Summary
Bacterial lipo-chitooligosaccharides (LCOs) are key mediators of the nitrogen-fixing root nodule symbiosis (RNS) in legumes. The isolation of LCOs from arbuscular mycorrhizal fungi suggested that LCOs are also signaling molecules in arbuscular mycorrhiza (AM). However, the corresponding plant receptors have remained uncharacterized. Here we show that petunia and tomato mutants in the LysM receptor-like kinases LYK10 are impaired in AM formation. Petunia and tomato LYK10 proteins have a high affinity for LCOs (Kd in the nM range) comparable to that previously reported for a legume LCO receptor essential for the RNS. Interestingly, the tomato and petunia LYK10 promoters, when introduced into a legume, were active in nodules similarly to the promoter of the legume orthologous gene. Moreover, tomato and petunia LYK10 coding sequences restored nodulation in legumes mutated in their orthologs. This combination of genetic and biochemical data clearly pinpoints Solanaceous LYK10 as part of an ancestral LCO perception system involved in AM establishment, which has been directly recruited during evolution of the RNS in legumes.
Keywords: symbiosis, evolution, plant, petunia, lysin motif receptor-like kinase, lipochitooligosaccharide, symbiotic signal, arbuscular mycorrhiza, nodulation
Highlights
-
•
Mutants in Solanaceaous LysM receptors LYK10 are impaired in arbuscular mycorrhiza
-
•
LYK10 proteins have a high affinity for lipo-chitooligosaccharidic signal molecules
-
•
LYK10 promoter is expressed in arbuscule-containing cells in tomato roots
-
•
Solanaceaous LYK10 can restore nodulation in legumes mutated in their orthologs
Soil rhizobial bacteria and arbuscular mycorrhizal (AM) fungi produce lipo-chitooligosaccharidic (LCO) signal molecules. Girardin et al. show that plant LCO receptors are involved in establishment of the ancient AM symbiosis and have been recruited during evolution for establishment of the nitrogen-fixing root nodule symbiosis with rhizobia.
Introduction
Arbuscular mycorrhiza (AM) is an ancient mutualistic symbiosis between Glomeromycota fungi and the majority of land plants, in which fungi provide plants with nutrients acquired from the soil in exchange for carbohydrates and lipids [1, 2]. To colonize plant roots, arbuscular mycorrhizal fungi (AMFs) first cross epidermal and outer cortical cells and then spread inter- or intra-cellularly within roots. Inside inner root cortical cells, AMFs form highly branched structures called arbuscules, across which most nutrient exchange takes place. In the more recent nitrogen-fixing root nodule symbiosis (RNS) that occurs between legumes and rhizobia, the bacteria can fix gaseous nitrogen inside the root nodules. Although the microorganisms are different between these two endosymbioses, the RNS is thought to have evolved through recruitment of genes implicated in the more ancient AM [3].
Nodule organogenesis and bacterial colonization rely on the secretion of lipo-chitooligosaccharide (LCO) signaling molecules by rhizobia [4]. All the rhizobial LCOs have a core structure of 4/5 N-acetyl glucosamine (GlcNAc) units of which the terminal non-reducing sugar is substituted with an acyl chain. Additional substitutions, which are important for host specificity, are characteristic of each bacterial strain [5]. Rhizobial LCOs are perceived by Lysin motif receptor-like kinases (LysM-RLKs) that are encoded by a multigenic family, some of which have the ability to bind LCOs [6, 7, 8]. Members of the LysM-RLK LYRIA phylogenetic group (Figure S1A) [9], such as Medicago truncatula NFP (MtNFP) or Lotus japonicus NFR5 (LjNFR5), are required for activation of a signaling pathway leading to oscillations of the nuclear Ca2+ concentration (Ca2+ spiking), nodule organogenesis, and bacterial colonization [10, 11, 12].
Two lines of evidence suggest that AM establishment also involves LCO-mediated signaling. The first line is the identification of LCOs from AMFs, and the second is the identification of potential plant LCO receptors. LCOs isolated from AMFs by Maillet et al. (hereafter collectively referred to as Myc-LCOs) have a core structure similar to the rhizobial LCOs and can be sulfated or not on the reducing sugar [13]. Exogenous application of these Myc-LCOs both increases the level of AMF root colonization [13] and activates Ca2+ spiking in various plant species [14, 15]. Short-chain chitooligosaccharides (COs) produced by AMFs can also activate Ca2+ spiking [16], indicating that both LCOs and short-chain COs have the potential to be involved in partner recognition during AM. However, whether Myc-LCOs and/or short-chain COs are indeed involved in AM establishment is not known.
Several LysM-RLKs (Parasponia andersonii PanNFP1 and/or PanNFP2, tomato SlLYK10 and SlLYK12, Medicago truncatula MtLYK9, and rice OsCERK1) have been shown to be involved in AM [17, 18, 19, 20, 21, 22], but their LCO/CO binding properties have not been determined so far. SlLYK12, MtLYK9, and OsCERK1 belong to the LYKI phylogenetic group (Figure S1B [9]). These LysM-RLKs are likely co-receptors, since MtLYK9 and OsCERK1 have a dual function in AM and defense [19, 20, 23], and OsCERK1 is involved in perception of various ligands including short-chain COs, chitin, and peptidoglycan [24, 25, 26], the latter two being components of fungal and bacterial cell walls, respectively, known as plant defense elicitors. The other LysM-RLKs known to control AM belong to the LYRIA group that contains members only in plant species that establish AM and/or RNS [27, 28]. In tomato, virus-induced silencing of the unique LYRIA gene (SlLYK10) resulted in significantly lower levels of AM colonization [21].
Although the current hypothesis is that the RNS evolved by coopting genes involved in the AM [3], it is unclear how LCO receptors may have evolved to become key players in RNS establishment.
Here, we functionally characterize LCO receptors from Solanaceae, a plant family that establishes AM but not RNS. We use heterologous expression in legumes to infer an evolutionary scenario of LCO receptor recruitment for RNS. Our data suggest that non-legume LYRIA genes encode LCO receptors involved in AM and that the transcriptional regulation required for LCO receptor function in RNS has been directly co-opted from AM.
Results
The Petunia and Tomato LYRIA Genes Are Involved in AM Establishment
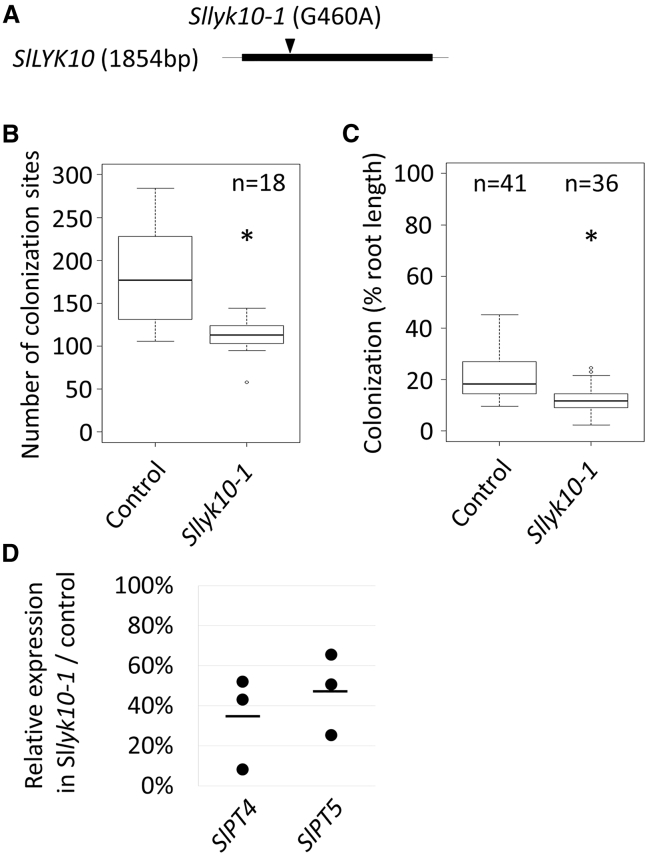
We have previously shown that knockdown of the LYRIA gene in tomato (SlLYK10) resulted in impaired AM establishment [21]. Because of the limitations of gene silencing, we screened an EMS-mutagenized tomato population and identified a line carrying a missense mutation in SlLYK10 affecting the second LysM (E154K) (Figure 1A). Segregants of this line with a homozygous mutation (Sllyk10-1) displayed reduced numbers of AMF colonization sites, root-length colonization, and expression of AM-marker genes (Figures 1B–1D) compared with segregants with a WT SlLYK10 allele (control).
Figure 1.
Sllyk10-1 Is Affected in AMF Colonization
(A) Schematic representation of SlLYK10. The thick line represents the single exon. Arrowhead indicates the position of the mutation in Sllyk10-1.
(B) Number of AMF colonization sites per root system. Boxplots represent the distribution between individuals from one experiment.
(C) Root-length colonization. Boxplots represent the distribution between root systems from three independent experiments.
(D) Relative expression of the plant AM-marker genes in Sllyk10-1 versus control roots measured by qRT-PCR. RNAs were extracted from pools of four root systems. The line represents the mean, and the dots represent each replicate.
Statistical differences were calculated using a Kruskal Wallis test in (B) and (C). See also Figures S1 and S2.
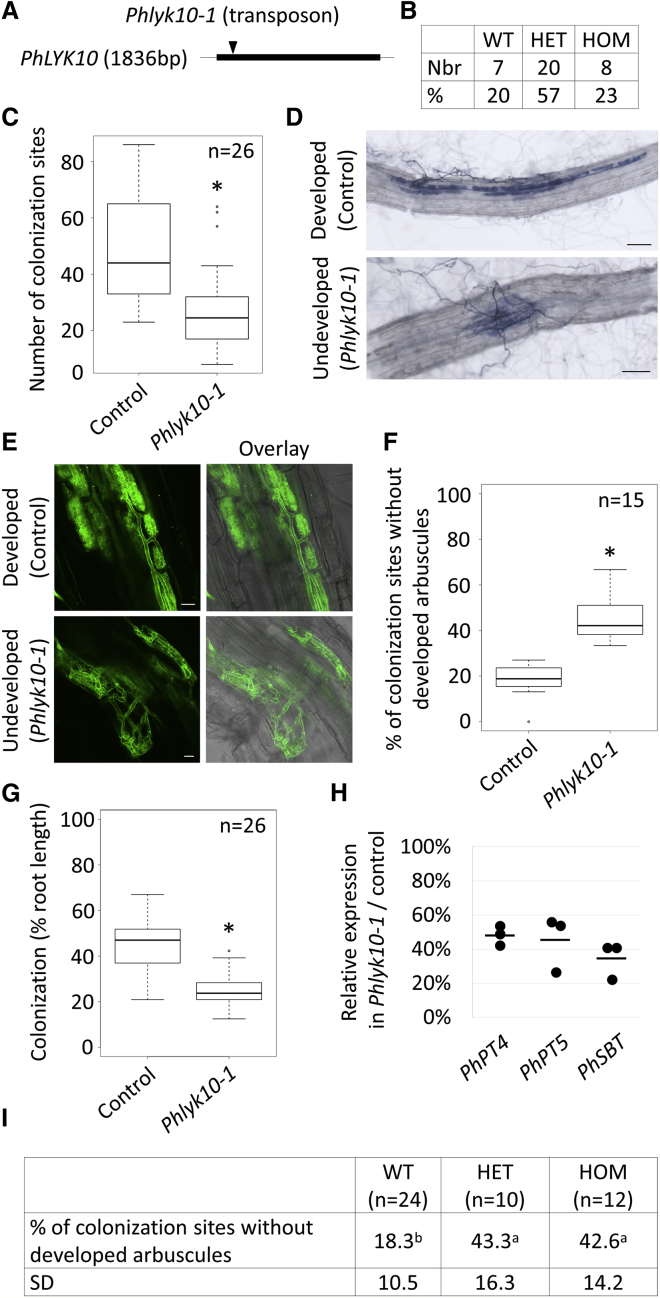
We also searched for knockout lines in a related Solanaceae species, Petunia hybrida, by screening a transposon-mutagenized population [29]. We identified a line with a dTPh1 insertion in the SlLYK10 ortholog PhLYK10 (Figures 2A and S2), which segregated with the expected 1:2:1 wild-type:heterozygous:homozygous ratio (Figure 2B). Segregants with a homozygous dTph1 insertion (Phlyk10-1) displayed a reduced number of AMF colonization sites (Figure 2C), many of them being impaired in arbuscule formation (Figure 2D), compared with segregants with a WT PhLYK10 allele (control). Confocal microscopy analysis of colonized cells showed hyphal coils instead of arbuscules (Figure 2E). The ratio of colonization sites with aberrant arbuscule development was significantly higher in Phlyk10-1 plants (Figure 2F). The Phlyk10-1 plants also displayed a reduced level of root-length colonization and expression of AM-marker genes (Figures 2G and 2H). Furthermore, in a segregating population, we found that increased numbers of colonization sites with aberrant arbuscule development correlated with the presence of the dTph1 insertion (Figure 2I). Unexpectedly, heterozygous individuals also showed impaired arbuscule development. This, together with the phenotypic similarity observed in SlLYK10-silenced plants [21] and the nature of the mutation (stop codon in dTPh1 close to the start codon of PhLYK10), suggests that PhLYK10 function is sensitive to gene dosage.
Figure 2.
Phlyk10-1 Is Affected in AMF Colonization and Arbuscule Formation
(A) Schematic representation of PhLYK10. The thick line represents the single exon. Arrowhead indicates the position of the dTph1 insertion in Phlyk10-1.
(B) Number of wild-type (WT), heterozygous (HET), and homozygous (HOM) individuals for the dTph1 insertion on progenies of HET F2 plants after a backcross. No significant difference with theoretical segregation was found.
(C) Number of AMF colonization sites per root system. Boxplots represent the distribution between individuals from three independent experiments.
(D) Images of ink-stained colonization sites.
(E) Images of WGA-CF488A-stained AMF.
(F) Percentage of colonization sites without developed arbuscules (as in D) versus the total number of colonization sites. Boxplots represent the distribution between root systems from one experiment.
(G) Root-length colonization. Boxplots represent the distribution between root systems from one experiment.
H) Relative expression of the plant AM-marker genes in Phlyk10-1 versus control roots measured by qRT-PCR. RNAs were extracted from pools of at least three root systems. The line represents the mean, and the dots represent each replicate.
(I) Same as in (F) except that measured on progenies of HET F2 plants after a backcross. Individual plants were genotyped and phenotyped. Means and SDs are shown in the table.
Statistical differences were calculated using a Xhi2 test in (B), a Student’s t test in (C), (F), and (G), or a Kruskal Wallis test in (I). Scale bars represent 100 μm in (D) and 20 μm in (E). See also Figures S1 and S2 and Table S1.
LCO Binding by LYRIA Proteins Predates the Evolution of RNS
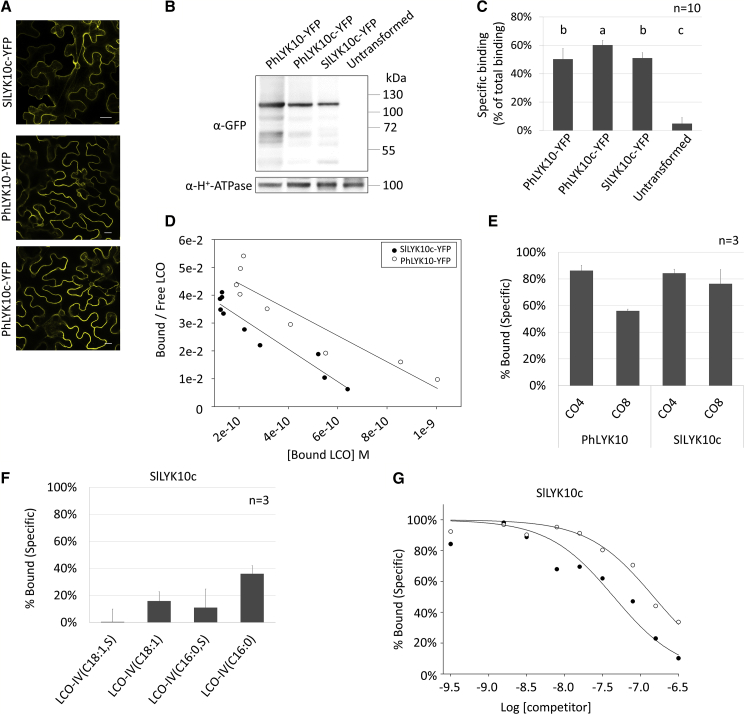
LCO-binding in legume LYRIA proteins may have originated from ancestral LCO-binding proteins, or it may have been gained in legumes as a key property in the evolution of the RNS. To discriminate between these two possibilities, we determined the LCO-binding properties of SlLYK10 and PhLYK10. We used Agrobacterium tumefaciens-mediated transient expression to produce SlLYK10-YFP and PhLYK10-YFP in leaves of Nicotiana benthamiana, a plant protein expression system which allows the formation of disulfide bridges essential for LysM-RLK function [30, 31]. SlLYK10-YFP was localized in undefined cytoplasmic structures in N. benthamiana leaf cells, although the protein was properly localized at the plasma membrane (PM) in transgenic tomato roots (Figures S3A–S3D). We previously observed that a chimeric LysM-RLK was well localized at the PM in N. benthamiana leaves and had LCO-binding properties similar to the corresponding full-length protein [6]. We thus generated a chimera (hereafter referred to as SlLYK10c) (Figure S4A) composed of SlLYK10 extracellular region (ECR) and MtNFP intracellular region. Although a fraction of SlLYK10c-YFP was localized to the endoplasmic reticulum (ER) of N. benthamiana leaf cells (Figure 3A), both co-localization with a PM marker and the analysis of N-glycan maturation indicated that a significant fraction of the proteins reached the PM (Figures S4B–S4D). Subcellular localization of PhLYK10 and a PhLYK10 chimera (PhLYK10c) was similar (Figures 3A and S4B).
Figure 3.
PhLYK10 and SlLYK10c Have a High Affinity for LCOs and Discriminate LCOs versus COs
(A) Confocal images of epidermal cells from N. benthamiana leaves expressing the indicated proteins. Scale bars represent 20 μm.
(B) Immunodetection of the YFP-fusion proteins in 10 μg of membrane fractions from N. benthamiana leaves.
(C) Binding of LCO-V(C18:1,NMe,35S) to membrane fractions containing the indicated proteins. Incubation with the radiolabeled ligand in the absence or in the presence of 1 μM unlabeled LCO-V(C18:1,NMe,S) allowed to determine the total and non-specific binding respectively and by difference the specific binding. The specific binding is expressed as a percentage of the total binding to normalize variations in protein expression level between biological replicates. Means and standard deviations between replicates are shown.
(D) Scatchard plot analysis of cold saturation experiments using a range of concentration of LCO-V(C18:1,NMe,S) as competitor. The plots are representative of experiments performed with three independent batches of membrane fractions.
(E) Selectivity of the PhLYK10 and SlLYK10c LCO-binding sites for LCOs versus COs. Membrane fractions were incubated with LCO-V(C18:1,NMe,35S) in the presence of 1 μM unlabeled CO4 or CO8 as competitors. Non-specific binding was determined with 1 μM LCO-V(C18:1,NMe,S). Bars represent the percentage of specific binding (means and standard deviations) obtained with independent batches of membrane fractions.
(F) Selectivity of the SlLYK10c LCO-binding sites for various Myc-LCO structures. This is the same as in (B) except that the unlabeled competitors are the indicated LCOs.
(G) Competitive inhibition using a range of concentration of Myc-LCO-IV(C16:0,S) (black circles) or Myc-LCO-IV(C16:0) (white circles).
See also Figures S3, S4, and S5.
SlLYK10c-YFP, PhLYK10-YFP, and PhLYK10c-YFP were all immunodetected in the membrane fractions extracted from N. benthamiana leaves (Figure 3B). Their affinity to LCOs was determined by radio-ligand binding assays using LCO-V(C18:1,NMe) labeled with 35S. Specific binding of LCOs to membrane fractions was detected in extracts of leaves expressing PhLYK10-YFP, PhLYK10c-YFP, or SlLYK10c-YFP but not in extracts of untransformed leaves (Figure 3C).
The affinity of PhLYK10-YFP and SlLYK10c-YFP for LCO-V(C18:1,NMe,S) was determined by a cold saturation experiment. Scatchard plot analysis revealed single class of binding sites (Figure 3D) with dissociation constants (Kd) of 22 nM ± 5 nM (n = 3) and 19 nM ± 4 nM (n = 3), for PhLYK10 and SlLYK10c, respectively, showing that both proteins exhibit high-affinity binding to this LCO. Their selectivity toward COs was then determined through competition assays between the 35S-LCO and an excess (1 μM) of unlabeled COs. CO4 and CO8 were much less efficient competitors of 35S-LCO binding (Figure 3E) with inhibitory constants (Ki) higher than 1 μM, showing that the LCO-binding site of PhLYK10 and SlLYK10c exhibits a low affinity for COs. We also determined the binding selectivity of SlLYK10c-YFP toward Myc-LCOs. All Myc-LCOs were able to compete the binding of the 35S-LCO (Figure 3F). The affinities of SlLYK10c-YFP for the sulfated and non-sulfated Myc-LCOs were further determined by competition assays. Ki of 192 nM ± 52 nM (n = 3) and 354 nM ± 60 nM (n = 3) were obtained for LCO-IV(C16:0,S) or LCO-IV(C16:0), respectively (Figure 3G). Finally, we found that affinity of PhLYK10c for LCOs (Kd of 60 nM ± 18 nM [n = 2]) and selectivity for LCOs versus COs were similar to that of PhLYK10 (Figures S5A and S5B), confirming that the LCO-binding properties of full-length proteins are conserved in our chimeric LysM-RLK.
Promoters from LYRIA Genes Did Not Neo-functionalize to Support RNS
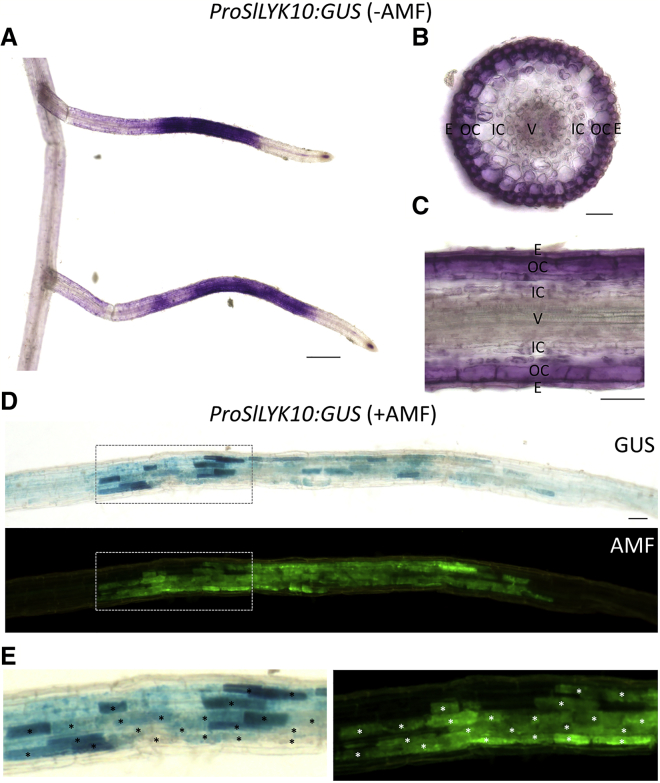
Evolutionary genetics in various eukaryotic models indicates that recruitment of existing pathways to new traits often involves the gain or loss of cis-regulatory elements in promoter regions [32, 33]. We tested whether change in the transcriptional regulation for the LYRIA gene occurred for advent of the RNS by analyzing the expression patterns of Solanaceae LYRIA promoters in AM and RNS. In un-inoculated transgenic tomato roots, a 1.8 kbp sequence of the SlLYK10 promoter region (ProSlLYK10) drove the expression of the GUS reporter primarily in lateral roots (Figure 4A), the preferred site for AMF penetration [34]. Transverse and longitudinal sections revealed GUS activity in the epidermis and outer cortex (Figures 4B and 4C). In transgenic roots maintained as root organ cultures (ROCs) and inoculated with AMF, GUS staining was observed in arbuscule-containing cells (Figures 4D and 4E). Strongest GUS expression was observed in cells at the border of colonization units. Interestingly, this is the site where young arbuscules develop [35].
Figure 4.
ProSlLYK10:GUS Is Expressed in Arbuscule-Containing Cells of Tomato Roots
(A) GUS activity (magenta) in tomato roots from chimeric plants in the absence of AMF.
(B–C) Transversal (B) and longitudinal (C) sections in a root segment showing GUS staining (E, epidermis; OC, outer cortex; IC, inner cortex; V, vessels).
(D) Tomato ROC line colonized by AMF (GUS staining, blue; AMF staining [WGA-CF488A], green).
(E) Close-up of (C). Arbuscule-containing cells are marked by an asterisk.
Scale bars represent 500 μm in (A) and 50 μm in (B–D).
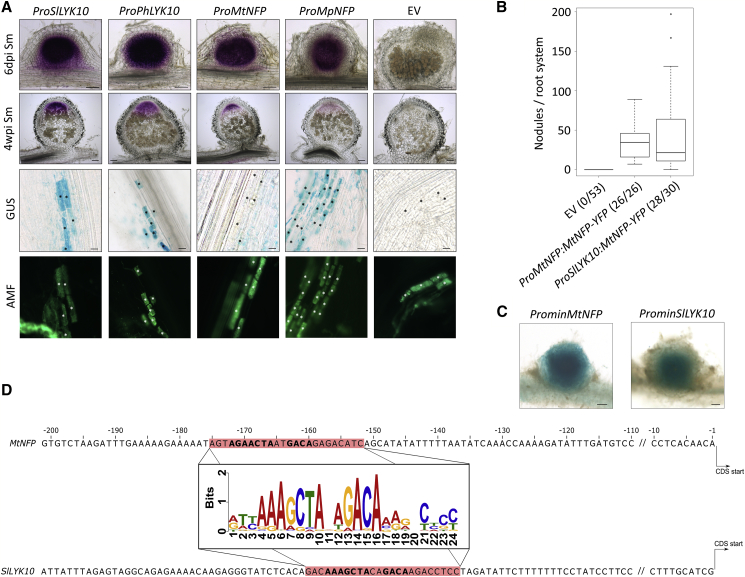
During nodulation, the M. truncatula LYRIA gene MtNFP is expressed in nodule primordia and later in the infection zone of mature nodules [10]. We analyzed the activity of the petunia and tomato LYRIA promoters during nodulation in M. truncatula. ProSlLYK10 and ProPhLYK10 exhibited an activity similar to ProMtNFP with GUS staining in the nodule primordia and in the apex of mature nodules (Figure 5A). This shows that the promoters of the two Solanaceae LYRIA genes contain all the information required for expression in legume nodules. We also compared the expression patterns of the three promoters in M. truncatula mycorrhizal roots. ProMtNFP showed a weak non-specific expression, while ProSlLYK10 and ProPhLYK10 were mostly active in arbuscule-containing cells (Figure 5A). These results suggest that ProSlLYK10 and ProPhLYK10 have the full symbiotic capacity required for expression during AM and RNS and that ProMtNFP has lost the ability to drive expression in mycorrhizal roots. In legumes, a whole-genome duplication at the base of the Papilionoideae gave rise to two paralogous LYRIA genes in Medicago, MtNFP, and MtLYR1. In contrast to MtNFP, MtLYR1 is expressed in mycorrhizal roots [36], but not in nodules (M. truncatula Gene Expression Atlas). The absence of ProMtNFP expression in mycorrhizal roots may reflect either a modification of the ancestral gene promoter required for its recruitment for RNS or the sub-functionalization following the gene duplication in the Papilionoideae. To test these possibilities, we analyzed the expression pattern of MpNFP, the LYRIA gene from Mimosa pudica, a legume from the Mimosoideae clade that did not undergo whole genome duplication [37]. We found that in M. truncatula, ProMpNFP drives a similar expression pattern to ProSlLYK10 and ProPhLYK10, with activity detected both in nodules and in arbuscule-containing cells (Figure 5A). This indicates that the evolution of RNS did not require the loss of LYRIA gene activation during AM. To determine whether Solanaceae LYRIA promoters are sufficient to provide LYRIA protein activity for RNS, we expressed the MtNFP coding sequence (CDS) under the control of ProSlLYK10 in a Mtnfp mutant line unable to form nodules. We observed a similar number of nodules in roots containing either the ProSlLYK10:MtNFP-YFP construct or the ProMtNFP:MtNFP-YFP construct (Figure 5B).
Figure 5.
ProSlLYK10:GUS and ProPhLYK10:GUS Are Expressed in Nodules and in Arbuscule-Containing Cells of M. truncatula Roots
(A) GUS staining (magenta) in young and mature nodules of M. truncatula transgenic roots containing the indicated constructs or the empty vector (EV) and inoculated with S. meliloti (Sm). GUS staining (blue) in arbuscule-containing cells (green) of M. truncatula transgenic roots inoculated with R. irregularis. Arbuscule-containing cells are marked by an asterisk.
(B) Number of nodules in Mtnfp roots complemented by the indicated constructs and inoculated with S. meliloti. Numbers in brackets indicate the numbers of root systems carrying nodules/root systems analyzed. Boxplots represent the distribution between individuals from at least two independent experiments. Scale bars represent 100 μm in the nodule sections and 20 μm the right panels.
(C) The GUS reporter (blue) under the control of a minimal MtNFP (ProminNFP, 240 bp before the start codon) or SlLYK10 (ProminSlLYK10, 185 bp before the start codon) promoters is expressed in young nodules of M. truncatula roots inoculated with S. meliloti. Scale bars represent 100 μm.
D) The putative cis-regulating element in MtNFP and SlLYK10 promoters is highlighted in red in the 200 bp sequences before the start codons. The most conserved positions are in bold. The logo shows the degree of conservation of the putative cis-regulating element among 71 dicotyledonous LYRIA genes.
These results suggest that cis-regulatory elements essential for expression in nodules are conserved between ProSlLYK10, ProPhLYK10, ProMtNFP, and ProMpNFP. To identify the region that contains these cis-regulatory elements, we first cloned a shorter version of the MtNFP promoter (240 bp before the start codon) and tested its activation during RNS. Similar to the 1.5 kb sequence, this shorter promoter was sufficient to drive expression of the GUS reporter in young nodules (Figure 5C). Through scanning the promoter region of orthologous LYRIA genes from nodulating and non-nodulating dicotyledonous species, we identified the AAAGCTANNGACA consensus sequence in the promoters of at least one LYRIA gene in 60% of 71 investigated species (Figure S6). This consensus sequence is located in the proximal region of MtNFP and SlLYK10 promoters (Figure 5D). A SlLYK10 promoter region starting 10 bp upstream of this consensus sequence (185 bp before the start codon) also exhibited activity in young nodules (Figure 5C).
Taken together, our results indicate that the recruitment of LYRIA genes for RNS did not require modification in the regulation of their expression.
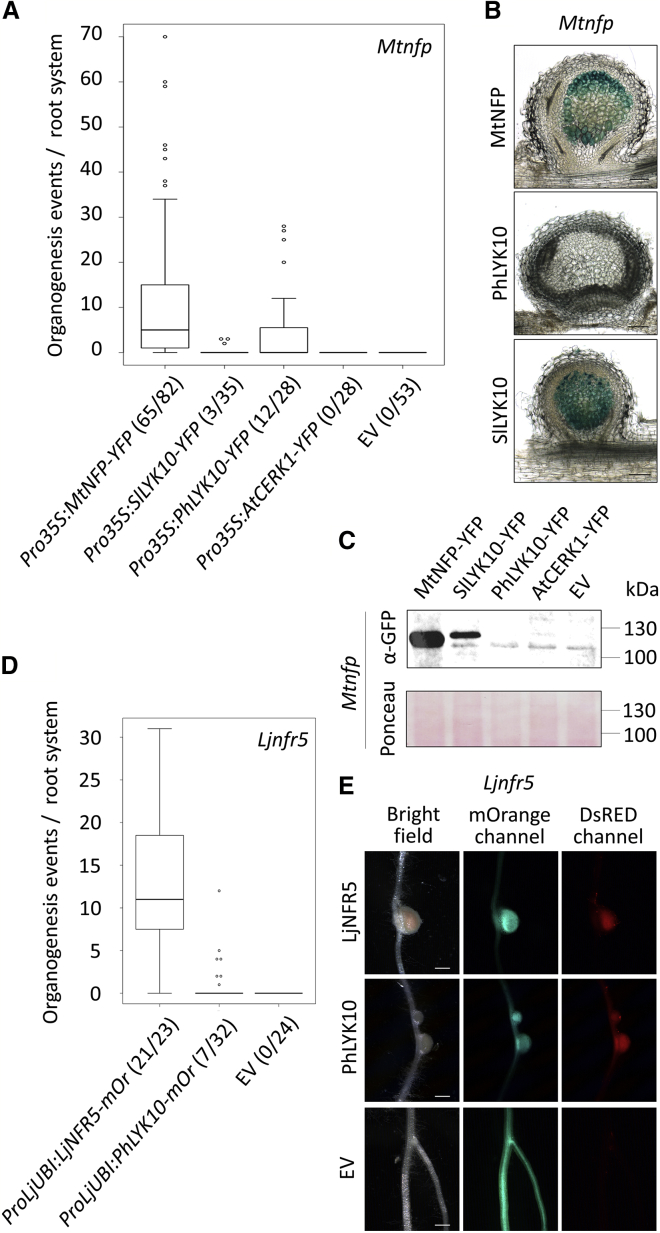
PhLYK10 Partially Complements the Lack of Nodules in Legume Mutants
Besides modifications in cis-regulatory elements, recruitment of a gene into a new trait may result from neo-functionalization of the encoded protein [32, 38]. To test whether the recruitment of LYRIA genes for RNS involved neofunctionalization, we performed complementation assays of Mtnfp and Ljnfr5 mutants with the CDS of PhLYK10 and SlLYK10. ProLjNFR5:SlLYK10 did not restore nodulation in Ljnfr5 mutant. This is similar to what was observed in Mtnfp mutant with the CDS of MtNFP ortholog in pea, PsSYM10, under the control of ProMtNFP [39]. However, we found that PsSYM10 under the control of the strong 35S promoter was able to complement Mtnfp for nodule formation and rhizobial colonization (Figures S7A and S7B). Strikingly, Pro35S:SlLYK10 and Pro35S:PhLYK10 were also able to restore the formation of nodules in Mtnfp (Figure 6A) while Mtnfp roots expressing AtCERK1, an A. thaliana LysM-RLK from the LYKI group (Figure S1B) did not produce any nodules. The nodules formed in roots expressing SlLYK10 were fully colonized by rhizobia, similarly to roots expressing MtNFP, while only a very weak rhizobial staining was observed in roots expressing PhLYK10 (Figure 6B). Immunodetection of proteins in Mtnfp roots revealed that MtNFP was expressed at the highest level (Figures 6C and S7C), whereas PhLYK10 was below the detection limit despite its ability to partially complement nodulation in Mtnfp. This may reflect differences in the stability of the orthologs in M. truncatula, which in turn may explain the different levels of complementation by the different LYRIA proteins. Nodulation was also restored in Ljnfr5 roots expressing PhLYK10 (Figure 6D), although, as in Mtnfp roots, fewer nodules were formed compared with complementation with the endogenous LYRIA gene. In this case, the nodules were fully colonized by rhizobia (Figure 6E). ProLjUBI:PhLYK10-mOrange also triggered spontaneous nodule formation in L. japonicus in the absence of rhizobia (Figures S7D and S7E) like overexpression of LjNFR5 [40].
Figure 6.
PhLYK10 Coding Sequence Complements the Lack of Nodulation in Mtnfp and Ljnfr5
(A) Number of organogenesis events (nodules and nodule primordia) 28 days post inoculation (dpi) with S. meliloti lacZ in Mtnfp roots complemented by the indicated constructs. Numbers in brackets indicate the numbers of root systems carrying organogenesis events/root systems analyzed. Boxplots represent the distribution among individuals from at least two independent experiments. Data for empty vector (EV) are the same as in Figure 5B.
(B) Sections of nodules from Mtnfp roots as in (A). S. meliloti LacZ were stained by X-Gal.
(C) Immunodetection of the YFP-fusion proteins in 20 mg of Mtnfp roots.
(D) Number of organogenesis events 26 dpi with M. loti DsRED in Ljnfr5 roots complemented by the indicated constructs. Numbers in brackets indicate the numbers of root systems carrying organogenesis events/root systems analyzed.
(E) Images of Ljnfr5 roots as in (D).
Scale bars represent 100 μm in (B) and 1 mm in (E). See also Figure S7.
Discussion
Myc-LCOs can induce gene transcription, Ca2+ spiking, and root branching [13, 14, 15, 41, 42]. However, until now it was not clear whether they are involved in AM establishment. Here, we demonstrate high-affinity LCO-binding properties of PhLYK10 and SlLYK10, which, together with the mycorrhizal phenotype of the Phlyk10-1 and Sllyk10-1 mutant lines, provide the strongest evidence to date that Myc-LCOs are directly involved in AM establishment.
Detailed characterization of PhLYK10 and SlLYK10 revealed that they are high-affinity LCO-binding proteins that discriminate LCOs versus COs; their affinity for LCOs being as high as that of the previously characterized legume LYRIA protein, LjNFR5, expressed in the same heterologous system [8]. SlLYK10 recognized the Myc-LCO structures described in [13] with similar affinity for sulfated and non-sulfated Myc-LCOs. However, SlLYK10 exhibited a higher affinity for LCO-V(C18:1,NMe,S) compared with the published Myc-LCO structures, indicating that such LCOs or related structures could potentially represent additional Myc-LCOs.
The similarity of the AM phenotype in the petunia line knockout for PhLYK10, the tomato line bearing a point mutation in SlLYK10, and the tomato SlLYK10-silenced plants [21] provides compelling evidence that the LYRIA gene is involved in AM establishment in Solanaceae. Reduction in the number of colonization sites in the above-mentioned plants suggests a role at early stages for AMF penetration in roots. Moreover, the aberrant arbuscule development observed in Phlyk10-1 and SlLYK10-silenced plants suggests an additional role in arbuscule development. The activity of the SlLYK10 promoter in tomato roots initially in the epidermis and upon colonization in arbuscule-containing cells further supports a role of the LYRIA gene at several steps of AM establishment in Solanaceae.
Although Phlyk10-1, Sllyk10-1, and the SlLYK10-silenced plants are affected in AM establishment, AMFs can still colonize roots and form arbuscules. In a mutant of the rice LYRIA gene OsNFR5, AM-marker gene expression was decreased, but the number of AMF colonization sites was not affected [18]. Mutants in MtNFP are also colonized normally by AMFs [19, 43] despite an almost complete block of symbiosis-related responses to both rhizobial LCOs and Myc-LCOs [13, 14, 43, 44]. Moreover, a double mutant in the two LYRIA genes LjNFR5 and LjLYS11 was not affected in AM establishment [45]. Altogether, this suggests redundancy at the level of LCO perception or that other signals could activate the LCO-mediated signaling pathway. Indeed, Ca2+ spiking can be measured in an Mtnfp mutant after treatment with CO4 [16], suggesting that short-chain CO receptors are also involved in AM establishment. Other signals such as karrikin-like molecules and effector proteins produced by AMFs are known to play important roles in plant-AMF communication [46], but the connection of their perception and/or mode of action to LCO-mediated signaling remains elusive.
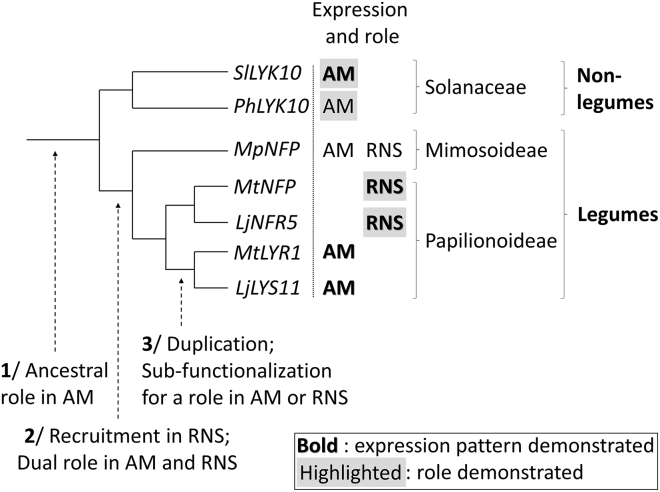
It has been postulated that RNS has evolved through recruitment of genes implicated in AM, but it is unclear how the LCO perception machinery may have been affected by the evolution of RNS. Our data are compatible with a scenario in which an ancestral LYRIA gene involved in LCO perception in AM was directly recruited for LCO perception for RNS in legumes (Figure 7). Because both symbiotic interfaces are intracellular, it can be proposed that LYRIA genes participate in these conserved accommodation mechanisms [47]. The promoters of the single LYRIA gene from the Solanaceae or from the legume M. pudica have the ability to drive dual expression both in mycorrhizal roots and in nodules of M. truncatula. In contrast, the LYRIA gene pairs in the legumes Medicago and Lotus, MtNFP/LjNFR5, and MtLYR1/LjLYS11 have retained transcriptional regulation only during nodulation or AM, respectively [10, 36, 45]. This is indicative of promoter sub-functionalization following the whole genome duplication that predated the radiation of the Papilionoideae, the legume clade to which Medicago and Lotus belong (Figure 7). Interestingly, RNS is evolutionarily more stable in Papilionoideae than in any other clade of RNS-forming plants, including the Mimosoideae to which Mimosa belongs [48]. In other words, the probability for a given species in the Papilionoideae to lose RNS is much lower than in other clades. Although the reason for this greater stability remains unknown, one possibility is that duplication and sub-functionalization of genes with a dual function in AM and RNS such as the LYRIA genes, for separated functions in AM and RNS, may have allowed stabilized symbiotic associations.
Figure 7.
Proposed Scenario for Evolution of the LYRIA Genes
(1/) Ancestral LYRIA genes were involved in AM.
(2/) When the RNS appeared, LYRIA genes had a dual function in AM and RNS in legumes.
(3/) After gene duplication, LYRIA genes were sub-functionalized for a role in RNS or in AM.
Shown are putative (observed in the M. truncatula heterologous system) or known (bold, demonstrated in the endogenous system) expression in mycorrhizal roots (AM) and/or nodules (RNS). Putative or known (highlighted) role in AM and/or RNS are shown. Ph, Petunia hybrida; Sl, Solananum lycopersicum (tomato); Mp, Mimosa pudica; Mt, Medicago truncatula; and Lj, Lotus japonicus.
The AAAGCTANNGACA sequence conserved in LYRIA promoters could represent an ancestral cis-regulatory element involved in transcriptional regulation during AM that has been recruited for transcriptional regulation during RNS. This putative cis-regulatory element is, however, conserved in the promoters of both paralogous LYRIA genes from the Papilionoideae, suggesting that sub-functionalization of the LYRIA promoter pairs has not occurred through divergence in this sequence. Further studies are required to validate the function of this putative cis-regulatory element and to identify the mechanism of LYRIA promoter sub-functionalization in Papilionoideae.
Strikingly, the Solanaceae LYRIA proteins PhLYK10 and SlLYK10 can restore the full nodulation program in the legume LYRIA mutants Mtnfp and Ljnfr5, although with lower efficiency than the respective endogenous LYRIA genes MtNFP and LjNFR5. This suggests that the legume and non-legume LYRIA proteins can fulfill the function of endogenous LYRIA proteins for both nodule formation and rhizobial colonization. Lower complementation efficiency of SlLYK10, PhLYK10, and PsSYM10 compared with MtNFP correlated with lower levels of protein detected in complemented Mtnfp roots. However, lower complementation efficiency of heterologous LYRIA proteins in Mtnfp and Ljnfr5 may also be due to inefficient interactions with the respective co-receptors MtLYK3 and LjNFR1, two LysM-RLKs belonging the LYKI group. It has been suggested that evolution of the LYRIA gene for a new role in RNS may have involved a tandem gene duplication (preceding the advent of RNS) followed by neofunctionalization of one copy for RNS and loss of other copy in the species that acquired the RNS [49]. However, our results suggest that both the promoter and the CDS of the ancestral non-duplicated LYRIA gene were already fully competent for both symbioses.
Intriguingly, our results raise the question of how signal specificity in AM and RNS may be encoded. The fact that PhLYK10 can complement both Mtnfp and Ljnfr5 for nodule formation while M. truncatula and L. japonicus can specifically recognize the respective major LCOs produced by Sinorhizobium meliloti (LCO-IV(C16:2,S) [50] and Mesorhizobium loti (LCO-V(C16:1,Cb,Fuc,Ac) [51] argues for limited LCO selectivity of MtNFP, LjNFR5, and their Solanaceous orthologs PhLYK10 and SlLYK10. This questions the hypothesis that MtNFP and LjNFR5 recognize specific LCO structures and suggests that co-receptors such as MtLYK3/LjNFR1, or yet unidentified proteins, may interact with MtNFP and LjNFR5 to confer LCO binding specificity to LCO receptor complexes. Consistent with such a scenario, the number of LysM-RLKs in the LYKI group has dramatically increased in legumes compared with non-legumes and contains a legume-specific subgroup to which MtLYK3 and LjNFR1 belong [52].
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal GFP antibodies | AMSBIO | TP401; RRID: AB_10890443 |
| monoclonal HSC70 (BIP) antibody | Enzo Life Sciences | ADI-SPA-818; RRID: AB_10617235 |
| Rabbit polyclonal H+-ATPase antibodies | [53] | N/A |
| Fungal and Bacterial Strains | ||
| Agrobacterium rhizogenes ARquA1 | [54] | N/A |
| Sinorhizobium meliloti 2011 pXLGD4 (lacZ reporter) | [55] | N/A |
| Mesorhizobium loti MAFF303099 DsRED | [56] | N/A |
| Agrobacterium tumefaciens LBA4404 VirGN54D | [57] | N/A |
| Rhizophagus irregularis DAOM 197198 | Agronutrition | AP2007-A |
| Gigaspora gigantea | [58] | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FM4-64 | Invitrogen | T3320 |
| DAPI | SIGMA | D9542 |
| PNGaseF | Roche Diagnostics | 11365169001 |
| LCO-V(C18:1Δ11,NMe) purified from Rhizobium tropici | [59] | N/A |
| LCO-V(C18:1Δ11,NMe,S) purified from Rhizobium tropici | [59] | N/A |
| Myc-LCOs, LCO-IV(C18:1Δ9) | [13] | N/A |
| LCO-IV(C18:1Δ9,S) | [13] | N/A |
| LCO-IV(C16:0) | [13] | N/A |
| LCO-IV(C16:O,S) | [13] | N/A |
| CO4 | CERMAV Grenoble, France | N/A |
| CO8 | CERMAV Grenoble, France | N/A |
| Critical Commercial Assays | ||
| Gateway BP mix | Invitrogen | 11789-100 |
| Gateway LR mix | Invitrogen | 11791-100 |
| Bsa I enzyme for Golden Gate reactions | New England BIOLABS | R0535S |
| X-Gluc | Biosynth | B7300 |
| Magenta-Gluc | Biosynth | B7350 |
| X-Gal substrate | Thermofisher | 10113253 |
| WGA CF488A conjugate | Biotum | BTM29022 |
| Macherey-Nagel NUCLEOSPIN RNA kit | Macherey-Nagel | 740955.250 |
| Agilent RNA Nano Chip and Reagents | Agilent Technologies | 5067-1511 |
| Superscript reverse transcriptase | Invitrogen | 18064071 |
| LightCycler480 Sybr Green I Master | Roche | 04707516001 |
| Attapulgite (American granules plain) | Oil-dri UK | GB100 |
| Experimental Models: Organisms/Strains | ||
| Solanum lycopersicum cv Marmande | NA | N/A |
| Petunia hybrida cv W138 | NA | N/A |
| Petunia hybrida cv W5 | [60] | N/A |
| Mimosa pudica | [37] | N/A |
| Solanum lycopersicum cv M82 Sllyk10-1 | This work | N/A |
| Petunia hybrida cv W138 Phlyk10-1 | This work | N/A |
| Medicago truncatula A17 | NA | N/A |
| Medicago truncatula A17 Mtnfp-2 | [10] | N/A |
| Lotus japonicus Gifu | NA | N/A |
| Lotus japonicus Ljnfr5-2 | [61] | N/A |
| Oligonucleotides | ||
| ProSlLYK10 for GGTCTCTAAATGGGTTATAGAGCTGTAATGC | This work | N/A |
| ProSlLYK10 rev GGTCTCATTTGCGATGCAAAGCTTAGATAAC | This work | N/A |
| ProPhLYK10 for ATCGGTCTCCAAATGAGCTGCAGGGCTTTTCTACG | This work | N/A |
| ProPhLYK10 rev ATCGGTCTCCTTTGTGCTGCAAAGCTCAGATGGC | This work | N/A |
| ProMpNFP for ATCGGTCTCCAAATAGAAAGTTTTCTGTTGTCCGG | This work | N/A |
| ProMpNFP rev: ATCGGTCTCCTTTGCTAATGAGAGTTTAGCAGAGG | This work | N/A |
| PhLYK10ECR for: GGTCTCCCAAAATGGTAGCTCCTCTTGCCTCCT | This work | N/A |
| PhLYK10ECR rev: GGTCTCGTAAGAATACTTAAAACGACAATGAGA | This work | N/A |
| SlLYK10ECR for GGTCTCGCAAAATGGTAGTTCCTCTTGTGTCCTTG | This work | N/A |
| SlLYK10ECR rev GGTCTCGTAAGTCCATGCTTGGATTTTCTACTGCTTGC | This work | N/A |
| SlLYK10 Genotyping for: GTGGTGCAAGATATGAATCC | This work | N/A |
| SlLYK10 Genotyping rev: GAGCTAAGTTAGACCTCCTC | This work | N/A |
| PhLYK10 Genotyping for: GCAGACAGAGACTTTTTGTGCTCT | This work | N/A |
| PhLYK10 Genotyping rev: ACAGCTTCCGTACCAACTGTC | This work | N/A |
| Recombinant DNA | ||
| Pcambia-Pro35S:PhLYK10-YFP | This work | N/A |
| PcambiaGG-Pro35S:PhLYK10c-YFP | This work | N/A |
| PcambiaGG-Pro35S:SlLYK10-YFP | This work | N/A |
| PcambiaGG-Pro35S:SlLYK10c-YFP | This work | N/A |
| PbinGW-Pro35S:SlLYK10-YFP | This work | N/A |
| PcambiaGG-ProSlLYK10:GUS | This work | N/A |
| PcambiaGG-ProminSlLYK10:GUS | This work | N/A |
| PcambiaGG-ProPhLYK10:GUS | This work | N/A |
| PcambiaGG-ProMpNFP:GUS | This work | N/A |
| Pbin-ProMtNFP:GUS | [10] | N/A |
| PcambiaGG-ProminMtNFP:GUS | This work | N/A |
| Pbin-ProMtNFP:MtNFP-YFP | This work | N/A |
| PcambiaGG-ProSlLYK10:MtNFP-YFP | This work | N/A |
| Pcambia-Pro35S:AtCERK1-YFP | [62] | N/A |
| Pcambia-ProLjUBI:LjNFR5-mOrange | This work | N/A |
| Pcambia-ProLjUBI:PhLYK10-mOrange | This work | N/A |
| Pbin-PsSYM10-YFP | [63] | N/A |
| Pbin-pro35S:PMA4-GFP | [64] | N/A |
| Pbin-pro35S:HDEL-GFP | [64] | N/A |
| Software and Algorithms | ||
| LASX | Leica | N/A |
| Zen | Leica | N/A |
| ImageJ | http://imagej.nih.gov/ij | N/A |
| R | http://r-project.org | N/A |
| tBLASTn v2.9.0+ | [65] | N/A |
| MAFFT v7.407 | [66] | N/A |
| TrimAl v1.4 | [67] | N/A |
| ModelFinder | [68] | N/A |
| IQ-TREE v1.6.1 | [69] | N/A |
| SH-alrt | [70] | N/A |
| iTOL platform v4.4.2 | [71] | N/A |
| MEME v5.0.1 | [72] | N/A |
| Other | ||
| Medicago truncatula Gene Expression Atlas | http://mtgea.noble.org/v3 | N/A |
| Axiozoom V16 microscope | Zeiss | N/A |
| Axioplan 2 microscope | Zeiss | N/A |
| SP2 confocal microscope | Leica | N/A |
| SP8 confocal microscope | Leica | N/A |
| S6E microscope | Leica | N/A |
| vribratome VT 1000S | Leica | N/A |
Lead Contact and Materials Availability
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Benoit Lefebvre (benoit.lefebvre@inra.fr). All unique/stable reagents generated in this study are available from the Lead Contact without restriction.
Experimental Model and Subject Details
Cloning
1.8, 1.5 and 1.9 kbp corresponding to the non-coding region between SlLYK10, PhLYK10, and MpNFP and the preceding genes, including the 5′ UTR were amplified by PCR (with the primers listed in the key resources table) from genomic DNA isolated from S. lycopersicum, P. hybrida and M. pudica, respectively, and cloned in transcriptional fusion with a GUS reporter containing a plant intron, in a pCambia 2200 modified for Golden gate cloning and containing a ProUbi:DsRed reporter as in [73]. Note that the PhLYK10 sequence in P. hybrida originates from the P. axillaris parent. 240 and 185 bp sequences preceding the SlLYK10 or MtNFP start codons were synthesized and cloned as described previously. ProSlLYK10:MtNFP-YFP was made by Golden gate cloning in a pCambia 2200 modified for Golden gate as in [6]. ProMtNFP:MtNFP-YFP was made as in [30] excepted that MtNFP was in translation fusion with YFP instead that of FLAG.
SlLYK10 and PhLYK10 coding sequences were amplified by PCR from genomic DNA isolated from S. lycopersicum and P. hybrida respectively and cloned in translational fusion with YFP under the control of Pro35S in a pbin vector modified for gateway cloning as in [74] for SlLYK10 or in a pCambia 2200 modified for Golden gate cloning as in [6] for PhLYK10. For expression in M. truncatula, SlLYK10 sequence was optimized with a M. truncatula codon usage and cloned in translational fusion with YFP under the control of Pro35S in a pCambia 2200 modified for Golden gate cloning as in [6]. For expression in L. japonicus, PhLYK10 coding sequence was amplified by PCR from genomic DNA isolated from P. hybrida and cloned in translational fusion with mOrange under the control of LjUbiquitin promoter into a pCambia-based Golden Gate expression vector [75].
For SlLYK10c and PhLYK10c constructs, the sequences coding the extracellular region of SlLYK10 or PhLYK10 were amplified by PCR (with the primers listed in the key resources table) and cloned in translational fusion with the sequences coding TM/ICR of MtNFP and YFP under the control of Pro35S in a pCambia 2200 modified for Golden gate cloning as in [6].
S. lycopersicum and P. hybrida mutant identification and genotyping
The Sllyk10-1 mutant allele (line 1051, G460A) was identified by sequencing (NGS) an amplicon (key resources table) obtained on tomato (cv M82) EMS-mutagenized lines. Homozygous mutant or WT SlLYK10 alleles were identified by sequencing (Sanger) a similar amplicon on the progeny. The Phlyk10-1 mutant allele (line LY0882, dTph1 insertion 116 bp from the start codon) was identified by BLAST-searching in a Petunia dTPh1 transposon flanking sequence database [29] with the full PhLYK10 coding sequence. This line was crossed with the stabilizer line W5 [60], to segregate out the activator locus required for dTph1 transposition. Genotyping on different progenies was done by PCR with the primers listed in the key resources table.
Agrobacterium rhizogenes mediated transformation
Tomato (cv Marmande) seeds were surface sterilized and germinated in vitro for 7 to 10 days until cotyledons were fully expanded. Plantlets were cut at the hypocotyl level, immerged in a A. rhizogenes ARqua1 suspension at OD600nm = 0.3 and grown for 3 days at 25°C on MS, then on MS supplemented with 50 mg/l kanamycin and 200 mg/l cefotaxim until emergence of transgenic roots. Transgenic roots were selected by fluorescence microscopy. Plantlets were transferred in pots containing vermiculite as described in [21]. ROC lines derived from transformed roots were grown in dark, on MS medium supplemented with 50 mg/l kanamycin.
Chimeric M. truncatula A17 and Mtnfp-2 plants were produced as described in [76] for analysis of promoter expression pattern and for complementation experiment, respectively. Chimeric L. japonicus Gifu and Ljnfr5-2 plants were produced as described in [77].
Inoculation with AMF
For AM phenotyping, petunia seeds were germinated on a sterilized potting soil until cotyledons were fully expanded. Tomato seeds were surface sterilized and germinated in sterile water. Petunia and tomato plantlets were then transferred in 50 mL containers filled with attapulgite, watered with 20 mL of 0.5x modified Long ashton (7.5 μM NaH2PO4), and inoculated with 500 spores of R. irregularis DAOM 197198. Roots were harvested, washed and stained between 3 and 4 weeks post inoculation.
For analysis of GUS activity in tomato roots, sterilized Gigantea gigaspora spores, harvested from a leek nurse culture, were pre-germinated 5 days on M medium [78] in a 3% CO2 incubator at 32°C. Two spores and one fragment of a transgenic tomato ROC line were then co-cultured on a Petri dish containing M medium supplemented with 50 mg/l kanamycin. Petri dishes were placed vertically with ROC lines above the fungal spores for 4 weeks. For analysis of GUS activity in M. truncatula transgenic roots, chimeric plantlets were transferred in 50 mL containers filled with a mix 1:1 of attapulgite and sand, watered with 20 mL of 0.5x modified Long ashton medium and inoculated with 200 spores of R. irregularis DAOM 197198. Roots were harvested, washed and stained 2 weeks post inoculation.
Inoculation with rhizobia and spontaneous nodulation
M. truncatula chimeric plantlets were transferred in 250 mL containers filled with attapulgite, watered with 20 mL of Farhaeus medium supplemented with 1 mM NH4NO3. After 4 days, 2.5 mL of a suspension at OD600nm = 0.025 of a S. meliloti strains 2011 harboring the hemA-lacZ plasmid (pXLGD4) was added around the hypocotyl. Roots were harvested, washed and stained 4 weeks post inoculation.
For complementation experiments, L. japonicus chimeric plantlets were transferred to Weck jars containing 300 mL of a mix of sand and vermiculite and inoculated with 20 mL of a M. loti MAFF303099 DsRED suspension in FP medium (OD600 = 0.05). Plants were phenotyped 25 days post inoculation.
Spontaneous nodulation experiments on L. japonicus roots were performed as described previously [40]. L. japonicus chimeric plantlets were transferred to Fahraeus medium plates containing 0.1 μM of the ethylene biosynthesis inhibitor L-α-(2-aminoethoxyvinyl)-glycine 2.5 weeks after transformation. Root systems were analyzed 60 days post transformation.
Transient Expression in N. benthamiana
Leaves of N. benthamiana were infiltrated with A. tumefaciens LBA4404 virGN54D strains as described in [79]. Leaves were harvested 3 days after infiltration.
Method Details
Microscopy
Tomato ROC expressing SlLYK10-YFP were incubated at room temperature 5 min in water with 1 μg / ml DAPI or 20 μM FM4-64 before confocal imaging. For plasmolysis, ROC lines were incubated for 1 h in 0.8 M mannitol. Tomato ROC and chimeric M. truncatula plants expressing the GUS reporter were stained with 0.1% X-Gluc or Magenta-Gluc (20 min under vacuum followed by incubation at 37°C). AMF were stained by treating root tissues with 100% ethanol for 4 h, then with 10% KOH for 8 min at 95°C (tomato ROC and P. hybrida roots) or 1,5 days at room temperature (M. truncatula roots) and finally with 0.2 M PBS pH 7.2, Triton X-100 0.01%, 1 μg/mL WGA CF488A conjugate overnight at room temperature. For analysis of subcellular localization, tomato ROC and N. benthamiana leaves were imaged using a SP8 confocal microscope. Arbuscules in P. hybrida were imaged with a SP2 confocal microscope. Overlay corresponds to merge of green fluorescence channel images with differential interference contrast images. GUS and WGA staining were imaged using an Axiozoom V16 microscope (Figure 4) or an Axioplan 2 microscope (Figure 5). Automatic delimitation and drawing of cells strongly expressing GUS was performed with ImageJ (Figure 4).
Numbers of colonization sites and root length colonization were quantified on entire root systems using a S6E microscope after ink staining of the AMF as described in [21].
M. truncatula nodulated roots systems expressing the GUS reporter were stained with 0.1% mangenta-gluc and then fixed with glutaraldehyde 1.25% in 0.1 M PBS pH7.2 (30 min under vacuum). In case of Mtnfp-2 complementation, nodulated roots systems were first fixed with glutaraldehyde 1.25% and then stained with 2% X-Gal (30min under vacuum and followed by incubation at 28°C). Nodules were sectioned after inclusion in 6% agarose low gelling temperature using a vribratome VT 1000S and sections were imaged with an Axioplan 2 microscope. Nodules of M. truncatula roots expressing the GUS reporter under the control of the minimal promoters were stained 0.1% X-Gluc and directly imaged with an Axiozoom V16 microscope.
Western blotting and membrane fraction preparation
Immunobloting of YFP fusions in M. truncatula roots was performed on 20 mg of a total extract of a pool of 10 root systems inoculated by S. meliloti. For LCO binding assays, approximately 20 g of leaves were homogenized at 4°C in a blender in the presence of 40 mL of extraction buffer (25 mM Tris, pH 8.5, 0.47 M sucrose, 5 mM EDTA, 10 mM DTT, 0.6% PVPP and protease inhibitors (0.1 mM AEBSF, and 1 mg/mL each of leupeptin, aprotinin, antipain, chymostatin, and pepstatin). Samples were centrifuged for 15 min at 3000 g, and then the supernatant was recentrifuged for 30 min at 45000 g. The pellet (membrane fraction) was first washed in 5 mL and then resuspended in 2 mL of binding buffer (25 mM Na-Cacodylate pH 6, 250 mM sucrose, 1 mM CaCl2, 1 mM MgCl2 and protease inhibitors). After each extraction, amount of fusion proteins was quantified by immunoblotting in 10 μg of membrane fraction proteins. PhLYK10-YFP, PhLYK10c-YFP and SlLYK10c-YFP have expected molecular masses of about 104, 102 and 102 kDa respectively (including 6 predicted N-glycans). For Figure S4D, after homogenization samples were centrifuged for 20 min at 100000 g and resuspended in the same volume of extraction buffer. Proportional volumes of total extract, resuspended pellet and supernatant were loaded on SDS-PAGE.
LCO binding assays
LCO-V(C18:1Δ11,NMe) and LCO-V(C18:1Δ11,NMe,S) were purified from the rhizobial strain Rhizobium tropici. Labeling of LCO-V(C18:1Δ11,NMe) was performed as described in [80]. LCO binding assays on membrane fractions containing 20 μg or 40 μg of proteins were performed as in [6] using between 1 and 2 nM of radiolabeled LCO and ranges of unlabeled LCO between 1 nM to 1 μM. Similar amount of membrane fraction from leaves expressing PhLYK10-YFP, PhLYK10-YFPc, SlLYK10c-YFP or from untransformed leaves were used in each experiment. Competition with COs were performed with 1 μM of unlabeled pure CO4 and CO8.
PNGaseF treatment and immunoblotting
PNGaseF treatment, SDS-PAGE, transfer to nitrocellulose membranes and western blotting were performed as described in [30].
qRT-PCR
RNA extraction, cDNA synthesis was performed as described in [21]. Relative expression levels were calculated using glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a reference gene. Primers were as in [81] and [21].
Promoter investigation
MtNFP orthologs were retrieved from genomes of 71 dicotyledonous species (list in Table S2) using tBLASTn and an e-value threshold of 1e-10. Putative orthologs were aligned with MAFFT with default parameters and aligned positions with more than 50% of gaps were removed using TrimAl. The best-fitting evolutionary model was tested using ModelFinder and according to the Bayesian Information Criteria. The model TVM+F+R5 was further used for Maximum Likelihood (ML) analysis using IQ-TREE. Branch support was tested using 10,000 replicates of SH-alrt. The resulting tree was annotated using the iTOL platform. For each ortholog, 600 bp promoter sequences were extracted upstream of the gene start using a custom Python script. Promoters were searched for enriched motif using MEME with following parameters: zero or one occurrence of motif per site, motif length comprises between 5 and 25 bp and a minimum of 2 sites by motifs.
Quantification and Statistical Analysis
Number of independent biological replicates and individuals analyzed, as well as the statistical tests used to analyze the data are indicated in the figure legends. All statistical analyses were performed using the R software (http://r-project.org).
Data and Code Availability
This study did not generate any unique datasets or code.
Acknowledgments
We thank Marie Cumener, Fabienne Maillet, Mireille Chabaud, and Véréna Poinsot for technical help, Sébastien Fort for CO4 an CO8 production, and Julie Cullimore and Malick Mbengue for critical reading of the manuscript. This work was supported by the ANR “WHEATSYM” (ANR-16-CE20-0025-01), the “Laboratoire d’Excellence (LABEX)” TULIP (ANR-10-LABX-41), the Swiss National Science Foundation (31003A_169732), and the research project Engineering Nitrogen Symbiosis for Africa (ENSA), which is funded through a grant to the University of Cambridge by the Bill & Melinda Gates Foundation (OPP1172165). A.G.’s fellowship was funded by Région Occitanie and INRA Department of Plant Health and Environment (SPE). T.W.’s fellowship was funded by the Chinese Scholarship Council (CSC). Work in M.P.’s lab was funded by the ERC Advanced Grant ERC-2013-ADG: “Molecular inventions underlying the evolution of the nitrogen-fixing root nodule symbiosis” (EVOLVINGNODULES).
Author Contributions
Conceptualization, B.L., D.R., J.-J.B., M.P., and P.-M.D.; Investigation, A.G., C.R., J.K., L.B., M.-C.A., M.G., M.R., T.V., T.W., V.G., and Y.D.; Resources, A.B., M.S., M.V., and P.M.; Writing, A.G., B.L., D.R., J.-J.B., M.P., and P.-M.D.
Declaration of Interests
The authors declare no competing interests.
Published: December 5, 2019
Footnotes
Supplemental Information can be found online at https://doi.org/10.1016/j.cub.2019.11.038.
Supplemental Information
References
- 1.Delaux P.M., Radhakrishnan G.V., Jayaraman D., Cheema J., Malbreil M., Volkening J.D., Sekimoto H., Nishiyama T., Melkonian M., Pokorny L. Algal ancestor of land plants was preadapted for symbiosis. Proc. Natl. Acad. Sci. USA. 2015;112:13390–13395. doi: 10.1073/pnas.1515426112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rich M.K., Nouri E., Courty P.E., Reinhardt D. Diet of Arbuscular Mycorrhizal Fungi: Bread and Butter? Trends Plant Sci. 2017;22:652–660. doi: 10.1016/j.tplants.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 3.Parniske M. Arbuscular mycorrhiza: the mother of plant root endosymbioses. Nat. Rev. Microbiol. 2008;6:763–775. doi: 10.1038/nrmicro1987. [DOI] [PubMed] [Google Scholar]
- 4.Murray J.D. Invasion by invitation: rhizobial infection in legumes. Mol. Plant Microbe Interact. 2011;24:631–639. doi: 10.1094/MPMI-08-10-0181. [DOI] [PubMed] [Google Scholar]
- 5.Fliegmann J., Bono J.J. Lipo-chitooligosaccharidic nodulation factors and their perception by plant receptors. Glycoconj. J. 2015;32:455–464. doi: 10.1007/s10719-015-9609-3. [DOI] [PubMed] [Google Scholar]
- 6.Fliegmann J., Canova S., Lachaud C., Uhlenbroich S., Gasciolli V., Pichereaux C., Rossignol M., Rosenberg C., Cumener M., Pitorre D. Lipo-chitooligosaccharidic symbiotic signals are recognized by LysM receptor-like kinase LYR3 in the legume Medicago truncatula. ACS Chem. Biol. 2013;8:1900–1906. doi: 10.1021/cb400369u. [DOI] [PubMed] [Google Scholar]
- 7.Malkov N., Fliegmann J., Rosenberg C., Gasciolli V., Timmers A.C., Nurisso A., Cullimore J., Bono J.J. Molecular basis of lipo-chitooligosaccharide recognition by the lysin motif receptor-like kinase LYR3 in legumes. Biochem. J. 2016;473:1369–1378. doi: 10.1042/BCJ20160073. [DOI] [PubMed] [Google Scholar]
- 8.Broghammer A., Krusell L., Blaise M., Sauer J., Sullivan J.T., Maolanon N., Vinther M., Lorentzen A., Madsen E.B., Jensen K.J. Legume receptors perceive the rhizobial lipochitin oligosaccharide signal molecules by direct binding. Proc. Natl. Acad. Sci. USA. 2012;109:13859–13864. doi: 10.1073/pnas.1205171109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buendia L., Girardin A., Wang T., Cottret L., Lefebvre B. LysM Receptor-Like Kinase and LysM Receptor-Like Protein Families: An Update on Phylogeny and Functional Characterization. Front. Plant Sci. 2018;9:1531. doi: 10.3389/fpls.2018.01531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arrighi J.F., Barre A., Ben Amor B., Bersoult A., Soriano L.C., Mirabella R., de Carvalho-Niebel F., Journet E.P., Ghérardi M., Huguet T. The Medicago truncatula lysin [corrected] motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 2006;142:265–279. doi: 10.1104/pp.106.084657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Radutoiu S., Madsen L.H., Madsen E.B., Felle H.H., Umehara Y., Grønlund M., Sato S., Nakamura Y., Tabata S., Sandal N., Stougaard J. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature. 2003;425:585–592. doi: 10.1038/nature02039. [DOI] [PubMed] [Google Scholar]
- 12.Indrasumunar A., Kereszt A., Searle I., Miyagi M., Li D., Nguyen C.D., Men A., Carroll B.J., Gresshoff P.M. Inactivation of duplicated nod factor receptor 5 (NFR5) genes in recessive loss-of-function non-nodulation mutants of allotetraploid soybean (Glycine max L. Merr.) Plant Cell Physiol. 2010;51:201–214. doi: 10.1093/pcp/pcp178. [DOI] [PubMed] [Google Scholar]
- 13.Maillet F., Poinsot V., André O., Puech-Pagès V., Haouy A., Gueunier M., Cromer L., Giraudet D., Formey D., Niebel A. Fungal lipochitooligosaccharide symbiotic signals in arbuscular mycorrhiza. Nature. 2011;469:58–63. doi: 10.1038/nature09622. [DOI] [PubMed] [Google Scholar]
- 14.Sun J., Miller J.B., Granqvist E., Wiley-Kalil A., Gobbato E., Maillet F., Cottaz S., Samain E., Venkateshwaran M., Fort S. Activation of symbiosis signaling by arbuscular mycorrhizal fungi in legumes and rice. Plant Cell. 2015;27:823–838. doi: 10.1105/tpc.114.131326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Camps C., Jardinaud M.F., Rengel D., Carrère S., Hervé C., Debellé F., Gamas P., Bensmihen S., Gough C. Combined genetic and transcriptomic analysis reveals three major signalling pathways activated by Myc-LCOs in Medicago truncatula. New Phytol. 2015;208:224–240. doi: 10.1111/nph.13427. [DOI] [PubMed] [Google Scholar]
- 16.Genre A., Chabaud M., Balzergue C., Puech-Pagès V., Novero M., Rey T., Fournier J., Rochange S., Bécard G., Bonfante P., Barker D.G. Short-chain chitin oligomers from arbuscular mycorrhizal fungi trigger nuclear Ca2+ spiking in Medicago truncatula roots and their production is enhanced by strigolactone. New Phytol. 2013;198:190–202. doi: 10.1111/nph.12146. [DOI] [PubMed] [Google Scholar]
- 17.Op den Camp R., Streng A., De Mita S., Cao Q., Polone E., Liu W., Ammiraju J.S., Kudrna D., Wing R., Untergasser A. LysM-type mycorrhizal receptor recruited for rhizobium symbiosis in nonlegume Parasponia. Science. 2011;331:909–912. doi: 10.1126/science.1198181. [DOI] [PubMed] [Google Scholar]
- 18.Miyata K., Hayafune M., Kobae Y., Kaku H., Nishizawa Y., Masuda Y., Shibuya N., Nakagawa T. Evaluation of the Role of the LysM Receptor-Like Kinase, OsNFR5/OsRLK2 for AM Symbiosis in Rice. Plant Cell Physiol. 2016;57:2283–2290. doi: 10.1093/pcp/pcw144. [DOI] [PubMed] [Google Scholar]
- 19.Zhang X., Dong W., Sun J., Feng F., Deng Y., He Z., Oldroyd G.E., Wang E. The receptor kinase CERK1 has dual functions in symbiosis and immunity signalling. Plant J. 2015;81:258–267. doi: 10.1111/tpj.12723. [DOI] [PubMed] [Google Scholar]
- 20.Miyata K., Kozaki T., Kouzai Y., Ozawa K., Ishii K., Asamizu E., Okabe Y., Umehara Y., Miyamoto A., Kobae Y. The bifunctional plant receptor, OsCERK1, regulates both chitin-triggered immunity and arbuscular mycorrhizal symbiosis in rice. Plant Cell Physiol. 2014;55:1864–1872. doi: 10.1093/pcp/pcu129. [DOI] [PubMed] [Google Scholar]
- 21.Buendia L., Wang T., Girardin A., Lefebvre B. The LysM receptor-like kinase SlLYK10 regulates the arbuscular mycorrhizal symbiosis in tomato. New Phytol. 2016;210:184–195. doi: 10.1111/nph.13753. [DOI] [PubMed] [Google Scholar]
- 22.Liao D., Sun X., Wang N., Song F., Liang Y. Tomato LysM Receptor-Like Kinase SlLYK12 Is Involved in Arbuscular Mycorrhizal Symbiosis. Front. Plant Sci. 2018;9:1004. doi: 10.3389/fpls.2018.01004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibelin-Viala C., Amblard E., Puech-Pages V., Bonhomme M., Garcia M., Bascaules-Bedin A., Fliegmann J., Wen J., Mysore K.S., le Signor C. The Medicago truncatula LysM receptor-like kinase LYK9 plays a dual role in immunity and the arbuscular mycorrhizal symbiosis. New Phytol. 2019;223:1516–1529. doi: 10.1111/nph.15891. [DOI] [PubMed] [Google Scholar]
- 24.Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., Shibuya N. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ao Y., Li Z., Feng D., Xiong F., Liu J., Li J.F., Wang M., Wang J., Liu B., Wang H.B. OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 2014;80:1072–1084. doi: 10.1111/tpj.12710. [DOI] [PubMed] [Google Scholar]
- 26.Carotenuto G., Chabaud M., Miyata K., Capozzi M., Takeda N., Kaku H., Shibuya N., Nakagawa T., Barker D.G., Genre A. The rice LysM receptor-like kinase OsCERK1 is required for the perception of short-chain chitin oligomers in arbuscular mycorrhizal signaling. New Phytol. 2017;214:1440–1446. doi: 10.1111/nph.14539. [DOI] [PubMed] [Google Scholar]
- 27.Bravo A., York T., Pumplin N., Mueller L.A., Harrison M.J. Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat. Plants. 2016;2:15208. doi: 10.1038/nplants.2015.208. [DOI] [PubMed] [Google Scholar]
- 28.Delaux P.M., Varala K., Edger P.P., Coruzzi G.M., Pires J.C., Ané J.M. Comparative phylogenomics uncovers the impact of symbiotic associations on host genome evolution. PLoS Genet. 2014;10:e1004487. doi: 10.1371/journal.pgen.1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vandenbussche M., Janssen A., Zethof J., van Orsouw N., Peters J., van Eijk M.J., Rijpkema A.S., Schneiders H., Santhanam P., de Been M. Generation of a 3D indexed Petunia insertion database for reverse genetics. Plant J. 2008;54:1105–1114. doi: 10.1111/j.1365-313X.2008.03482.x. [DOI] [PubMed] [Google Scholar]
- 30.Lefebvre B., Klaus-Heisen D., Pietraszewska-Bogiel A., Hervé C., Camut S., Auriac M.-C., Gasciolli V., Nurisso A., Gadella T.W.J., Cullimore J. Role of N-glycosylation sites and CXC motifs in trafficking of medicago truncatula Nod factor perception protein to plasma membrane. J. Biol. Chem. 2012;287:10812–10823. doi: 10.1074/jbc.M111.281634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawaharada Y., Kelly S., Nielsen M.W., Hjuler C.T., Gysel K., Muszyński A., Carlson R.W., Thygesen M.B., Sandal N., Asmussen M.H. Receptor-mediated exopolysaccharide perception controls bacterial infection. Nature. 2015;523:308–312. doi: 10.1038/nature14611. [DOI] [PubMed] [Google Scholar]
- 32.Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 33.Jiang P., Rausher M. Two genetic changes in cis-regulatory elements caused evolution of petal spot position in Clarkia. Nat. Plants. 2018;4:14–22. doi: 10.1038/s41477-017-0085-6. [DOI] [PubMed] [Google Scholar]
- 34.Gutjahr C., Paszkowski U. Multiple control levels of root system remodeling in arbuscular mycorrhizal symbiosis. Front. Plant Sci. 2013;4:204. doi: 10.3389/fpls.2013.00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kobae Y., Hata S. Dynamics of periarbuscular membranes visualized with a fluorescent phosphate transporter in arbuscular mycorrhizal roots of rice. Plant Cell Physiol. 2010;51:341–353. doi: 10.1093/pcp/pcq013. [DOI] [PubMed] [Google Scholar]
- 36.Gomez S.K., Javot H., Deewatthanawong P., Torres-Jerez I., Tang Y., Blancaflor E.B., Udvardi M.K., Harrison M.J. Medicago truncatula and Glomus intraradices gene expression in cortical cells harboring arbuscules in the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 2009;9:10. doi: 10.1186/1471-2229-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Griesmann M., Chang Y., Liu X., Song Y., Haberer G., Crook M.B., Billault-Penneteau B., Lauressergues D., Keller J., Imanishi L. Phylogenomics reveals multiple losses of nitrogen-fixing root nodule symbiosis. Science. 2018;361:eaat1743. doi: 10.1126/science.aat1743. [DOI] [PubMed] [Google Scholar]
- 38.Monte I., Ishida S., Zamarreño A.M., Hamberg M., Franco-Zorrilla J.M., García-Casado G., Gouhier-Darimont C., Reymond P., Takahashi K., García-Mina J.M. Ligand-receptor co-evolution shaped the jasmonate pathway in land plants. Nat. Chem. Biol. 2018;14:480–488. doi: 10.1038/s41589-018-0033-4. [DOI] [PubMed] [Google Scholar]
- 39.Bensmihen S., de Billy F., Gough C. Contribution of NFP LysM domains to the recognition of Nod factors during the Medicago truncatula/Sinorhizobium meliloti symbiosis. PLoS ONE. 2011;6:e26114. doi: 10.1371/journal.pone.0026114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ried M.K., Antolín-Llovera M., Parniske M. Spontaneous symbiotic reprogramming of plant roots triggered by receptor-like kinases. eLife. 2014;3 doi: 10.7554/eLife.03891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Czaja L.F., Hogekamp C., Lamm P., Maillet F., Martinez E.A., Samain E., Dénarié J., Küster H., Hohnjec N. Transcriptional responses toward diffusible signals from symbiotic microbes reveal MtNFP- and MtDMI3-dependent reprogramming of host gene expression by arbuscular mycorrhizal fungal lipochitooligosaccharides. Plant Physiol. 2012;159:1671–1685. doi: 10.1104/pp.112.195990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Buendia L., Maillet F., O’Connor D., van de-Kerkhove Q., Danoun S., Gough C., Lefebvre B., Bensmihen S. Lipo-chitooligosaccharides promote lateral root formation and modify auxin homeostasis in Brachypodium distachyon. New Phytol. 2019;221:2190–2202. doi: 10.1111/nph.15551. [DOI] [PubMed] [Google Scholar]
- 43.Amor B.B., Shaw S.L., Oldroyd G.E.D., Maillet F., Penmetsa R.V., Cook D., Long S.R., Dénarié J., Gough C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003;34:495–506. doi: 10.1046/j.1365-313x.2003.01743.x. [DOI] [PubMed] [Google Scholar]
- 44.Hohnjec N., Czaja-Hasse L.F., Hogekamp C., Küster H. Pre-announcement of symbiotic guests: transcriptional reprogramming by mycorrhizal lipochitooligosaccharides shows a strict co-dependency on the GRAS transcription factors NSP1 and RAM1. BMC Genomics. 2015;16:994. doi: 10.1186/s12864-015-2224-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rasmussen S.R., Füchtbauer W., Novero M., Volpe V., Malkov N., Genre A., Bonfante P., Stougaard J., Radutoiu S. Intraradical colonization by arbuscular mycorrhizal fungi triggers induction of a lipochitooligosaccharide receptor. Sci. Rep. 2016;6:29733. doi: 10.1038/srep29733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lanfranco L., Fiorilli V., Gutjahr C. Partner communication and role of nutrients in the arbuscular mycorrhizal symbiosis. New Phytol. 2018;220:1031–1046. doi: 10.1111/nph.15230. [DOI] [PubMed] [Google Scholar]
- 47.Parniske M. Intracellular accommodation of microbes by plants: a common developmental program for symbiosis and disease? Curr. Opin. Plant Biol. 2000;3:320–328. doi: 10.1016/s1369-5266(00)00088-1. [DOI] [PubMed] [Google Scholar]
- 48.Werner G.D., Cornwell W.K., Sprent J.I., Kattge J., Kiers E.T. A single evolutionary innovation drives the deep evolution of symbiotic N2-fixation in angiosperms. Nat. Commun. 2014;5:4087. doi: 10.1038/ncomms5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.van Velzen R., Holmer R., Bu F., Rutten L., van Zeijl A., Liu W., Santuari L., Cao Q., Sharma T., Shen D. Comparative genomics of the nonlegume Parasponia reveals insights into evolution of nitrogen-fixing rhizobium symbioses. Proc. Natl. Acad. Sci. USA. 2018;115:E4700–E4709. doi: 10.1073/pnas.1721395115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lerouge P., Roche P., Faucher C., Maillet F., Truchet G., Promé J.C., Dénarié J. Symbiotic host-specificity of Rhizobium meliloti is determined by a sulphated and acylated glucosamine oligosaccharide signal. Nature. 1990;344:781–784. doi: 10.1038/344781a0. [DOI] [PubMed] [Google Scholar]
- 51.Bek A.S., Sauer J., Thygesen M.B., Duus J.Ø., Petersen B.O., Thirup S., James E., Jensen K.J., Stougaard J., Radutoiu S. Improved characterization of nod factors and genetically based variation in LysM Receptor domains identify amino acids expendable for nod factor recognition in Lotus spp. Mol. Plant Microbe Interact. 2010;23:58–66. doi: 10.1094/MPMI-23-1-0058. [DOI] [PubMed] [Google Scholar]
- 52.De Mita S., Streng A., Bisseling T., Geurts R. Evolution of a symbiotic receptor through gene duplications in the legume-rhizobium mutualism. New Phytol. 2014;201:961–972. doi: 10.1111/nph.12549. [DOI] [PubMed] [Google Scholar]
- 53.Maudoux O., Batoko H., Oecking C., Gevaert K., Vandekerckhove J., Boutry M., Morsomme P. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J. Biol. Chem. 2000;275:17762–17770. doi: 10.1074/jbc.M909690199. [DOI] [PubMed] [Google Scholar]
- 54.Quandt H.J., Puhler A., Broer I. Transgenic root-nodules of Vicia-hirsuta: a fast and efficient system for the study of gene-expression in indeterminate-type nodules. Mol. Plant Microbe Interact. 1993;6:699–706. [Google Scholar]
- 55.Ardourel M., Demont N., Debellé F., Maillet F., de Billy F., Promé J.C., Dénarié J., Truchet G. Rhizobium meliloti lipooligosaccharide nodulation factors: different structural requirements for bacterial entry into target root hair cells and induction of plant symbiotic developmental responses. Plant Cell. 1994;6:1357–1374. doi: 10.1105/tpc.6.10.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maekawa-Yoshikawa M., Müller J., Takeda N., Maekawa T., Sato S., Tabata S., Perry J., Wang T.L., Groth M., Brachmann A., Parniske M. The temperature-sensitive brush mutant of the legume Lotus japonicus reveals a link between root development and nodule infection by rhizobia. Plant Physiol. 2009;149:1785–1796. doi: 10.1104/pp.108.135160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Fits L., Deakin E.A., Hoge J.H.C., Memelink J. The ternary transformation system: constitutive virG on a compatible plasmid dramatically increases Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;43:495–502. doi: 10.1023/a:1006440221718. [DOI] [PubMed] [Google Scholar]
- 58.Sejalon-Delmas N., Magnier A., Douds D., Becard G. Cytoplasmic autofluorescence of an arbuscular mycorrhizal fungus Gigaspora gigantea and nondestructive fungal observations in planta. Mycologia. 1998;90:921–926. [Google Scholar]
- 59.Poupot R., Martinez-Romero E., Promé J.C. Nodulation factors from Rhizobium tropici are sulfated or nonsulfated chitopentasaccharides containing an N-methyl-N-acylglucosaminyl terminus. Biochemistry. 1993;32:10430–10435. doi: 10.1021/bi00090a019. [DOI] [PubMed] [Google Scholar]
- 60.Stuurman J., Kuhlemeier C. Stable two-element control of dTph1 transposition in mutator strains of Petunia by an inactive ACT1 introgression from a wild species. Plant J. 2005;41:945–955. doi: 10.1111/j.1365-313X.2005.02340.x. [DOI] [PubMed] [Google Scholar]
- 61.Madsen E.B., Madsen L.H., Radutoiu S., Olbryt M., Rakwalska M., Szczyglowski K., Sato S., Kaneko T., Tabata S., Sandal N., Stougaard J. A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature. 2003;425:637–640. doi: 10.1038/nature02045. [DOI] [PubMed] [Google Scholar]
- 62.Pietraszewska-Bogiel A., Lefebvre B., Koini M.A., Klaus-Heisen D., Takken F.L.W., Geurts R., Cullimore J.V., Gadella T.W.J. Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS ONE. 2013;8:e65055. doi: 10.1371/journal.pone.0065055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kirienko A.N., Porozov Y.B., Malkov N.V., Akhtemova G.A., Le Signor C., Thompson R., Saffray C., Dalmais M., Bendahmane A., Tikhonovich I.A., Dolgikh E.A. Role of a receptor-like kinase K1 in pea Rhizobium symbiosis development. Planta. 2018;248:1101–1120. doi: 10.1007/s00425-018-2944-4. [DOI] [PubMed] [Google Scholar]
- 64.Lefebvre B., Batoko H., Duby G., Boutry M. Targeting of a Nicotiana plumbaginifolia H+ -ATPase to the plasma membrane is not by default and requires cytosolic structural determinants. Plant Cell. 2004;16:1772–1789. doi: 10.1105/tpc.022277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Katoh K., Standley D.M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Capella-Gutiérrez S., Silla-Martínez J.M., Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kalyaanamoorthy S., Minh B.Q., Wong T.K.F., von Haeseler A., Jermiin L.S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nguyen L.T., Schmidt H.A., von Haeseler A., Minh B.Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Guindon S., Dufayard J.F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 71.Letunic I., Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1) doi: 10.1093/nar/gkw290. W242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bailey T.L., Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 73.Sevin-Pujol A., Sicard M., Rosenberg C., Auriac M.C., Lepage A., Niebel A., Gough C., Bensmihen S. Development of a GAL4-VP16/UAS trans-activation system for tissue specific expression in Medicago truncatula. PLoS ONE. 2017;12:e0188923. doi: 10.1371/journal.pone.0188923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lefebvre B., Timmers T., Mbengue M., Moreau S., Hervé C., Tóth K., Bittencourt-Silvestre J., Klaus D., Deslandes L., Godiard L. A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. USA. 2010;107:2343–2348. doi: 10.1073/pnas.0913320107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Binder A., Lambert J., Morbitzer R., Popp C., Ott T., Lahaye T., Parniske M. A modular plasmid assembly kit for multigene expression, gene silencing and silencing rescue in plants. PLoS ONE. 2014;9:e88218. doi: 10.1371/journal.pone.0088218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chabaud M., Boisson-Dernier A., Zhang J., Taylor C.G., Yu O., Barker D.G. Agrobacterium rhizogenes-mediated root transformation. In The Medicago truncatula handbook, U. Mathesius, In: Journet E.P., Sumner L.W., editors. Noble Research Institute; 2006. pp. 1–8. [Google Scholar]
- 77.Charpentier M., Bredemeier R., Wanner G., Takeda N., Schleiff E., Parniske M. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell. 2008;20:3467–3479. doi: 10.1105/tpc.108.063255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Becard G., Fortin J.A. Early events of vesicular-arbuscular mycorrhiza formation on Ri T-DNA transformed roots. New Phytol. 1988;108:211–218. doi: 10.1111/j.1469-8137.1988.tb03698.x. [DOI] [PubMed] [Google Scholar]
- 79.Mbengue M., Camut S., de Carvalho-Niebel F., Deslandes L., Froidure S., Klaus-Heisen D., Moreau S., Rivas S., Timmers T., Hervé C. The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell. 2010;22:3474–3488. doi: 10.1105/tpc.110.075861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gressent F., Cullimore J.V., Ranjeva R., Bono J.J. Radiolabeling of lipo-chitooligosaccharides using the NodH sulfotransferase: a two-step enzymatic procedure. BMC Biochem. 2004;5:4. doi: 10.1186/1471-2091-5-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Rich M.K., Schorderet M., Bapaume L., Falquet L., Morel P., Vandenbussche M., Reinhardt D. The Petunia GRAS Transcription Factor ATA/RAM1 Regulates Symbiotic Gene Expression and Fungal Morphogenesis in Arbuscular Mycorrhiza. Plant Physiol. 2015;168:788–797. doi: 10.1104/pp.15.00310. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
This study did not generate any unique datasets or code.