Summary
Humans have used yeasts to make cheese and kefir for millennia, but the ability to ferment the milk sugar lactose is found in only a few yeast species, of which the foremost is Kluyveromyces lactis [1]. Two genes, LAC12 (lactose permease) and LAC4 (lactase), are sufficient for lactose uptake and hydrolysis to glucose and galactose [2]. Here, we show that these genes have a complex evolutionary history in the genus Kluyveromyces that is likely the result of human activity during domestication. We show that the ancestral Lac12 was bifunctional, able to import both lactose and cellobiose into the cell. These disaccharides were then hydrolyzed by Lac4 in the case of lactose or Cel2 in the case of cellobiose. A second cellobiose transporter, Cel1, was also present ancestrally. In the K. lactis lineage, the ancestral LAC12 and LAC4 were lost and a separate upheaval in the sister species K. marxianus resulted in loss of CEL1 and quadruplication of LAC12. One of these LAC12 genes became neofunctionalized to encode an efficient lactose transporter capable of supporting fermentation, specifically in dairy strains of K. marxianus, where it formed a LAC4-LAC12-CEL2 gene cluster, although another remained a cellobiose transporter. Then, the ability to ferment lactose was acquired very recently by K. lactis var. lactis by introgression of LAC12 and LAC4 on a 15-kb subtelomeric region from a dairy strain of K. marxianus. The genomic history of the LAC genes shows that strong selective pressures were imposed on yeasts by early dairy farmers.
Keywords: introgression, domestication, lactose, fermentation, yeast, neofunctionalization, cluster, lactis, marxianus, Kluyveromyces
Graphical Abstract
Highlights
-
•
Early farmers domesticated the milk yeast Kluyveromyces lactis
-
•
A dairy lineage of Kluyveromyces marxianus donated the LAC genes to K. lactis
-
•
A new Kluyveromyces-specific gene cluster for utilizing cellobiose was identified
-
•
K. marxianus carries an integrated cellobiose and lactose utilization gene cluster
It is well known that humans domesticated brewer’s yeast, and now, Varela et al. report that another yeast is also the product of human activity. They show that an insect-associated, lactose-negative progenitor of the milk yeast Kluyveromyces lactis acquired the genes that enable lactose fermentation from a dairy-adapted population of K. marxianus.
Results and Discussion
Genomes, Phylogeny, and Phenotypes of Kluyveromyces Species
The yeast genera Kluyveromyces and Saccharomyces, which diverged about 150 mya, both contain species that are important producers of fermented foods or beverages or that serve as hosts for production of metabolites and proteins for biotechnology [2, 3]. The capacity to grow on lactose as a sole carbon source is a defining trait in the food yeasts Kluyveromyces lactis and Kluyveromyces marxianus.
The lactose utilization system was elucidated in K. lactis and depends primarily on two neighboring genes, LAC12 and LAC4 [2, 4, 5]. Lac12 is a membrane permease that imports lactose into the cell, and Lac4 is an intracellular lactase (β-galactosidase) that hydrolyzes lactose into the easily catabolized monosaccharides glucose and galactose. The kinetics of uptake of lactose by Lac12 and its hydrolysis by Lac4 are sufficient to allow fermentative growth of K. lactis [6] and dairy isolates of K. marxianus [7] in oxygen-limiting conditions. K. lactis and K. marxianus are often associated with fermented dairy products, such as artisan cheese and kefir. K. marxianus can also be isolated from plants and other habitats [7, 8]. Two varieties of K. lactis are recognized: K. lactis var. lactis, which is milk associated, and K. lactis var. drosophilarum, which is insect associated [1]. Our previous population studies of K. marxianus found three distinct genomic haplotypes (A, B, and C). Haplotype B is dairy associated and carries a LAC12 allele that encodes a protein variant, Lac12L, with enhanced capacity to transport lactose [7, 9]. There are four LAC12 genes in the K. marxianus genome, but only the Lac12L variant has efficient lactose-uptake properties [9].
The six currently recognized species in the genus Kluyveromyces vary widely in their ability to metabolize lactose [1]. Three phenotypic groups can be described. First, only K. lactis var. lactis and dairy strains of K. marxianus (B haplotype) can ferment lactose. Second, K. dobzhanskii and K. lactis var. drosophilarum are lactose negative, unable to utilize this sugar at all. A third phenotypic group is formed by K. aestuarii, K. nonfermentans, and K. wickerhamii and non-dairy isolates of K. marxianus (A and C haplotypes), which are Kluyver effect positive for lactose—meaning that they can respire, but not ferment the sugar [10]. Because these three phenotypic groups do not correspond to phylogenetic clades, and because the trait of biotechnological interest (lactose fermentation) is polymorphic for presence/absence in both K. lactis and K. marxianus, we were motivated to investigate the origin and evolution of the LAC genes.
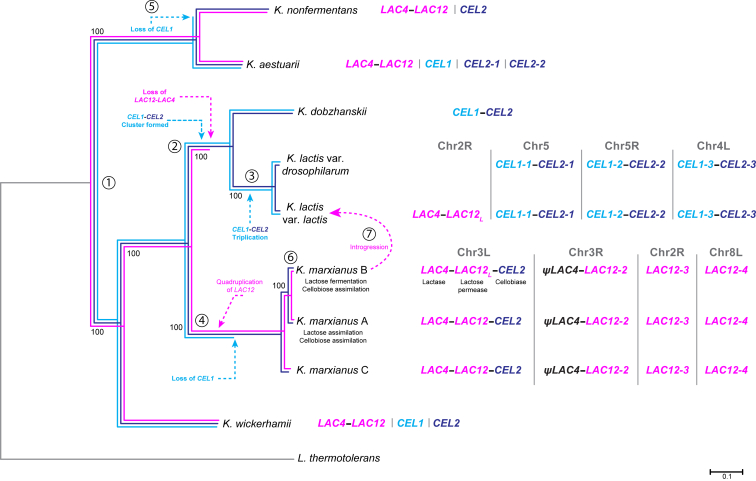
Genome sequences are available for all six Kluyveromyces species, including K. marxianus haplotypes A, B, and C and the type strain of K. lactis var. lactis (CBS2359). We sequenced the type strain of K. lactis var. drosophilarum (CBS2105) and assembled it into six complete chromosome sequences. Comparison to CBS2359 reveals genome-wide nucleotide sequence identity of 95.4% and karyotypes that differ by two inversions and four reciprocal translocations. A phylogenomic tree of the six species (Figure S1) gives a topology that agrees with a recent phylogenomic study [11]. The two varieties of K. lactis are seen to be very closely related, as are the three haplotypes of K. marxianus (Figure 1).
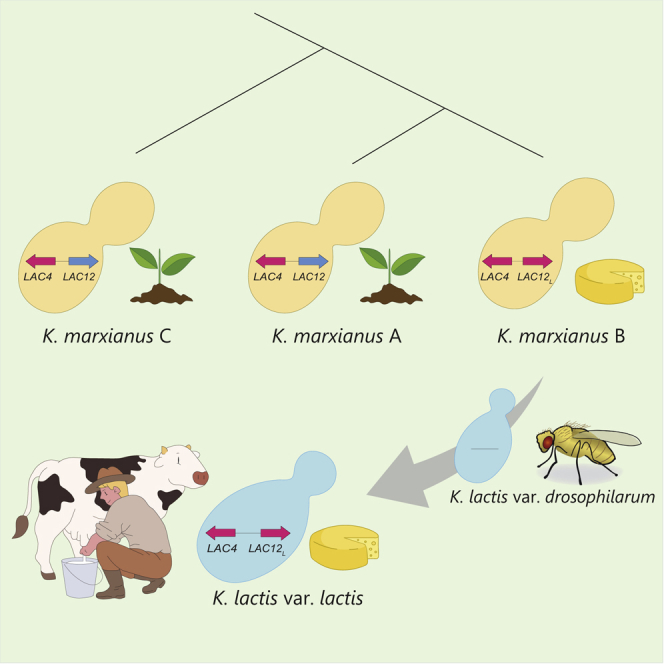
Figure 1.
Summary of Evolutionary Steps in Lactose and Cellobiose Utilization in the Genus Kluyveromyces
Colored branches on the phylogenetic tree trace the history of the LAC4-LAC12 gene cluster (magenta), CEL1 (cyan), and CEL2 (dark blue). Dashed arrows mark key evolutionary steps, including gene duplications, losses, and relocations. Numbers refer to specific events that are discussed in the main text. The dashed magenta arrow shows the introgression of the LAC12L gene and the neighboring LAC4 from a K. marxianus haplotype B strain into a lactose negative progenitor of K. lactis var. lactis, leading to the modern species that is now able to assimilate and ferment lactose. The LAC and CEL genotype is shown for every species. The chromosome numbers for the K. lactis species refers to chromosomes in K. lactis var. lactis CBS2359. Dashes between gene names indicate genes that are clustered in the genome.
The LAC Genes of K. lactis var. lactis Were Acquired by Introgression from K. marxianus
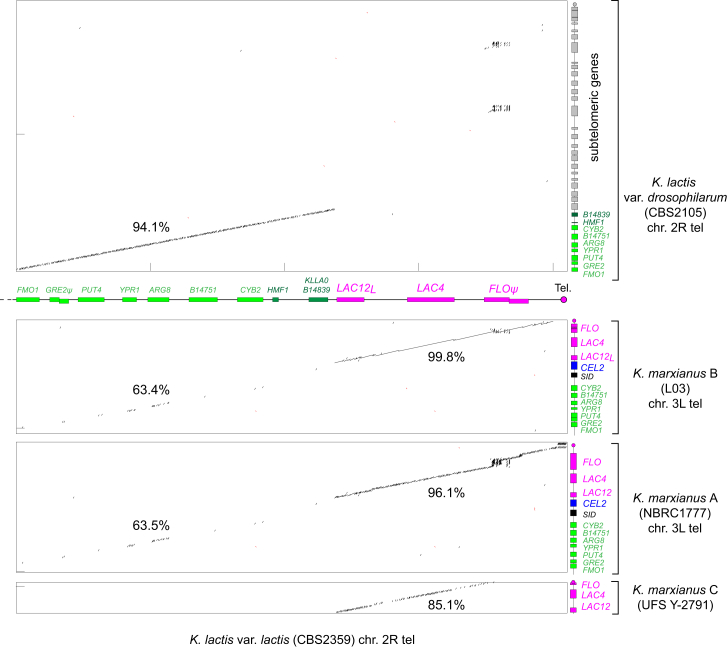
The genome sequence of K. lactis var. drosophilarum does not contain any LAC12 or LAC4 genes, consistent with a previous report based on Southern blotting [12] and with the inability of this strain to grow on lactose. The LAC12-LAC4 gene cluster is located in a subtelomeric region in both K. lactis and K. marxianus, so we used dot matrix plots to compare this region between K. lactis var. lactis and either K. lactis var. drosophilarum or K. marxianus haplotypes A, B, and C (Figure 2). The genomic region between FMO1 and CYB2 (green in Figure 2) is orthologous between K. lactis and K. marxianus, even though its DNA sequence identity is low (63%). Between the two K. lactis varieties, the high DNA sequence identity (94.1%), which extends over most of the chromosome, terminates abruptly after the gene (KLLA0B14839) located immediately before the LAC12-LAC4 gene cluster and the telomere (Figure 2, upper panel). Instead, the region from LAC12 to the telomere in K. lactis var. lactis has very high similarity to the corresponding region of K. marxianus (Figure 2, lower panels). Moreover, it has higher sequence identity to the B haplotype of K. marxianus (99.8% identity; only 31-nt differences in 13,961 bp aligned) than to the A and C haplotypes (96.1% and 85.1% identity, respectively). Phylogenetic analysis of the proteins encoded by LAC12 and LAC4 confirms that these genes in K. lactis var. lactis are more closely related to their homologs in a K. marxianus B-haplotype strain than to A- or C-haplotype strains (Figures S2 and S3).
Figure 2.
The LAC4-LAC12 Gene Region in K. lactis var. lactis Was Formed by Introgression of a 15-kb Subtelomeric Region from K. marxianus
Four dot matrix plots are shown. The x axis in all the plots is a 35-kb region beside the telomere of chromosome 2R of K. lactis var. lactis, including the genes LAC12, LAC4, and a flocculin pseudogene (FLOψ). This region is compared to K. lactis var. drosophilarum in the uppermost plot and to K. marxianus (haplotypes A, B, and C) in the three lower plots. The introgression into K. lactis var. lactis replaced a subtelomeric region containing approximately 26 genes (gray) with one containing 3 genes (magenta). Numbers show percent DNA sequence identity in CLUSTALΩ alignments of the regions from FMO1 to CYB2 and from LAC12 to the telomere between genomes. The genes HMF1 and KLLA0B14839 are present only in K. lactis in this region, and the genes CEL2 and SID (a putative siderophore transporter) are present only in K. marxianus. The plots were made using DNAMAN (https://www.lynnon.com) with a criterion of 17 matches per 20-bp window.
See also Table S1.
These results show that the telomere-proximal region, including the LAC genes, was transferred between the two Kluyveromyces species, confirming a hypothesis by Naumov that K. lactis might have obtained its LAC genes by horizontal gene transfer [13]. The donor was a B-haplotype (dairy) strain of K. marxianus, and the recipient was a K. lactis strain that became the progenitor of K. lactis var. lactis. The transferred region (magenta in Figure 2) was approximately 15 kb and included LAC12L, LAC4, and a flocculin (FLO) gene that has since acquired frameshift mutations and is thus a pseudogene. The telomere was also transferred during the introgression event. We infer that the transfer replaced a previous subtelomeric region in K. lactis that resembled the current subtelomere of K. lactis var. drosophilarum, which consists of a 70-kb region containing 26 genes (gray in Figure 2). The most likely mechanism of DNA transfer was introgression, i.e., interspecies mating between K. marxianus and K. lactis, followed by repeated backcrossing to K. lactis, although the rearrangement was more complex than a meiotic crossover in the last orthologous gene, CYB2 (Figure 2).
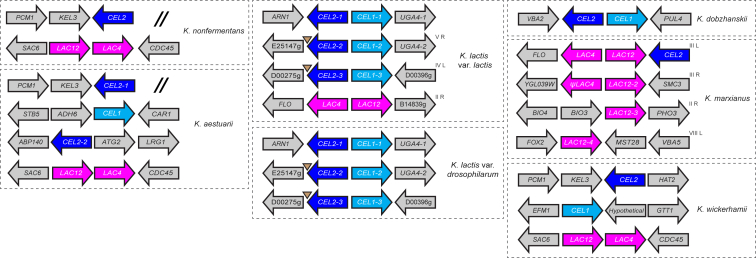
The CEL1-CEL2 Gene Cluster Encodes a Newly Identified Cellobiose Utilization System in Kluyveromyces
The gene immediately beside the LAC12-LAC4 gene cluster in K. marxianus has features suggesting that it codes for a β-glucosidase. There are no other candidate β-glucosidase genes in the K. marxianus genome, so we hypothesized that this gene, which we named CEL2, encodes a cellobiase that enables K. marxianus to grow on the disaccharide cellobiose by hydrolyzing it to glucose. It has previously been shown that K. lactis Lac12 (Lac12L) is able to transport cellobiose when expressed in S. cerevisiae [14, 15], so we wondered whether the juxtaposition of LAC12 and CEL2 in the K. marxianus genome, coding respectively for proteins that can potentially import and hydrolyze cellobiose, constitutes a functional gene cluster. But if this is the case, what is the system for utilizing cellobiose in species such as K. lactis var. drosophilarum and K. dobzhanskii that can grow on this sugar but do not carry LAC12?
CEL2 genes are present in all six Kluyveromyces species, with some having multiple paralogs (Figure 3). Phylogenetic analysis shows that the putative cellobiase Cel2 groups with fungal β-glucosidases, whereas the lactase Lac4 forms a clade with β-galactosidases from other fungi and bacteria (Figure S2). In the K. dobzhanskii/K. lactis clade, we also found a putative sugar transporter gene that we named CEL1. This gene is adjacent to CEL2, which is suggestive of another functional gene cluster. K. wickerhamii and K. aestuarii also have CEL1 genes, but they are not linked to CEL2. In contrast, CEL1 is absent in K. marxianus and K. nonfermentans. Comparison to known cellobiose and lactose transporters from other fungi shows that Cel1 groups with cellobiose transporters, whereas Lac12 groups with fungal lactose permeases (Figure S3). Based on these phylogenetic analyses and the previous report that Lac12 can transport cellobiose as well as lactose [14, 15], we hypothesized that (1) Cel1 and Cel2 together constitute a system for import and hydrolysis of cellobiose and (2), in some species, Lac12 serves as cellobiose transporter that is an alternative to Cel1.
Figure 3.
Organization of LAC and CEL Genes in the Genomes of Kluyveromyces Species and Varieties
Dashed lines delimit the genes that belong to each species. The LAC genes are shown in magenta. CEL1 and CEL2 are shown in cyan and dark blue, respectively. Missing information due to incomplete genome assembly is indicated with a double slash symbol in the K. nonfermentans and K. aestuarii panels. LTR elements are represented by inverted triangles. The organization of the K. marxianus genes is based on the CBS397 assembly [7]. ψLAC4 is a conserved pseudogene and LAC12-4 is a pseudogene only in some strains, and variability both in the sequence of Lac4 and in the genomic organization in this telomere of chromosome 8 is seen between strains [9]. Superscript text indicates the position (chromosome number and arm) of the LAC and CEL genes in K. marxianus and the K. lactis var. lactis variants. Loci in K. lactis var lactis and K. lactis var. drosophilarum are syntenic, but reciprocal translocations may have changed chromosome numbers.
See also Figures S2 and S3 and Table S1.
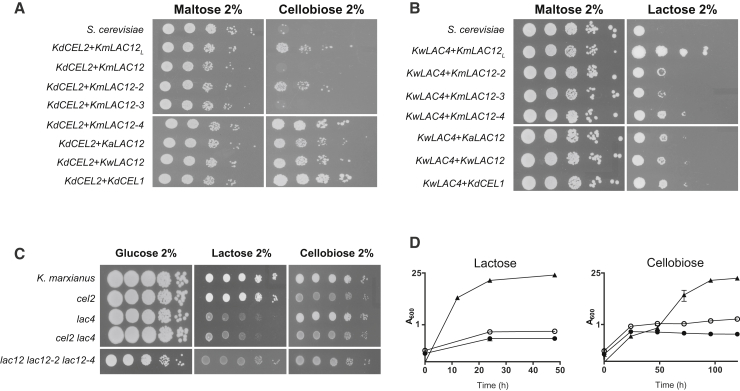
To test these hypotheses, we cloned candidate transporter genes (LAC12 or CEL1) and expressed them in a S. cerevisiae strain co-expressing CEL2 from K. dobzhanskii (Figure 4A). Co-expression of K. dobzhanskii CEL1 and CEL2 conferred growth on cellobiose, proving that these genes encode a functional cellobiose utilization system: Cel1 can transport cellobiose and Cel2 is a cellobiase. Co-expression of CEL2 with the single LAC12 gene from either K. aestuarii or K. wickerhamii also enabled S. cerevisiae to grow on cellobiose, demonstrating that these Lac12 proteins can transport cellobiose. This is also the case for K. marxianus LAC12L, LAC12-2, and LAC12-4, but neither LAC12-3 nor LAC12 from an A-haplotype strain are efficient cellobiose transporters (Figure 4A).
Figure 4.
Functional Analyses Confirm that the LAC and CEL Genes Encode Functional Sugar Assimilation Systems
The function of the putative permeases and hydrolases was assessed by heterologous expression in S. cerevisiae and CRISPR-Cas9-induced mutagenesis in K. marxianus.
(A and B) S. cerevisiae strains transformed with different combinations of putative hydrolyses (LAC4 or CEL2) and permeases (LAC12 alleles or CEL1) were assessed for growth on SC medium with cellobiose (A) or lactose (B) as the carbon source. The control strain is S. cerevisiae transformed with the empty pGREG505 and p426 plasmids and growth on maltose assessed as a positive control. The function of the LAC and CEL genes was also evaluated in K. marxianus. Targeted mutants of the LAC12 genes, LAC4 and CEL2, were constructed using the CRISPR-Cas9 system in K. marxianus NBRC 1777.
(C and D) These mutants were assessed for growth on plates (C) and in liquid (D) in mineral medium with the sole carbon sources indicated. Curves are the mean of three replicates. Legend: close triangles, wild-type; closed circle, cel2 lac4; open circle, lac12 lac12-2 lac12-4.
To assess lactose transport, K. wickerhamii LAC4 was co-expressed in S. cerevisiae with putative transporters (Figure 3B). The lactase activity of K. wickerhamii Lac4 was confirmed by co-expression with K. marxianus LAC12L, but co-expression with other LAC12 genes or with CEL1 led to little growth. This result agrees with previous data that, in K. marxianus, only the B-haplotype allele LAC12L encodes a functional lactose transporter [9] and suggests that the protein encoded by LAC12 in other Kluyveromyces species is a poor lactose transporter. This observation is also consistent with data that K. aestuarii and K. wickerhamii grow slowly on lactose by respiration and cannot ferment it (Kluyver effect positive) [16, 17]. The data also demonstrate that Cel1 is specific for cellobiose and is unable to transport lactose.
Cellobiose and Lactose Uptake Is Mediated by Lac12 Transporters in K. marxianus
Our data indicate that there are two ancestral disaccharide utilization systems in the genus Kluyveromyces: Cel1 and Cel2, which import and hydrolyze cellobiose, and Lac12 and Lac4, which import and hydrolyze lactose. Lac12, however, is bifunctional, able to transport both lactose and cellobiose in heterologous expression assays (Figure 4A). Because the Cel1 transporter has been lost in K. marxianus, we hypothesized that cellobiose transport in this species is carried out exclusively by Lac12. To investigate whether K. marxianus Lac12 has this proposed bifunctionality and to confirm the separate enzymatic functions of Lac4 and Cel2, K. marxianus mutants were constructed using CRISPR-Cas9-induced nonsense mutation and analyzed for growth on lactose or cellobiose (Figure 4C). Comparing growth first on solid medium, the cel2 mutant grows on lactose, but not cellobiose; the lac4 mutant grows on cellobiose, but not lactose; and the cel2 lac4 mutant grows on neither lactose nor cellobiose (Figure 4C). The faint apparent growth in these plate assays is background due to dead cells, a point confirmed by the failure of the cel2 lac4 double mutant to grow on either disaccharide in liquid medium (Figure 4D). Because heterologous expression (Figure 4A) indicated that any of the Lac12L, Lac12-2, or Lac12-4 proteins was able to transport cellobiose, a K. marxianus triple mutant lacking all three genes was assessed (Figures 4C and 4D). The triple mutant was unable to grow on either lactose or cellobiose, confirming the dual function of the K. marxianus LAC12 gene family and establishing that there is no other unidentified cellobiose transporter in K. marxianus. Thus, in K. marxianus, Lac12 is a bifunctional transporter that can transport either lactose or cellobiose, which is then cleaved into monosaccharides by the dedicated hydrolytic enzyme, Lac4 or Cel2 (Figure S4).
Reconstruction of the Evolutionary Trajectory of the LAC and CEL Genes in Kluyveromyces Species
Our analyses enable us to reconstruct the evolutionary history of the genes for lactose and cellobiose utilization in the Kluyveromyces genus (Figure 1). Because K. wickerhamii and K. aestuarii are located on branches that diverged at the base of the genus, it is likely that the similar genomic organization of the CEL and LAC genes in these species (Figure 3) represents the ancestral state in the genus. Thus, LAC12 and LAC4 were already contiguous in the genome of the ancestor (beside SAC6; Figure 3), but CEL1 and CEL2 were not contiguous. This ancestor is labeled as point 1 in Figure 1. On the branch that led to the K. dobzhanskii/K. lactis clade, there were several reorganizations of the CEL genes (point 2). One of these brought CEL1 and CEL2 together to assemble a gene cluster for cellobiose utilization. This CEL gene cluster became located in a subtelomere in K. lactis and was later triplicated during a large-scale amplification of K. lactis subtelomeres (point 3) [18]. In contrast, CEL1 was lost from the K. marxianus branch (point 4) and also from K. nonfermentans (point 5). Separate reorganization of the LAC genes took place. The LAC cluster was lost from the K. dobzhanskii/K. lactis branch (point 2). In the K. marxianus branch (point 4), it was rearranged such that LAC12 and LAC4 are now divergently transcribed from a common promoter (Figure 3). The LAC gene cluster became quadruplicated onto multiple K. marxianus subtelomeres, but LAC4 was lost or degenerated into a pseudogene in all the copies except the one at the subtelomere of chromosome 3L. Also at point 4, CEL2 was relocated to the same subtelomere, forming the three-gene cluster LAC4-LAC12-CEL2. Within the species K. marxianus, there was divergence into distinct haplotypes and the LAC12 allele in haplotype B (LAC12L) acquired amino acid changes that improved its ability to transport lactose (point 6). Then, recently, a 15-kb subtelomeric region containing the LAC genes introgressed from a K. marxianus B-haplotype strain into K. lactis (point 7), splitting the latter species into two varieties and restoring lactose utilization (now fermentable because of efficient uptake) to K. lactis var. lactis. The transferred region contained LAC12L and LAC4 from the three-gene cluster, but not K. marxianus CEL2 (K. lactis already had native CEL2 genes in its CEL1-CEL2 clusters; Figure 3).
From the genomic data, it is possible to say that the host for the introgression was a K. lactis strain resembling the insect-associated species K. lactis var. drosophilarum. We expect that further analysis of genetic diversity in K. lactis will show that strains containing the introgressed LAC genes form a subclade that lies within a more diverse set of insect-associated K. lactis strains that cannot grow on lactose. The introgression can be inferred to be a recent event on evolutionary timescales because of the low number of nucleotide differences between the donor and recipient. Using Rolland and Dujon’s method for yeast molecular clocks [19], we estimate that the introgressed DNA diverged from the K. marxianus B-haplotype approximately 3.7 million generations ago, which corresponds to between 3,700 and 37,000 years ago, depending on the number of yeast generations assumed per year. This is close to or within the timescale for the emergence of agriculture. The milk-producing animals cow, sheep, and goat were all domesticated between 8,000 and 10,000 years ago [20], and paleoproteomic analysis of dental calculus has shown that humans were consuming milk, most likely as cheese or other fermented products, by 5,500 years ago [21]. Because our estimate for the age of the LAC gene introgression extends into the period of milk animal domestication, it is plausible that selection for the introgression was the result of human activity during production of a fermented milk product, such as cheese or kefir. There are some parallels between this event and the recently reported trans-species introgression of GAL genes from an unknown donor Saccharomyces species into milk- or cheese-associated strains of S. cerevisiae, which utilize the galactose and glucose formed by bacterial hydrolysis of lactose [22, 23]. Together, the introgressions in both Kluyveromyces and Saccharomyces point to strong selective pressures imposed on yeasts by early farmers for the ability to ferment animal milk.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Deposited Data | ||
| K. lactis var. drosophilarum genome assembly | This paper | GenBank: GCA_007993695.1 |
| K. marxianus L03 genome assembly | This paper | GenBank: GCA_008000265.1 |
| Experimental Models: Organisms/Strains | ||
| Kluyveromyces and S. cerevisiae strains | N/A | See Table S1 |
| Genotype of K. marxianus mutants | This study | See Table S4 |
| Oligonucleotides | ||
| Oligonucleotides | Sigma | See Table S3 |
| Recombinant DNA | ||
| Plasmids | N/A | See Table S2 |
| Software and Algorithms | ||
| SPAdes v3.11.1 | N/A | http://cab.spbu.ru/files/release3.11.1/ |
| RAxML-NG v0.8.1 | N/A | https://github.com/amkozlov/raxml-ng/releases |
| MUMmer | N/A | http://mummer.sourceforge.net/ |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, John P. Morrissey (j.morrissey@ucc.ie).
Experimental Model and Subject Details
Experiments were conducted using S. cerevisiae and Kluyveromyces yeasts. All the yeast strains used in this study are listed in Table S1. Kluyveromyces strains were purchased from the Westerdijk Fungal Biodiversity Institute and S. cerevisiae EBY.VW4000 was kindly provided by Dr Eckhard Boles, Goethe University Frankfurt, Germany. This strain is deleted for 17 hexose transporter genes but can grow on the disaccharide, maltose, which is therefore used as a control in the experiments. S. cerevisiae EBY.VW4000 was used for heterologous expression of Kluyveromyces genes. K. marxianus NBRC1777 (haploid, haplotype A) was used to construct mutants for transporters and hydrolases and K. marxianus CBS397 (diploid, haplotype AB) was used for phenotypic tests and as the source of template DNA to clone LAC and CEL genes. Genome sequences of K. marxianus NBRC 1777 (haploid, haplotype A), K. marxianus L03 (triploid, haplotype BBB), and K. marxianus Y-UFS 2791 (haploid, haplotype C) were used for comparison of genomic structure between K. marxianus and K. lactis spp.
Method Details
Strains and culture conditions
The Kluyveromyces species were routinely grown in YPD broth (1% yeast extract, 2% peptone, 2% glucose). For sugar utilization experiments the strains were grown on mineral medium [24] supplemented with 2% glucose, then washed twice with sterile water, diluted to A600 1, diluted serially (10-fold) and spotted onto minimal medium plates containing 2% glucose, lactose, raffinose or cellobiose (Thermo-Fisher Scientific, MA, USA) as the sole carbon source. For growth curves, the K. marxianus mutants were grown overnight on minimal medium and transferred to fresh medium containing the same carbon source to an A600 of 0.1. Growth was monitored by measuring A600 over time. For selecting transformants, K. marxianus was grown on YPD plates supplemented with 200 μg/mL-1. Hygromycin B (Sigma-Aldrich, MI., USA). S. cerevisiae EBY.VW4000 was used in heterologous expression experiments. The strain was grown on synthetic complete (SC) medium (1.7 g L-1 yeast nitrogen base, 5 g L-1 ammonium plus synthetic complete drop-out lacking uracil and L-leucine) supplemented with 2% maltose. In experiments testing the function of putative transporters, the S. cerevisiae strains containing different plasmids were grown on SC maltose, serially diluted and spotted on SC plates containing maltose, lactose or cellobiose to a final concentration of 2%. All the yeast strains in this study were grown at 30°C with 200 rpm agitation. E. coli, used for cloning experiments, was grown in LB medium (0.5% yeast extract, 1% bactopeptone, 1% NaCl) supplemented with 100 μg/mL ampicillin.
Heterologous expression
The CEL and LAC genes were cloned and expressed in S. cerevisiae as described previously [9]. Genes encoding enzymes and transporters were cloned into the p426 (2 μ) and pGREG (CEN/ARS) plasmids, respectively (Table S2). All the genes were expressed under the control of the TEF1 promoter from S. cerevisiae. Plasmids were introduced by transformation into S. cerevisiae EBY.VW4000 following the LiAC/SS carrier DNA/PEG protocol [25]. Cultures transformed with these plasmids were plated onto SC plates lacking uracil and leucine. Primers used to construct the expression plasmids are listed in Table S3.
Construction of K. marxianus mutants
The CRISPR-Cas9 system was used for the construction of cel2 and lac4 K. marxianus mutants. Target sequence identification was carried out using the sgRNAcas9 [26]. Primers encoding the target sequences and specific overhangs (Table S3) were annealed and cloned into pUCC001, as described previously [27, 28]. In brief, the two complementary oligonucleotides encoding the target sequence were combined and phosphorylated using T4 polynucleotide kinase. The resulting DNA duplex was then cloned into pUCC001 via Golden Gate assembly. The BSA-R primer was used in combination with the target forward primer to check for correct assembly of the plasmids in E. coli. Plasmids containing the target sequences were purified and introduced into K. marxianus NBRC1777 by transformation. Transformants were checked by PCR using diagnosis primers and plasmid curing was performed by growing the strains in YPD without Hygromycin B for 16 hours. The strains were then plated in YPD and three colonies were streaked in YPD + Hygromycin B to confirm plasmid loss. The genotype of the mutants constructed is shown in Table S4.
Genome sequencing
The type strain of K. lactis var. drosophilarum (CBS2105) was purchased from the Westerdijk Institute (the Netherlands) and its genome was sequenced using Illumina and Pacific Biosciences technology. For Illumina sequencing, genomic DNA was harvested from stationary-phase cultures by homogenization with glass beads followed by phenol-chloroform extraction and ethanol precipitation, and concentrated with the Genomic DNA Clean & Concentrator-10 (Zymo Research, catalog D4010). Illumina sequencing was done by BGI Tech Solutions (Hong Kong) on a HiSeq2500 instrument generating 150 bp paired-end reads. Illumina data was assembled using SPAdes version v3.11.1 [29], giving 65x coverage of the genome. Genomic DNA for PacBio sequencing was prepared as in [30]. PacBio sequencing was done by the Earlham Institute (UK) using a PacBio Sequel instrument (1 SMRT cell), and assembled by them using HGAP4 [31], producing 224x coverage. BLASTN alignments to the Illumina contigs were used to detect and correct single-base indel errors in the PacBio scaffolds. The K. lactis var. drosophilarum CBS2105 genome sequence was annotated using YGAP [32] and submitted to the NCBI/ENA/DDBJ database (GenBank: GCA_007993695.1). The K. marxianus L03 genome was sequenced and assembled as described in [7] and was filtered using the CVL method [33] with 20x kmer coverage and 5.5 kb length cut-offs before deposition in the NCBI/ENA/DDBJ database (GenBank: GCA_008000265.1).
Bioinformatic analysis
The phylogenetic tree in Figure 1 was produced using a set of 1,515 single-copy orthologous amino acid sequences obtained from genome assemblies of 9 Kluyveromyces type strains plus Lachancea thermotolerans using the saccharomyceta_odb9 dataset with BUSCO v3.1.0 [34]. Orthologous sequences were obtained from K. marxianus NBRC1777 (GenBank assembly accession: GCA_001417835.1 [35]), L. thermotholerans CBS6340T (GCF_000142805.1), K. aestuarii CBS4438T (GCA_003707555.1), K. nonfermentans CBS8778T (GCA_003670155.1), K. wickerhamii CBS2745T (GCA_000179415.1), K. dobzhanskii CBS2104T (GCA_000820885.1), K. lactis var. lactis CBS2359T (GCF_000002515.2), K. lactis var. drosophilarum CBS2105T (GCA_007993695.1), K. marxianus L03 (GCA_008000265.1), and K. marxianus UFS-Y2791 (GCA_001692465.1). Each group of 10 orthologous sequences were aligned using CLUSTALΩ (v1.2.4 [36];) removing gaps with trimAl v1.2rev59 in -nogaps mode [37]. Trimmed multiple sequence alignments were then concatenated and used to calculate a phylogenetic tree with RAxML-NG v0.8.1 [38] using 10 random and 10 parsimony-based starting trees, to pick the best-scoring topology to then do 100 bootstrap replicates with the PROTGTR substitution model.
Global nucleotide sequence identity between the K. lactis var. lactis CBS2359 and K. lactis var. drosophilarum CBS2105 genomes was calculated using the nucmer program of the MUMmer package (version 3.23) [39], filtered to retain only alignments longer than 10 kb (option delta-filter –g –l 10000). The divergence time of the introgressed region was calculated assuming a mutation rate of 3e-10 per site per generation and 10-100 yeast generations per year, as in [19].
Data and Code Availability
The accession numbers for the K. lactis var. drosophilarum CBS2105 and K. marxianus L03 genome assemblies reported in this paper are GenBank: GCA_007993695.1 and GenBank: GCA_008000265.1, respectively.
Acknowledgments
This work was performed with support from the YEASTCELL Marie Curie ITN project, which received funding from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme FP7/2007-2013/ under REA grant agreement no. 606795, and the CHASSY project, which has received funding from the European Union’s Horizon 2020 Framework Programme for Research and Innovation—grant agreement no. 720824. M.P. and R.G. were supported by the EU ERASMUS Plus Programme. K.H.W. is supported by the European Research Council (grant number 789341). R.A.O.-M. was supported by European Research Council award no. 694633 to Jack Pronk.
Author Contributions
J.P.M., K.H.W., and J.A.V. conceived the project and designed and interpreted the experiments. M.P., J.A.V., R.G., and S.B.-G. performed the experiments. R.A.O.-M., K.H.W., and J.A.V. performed bioinformatics analyses and analyzed the data. J.P.M., K.H.W., J.A.V., and R.A.O.-M. wrote the manuscript.
Declaration of Interests
The authors declare no competing interests
Published: December 5, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.10.044.
Supplemental Information
References
- 1.Lachance M.A. Kluyveromyces van der Walt (1971) In: Kurtzman C.P., Fell J.W., Boekhout T., editors. The Yeasts: A Taxonomic Study. Elsevier; 2011. pp. 471–481. [Google Scholar]
- 2.Rodicio R., Heinisch J.J. Yeast on the milky way: genetics, physiology and biotechnology of Kluyveromyces lactis. Yeast. 2013;30:165–177. doi: 10.1002/yea.2954. [DOI] [PubMed] [Google Scholar]
- 3.Nielsen J. Yeast systems biology: model organism and cell factory. Biotechnol. J. 2019;14:e1800421. doi: 10.1002/biot.201800421. [DOI] [PubMed] [Google Scholar]
- 4.Fukuhara H. Kluyveromyces lactis- a retrospective. FEMS Yeast Res. 2006;6:323–324. doi: 10.1111/j.1567-1364.2005.00012.x. [DOI] [PubMed] [Google Scholar]
- 5.Schaffrath R., Breunig K.D. Genetics and molecular physiology of the yeast Kluyveromyces lactis. Fungal Genet. Biol. 2000;30:173–190. doi: 10.1006/fgbi.2000.1221. [DOI] [PubMed] [Google Scholar]
- 6.González Siso M.I., Ramil E., Cerdán M.E., Freire-Picos M.A. Respirofermentative metabolism in Kluyveromyces lactis: ethanol production and the Crabtree effect. Enzyme Microb. Technol. 1996;18:585–591. doi: 10.1016/s0141-0229(00)00161-7. [DOI] [PubMed] [Google Scholar]
- 7.Ortiz-Merino R.A., Varela J.A., Coughlan A.Y., Hoshida H., da Silveira W.B., Wilde C., Kuijpers N.G.A., Geertman J.-M., Wolfe K.H., Morrissey J.P. Ploidy variation in Kluyveromyces marxianus separates dairy and non-dairy isolates. Front. Genet. 2018;9:94. doi: 10.3389/fgene.2018.00094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lane M.M., Morrissey J.P. Kluyveromyces marxianus: a yeast emerging from its sister’s shadow. Fungal Biol. Rev. 2010;24:17–26. [Google Scholar]
- 9.Varela J.A., Montini N., Scully D., Van der Ploeg R., Oreb M., Boles E., Hirota J., Akada R., Hoshida H., Morrissey J.P. Polymorphisms in the LAC12 gene explain lactose utilisation variability in Kluyveromyces marxianus strains. FEMS Yeast Res. 2017;17:fox021. doi: 10.1093/femsyr/fox021. [DOI] [PubMed] [Google Scholar]
- 10.Fukuhara H. The Kluyver effect revisited. FEMS Yeast Res. 2003;3:327–331. doi: 10.1016/S1567-1356(03)00112-0. [DOI] [PubMed] [Google Scholar]
- 11.Krause D.J., Kominek J., Opulente D.A., Shen X.-X., Zhou X., Langdon Q.K., DeVirgilio J., Hulfachor A.B., Kurtzman C.P., Rokas A., Hittinger C.T. Functional and evolutionary characterization of a secondary metabolite gene cluster in budding yeasts. Proc. Natl. Acad. Sci. USA. 2018;115:11030–11035. doi: 10.1073/pnas.1806268115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naumov G.I., Naumova E.S., Barrio E., Querol A. [Genetic and molecular study of inability of the yeast Kluyveromyces lactis var Drosophilarum to ferment lactose] Mikrobiologiia. 2006;75:299–304. [PubMed] [Google Scholar]
- 13.Naumov G.I. Domestication of dairy yeast Kluyveromyces lactis: transfer of the beta-galactosidase (LAC4) and lactose permease (LAC12) gene cluster? Dokl. Biol. Sci. 2005;401:120–122. doi: 10.1007/s10630-005-0061-6. [DOI] [PubMed] [Google Scholar]
- 14.Rubio-Texeira M., Arévalo-Rodríguez M., Lequerica J.L., Polaina J. Lactose utilization by Saccharomyces cerevisiae strains expressing Kluyveromyces lactis LAC genes. J. Biotechnol. 2001;84:97–106. doi: 10.1016/s0168-1656(00)00350-3. [DOI] [PubMed] [Google Scholar]
- 15.Sadie C.J., Rose S.H., den Haan R., van Zyl W.H. Co-expression of a cellobiose phosphorylase and lactose permease enables intracellular cellobiose utilisation by Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 2011;90:1373–1380. doi: 10.1007/s00253-011-3164-z. [DOI] [PubMed] [Google Scholar]
- 16.Naumov G.I. Why does the yeast Kluyveromyces wickerhamii assimilates but not ferments lactose? Dokl. Biol. Sci. 2005;403:310–312. doi: 10.1007/s10630-005-0121-y. [DOI] [PubMed] [Google Scholar]
- 17.Kurtzman C.P. Molecular taxonomy of the yeasts. Yeast. 1994;10:1727–1740. doi: 10.1002/yea.320101306. [DOI] [PubMed] [Google Scholar]
- 18.Fairhead C., Dujon B. Structure of Kluyveromyces lactis subtelomeres: duplications and gene content. FEMS Yeast Res. 2006;6:428–441. doi: 10.1111/j.1567-1364.2006.00033.x. [DOI] [PubMed] [Google Scholar]
- 19.Rolland T., Dujon B. Yeasty clocks: dating genomic changes in yeasts. C. R. Biol. 2011;334:620–628. doi: 10.1016/j.crvi.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 20.Larson G., Fuller D.Q. The evolution of animal domestication. Annu. Rev. Ecol. Evol. Syst. 2014;45:115–136. [Google Scholar]
- 21.Charlton S., Ramsøe A., Collins M., Craig O.E., Fischer R., Alexander M., Speller C.F. New insights into Neolithic milk consumption through proteomic analysis of dental calculus. Archaeol. Anthropol. Sci. 2019 Published online September 9, 2019. [Google Scholar]
- 22.Legras J.-L., Galeote V., Bigey F., Camarasa C., Marsit S., Nidelet T., Sanchez I., Couloux A., Guy J., Franco-Duarte R. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol. Biol. Evol. 2018;35:1712–1727. doi: 10.1093/molbev/msy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Duan S.F., Shi J.Y., Yin Q., Zhang R.P., Han P.J., Wang Q.M., Bai F.Y. Reverse evolution of a classic gene network in yeast offers a competitive advantage. Curr. Biol. 2019;29:1126–1136.e5. doi: 10.1016/j.cub.2019.02.038. [DOI] [PubMed] [Google Scholar]
- 24.Verduyn C., Postma E., Scheffers W.A., Van Dijken J.P. Effect of benzoic acid on metabolic fluxes in yeasts: a continuous-culture study on the regulation of respiration and alcoholic fermentation. Yeast. 1992;8:501–517. doi: 10.1002/yea.320080703. [DOI] [PubMed] [Google Scholar]
- 25.Gietz R.D., Schiestl R.H. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat. Protoc. 2007;2:31–34. doi: 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- 26.Xie S., Shen B., Zhang C., Huang X., Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PLoS ONE. 2014;9:e100448. doi: 10.1371/journal.pone.0100448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Varela J.A., Puricelli M., Montini N., Morrissey J.P. Expansion and diversification of MFS transporters in Kluyveromyces marxianus. Front. Microbiol. 2019;9:3330. doi: 10.3389/fmicb.2018.03330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juergens H., Varela J.A., Gorter de Vries A.R., Perli T., Gast V.J.M., Gyurchev N.Y., Rajkumar A.S., Mans R., Pronk J.T., Morrissey J.P., Daran J.G. Genome editing in Kluyveromyces and Ogataea yeasts using a broad-host-range Cas9/gRNA co-expression plasmid. FEMS Yeast Res. 2018;18:foy012. doi: 10.1093/femsyr/foy012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bankevich A., Nurk S., Antipov D., Gurevich A.A., Dvorkin M., Kulikov A.S., Lesin V.M., Nikolenko S.I., Pham S., Prjibelski A.D. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ortiz-Merino R.A., Kuanyshev N., Braun-Galleani S., Byrne K.P., Porro D., Branduardi P., Wolfe K.H. Evolutionary restoration of fertility in an interspecies hybrid yeast, by whole-genome duplication after a failed mating-type switch. PLoS Biol. 2017;15:e2002128. doi: 10.1371/journal.pbio.2002128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chin C.S., Alexander D.H., Marks P., Klammer A.A., Drake J., Heiner C., Clum A., Copeland A., Huddleston J., Eichler E.E. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat. Methods. 2013;10:563–569. doi: 10.1038/nmeth.2474. [DOI] [PubMed] [Google Scholar]
- 32.Proux-Wéra E., Armisén D., Byrne K.P., Wolfe K.H. A pipeline for automated annotation of yeast genome sequences by a conserved-synteny approach. BMC Bioinformatics. 2012;13:237. doi: 10.1186/1471-2105-13-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Douglass A.P., O’Brien C.E., Offei B., Coughlan A.Y., Ortiz-Merino R.A., Butler G., Byrne K.P., Wolfe K.H. Coverage-versus-length plots, a simple quality control step for de novo yeast genome sequence assemblies. G3. 2019;9:879–887. doi: 10.1534/g3.118.200745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Waterhouse R.M., Seppey M., Simão F.A., Manni M., Ioannidis P., Klioutchnikov G., Kriventseva E.V., Zdobnov E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018;35:543–548. doi: 10.1093/molbev/msx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Inokuma K., Ishii J., Hara K.Y., Mochizuki M., Hasunuma T., Kondo A. Complete genome sequence of Kluyveromyces marxianus NBRC1777, a nonconventional thermotolerant yeast. Genome Announc. 2015;3:e00389-15. doi: 10.1128/genomeA.00389-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sievers F., Wilm A., Dineen D., Gibson T.J., Karplus K., Li W., Lopez R., McWilliam H., Remmert M., Söding J. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozlov A.M., Darriba D., Flouri T., Morel B., Stamatakis A. RAxML-NG: a fast, scalable and user-friendly tool for maximum likelihood phylogenetic inference. Bioinformatics. 2019;35:4453–4455. doi: 10.1093/bioinformatics/btz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Delcher A.L., Kasif S., Fleischmann R.D., Peterson J., White O., Salzberg S.L. Alignment of whole genomes. Nucleic Acids Res. 1999;27:2369–2376. doi: 10.1093/nar/27.11.2369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The accession numbers for the K. lactis var. drosophilarum CBS2105 and K. marxianus L03 genome assemblies reported in this paper are GenBank: GCA_007993695.1 and GenBank: GCA_008000265.1, respectively.