Abstract
In cancer, aberrant growth factor receptor signaling reprograms cellular metabolism and global gene transcription to drive aggressive growth, but the underlying mechanisms are not well-understood. Here we show that in the highly lethal brain tumor glioblastoma (GBM), mTOR complex 2 (mTORC2), a critical core component of the growth factor signaling system, couples acetyl-CoA production with nuclear translocation of histone-modifying enzymes including pyruvate dehydrogenase and class IIa histone deacetylases to globally alter histone acetylation. Integrated analyses in orthotopic mouse models and in clinical GBM samples reveal that mTORC2 controls iron metabolisms via histone H3 acetylation of the iron-related gene promoter, promoting tumor cell survival. These results nominate mTORC2 as a critical epigenetic regulator of iron metabolism in cancer.
Keywords: mammalian target of rapamycin (mTOR), histone acetylation, glucose metabolism, acetyl coenzyme A (acetyl-CoA), iron metabolism, glioblastoma
Introduction
Metabolic reprogramming is a core hallmark of cancer (1, 2), enabling cancer cells to meet the coordinately elevated anabolic and energetic demands imposed by rapid tumor growth (3). Aberrant growth factor receptor signaling, frequently as a consequence of receptor tyrosine kinase (RTK)3 amplification and/or gain of function mutations, promotes tumor growth by coupling proliferative signaling with enhanced nutrient uptake and utilization. This process also alters gene expression, including via epigenetic regulation (4), although the molecular mechanisms are still incompletely understood. Persistent growth factor receptor signaling can enhance the cellular pool of acetyl-CoA, which provides the acetyl groups needed for histone acetylation (5, 6). At present, the components of the growth factor signaling system that link oncogenic mutations with epigenetic regulation, and the specific genes that may be regulated by this process, mandate further study.
We focus on mTOR complex 2 (mTORC2), a core component of the growth factor signaling in glioblastoma (GBM), the highly lethal form of brain tumor. Epidermal growth factor receptor (EGFR) is amplified in over 50% of GBMs (7), commonly in association with the gain of function mutation EGFRvIII. We have previously shown that EGFRvIII potently activates mTORC2, which is required for GBM pathogenesis via its effect on glucose, amino acid, and lipid metabolism (8–11). We have also shown that mTORC2 integrates aberrant growth factor signaling with glucose and amino acid availability, ensuring that aberrant growth factor receptor signaling is coupled with sufficient environmental nutrients to support rapid tumor growth (9, 11, 12). We hypothesized that mTORC2 could also play a role in gene regulation, by controlling histone acetylation. Here, we report a previously unanticipated role for mTORC2 in driving GBM growth by promoter histone acetylation of iron metabolism genes.
Results
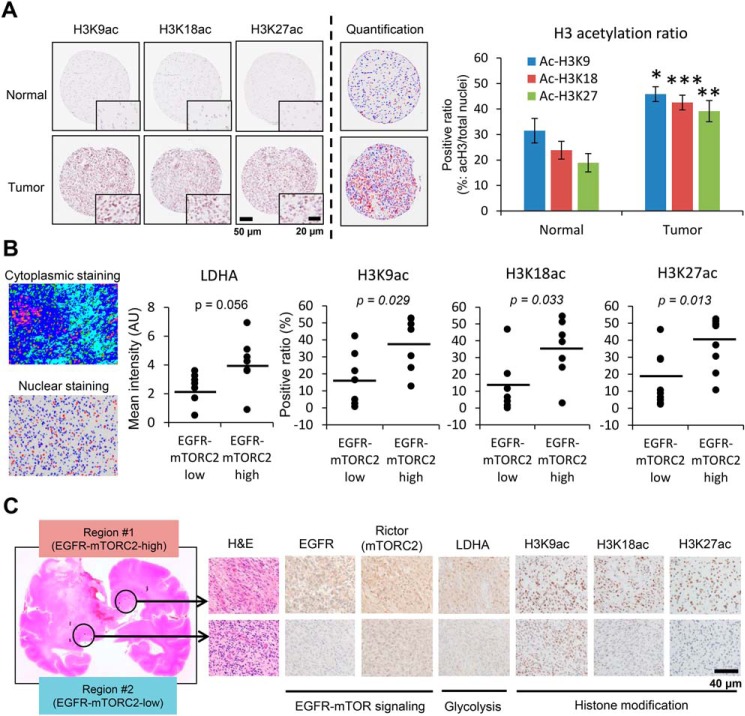
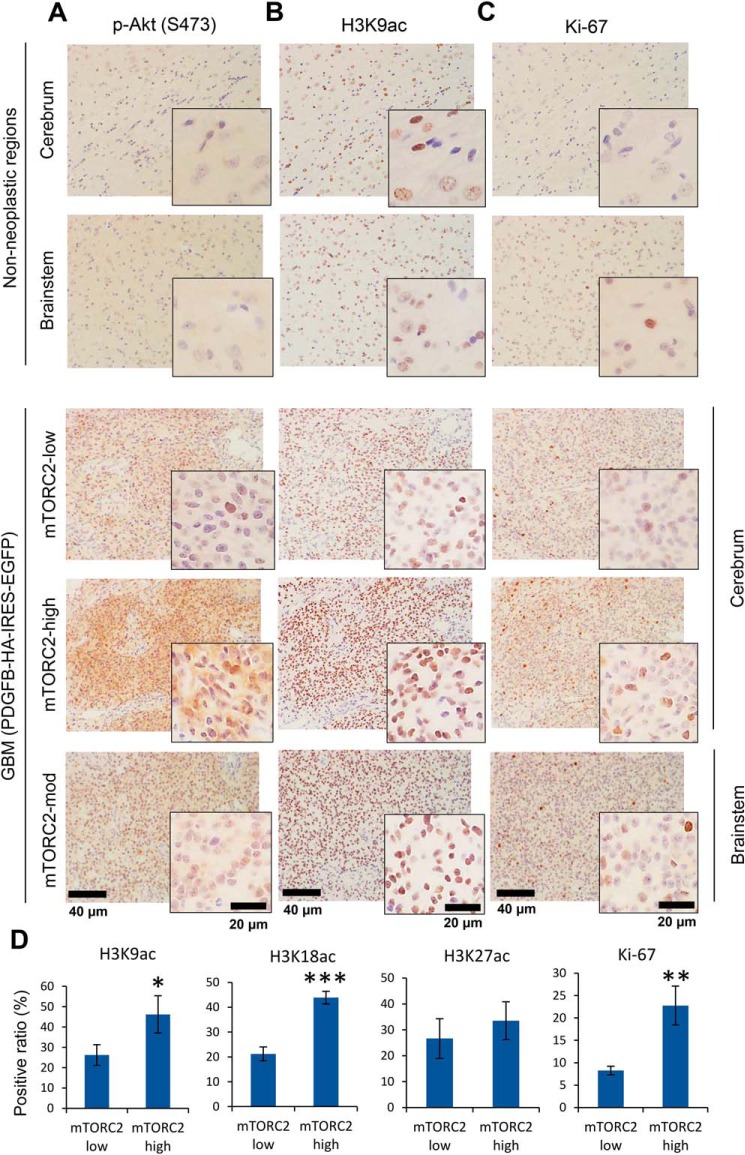
To examine the histone acetylation status in human GBM, we performed quantitative immunohistochemical analyses of EGFR/mTORC2 signaling, glycolytic enzyme lactate dehydrogenase A (LDHA) expression, and acetylation of histone H3 tails including lysines 9, 18, and 27 (H3K9ac, K18ac, and K27ac) in surgical and autopsied cases of human GBM, revealing increased H3K9ac, K18ac, and K27ac in EGFR/mTORC2-activated GBM clinical samples (Fig. 1, A–C, and Fig. S1, B and D). Notably, GBM and breast cancer specimens display that EGFR highly expressing tumor cells increase histone acetylation within tumor (Fig. S1C), suggesting that genetic and epigenetic interaction may be a phenomenon observed in a broader range of cancer types. Similar to persistent EGFR signaling, constitutive platelet-derived growth factor (PDGF)-mediated activation in vivo, using a retrovirus-mediated PDGF ligand overexpression to activate PDGF receptor (PDGFR) signaling induced GBM tumors including cerebrum as well as brainstem, with tumor cells that similarly displayed high levels of acetylated histone H3 compared with the control non-neoplastic brain regions (Fig. 2B and Fig. S2A). The expression of H3 acetylation was correlated with a proliferative marker Ki-67 (Fig. 2C). Interestingly, even in the control rat brain, H3 acetylation was observed in specific regions such as in the white matter and the subventricular zone (Fig. S2B), and these might reflect the glioma-initiating cells (13, 14). We next interrogated signaling pathways downstream of PDGF to identify those responsible for promoting histone acetylation. The level of mTORC2 signaling markers was compatible with the histone acetylation level in the tumor (Fig. 2, A and D, and Fig. S2A). These data indicate that RTK-mTORC2–dependent histone acetylation is prevalent among GBM driven either by EGFR or PDGFR signaling.
Figure 1.
EGFR-mTORC2 signaling correlates with histone acetylation in human GBM. A, histone H3K9, K18, and K27 acetylation immunostaining of GBM tissue (15 normal brain tissue and 17 GBM tissue). A bar graph showing the positive ratio of each histone mark (acetylated histone H3 (red)/total nuclei (red + blue)). Scale bars, 50 μm (20 μm for insets). B, quantitative immunohistochemical analysis of LDHA (a glycolytic enzyme) and histone H3 acetylation (K9ac, K18ac, and K27ac), based on low and high expression regions in EGFR-mTORC2 signaling, obtained at autopsy from 7 patients with GBM or surgically resected 10 GBM tissue. The averaged mean saturation of Rictor for the mTORC2-low group is 5.39, and the mTORC2-high group is 8.83. Also shown are representative images of immunohistochemical quantification: cytoplasmic staining with Visiopharm and nuclear staining with ImageJ. AU, arbitrary unit. C, representative images for differential expression in EGFR-mTOR signaling, glycolytic metabolism and histone acetylation in autopsied GBM samples. Scale bar, 40 μm.
Figure 2.
mTORC2 signaling and histone acetylation are promoted in PDGF-induced GBM animal models. A–C, cerebral and brainstem tissue with GBM tumors was harvested from rats infected with PDGFB-HA-IRES-EGFP retroviral vectors (n = 3 for each region). Immunohistochemistry was performed on paraffin-embedded tissue sections against an mTORC2 marker (p-Akt S473, A), a histone acetylation mark (H3K9ac, B), and a proliferation marker (Ki-67, C). mTORC2-high, -moderate, and -low regions in GBM tissue was compared with contralateral non-neoplastic regions. Representative images are demonstrated with higher magnification of regions of interest. Scale bars, 40 μm (20 μm for insets). D, quantification of histone H3 acetylation (K9ac, K18ac, K27ac) and Ki-67, based on low and high activity regions in mTORC2 signaling.
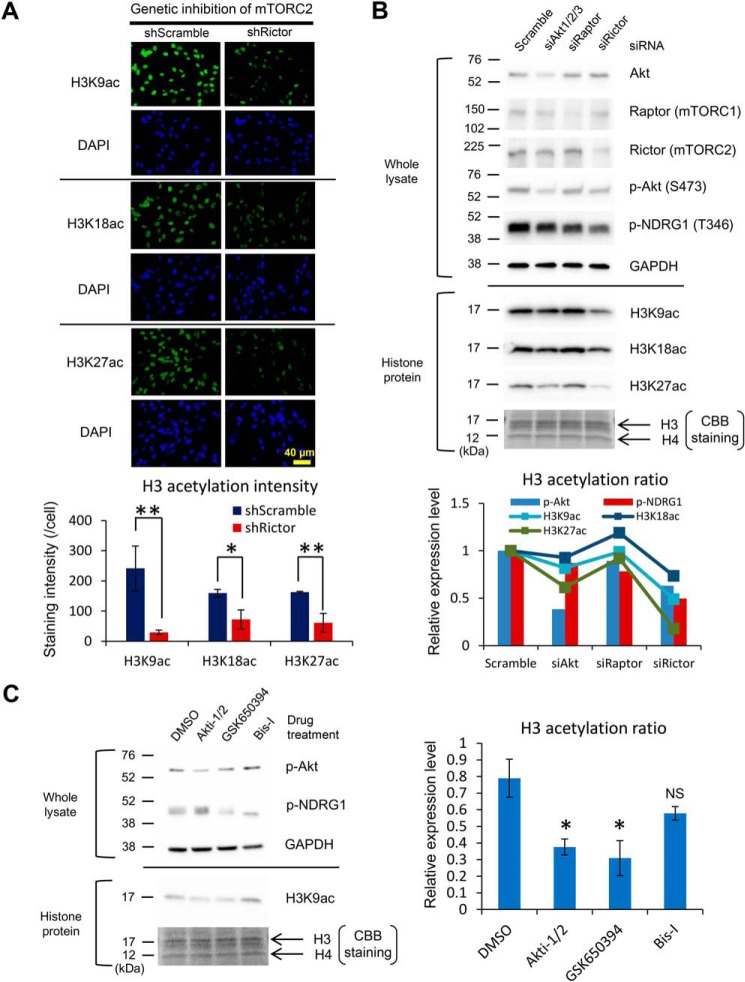
We next examined whether mTORC2 directly regulates histone acetylation. Lentivirus-mediated genetic knockdown of RICTOR (a core component of mTORC2) by shRNA decreases nuclear expression of histone acetylation in GBM cells (Fig. 3A). Of note, siRNA-mediated knockdown studies on signaling components downstream of EGFR demonstrated that mTORC2 inhibition potently decreases histone acetylation, whereas Akt, one of the effectors downstream of mTORC2, has the mild effect on it (Fig. 3B). We further investigated the effector to regulate histone acetylation downstream of mTORC2 and found that both Akt and SGK1 are the potential regulators of histone acetylation downstream of mTORC2 (Fig. 3C). We and others speculate that the level of intermediary metabolites derived from metabolic reactions could have a profound effect on epigenetic regulation (5, 6). Indeed, metabolomic analyses demonstrated the drastic change in intermediary metabolites by mTORC2 inhibition (Fig. S3, A and B). To examine the effect of metabolic reprogramming on histone acetylation, we analyzed the role of acetyl-CoA production on the acetylation of histone protein. Glucose and acetate are two major nutrients for acetyl-CoA production (15), and we found that glucose deprivation from the culture medium or PP242 treatment (mTORC2 inhibition) significantly decreased global histone H3 acetylation, which was restored by the addition of exogenous acetate (Fig. S3, C and D). Furthermore, RICTOR overexpression augmented the histone H3K9 acetylation, and glucose withdrawal abrogated the effect of RICTOR overexpression on histone acetylation, which was further rescued by the addition of acetate (Fig. S3E). Together, these findings indicate that mTORC2 facilitates histone acetylation via metabolic reprogramming and the production of a metabolite acetyl-CoA.
Figure 3.
mTORC2 facilitates histone acetylation through its downstream effector Akt and SGK1. A, immunofluorescent staining of H3K9ac, K18ac, and K27ac (green) in U87-EGFRvIII cells with shRNA against Scramble sequence or Rictor. DAPI staining shows the counter nuclear staining (blue). Bar graphs demonstrate quantification of histone H3K9, K18, and K27 acetylation in U87-EGFRvIII cells with Scramble or Rictor shRNA (n = 3). Scale bar, 40 μm. B, immunoblot detection and quantification of histone H3 acetylation (H3K9ac, H3K18ac, H3K27ac) in U87-EGFRvIII cells with the indicated siRNAs targeting the effector molecules in EGFR-mTOR signaling including Akt, Raptor (mTORC1), and Rictor (mTORC2). Bar graphs showing the activation markers for Akt and mTORC2 pathways (p-Akt S473 and p-NDRG1 T346), and line graphs for H3 acetylation ratio. CBB, Coomassie Brilliant Blue staining. C, immunoblot detection and quantification of histone H3K9 acetylation in U87-EGFRvIII cells treated with drugs targeting mTORC2 substrates including Akt (Akti-1/2: 2.5 μm), SGK1 (GSK650394: 2.0 μm), and PKC-α (Bis-I: 10 μm) for 48 h. NS, not significant.
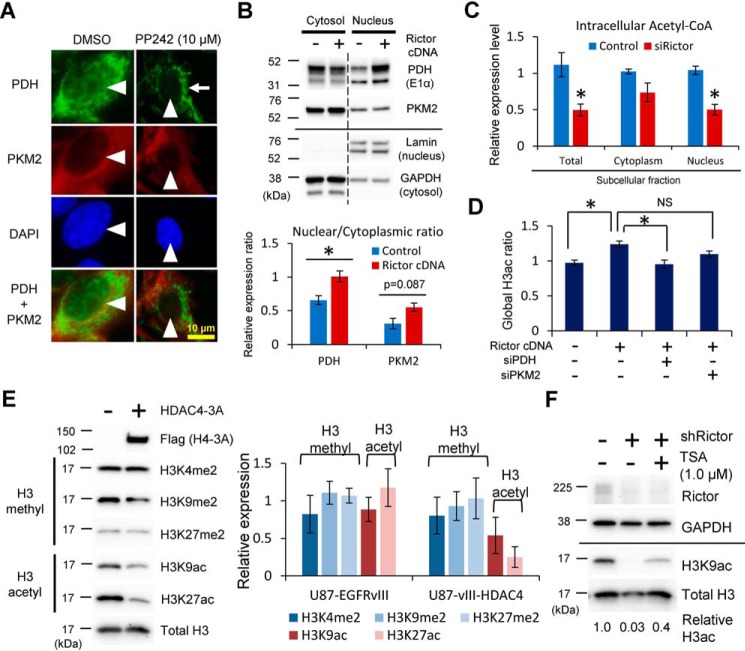
Acetyl-CoA–producing enzymes were reported to promote histone acetylation via metabolic reprogramming (16, 17). A recent report demonstrated that nuclear translocation of pyruvate dehydrogenase (PDH) complexes from mitochondria along with pyruvate kinase M2 (PKM2) isoform increase histone acetylation (20, 21). We found that mTORC2 activity significantly regulated the subcellular localization of PDH as well as mildly that of PKM2 (Fig. 4, A and B). Supporting our hypothesis, knockdown of RICTOR decreased the total, cytoplasmic as well as nuclear level of acetyl-CoA (Fig. 4C), important in the acetylation of nuclear histone (16, 21). Consistent with this, an increase in global H3 histone acetylation by RICTOR overexpression was mitigated by the concurrent knockdown of PDH (Fig. 4D). Another important source of acetyl-CoA was from glutamine via anaplerotic reactions (i.e. glutaminolysis) (22), and it may be related to mTORC2-dependent histone acetylation. However, the effect of glutaminolysis inhibitor (BPTES) on mTORC2-mediated histone acetylation was at least partly independent of the status of Rictor (Fig. S4), suggesting that PDH is the main enzyme to control acetyl-CoA and histone acetylation downstream of mTORC2. In addition to the acetyl-donor substrate (acetyl-CoA), histone tail acetylation necessitates the balance between histone acetyltransferases (HATs) and histone deacetylases (HDACs) (18, 19). Considering the previous reports that mTORC2 mainly affects the status of class IIa HDACs among various HATs and HDACs (8), mTORC2 may promote the histone acetylation through the regulation of class IIa HDACs. We thus examined the additional effect of class IIa HDACs on mTORC2-mediated histone acetylation. Overexpression of class IIa HDACs in GBM cells with mTORC2 activation by EGFRvIII reduced histone acetylation, but had no effect on the methylation of histone H3 tails (Fig. 4E). Furthermore, reduction of histone acetylation by RICTOR knockdown was partially rescued by the pharmacological inhibition of class IIa HDACs with tricostatin A (TSA) (Fig. 4F). Taken together, mTORC2 promotes histone acetylation both by increasing the acetyl-donor substrate via shifting the subcellular localization of acetyl-CoA producing enzymes PDH as well as by affecting the activity of histone modifying enzymes, class IIa HDACs.
Figure 4.
mTORC2 regulates histone acetylation through nuclear translocation of PDH/PKM2 and suppression of class IIa HDACs. A, immunofluorescent staining of PDH and PKM2 in U87-EGFRvIII cells treated by PP242 (mTORC1/C2 inhibitors) for 24 h. Arrowheads indicate each nucleus, and an arrow shows the mitochondrial distribution. Green, PDH staining; red, PKM2 staining; blue, 4′,6-diamidino-2-phenylindole (DAPI) staining. Scale bar, 10 μm. B, subcellular fractionation of cytosolic and nuclear protein from U87 cells with or without Rictor cDNA overexpression, followed by immunoblotting against PDH (E1α) and PKM2. Lamin and GAPDH are loading controls for nuclear and cytoplasmic proteins, respectively. Bar graphs showing the nuclear/cytoplasmic ratio of each protein in control and Rictor-overexpressing U87 cells. C, bar graphs showing the total, cytoplasmic, and nuclear amount of acetyl-CoA from U87-EGFRvIII cells with Scramble or Rictor knockdown. D, ELISA-based detection of global histone H3 acetylation in U87 cells overexpressing Rictor, combined with siRNA-mediated knockdown of PDH or PKM2. E, quantitative immunoblot analyses of histone modifications including methylation and acetylation in U87-EGFRvIII cells with the overexpression of class IIa HDAC (HDAC4–3A: phosphorylation-resistant mutant form). F, immunoblot analysis demonstrates that H3K9ac reduction in Rictor-depleted U87-EGFRvIII cells was partially rescued by the concurrent inhibition of HDACs with TSA for 24 h.
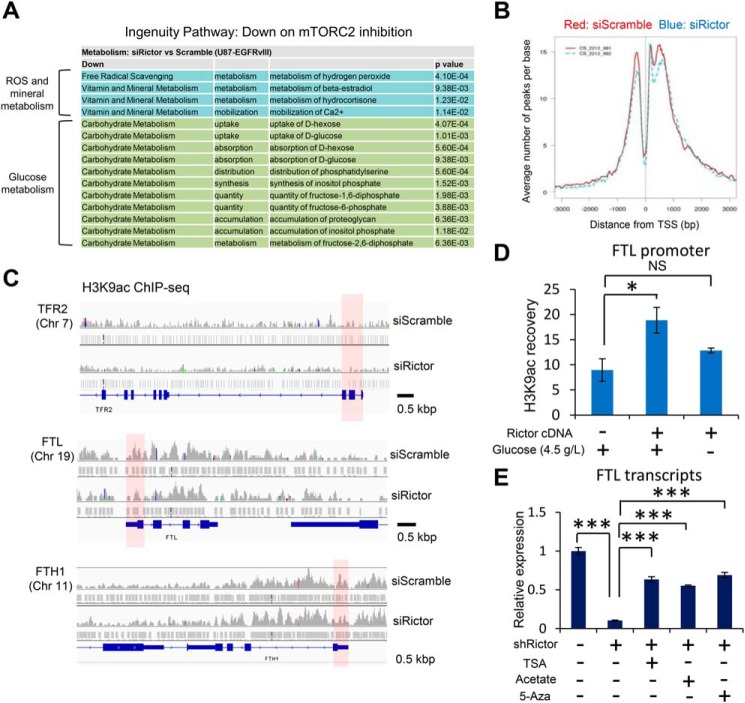
Having shown that histone acetylation is dynamically controlled by mTORC2 through metabolic reprogramming, we next examined its epigenetic effect on the expression of tumor-promoting genes because histone acetylation is generally considered to relax the chromatin configuration and facilitate the gene transcription (23, 24). Notably, pathway analyses on RNA-Seq–based transcriptome data demonstrated that mTORC2 inhibition affected the expression of genes related to not only RTK signaling, glioma, and metabolism (Fig. S5A), but also reactive oxygen species and mineral metabolism as well as glucose metabolism (Fig. 5A), suggesting that mTORC2 may regulate the genes related to iron metabolism in cancer cells through the epigenetic shift. Consistent with this idea, genes related to iron metabolism were up-regulated in GBM cells treated with TSA and acetate, which potentially increase the histone acetylation (Fig. S5B), further supporting the idea that iron-related genes could be epigenetically up-regulated in cancer cells. On the contrary, RNA-seq data demonstrated that the expression of transcription factors to potentially regulate the ferritin genes, such as SP1 (promotion) and ATF1 (repression), was not significantly different between control and RICTOR-knockdown GBM cells (Fig. S5C) (25, 26). The histone mark of H3K9ac is a known modification of promoted transcription (27, 28), and we performed the ChIP-sequencing analyses to determine the effect of mTORC2 on gene regulation through this histone modification. The twin peaks of H3K9ac were observed within 1,000 bp around the transcription start site (TSS) (Fig. 5B), indicating its association with the gene promoter regions. Interestingly, mTORC2 suppression by RICTOR knockdown reduced these dual peaks (Fig. 5B). We then sought the specific groups of the genes regulated by this epigenetic shift, and unexpectedly, mTORC2 inhibition clearly reduced the H3K9ac peaks around TSS along with the expression of the genes relevant to the iron metabolism such as transferrin receptor (TFR), ferritin light chain (FTL), and heavy chain (FTH1) (Fig. 5C), which are essential in maintaining iron metabolism (29). Notably, mTORC2 activation by overexpression of RICTOR promoted the H3K9 acetylation marks at the promoter region of FTL, which was abrogated by the concurrent withdrawal of glucose (Fig. 5D). Consistent with this, decreased expression of FTL transcripts by mTORC2 inhibition was recovered by the treatment with TSA (HDAC inhibition), acetate, or 5-aza-2′-deoxycytidine (DNA methylation inhibitor) (Fig. 5E and Fig. S5D), and knockdown of PDH and PKM2 reduced the level of FTL and FTH1 transcripts (Fig. S5E), suggesting the involvement of histone acetylation and potentially subsequent DNA demethylation on this process. These findings indicate that mTORC2 increases the expression of iron-related genes through the regulation of histone acetylation at their promoters.
Figure 5.
mTORC2-dependent histone acetylation promotes the expression of iron-related genes. A, ingenuity canonical pathway annotations that were down-regulated, but not up-regulated on mTORC2 inhibition by Rictor knockdown. B, average Peak densities of H3K9ac around the TSS in U87-EGFRvIII cells with siRNA against Scramble sequences (red) or Rictor (blue). C, University of California, Santa Cruz, browser image depicting H3K9ac peaks around the TSS of TER2 gene (locus on chromosome 3), FTL gene (on chromosome 19), and FTH1 gene (on chromosome 11) in U87-EGFRvIII cells with Scramble or Rictor siRNAs. Red shades, potential promoter regions of each gene. D, ChIP-qPCR analysis on U87-EGFRvIII cells transfected with Control (GFP-NTD) or Rictor cDNA combined with glucose withdrawal for 24 h, assessing H3K9ac recovery on binding elements in FTL promoter regions. E, RT-qPCR analyses on FTL transcripts in U87-EGFRvIII cells transfected with shRNA, combined with supplementation of TSA (1.0 μm), acetate (10 mm), or 5-aza-2′-deoxycytidine (100 nm) for 3 days.
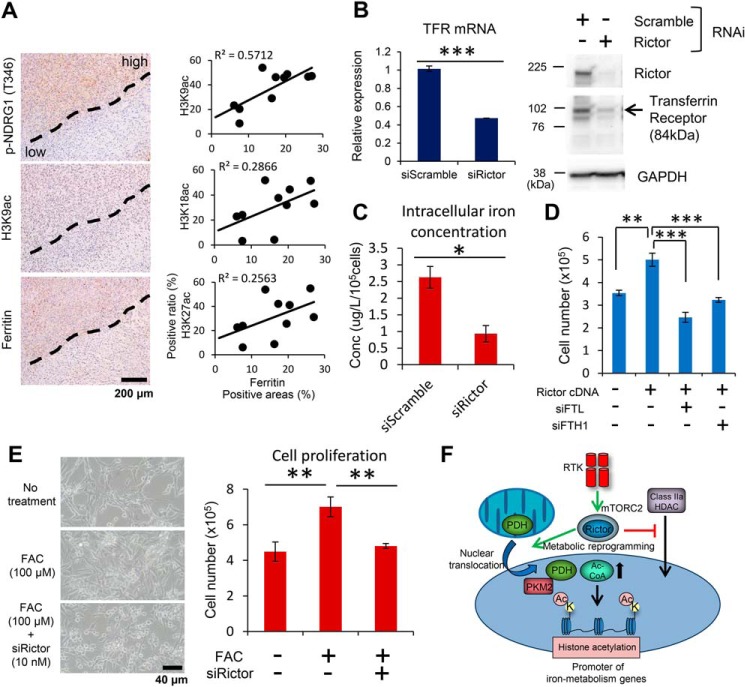
We last determined the functional role of mTORC2-dependent regulation of the components in iron metabolism. TCGA (The Cancer Genome Atlas) data sets (30, 31) demonstrated that the expression of iron metabolism-related genes such as TFRC, FTL, and FTH1 was up-regulated in GBM compared with lower grade gliomas, which usually exhibit less activity of the mTOR signaling pathway (Fig. S6A). Human GBM (IDH-WT) tissue showed higher staining of ferritin molecules and iron compared with normal brain (Fig. S6B). Furthermore, expression of ferritin in surgical GBM specimens was concordant with the mTORC2 activation marker and the acetylated histone mark (Fig. 6A). Consistent with the clinical findings, GBM cells with RICTOR knockdown markedly down-regulated the expression of iron transporters such as TFR and divalent metal transporter 1 (Fig. 6B and Fig. S6C). More importantly, the intracellular concentration of total iron was significantly reduced by RICTOR knockdown (Fig. 6C and Fig. S6D). We then hypothesized that mTORC2 signaling may render GBM cells addicted to iron metabolism for their survival. Of note, promotion of GBM cell proliferation by RICTOR overexpression was mitigated by the simultaneous knockdown of ferritin genes (FTL and FTH1) (Fig. 6D and Fig. S6E). Furthermore, exogenous addition of iron promotes the proliferation of GBM cells in an mTORC2-dependent manner (Fig. 6E). Taken together, these results indicate that GBM cells with activated mTORC2 signaling facilitate cellular survival by enhancing iron metabolism through the regulation of acetylated histones in their promoters (Fig. 6F).
Figure 6.
mTORC2-dependent iron metabolism regulates GBM cell survival. A, histological analyses for differential expression in mTORC2 signaling (p-NDRG1 T346), histone acetylation (H3K9ac, K18ac, and K27ac), and ferritin expression in surgical GBM samples (n = 10). Scatter plots showing the association of each histone mark and ferritin expression with H3K9ac showing the strongest correlation with ferritin expression. Scale bar, 200 μm. B, transcript and protein expression of TFR in U87-EGFRvIII cells with siRNAs against Rictor. C, bar graph showing the difference in total intracellular concentration of iron between U87-EGFRvIII cells with siScramble and siRictor knockdown (n = 3), quantified by atomic absorption spectrometry. D, bar graph showing the difference in cell proliferation (cell number) of U87 cells overexpressing Rictor with concurrent knockdown of FTL or FTH1 for 2 days (n = 3). E, quantitative analyses of the proliferation in U87-EGFRvIII cells treated with exogenous addition of 100 μm FAC, associated with siRNA-mediated knockdown of Rictor for 2 days (n = 3). Scale bar, 40 μm. F, RTK signaling activates mTORC2, which in turn promotes histone acetylation via the regulation of both the substrate and modifying enzymes, leading to the up-regulation of iron-related genes, a key downstream effector of cell survival in GBM.
Discussion
Amplification and/or gain of function mutations in cancer are prevalent because they sit at the top of a cascade of interlacing events that integrate proliferative signaling and nutrient uptake and utilization with epigenetic regulation and gene transcription. Dissecting the critical nodes that integrate environmental nutrient levels with genetic alterations will be critical for better understanding cancer pathogenesis and for identifying potential therapeutic targets (32). Here, we have found that mTORC2 promotes GBM growth by altering iron metabolism, and we demonstrate that this activity is mediated by promoter histone modification. These results further nominate mTORC2 as an important regulator of oncogenic growth factor signaling in GBM, integrating the local tissue microenvironmental nutrient levels with tumor growth. Further studies will be needed to better understand its potential role in other cancer types.
Histone modifications are a dynamic chromatin mark with various important roles in gene regulation (33). In particular, histone H3 acetylation is tightly associated with the transcriptional start sites of genes, highly predictive of gene activity (27, 28). H3K9ac is one of the major marks reported to be found in actively transcribed promoters, and mTORC2 surprisingly increases H3K9ac at the promoter regions of the components in iron metabolism. Iron had been thought to be associated with cancer incidence, and a recent report demonstrated that GBM stem-like cells promoted iron trafficking, driving tumor growth (34). Although the previous comprehensive studies of lymphocytes with RICTOR null mouse by Ye et al. (47) did not demonstrate the significant transcriptome changes in iron metabolism pathways (GSE111536: Gene expression data from LCMV-specific Rictor−/− and WT follicular T helper cells), the present study demonstrates that mTORC2 affects the cancer cell survival at least partly through the epigenetic regulation of iron metabolism. The findings could be consistent with the idea that iron metabolism is regulated mainly according to the status of the environmental amount of iron in normal cells, whereas that may not be the case for cancer cells. Additionally, intriguing speculation would be that the central carbon metabolism and mineral metabolism including iron might be epigenetically interconnected in cancer cells and iron metabolism could be actively and autonomously regulated in cancer cells.
Intensive effort is currently aimed at targeting cancer cell metabolism and epigenetics. Unfortunately, however, therapeutics targeting the metabolism and epigenetics have not been a success thus far because rapidly dividing normal constituents, such as hematopoietic cells, also harness the same kind of metabolic and epigenetic systems as cancer cells (2, 35). This study provides strong evidence that mTORC2 could reside at the intersection of metabolism and epigenetics, which could be a therapeutic target against cancer metabolism as well as epigenetics. Our data suggest that GBM cells with mTORC2 activation are highly dependent on iron metabolism, suggesting that the autonomous iron metabolism in cancer cells may be therapeutically exploitable.
Experimental procedures
Cell culture and human samples
U87 and U87-EGFRvIII GBM cell lines were obtained as described previously (36). Cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Omega Scientific, Tarzana, CA) in a humidified 5% CO2 incubator at 37 °C. Autopsied cases of human GBM were a part of the collections from the Tokyo Metropolitan Institute of Medical Science, and surgical tissues of glioma and breast cancer samples were obtained from the Tokyo Women's Medical University Hospital and the UCLA-affiliated hospitals. Physicians obtained informed consent from the patients. Gene expression analysis for TFRC, FTL, and FTH1 in lower grade gliomas and GBMs were performed using TCGA data sets with cBioPortal for Cancer Genomics (45, 46) (http://www.cbioportal.org/).4 All methods and experimental protocols related to human subjects were approved by each institutional review board Ethics Committee, and the procedures related to human subjects were carried out in accordance with each institutional review board-approved protocol and Declaration of Helsinki, 2013.
Antibodies and reagents
Cell Signaling antibodies used were: EGF receptor vIII (D6T2Q, catalog number 64952), Akt (catalog number 9272), p-Akt (S473, catalog number 4060), p-NDRG1 (T346, catalog number 5482), Raptor (catalog number 2280), Rictor (catalog number 2114), LDHA (catalog number 3582), H3K9ac (catalog number 9649), H3K18ac (catalog number 9675), H3K27ac (catalog number 4353), H3K4me2 (catalog number 9725), H3K9me2 (catalog number 4658), H3K27me2 (catalog number 9755), Histone H3 (catalog number 4499), GAPDH (catalog number 5174), PKM2 (catalog number 4053), Lamin A/C (catalog number 4777), Myc tag (catalog number 2276), and β-actin (catalog number 3700), horseradish peroxidase-linked anti-rabbit IgG (catalog number 7074), and horseradish peroxidase-linked anti-mouse IgG (catalog number 7076). Santa Cruz (Dallas, TX) antibodies were: PDH (E1α, catalog number sc-377092). The Sigma antibodies used was: EGFR (catalog number SAB4300352) and FLAG tag (catalog number F1804). The MBL International (Woburn, MA) antibody was H3K9ac (catalog number MABI0305). DAKO (Glostrup, Denmark) antibodies were: Ki-67 (catalog number M7240) and ferritin (catalog number A113). Thermo Fisher antibodies were: transferrin receptor (H68.4) (catalog number 13-6890)). Reagents used were sodium acetate (Sigma-Aldrich, catalog number S5636), PP242 (Cayman Chemical, Ann Arbor, MI, catalog number 13643), Trichostatin A (Sigma, catalog number T1952), 5-aza-2′-deoxycytidine (WAKO, Osaka, Japan; catalog number 018-20941), ferric ammonium citrate (FAC; Sigma, catalog number 5879), BPTES (Selleck, Houston, TX, catalog number S7753), Akti-1/2 (Calbiochem, La Jolla, CA, catalog number 124018), Bisindolylmaleimide I (Bis-I; Santa Cruz, catalog number sc-24003) and GSK 650394 (Tocris Bioscience, Bristol, UK, catalog number 3572/10).
DNA plasmid, siRNA, and shRNA transfection
GFP-NTD, Myc-Rictor and FLAG-HDAC4–3A DNA plasmids were obtained from Addgene. Lentiviral shRNA vectors targeting human Rictor and scramble sequences were also obtained from Addgene. Transfections of DNA plasmids were performed using X-tremeGENE HP (Roche Applied Science) in full serum, with medium change after 24 h, and cells were typically harvested 48 h post-transfection. Transfection of siRNA into cell lines was carried out using Lipofectamine RNAiMAX (Invitrogen) in full serum, with medium change after 24 h. siRNAs were used at 10 nm, and cells were harvested 48 h post-transfection. Lentivirus-mediated delivery of shRNA was performed as described previously (37). Cells were infected in the presence of 6 μg/ml of protamine sulfate, selected for puromycin resistance, and analyzed on the seventh day after infection.
Antibody validation, immunostaining, and image analysis-based scoring
Immunostaining was performed as previously described (38). Antibodies against Rictor (at a dilution of 1:50) and H3K27ac (at a dilution of 1:100) were validated for the immunostaining applicability with formalin-fixed, paraffin-embedded (FFPE) cell blocks from Rictor KD U87-EGFRvIII cells and TSA-treated U87 cells, respectively (Fig. S1A). Additionally, the antibody against ferritin (at a dilution of 1:500) was also immunohistochemically validated with the use of negative and positive staining controls in the study of human ALS cases.5 Prior to staining for Rictor and H3K27ac, sections were pre-treated with microwaving in citrate buffer, pH 6.0, for 10 min at 500 watts for antigen retrieval. Slides were counterstained with hematoxylin or DAPI (Invitrogen) to visualize nuclei. Immunostained sections underwent immunohistochemical analysis in which the results were evaluated independently by two pathologists who were unaware of the findings of the molecular analyses. Immunofluorescent samples were analyzed with a fluorescent microscope (Olympus BX53 Digital Fluorescence Microscope). Quantitative image analysis was performed with Soft Imaging System software (Visiopharm, Hørsholm, Denmark) for cytoplasmic proteins. Images from each immunostained section were captured from representative regions of the tumor with sufficiently high tumor cell content based on H&E staining evaluation. Negative control staining was also performed for each section without primary antibodies to determine the threshold for immunopositivity. Nuclear scoring was performed by ImageJ-based scoring systems (ImageJ version 1.49, NIH). Quantification of the immunofluorescent staining for the cultured cells was performed with WinROOF V6.5 (MITANI Co., Tokyo, Japan) according to the manufacturer's instructions.
Metabolome analysis
Metabolome analysis was conducted by C-SCOPE package of Human Metabolome Technologies (HMT, Yamagata, Japan) using capillary electrophoresis TOF-MS (CE-TOFMS) for cation analysis and CE-tandem MS (CE-MS/MS) for anion analysis based on the methods described previously (39, 40). Peaks were extracted using MasterHands, automatic integration software (Keio University, Tsuruoka, Yamagata, Japan) (41) and MassHunter Quantitative Analysis B.04.00 (Agilent Technologies) to obtain peak information including m/z, peak area, and migration time. Signal peaks were annotated according to the HMT metabolite database based on their m/z values with the migration times. Concentrations of metabolites were calculated by normalizing the peak area of each metabolite with respect to the area of the internal standard and using standard curves with three-point calibrations. Hierarchical cluster analysis and principal component analysis were performed by HMT proprietary software, PeakStat and SampleStat, respectively. Detected metabolites were plotted on metabolic pathway maps using VANTED software (42).
Western blot and immunoprecipitation
Immunoblotting was performed according to a modified version of the previously reported methods (8). Cell lines or snap-frozen tissue samples were lysed and homogenized with radioimmunoprecipitation assay (RIPA) lysis buffer (50 mm Tris-HCl, 150 mm NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) from Boston BioProducts (Boston, MA). Protein concentration of each sample was determined using the BCA kit (Thermo Fisher Scientific) according to the manufacturer's instructions. Equal amounts of protein extracts were separated by electrophoresis on 4–20% Mini-PROTEAN TGX Precast Gels (Bio-Rad), and then transferred to a nitrocellulose membrane with Trans-Blot Turbo Transfer System (Bio-Rad). The membrane was probed with the primary antibodies, followed by secondary antibodies conjugated to horseradish peroxidase. The immunoreactivity was detected with Super Signal West Pico Chemiluminescent Substrate or West Femto Trial kit (Thermo Fisher Scientific). Quantitative densitometry analysis was performed with an image analysis software (ImageJ version 1.49, NIH). For immunoprecipitation analyses, cells were lysed with the Pierce IP Lysis Buffer (25 mm Tris-HCl, pH 7.4, 150 mm NaCl, 1 mm EDTA, 1% Nonidet P-40, and 5% glycerol), supplemented with phosphatase and protease inhibitors (Thermo Scientific). Cell lysates were incubated overnight at 4 °C with 50 μl of the Dynabeads Protein A (Invitrogen) conjugated with 5 μl of each antibody. After washing 3 times with ice-cold PBS with Tween 20, the beads were boiled with denaturing elution buffer, and the eluted protein was analyzed by SDS-PAGE and immunoblotting.
Quantitative reverse transcription-polymerase chain reaction analysis
Total RNA was extracted by the use of RNeasy Plus Micro Kit (Qiagen). Firststrand cDNA was synthesized by the use of iScript™ RT Supermix for RT-quantitative PCR (Bio-Rad). Real-time RT-PCR was performed with the SYBR® Premix Ex TaqTM II (Tli RNaseH Plus) (Takara, Kyoto, Japan) on Thermal Cycler Dice Real Time System TP800 (Takara) following the manufacturer's instructions. β-Actin was used as an endogenous control. Primer sequences were available upon request.
RNA-sequencing and functional/canonical pathway analyses
U87-EGFRvIII cells were treated with siRNA against Scramble sequence or Rictor for 48 h (n = 2 for each cell line). Total RNA was isolated by RNeasy Plus Mini Kit (Qiagen, Venlo, The Netherlands) and submitted to Eurofins Genomics (Kanagawa, Japan) for library preparation and sequencing. Gene expression data were analyzed by Chemicals Evaluation and Research Institute (CERI) (Tokyo, Japan), using Ingenuity Pathway Analysis software (IPA; Ingenuity Systems, Redwood City, CA). The data have been deposited in Gene Expression Omnibus (GEO accession number GSE138475).
Chromatin immunoprecipitation (ChIP)-sequencing and qPCR
ChIP experiment was performed using SimpleChIPTM Enzymatic Chromatin IP Kit (Cell Signaling Technology, Beverly, MA) according to the manufacturer's instruction. H3K9ac ChIP was performed from 5 × 106 cross-linked U87-EGFRvIII cells treated with siRNA against Scramble sequence or Rictor for 48 h (n = 2 for each cell line: ChIPed and input DNAs). ChIPed DNA was submitted to RIKEN GENESIS CO., LTD. (Tokyo, Japan) for library preparation and sequencing. Model-based analysis of ChIP-seq (MACS) normalizes the total tag counts between ChIPed and input DNAs. The data have been deposited in Gene Expression Omnibus (GEO accession number GSE133117). For ChIP-qPCR analyses, H3K9ac ChIP was performed from 5 × 106 cross-linked U87-EGFRvIII cells overexpressing Rictor in combination with glucose withdrawal for 24 h. Immunoprecipitated chromatin was washed and de-cross-linked, and purified DNA was quantified by SYBR Green real-time quantitative PCR. Recoveries were calculated as percent of input according to the previously reported methods (8).
Global histone H3 detection assay
Global histone H3 detection assay was performed using EpiQuikTM Global Histone H3 Acetylation Assay Kit (Epigentek, Farmingdale, NY) according to the manufacturer's instructions. Histone extraction was performed with Histone Extraction Kit (Abcam) and 2 μg of each histone sample was applied to each well of the assay. The absorbance was read on a microplate reader (Multiskan GO Microplate Spectrophotometer; Thermo Fisher) at 450 nm.
Subcellular fractionation and acetyl-CoA measurement
Nuclear fractionation was prepared from subconfluent 10-cm plates using a kit according to the manufacturer's instructions (Active Motif, Carlsbad, CA). Intracellular acetyl-CoA level was calculated in a picomole range using PicoProbe Acetyl-CoA Assay Kit (Abcam). Fluorescence was measured using excitation/emission = 535/589 nm with a microplate reader (Thermo Fisher). After correcting background from all readings, values for each sample were determined and normalized by the protein concentration of each sample.
Quantification of total intracellular iron by atomic absorption spectrometry
U87-EGFRvIII cells (1.0 × 106 cells per well) were seeded with siRNA against scramble (control) or Rictor for 2 days prior to use. The cells were collected and counted by trypsin treatment. The cell pellets were re-suspended in concentrated HNO3 (400 μl) and incubated for 6 h. The lysates were diluted to 400 μl with distilled water. The concentrations of iron in the samples were measured by furnace atomic absorption spectroscopy with a Shimadzu AA-7000 atomic absorption spectrometer (43). The obtained values were normalized with the cell numbers.
Quantitative measurement of soluble iron in the media
U87-EGFRvIII with siRNA against Scramble sequence or Rictor were plated in 6-well plates and cultured with supplementation of 100 μm ferric ammonium citrate (FAC) for 48 h (n = 3 for each cell lines). Medium was collected and the amount of soluble iron content of each sample was determined by the Ferrozine method using a Metalloassay FeII/FeIII kit (Metallogenics, Chiba, Japan) according to the manufacturer's instructions. The iron concentration was determined by the formation of a chelate complex between ferrozine and iron. Iron bound to transport proteins such as transferrin is dissociated from the protein by the action of weak acid, denaturant and reducing agent, and Fe3+ is reduced to Fe2+ to form Fe2+-ferrozine complex. The iron concentration can be determined by measuring this complex at a wavelength of 560 nm. A value of soluble iron content on a microplate was normalized by protein concentration of each sample detected with a BCA protein assay.
Animal studies
U87 and U87-EGFRvIII cell lines were implanted into immunodeficient SCID/Beige mice for subcutaneous xenograft studies as previously reported (8). Proneural GBM rat models were stereotactically induced by injecting pQ-PDGFB-HA-IRES-EGFP (PDGF-GFP) retroviruses into the subventricular zone of the lateral ventricle in the cerebrum or tegmentum of the brainstem as described previously (44). The procedures related to animals were in accordance with the Guidelines for Animal Experiments of each institutional review board and the Law and Notification of the Japanese Government.
Statistical analysis
Statistical differences between the two groups were analyzed using Student's two-tailed unpaired t test, and those among three or more groups using one-way analysis of variance, followed by a Turkey test and Dunnett's test. Error bars represented mean ± S.D. unless otherwise noted, and statistical significance was indicated as: *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
Author contributions
K. M., W. K. C., P. S. M., and N. S. conceptualization; K. M. data curation; K. M., M. H., S. I., H. Y., and H. O. formal analysis; K. M. and P. S. M. funding acquisition; K. M., M. H., S. I., H. Y., and H. O. investigation; K. M. and P. S. M. writing-original draft; S. I. software; W. H. Y., T. F. C., Y. M., T. Kawamata, N. A., and T. Komori resources; W. K. C., P. S. M., and N. S. supervision; W. K. C., P. S. M., and N. S. writing-review and editing; P. S. M. and N. S. project administration.
Supplementary Material
Acknowledgments
We thank Noriko Sakayori (Tokyo Women's Medical University) and Erika Seki (Tokyo Metropolitan Institute of Medical Science) for helpful assistance and the UCLA Brain Tumor Translational Resource for biospecimen and biorepository. Intracellular iron measurement with furnace atomic absorption spectroscopy was performed with the help of Drs. Tasuku Hirayama and Hideko Nagasawa (Gifu Pharmaceutical University).
This work was supported by National Institutes of Health NINDS Grant NS73831, the Defeat GBM Research Collaborative, a subsidiary of National Brain Tumor Society, National Institutes of Health NCI Grant CA119347, the Ben and Catherine Ivy Foundation, the Ziering Family Foundation in memory of Sigi Ziering, a Grant-in-Aid from Takeda Science Foundation, and Japan Society for the Promotion of Science KAKENHI Grant 19K07649. P.S.M. is a scientific co-founder and consultant for Pretzel Therapeutics, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
The next-generation sequencing (NGS) data reported in this paper have been submitted to the Gene Expression Omnibus (GEO) database under GEO accession nos. GSE138475 and GSE133117.
This article contains Figs. S1–S6.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
M. Niida-Kawaguchi, A. Kakita, N. Noguchi, M. Kazama, K. Masui, Y. Kato, T. Yamamoto, T. Sawada, K. Kitagawa, K. Watabe, and N. Shibata, unpublished data.
- RTK
- receptor tyrosine kinase
- mTOR
- mammalian target of rapamycin
- mTORC2
- mTOR complex 2
- GBM
- glioblastoma
- EGFR
- epidermal growth factor receptor
- LDHA
- lactate dehydrogenase A
- PDGF
- platelet-derived growth factor
- shRNA
- short hairpin RNA
- PDH
- pyruvate dehydrogenase
- PKM2
- pyruvate kinase M2
- HAT
- histone acetyltransferase
- HDAC
- histone deacetylase
- TSA
- tricostatin A
- TSS
- transcription start site
- TFR
- transferrin receptor
- FTL
- ferritin light chain
- FTH1
- ferritin heavy chain 1
- FAC
- ferric ammonium citrate
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
References
- 1. Hanahan D., and Weinberg R. A. (2011) Hallmarks of cancer: the next generation. Cell 144, 646–674 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 2. Pavlova N. N., and Thompson C. B. (2016) The emerging hallmarks of cancer metabolism. Cell Metab. 23, 27–47 10.1016/j.cmet.2015.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Vander Heiden M. G., and DeBerardinis R. J. (2017) Understanding the intersections between metabolism and cancer biology. Cell 168, 657–669 10.1016/j.cell.2016.12.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Liu F., Hon G. C., Villa G. R., Turner K. M., Ikegami S., Yang H., Ye Z., Li B., Kuan S., Lee A. Y., Zanca C., Wei B., Lucey G., Jenkins D., Zhang W., et al. (2015) EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol. Cell 60, 307–318 10.1016/j.molcel.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaelin W. G. Jr., and McKnight S. L. (2013) Influence of metabolism on epigenetics and disease. Cell 153, 56–69 10.1016/j.cell.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kinnaird A., Zhao S., Wellen K. E., and Michelakis E. D. (2016) Metabolic control of epigenetics in cancer. Nat. Rev. Cancer 16, 694–707 10.1038/nrc.2016.82 [DOI] [PubMed] [Google Scholar]
- 7. Cloughesy T. F., Cavenee W. K., and Mischel P. S. (2014) Glioblastoma: from molecular pathology to targeted treatment. Annu. Rev. Pathol. 9, 1–25 10.1146/annurev-pathol-011110-130324 [DOI] [PubMed] [Google Scholar]
- 8. Masui K., Tanaka K., Akhavan D., Babic I., Gini B., Matsutani T., Iwanami A., Liu F., Villa G. R., Gu Y., Campos C., Zhu S., Yang H., Yong W. H., Cloughesy T. F., et al. (2013) mTOR complex 2 controls glycolytic metabolism in glioblastoma through FoxO acetylation and upregulation of c-Myc. Cell Metab. 18, 726–739 10.1016/j.cmet.2013.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Masui K., Tanaka K., Ikegami S., Villa G. R., Yang H., Yong W. H., Cloughesy T. F., Yamagata K., Arai N., Cavenee W. K., and Mischel P. S. (2015) Glucose-dependent acetylation of Rictor promotes targeted cancer therapy resistance. Proc. Natl. Acad. Sci. U.S.A. 112, 9406–9411 10.1073/pnas.1511759112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Villa G. R., Hulce J. J., Zanca C., Bi J., Ikegami S., Cahill G. L., Gu Y., Lum K. M., Masui K., Yang H., Rong X., Hong C., Turner K. M., Liu F., Hon G. C., et al. (2016) An LXR-cholesterol axis creates a metabolic co-dependency for brain cancers. Cancer Cell 30, 683–693 10.1016/j.ccell.2016.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gu Y., Albuquerque C. P., Braas D., Zhang W., Villa G. R., Bi J., Ikegami S., Masui K., Gini B., Yang H., Gahman T. C., Shiau A. K., Cloughesy T. F., Christofk H. R., Zhou H., et al. (2017) mTORC2 regulates amino acid metabolism in cancer by phosphorylation of the cystine-glutamate antiporter xCT. Mol. Cell 67, 128–138.7 10.1016/j.molcel.2017.05.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Harachi M., Masui K., Okamura Y., Tsukui R., Mischel P. S., and Shibata N. (2018) mTOR complexes as a nutrient sensor for driving cancer progression. Int. J. Mol. Sci. 19, e3267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Assanah M., Lochhead R., Ogden A., Bruce J., Goldman J., and Canoll P. (2006) Glial progenitors in adult white matter are driven to form malignant gliomas by platelet-derived growth factor-expressing retroviruses. J. Neurosci. 26, 6781–6790 10.1523/JNEUROSCI.0514-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jackson E. L., Garcia-Verdugo J. M., Gil-Perotin S., Roy M., Quinones-Hinojosa A., VandenBerg S., and Alvarez-Buylla A. (2006) PDGFR α-positive B cells are neural stem cells in the adult SVZ that form glioma-like growths in response to increased PDGF signaling. Neuron 51, 187–199 10.1016/j.neuron.2006.06.012 [DOI] [PubMed] [Google Scholar]
- 15. Pietrocola F., Galluzzi L., Bravo-San Pedro J. M., Madeo F., and Kroemer G. (2015) Acetyl coenzyme A: a central metabolite and second messenger. Cell Metab. 21, 805–821 10.1016/j.cmet.2015.05.014 [DOI] [PubMed] [Google Scholar]
- 16. Wellen K. E., Hatzivassiliou G., Sachdeva U. M., Bui T. V., Cross J. R., and Thompson C. B. (2009) ATP-citrate lyase links cellular metabolism to histone acetylation. Science 324, 1076–1080 10.1126/science.1164097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee J. V., Carrer A., Shah S., Snyder N. W., Wei S., Venneti S., Worth A. J., Yuan Z. F., Lim H. W., Liu S., Jackson E., Aiello N. M., Haas N. B., Rebbeck T. R., Judkins A., et al. (2014) Akt-dependent metabolic reprogramming regulates tumor cell histone acetylation. Cell Metab. 20, 306–319 10.1016/j.cmet.2014.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yang X. J., and Seto E. (2007) HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene 26, 5310–5318 10.1038/sj.onc.1210599 [DOI] [PubMed] [Google Scholar]
- 19. Peserico A., and Simone C. (2011) Physical and functional HAT/HDAC interplay regulates protein acetylation balance. J. Biomed. Biotechnol. 371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yang W., Xia Y., Hawke D., Li X., Liang J., Xing D., Aldape K., Hunter T., Alfred Yung W. K., and Lu Z. (2012) PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell 150, 685–696 10.1016/j.cell.2012.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sutendra G., Kinnaird A., Dromparis P., Paulin R., Stenson T. H., Haromy A., Hashimoto K., Zhang N., Flaim E., and Michelakis E. D. (2014) A nuclear pyruvate dehydrogenase complex is important for the generation of acetyl-CoA and histone acetylation. Cell 158, 84–97 10.1016/j.cell.2014.04.046 [DOI] [PubMed] [Google Scholar]
- 22. Yang C., Ko B., Hensley C. T., Jiang L., Wasti A. T., Kim J., Sudderth J., Calvaruso M. A., Lumata L., Mitsche M., Rutter J., Merritt M. E., and DeBerardinis R. J. (2014) Glutamine oxidation maintains the TCA cycle and cell survival during impaired mitochondrial pyruvate transport. Mol. Cell 56, 414–424 10.1016/j.molcel.2014.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Eberharter A., and Becker P. B. (2002) Histone acetylation: a switch between repressive and permissive chromatin: second in review series on chromatin dynamics. EMBO Rep. 3, 224–229 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Verdin E., and Ott M. (2015) 50 years of protein acetylation: from gene regulation to epigenetics, metabolism and beyond. Nat. Rev. Mol. Cell Biol. 16, 258–264 10.1038/nrm3931 [DOI] [PubMed] [Google Scholar]
- 25. Torti F. M., and Torti S. V. (2002) Regulation of ferritin genes and protein. Blood 99, 3505–3516 10.1182/blood.V99.10.3505 [DOI] [PubMed] [Google Scholar]
- 26. Hailemariam K., Iwasaki K., Huang B. W., Sakamoto K., and Tsuji Y. (2010) Transcriptional regulation of ferritin and antioxidant genes by HIPK2 under genotoxic stress. J. Cell Sci. 123, 3863–3871 10.1242/jcs.073627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Strahl B. D., and Allis C. D. (2000) The language of covalent histone modifications. Nature 403, 41–45 10.1038/47412 [DOI] [PubMed] [Google Scholar]
- 28. Yan C., and Boyd D. D. (2006) Histone H3 acetylation and H3 K4 methylation define distinct chromatin regions permissive for transgene expression. Mol. Cell. Biol. 26, 6357–6371 10.1128/MCB.00311-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bogdan A. R., Miyazawa M., Hashimoto K., and Tsuji Y. (2016) Regulators of iron homeostasis: new players in metabolism, cell death, and disease. Trends Biochem. Sci. 41, 274–286 10.1016/j.tibs.2015.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cancer Genome Atlas Research Network. (2008) Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brennan C. W., Verhaak R. G., McKenna A., Campos B., Noushmehr H., Salama S. R., Zheng S., Chakravarty D., Sanborn J. Z., Berman S. H., Beroukhim R., Bernard B., Wu C. J., Genovese G., Shmulevich I., et al. (2013) The somatic genomic landscape of glioblastoma. Cell 155, 462–477 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Masui K., Onizuka H., Cavenee W. K., Mischel P. S., and Shibata N. (2019) Metabolic reprogramming in the pathogenesis of glioma: update. Neuropathology 39, 3–13 10.1111/neup.12535 [DOI] [PubMed] [Google Scholar]
- 33. Martinez-Pastor B., Cosentino C., and Mostoslavsky R. (2013) A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov. 3, 497–501 10.1158/2159-8290.CD-13-0059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Schonberg D. L., Miller T. E., Wu Q., Flavahan W. A., Das N. K., Hale J. S., Hubert C. G., Mack S. C., Jarrar A. M., Karl R. T., Rosager A. M., Nixon A. M., Tesar P. J., Hamerlik P., Kristensen B. W., et al. (2015) Preferential iron trafficking characterizes glioblastoma stem-like cells. Cancer Cell 28, 441–455 10.1016/j.ccell.2015.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vernieri C., Casola S., Foiani M., Pietrantonio F., de Braud F., and Longo V. (2016) Targeting cancer metabolism: dietary and pharmacologic interventions. Cancer Discov. 6, 1315–1333 10.1158/2159-8290.CD-16-0615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tanaka K., Babic I., Nathanson D., Akhavan D., Guo D., Gini B., Dang J., Zhu S., Yang H., De Jesus J., Amzajerdi A. N., Zhang Y., Dibble C. C., Dan H., Rinkenbaugh A., et al. (2011) Oncogenic EGFR signaling activates an mTORC2-NF-κB pathway that promotes chemotherapy resistance. Cancer Discov. 1, 524–538 10.1158/2159-8290.CD-11-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Guo D., Prins R. M., Dang J., Kuga D., Iwanami A., Soto H., Lin K. Y., Huang T. T., Akhavan D., Hock M. B., Zhu S., Kofman A. A., Bensinger S. J., Yong W. H., Vinters H. V., et al. (2009) EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci. Signal. 2, ra82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Mellinghoff I. K., Wang M. Y., Vivanco I., Haas-Kogan D. A., Zhu S., Dia E. Q., Lu K. V., Yoshimoto K., Huang J. H., Chute D. J., Riggs B. L., Horvath S., Liau L. M., Cavenee W. K., Rao P. N., et al. (2005) Molecular determinants of the response of glioblastomas to EGFR kinase inhibitors. N. Engl. J. Med. 353, 2012–2024 10.1056/NEJMoa051918 [DOI] [PubMed] [Google Scholar]
- 39. Ooga T., Sato H., Nagashima A., Sasaki K., Tomita M., Soga T., and Ohashi Y. (2011) Metabolomic anatomy of an animal model revealing homeostatic imbalances in dyslipidaemia. Mol. Biosyst. 7, 1217–1223 10.1039/c0mb00141d [DOI] [PubMed] [Google Scholar]
- 40. Ohashi Y., Hirayama A., Ishikawa T., Nakamura S., Shimizu K., Ueno Y., Tomita M., and Soga T. (2008) Depiction of metabolome changes in histidine-starved Escherichia coli by CE-TOFMS. Mol. BioSyst. 4, 135–147 10.1039/B714176A [DOI] [PubMed] [Google Scholar]
- 41. Sugimoto M., Wong D. T., Hirayama A., Soga T., and Tomita M. (2010) Capillary electrophoresis mass spectrometry-based saliva metabolomics identified oral, breast and pancreatic cancer-specific profiles. Metabolomics 6, 78–95 10.1007/s11306-009-0178-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Junker B. H., Klukas C., and Schreiber F. (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7, 109 10.1186/1471-2105-7-109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hirayama T., Miki A., and Nagasawa H. (2019) Organelle-specific analysis of labile Fe(II) during ferroptosis by using a cocktail of various color organelle-targeted fluorescent probes. Metallomics 11, 111–117 10.1039/C8MT00212F [DOI] [PubMed] [Google Scholar]
- 44. Masui K., Suzuki S. O., Torisu R., Goldman J. E., Canoll P., and Iwaki T. (2010) Glial progenitors in the brainstem give rise to malignant gliomas by platelet-derived growth factor stimulation. Glia 58, 1050–1065 10.1002/glia.20986 [DOI] [PubMed] [Google Scholar]
- 45. Cerami E., Gao J., Dogrusoz U., Gross B. E., Sumer S. O., Aksoy B. A., Jacobsen A., Byrne C. J., Heuer M. L., Larsson E., Antipin Y., Reva B., Goldberg A. P., Sander C., and Schultz N. (2012) The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2, 401–404 10.1158/2159-8290.CD-12-0095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao J., Aksoy B. A., Dogrusoz U., Dresdner G., Gross B., Sumer S. O., Sun Y., Jacobsen A., Sinha R., Larsson E., Cerami E., Sander C., and Schultz N. (2013) Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 6, pl1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hao Y., Wang Y., Liu X., Yang X., Wang P., Tian Q., Bai Q., Chen X., Li Z., Wu J., Xie Z., Zhou X., Zhou Y., Yin Z., Wu Y., et al. (2018) The kinase complex mTOR complex 2 promotes the follicular migration and functional maturation of differentiated follicular helper CD4+ T cells during viral infection. Front. Immunol. 9, 1127 10.3389/fimmu.2018.01127 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.