.1093/bioinformatics/btw024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sebastian A., and Contreras-Moreira B. (2014) footprintDB: a database of transcription factors with annotated cis elements and binding interfaces. Bioinformatics 30, 258–265. 10.1093/bioinformatics/btt663 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.