Summary
Background
The Middle East respiratory syndrome coronavirus (MERS-CoV) is a lethal zoonotic pathogen endemic to the Arabian Peninsula. Dromedary camels are a likely source of infection and the virus probably originated in Africa. We studied the genetic diversity, geographical structure, infection prevalence, and age-associated prevalence among camels at the largest entry port of camels from Africa into the Arabian Peninsula.
Methods
In this prospective genomic study, we took nasal samples from camels imported from Sudan and Djibouti into the Port of Jeddah in Jeddah, Saudi Arabia, over an almost 2-year period and local Arabian camels over 2 months in the year after surveillance of the port. We determined the prevalence of MERS-CoV infection, age-associated patterns of infection, and undertook phylogeographical and migration analyses to determine intercountry virus transmission after local lineage establishment. We compared all virological characteristics between the local and imported cohorts. We compared major gene deletions between African and Arabian strains of the virus. Reproductive numbers were inferred with Bayesian birth death skyline analyses.
Findings
Between Aug 10, 2016, and May 3, 2018, we collected samples from 1196 imported camels, of which 868 originated from Sudan and 328 from Djibouti, and between May 1, and June 25, 2018, we collected samples from 472 local camels, of which 189 were from Riyadh and 283 were from Jeddah, Saudi Arabia. Virus prevalence was higher in local camels than in imported camels (224 [47·5%] of 472 vs 157 [13·1%] of 1196; p<0·0001). Infection prevalence peaked among camels older than 1 year and aged up to 2 years in both groups, with 255 (66·9%) of 381 positive cases in this age group. Although the overall geographical distribution of the virus corresponded with the phylogenetic tree topology, some virus exchange was observed between countries corresponding with trade routes in the region. East and west African strains of the virus appear to be geographically separated, with an origin of west African strains in east Africa. African strains of the virus were not re-sampled in Saudi Arabia despite sampling approximately 1 year after importation from Africa. All local Arabian samples contained strains of the virus that belong to a novel recombinant clade (NRC) first detected in 2014 in Saudi Arabia. Reproduction number estimates informed by the sequences suggest sustained endemicity of NRC, with a mean Re of 1·16.
Interpretation
Despite frequent imports of MERS-CoV with camels from Africa, African lineages of MERS-CoV do not establish themselves in Saudi Arabia. Arabian strains of the virus should be tested for changes in virulence and transmissibility.
Funding
German Ministry of Research and Education, EU Horizon 2020, and Emerging Diseases Clinical Trials Partnership.
Introduction
The Middle East respiratory syndrome coronavirus (MERS-CoV) is a priority zoonotic pathogen listed in the WHO R&D Blueprint for 2018 because of its epidemic potential, high case fatality rate, and no available treatment or vaccine.1
As of Aug 2, 2019, 2468 laboratory-confirmed cases of MERS, with 851 deaths (34·5% mortality) had been reported to WHO since September, 2012, globally.2 2090 (84%) of these cases occurred in Saudi Arabia and the largest outbreak outside of Saudi Arabia occurred in South Korea in May, 2015, with 186 cases and 36 deaths reported.2 The number of cases in Saudi Arabia and Oman has recently increased, with 126 cases reported in January–March, 2019, compared with 189 cases reported from July, 2017, to June, 2018.2 Discovered in 2012, MERS-CoV continues to circulate in the Middle East and remains a threat to global health security. Despite many WHO scoping reviews and stakeholder meetings defining urgent priority research needs, major knowledge gaps in the epidemiology, transmission, pathogenesis, and evolution of MERS-CoV remain.1, 3
Research in context.
Evidence before this study
We searched PubMed, Web of Science, and Google Scholar for studies on the prevalence and diversity of Middle East respiratory syndrome coronavirus (MERS-CoV) infection from database inception until May 30, 2019, without language restrictions. We used the term “MERS*” combined with any single other term from the following list: “coronavirus*”, “camels*”, “dromedaries*”, “recombinant*”, “phylogeny*”, “phylogeography”, “Africa*”, “sequenc*”, “prevalence”, “age”, and “transmission”. Since the discovery of MERS-CoV in 2012, multiple sequencing studies have been done on viruses from camels and humans mainly in the Arabian Peninsula. Few studies exist on sequences from Africa, but all of these sequences are from camels rather than from humans, whereas most of the sequences from the Arabian Peninsula are from humans. Sampling bias is likely to affect all studies. The number of studies, and hence samples collected, from Africa is small compared with those from the Arabian Peninsula.
Added value of this study
We took advantage of sampling opportunities at the Port of Jeddah, in Jeddah, Saudi Arabia, where large numbers of camels are continuously imported from Africa. By sampling before offloading from ships we made sure to take samples from animals that came directly from Africa and had no contact with local camels in Saudi Arabia. To our knowledge, the resulting sample of African camel-borne MERS-CoV is the largest so far from the African continent. Our data enhance the overall picture of African strains of the virus, including the phylogenetic and geographical associations, which has enabled us to undertake comparisons of diversity against representatively large samples from the Arabian Peninsula. Our comparisons take sampling dates into account. We infer that Arabian and African strains of the virus have been separated for a time that exceeds the present observation period in all studies (ie, the virus strains have been separated since before 2012). We suggest that African strains of the virus are not transmitted in Arabian camels, despite large numbers of African camels frequently being imported into Saudi Arabia; and that Arabian strains of the virus might be self-sufficient in terms of their reproductive rate to maintain endemic circulation. By sampling from local camels in Saudi Arabia, we found that a novel clade of MERS-CoV that emerged in 2014 remains the only detected viral variant in camels from Saudi Arabia, suggesting that strains of this clade might be dominant over other viral strains that were co-endemic in camels before 2014.
Implications of all the available evidence
Our results suggest that a recently emerged clade of MERS-CoV in Saudi Arabia should be compared with African strains of the virus and with older Arabian strains to address the question of a potential increase in transmissibility and virulence. Change in transmissibility and virulence would affect the general assessment of pandemic risks emanating from MERS-CoV.
MERS-CoV seroprevalence among the general human population in Saudi Arabia is less than 0·5%, although it is substantially higher in camel shepherds (2·3%) and slaughterhouse workers (3·6%).4 MERS-CoV is highly prevalent in dromedary camels on the Arabian Peninsula and dromedary camels are the likely source of primary human MERS-CoV infections.5
Serological and nucleic acid-based evidence suggests that dromedary camels from Africa and Asia have harboured MERS-CoV for more than 35 years.6, 7, 8 The high diversity of MERS-CoV in African camels and the existence of a conspecific virus in African bats point to its geographical roots in Africa.8, 9, 10 However, the geographical structure (in terms of phylogeography) of African MERS-CoV remains understudied.
Farming and trade of dromedary camels has increased over the past three decades in and between the Middle East and Africa. A large proportion of dromedary camels in the Middle East are imported from east African countries, where 19 million of the world's estimated population of 30 million dromedary camels reside.11 MERS-CoV strains identified in African camels are genetically distinct from strains detected in camels in the Arabian Peninsula.8, 9 In Africa, east and west African strains of MERS-CoV can be discriminated by their genetic markers that include accessory gene deletions that might affect the extent of virus replication or virulence.8
Despite the continuous, extensive, and unidirectional export of African dromedaries, whether African MERS-CoV lineages reach the Arabian Peninsula and can be transmitted onward remains unknown. An important aspect of potential transmission is the age structure of imported camels, which are usually adult animals but can also include animals shipped towards the end of their first year of life. In large husbandries on the Arabian Peninsula, acute MERS-CoV infection mostly occurs in young camels younger than 1 year.12 Age at the time of infection might differ among imported camels because of different husbandry practices across the distribution area, with additional effects due to cohorting and mixing during animal transportation.11, 13
The seaport at Port of Jeddah, in Jeddah, Saudi Arabia, receives animals from east Africa linked with the major trading routes in the Horn of Africa, Egypt, and from the trading route in the Sahel region connected with regions in west Africa.11 We undertook a study of the pattern of infection age, infection prevalence, and genetic diversity of MERS-CoV in camels being imported from Africa (Sudan and Djibouti) into the Port of Jeddah, the largest entry port of camels into Saudi Arabia. We also analysed the genetic diversity of the virus strains identified, and compared all virological characteristics of imported camels with local dromedaries sampled during the year after surveillance at the port.
Methods
Study design and cohorts
In this prospective genomic study, we obtained respiratory samples from two different cohorts of dromedary camels. The first cohort comprised dromedary camels imported into Saudi Arabia on incoming vessels from Sudan or Djibouti at the Port of Jeddah. Samples were obtained over an almost 2-year period and were collected from about 15% of all camels aboard a ship. Sampling took place by entering the animal compartment and sampling accessible animals at random. Often ships held camels in two separate compartments, and so for our study we took samples from camels in both compartments. The second cohort comprised local camels from herds in Jeddah and Riyadh, Saudi Arabia. Local samples were collected after the surveillance at the Port of Jeddah. In Jeddah, we collected 30% of samples from three camel farms approximately 45 km south of Jeddah and one farm approximately 50 km north of Jeddah, while we collected the other 70% of samples at an abattoir in the city before the camels were slaughtered. In Riyadh, we collected samples from camel farms and a camel market in a circle with a 60 km radius in and around the city.
Permission for this research was granted by the Indian Ministry of Environment, Water and Agriculture.
Procedures
We took nasal swabs from all camels using dacron swabs in viral transport medium from Vircell (Granada, Spain) and stored them on ice during transportation to the Special Infectious Agents Unit laboratory in Jeddah, Saudi Arabia, where they were stored at −80°C until they were thawed and used for RNA extraction and MERS-CoV testing.
Viral RNA was extracted with the MagnaNApure compact system (Roche, Penzberg, Germany) using 200 μL of the viral transport medium sample and eluted in 50 μL of elution buffer. Screening for MERS-CoV was done in accordance with the WHO interim guidelines for laboratory testing for MERS-CoV case definition, with RT-PCR assays targeting two different genomic targets (the upE region in the E gene and open reading frame [ORF1A] in the ORF1a gene) as described before.14, 15
Three genome regions upstream and downstream of known recombination breakpoints,16, 17 including the ORF4b region carrying deletions in African viruses,8 were amplified with established protocols (full list of genome regions is in the appendix [p 1]).10 After initial phylogenetic analyses for preliminary genotyping, several samples representing all African clades of the virus and several samples from Saudi Arabia were chosen on the basis of initial viral load quantification (appendix p 1) for full genome analysis using a combined RT-PCR and unbiased, non-amplified sequencing approach in an Illumina MiSeq instrument using 2 × 300 bp paired-end reads chemistry, as outlined in the appendix (p 1). The resulting sequences were assembled into full or near-full genome scaffolds and analysed for recombination.
Novel strains of African viruses identified in our cohort were combined with representative members of MERS-CoV Arabian clades A and B17 and all African clade C (non-A and non-B) MERS-CoV complete genomes that had been published as of Feb 1, 2019.8
Statistical analysis
We undertook phylogenetic analyses using Beast 1.10.4 software, with time-stamping based on the sampling dates of viral sequences; parameters used are listed in the appendix (pp 1–2).18 We applied discrete phylogeographic diffusion models to the full dataset of viral strains to identify signs of geographical migration after the establishment of viral phylogenetic lineages, as has been previously published.19 We undertook analyses and visualisations of the geographical association of phylogenetic lineages and the geographical diffusion process using SpreaD3,20 and we used RDP version 4.95 for recombination analyses.21 To study viral transmission dynamics, we did an analysis of reproductive number on a curated dataset of Arabian viral genome sequences listed in the appendix (pp 7–9). This analysis involved Bayesian birth death skyline analyses in Beast2,22 using previous assumptions of parameters in accordance with the methods of Dudas and colleagues (full details are in the appendix [pp 1–3]).23 We repeated our phylogenetic analysis to determine the robustness of our model.
To test the potential uncertainty introduced by use of recombinant sequences and the additional sequence information contributed to the phylogenetic tree, we repeated the phylogenetic analysis using only a non-recombinant sequence fragment (genome positions 91–11 343 in full genomes and 10 224–11 343 in partial sequences). This additional analysis generated essentially the same result with somewhat less statistical support for tree nodes at higher tree nodes but had no influence on main lineage separation (data not shown). Therefore, for the rest of analyses, we used our original full dataset.
We did all other statistical analyses using GraphPad Prism version 7. For comparison of detection rates in different groups we used the χ2 test without Yates' correction.
Role of the funding source
The funders of the study had no role in study design, data collection, data analysis, data interpretation, or writing of the report. VMC, SAE-K, CD, AZ, and EIA had access to all data and had final responsibility for the decision to submit for publication.
Results
Between Aug 10, 2016, and May 3, 2018, 1196 samples were obtained from imported camels before being offloaded at the Port of Jeddah, of which 868 originated from Sudan and 328 from Djibouti. And between May 1, and June 25, 2018, we obtained samples from 472 local camels, 189 in Riyadh and 283 in Jeddah, Saudi Arabia (table ). Testing by RT-PCR identified MERS-CoV in 381 (22·8%) of 1668 animals sampled, and the prevalence of MERS-CoV was significantly higher among local camels than imported camels (224 [47·5%] of 472 local camels vs 157 [13·1%] of 1196 imported camels; χ2 test p<0·0001).
Table.
MERS-CoV detection per cohort and by age group of dromedary camels
| Sudan (n=868) | Djibouti (n=328) | Riyadh, Saudi Arabia (n=189) | Jeddah, Saudi Arabia (n=283) | |||
|---|---|---|---|---|---|---|
| Tested positive | 120 (13·8%) | 37 (11·3%) | 133 (70·4%) | 91 (32·2%) | ||
| Age | ||||||
| ≤1 year | 17 (2·0%) | 0 | 0 | 31 (11·0%) | ||
| Tested positive | 1/17 (6%) | .. | .. | 3/31 (10%) | ||
| >1–2 years | 322 (37·1%) | 25 (7·6%) | 189 (100%) | 85 (30·0%) | ||
| Tested positive | 86/322 (27%) | 0 | 133/189 (70%) | 36/85 (42%) | ||
| >2–5 years | 298 (34·3%) | 280 (85·4%) | 0 | 167 (59·0%) | ||
| Tested positive | 33/298 (11%) | 37/280 (13%) | .. | 52/167 (31%) | ||
| Unknown | 231 (26·6%) | 23 (7·0%) | 0 | 0 | ||
| Tested positive | 0 | 0 | .. | .. | ||
Data are n (%) or n/N (%). Camels older than 5 years might be included in the unknown age category. MERS-CoV=Middle East respiratory syndrome coronavirus.
The age range of the animals was between 6 months and 5 years. MERS-CoV RNA detection was pronounced between those older than 1 year and up to age 2 years (χ2 test p<0·0001). This difference was significant for both the imported and local cohorts in which samples from all the different age groups were available—ie, Sudan (χ2 test p<0·0001) and Jeddah (χ2 test p<0·0035; table).
We sequenced 124 viruses via RT-PCR and initial sequencing of the RT-PCR products suggested diverse clade associations. Based on preliminary phylogenetic analyses, we chose 46 individual samples (22 from the imported cohort and 24 of the Arabian samples) were chosen for full genome sequencing. All sequences were deposited under GenBank accession numbers MN541181–1304 (appendix pp 1–2, 5–6). Analysis of the assembled full or near-full genome scaffolds for recombination identified recombination signals for only one lineage resulting from recombination between previously circulating MERS-CoV lineages 3 and 5 (termed as a novel recombinant clade [NRC]) that emerged in Saudi Arabia in 2014–15.17 All 24 MERS-CoV strains sampled from domestic dromedary camels in Saudi Arabia belonged to this NRC.
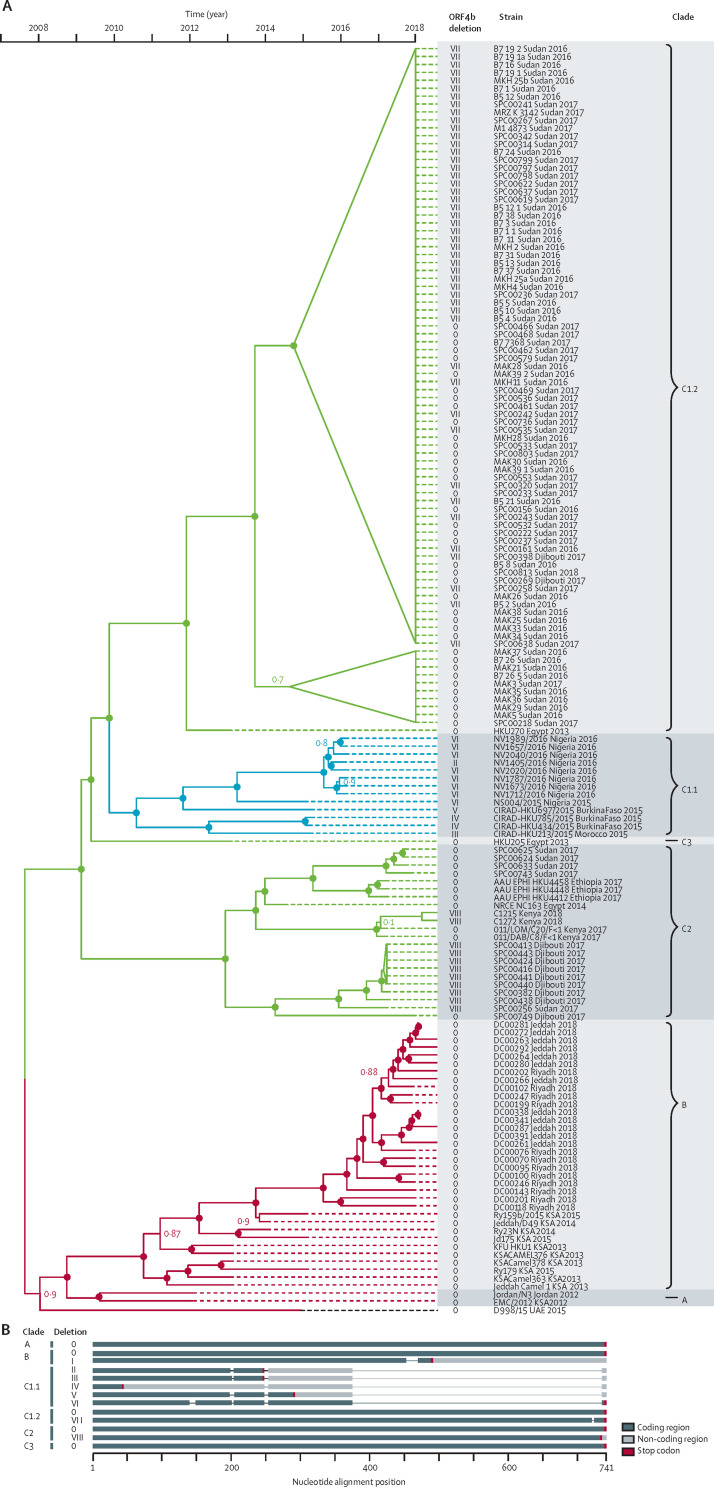
All viruses from imported camels belonged to clade C, which was exclusively found in Africa in previous studies.8, 9, 24 A time-stamped phylogenetic tree including all novel sequences from imported camels is shown in figure 1 .
Figure 1.
Phylogenetic and geographical attribution of MERS-CoV strains in camels imported into Saudi Arabia
(A) Time-stamped phylogenetic tree indicating ORF4b region deletion types. Dots at tree nodes indicate statistically high (>95%) node support and numbers indicate statistically lower (<95%) node support, as indicated by the number. GenBank accession numbers are given in the appendix (pp 5–6). (B) Deletion patterns in ORF4b region. The diagram shows the variants found in clade C sequences (C1.1, C1.2, C2, and C3) of ORF4b compared with clade A and B sequences. MERS-CoV=Middle East respiratory syndrome coronavirus.
Our phylogenetic results support subclassification of African MERS-CoV lineages. Our results support the designation of clade C for all African strains of the virus. One subclade, designated C1.1, was previously found to contain strains from west Africa including from Nigeria, Burkina Faso, and Morocco (designated as C1 by Chu and colleagues8). This clade has a novel sister clade here designated as C1.2, including sequences from Sudan and Djibouti found in the present study. With a similar internal diversity as clades C1.1 and C1.2, a novel subclade is identified, C2, that contains multiple novel viruses from the present study derived from Sudan and Djibouti, and viruses previously detected by us and others in Egypt, Ethiopia, and Kenya.8, 9, 25 A strain previously detected in Egypt is highly distinct and designates a novel clade C3.25
Deletions in the ORF4b region have occurred recently in parallel lineages of African strains of the virus but not Arabian strains (figure 1). Deletion types are specific for their respective clades, cluster with viral strains that carry wild-type ORF4b genes, and are highly region specific, indicating that geographical spread of these recent variants has not occurred (the topology of deletion type VII does not indicate convergence because it is merely deletion of one amino acid at the C-terminus). Viruses from Nigeria (C1.1) show particularly large deletions (types II–VI) that are not observed in other clades and suggest the geographical isolation of west African clades. Two sequences described in Saudi Arabia in 2012 (Bisha_1_2012; KF600620 and Riyadh_1_2012; KF600612.1) both belonging to clade B also show deletions (type 1) in ORF4b region, but with a distinct location.
In the ORF3 region, we also observed some variations in the sequence of clade C3. Four of six sequences originating in Djibouti showed a 13 nucleotide deletion at the 3′-terminal part of ORF3, resulting in a new stop codon and an ORF3 predicted to be extended by six amino acids. In the same genomic region, one sequence also from Djibouti showed deletion of two nucleotides, resulting in a premature stop codon and a predicted truncated ORF3 (six amino acids shorter). Deletions and insertions at the 3′-terminus of ORF3 have been described before in viral strains from Nigeria, Burkina Faso (belonging to clade C1.1),8 and the United Arab Emirates,26 and sequences obtained during a hospital outbreak of MERS-CoV in Jordan in 2015 (clade B).27
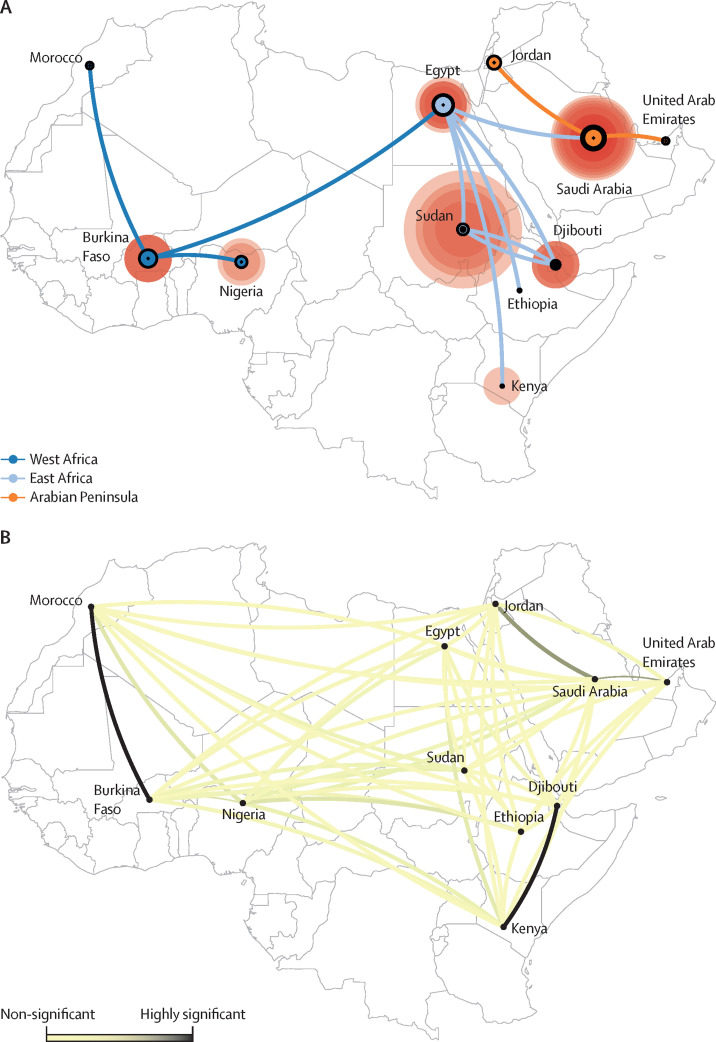
Having inferred the structure of the viral phylogenetic tree, we aimed to establish the extent of virus transmission between countries and how it has affected the present geographical distribution of the virus. We did a phylogeographical analysis of between-location migration in discrete space with determination of significant migration rate parameters based on a Bayesian stochastic search variable selection procedure (figure 2 ).19 The most significant indications for migration were obtained between Saudi Arabia and Jordan, Saudi Arabia and the United Arab Emirates, and Djibouti and Kenya. A high migration rate was also inferred between Burkina Faso and Morocco, but this rate was based on only one sequence and is therefore uncertain (appendix p 10). The overall result of the analysis suggests a tree structure that adheres to geographical distribution of most, if not all, viral strains.
Figure 2.
Phylogeographic and social network structure of MERS-CoV strains
(A) Maximum clade credibility tree projected on a map of the study region. Outer circle sizes represent number of taxa associated with the country and inner circle sizes indicate deepest node per country, with a larger circle indicating a deeper node. (B) Social network inferred by Bayesian stochastic search variable selection approach. Only one network connection (Burkina Faso to Morocco) is identified as highly significant migration that does not adhere to the tree structure, based on Bayes factor. Significant indications for migration were obtained for Djibouti to Kenya, Saudi Arabia to the United Arab Emirates, and Saudi Arabia to Jordan. MERS-CoV=Middle East respiratory syndrome coronavirus.
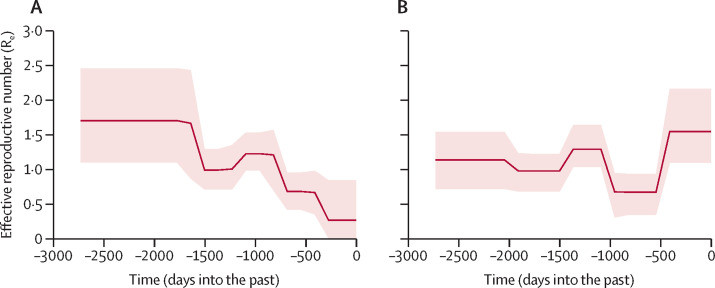
Despite the obvious importation of African virus lineages into Saudi Arabia, these viruses were not observed in the local camels under study in Saudi Arabia. Because previous studies have shown that serotype discrimination does not exist between MERS-CoV clades, our findings suggests that viruses in Saudi Arabia can remain endemic without introduction of new strains from Africa.8 Therefore, we focused on the reproductive rate of viruses in Saudi Arabia. Using a curated dataset of Arabian strains of the virus, we subjected all strains of the virus belonging to clade B, whether derived from humans or camels, to analyses of Re based on Bayesian birth death skyline analyses. We used this approach to obtain a dataset as complete as possible from a virus population that is close to the requirements of coalescent analysis—namely, panmixis and contingency as a population. Human-derived viral sequences were selected in such a way that they most likely represent primary cases; human-derived viruses were thus regarded as sentinels for enzootic evolution in camels, as previously reported.23 We only used the non-recombinant part of clade A and B virus genomes for this analysis because all recent isolates belonged to the NRC. The overall estimate of Re had a mean of 1·16, compatible with sustained endemicity. Re increased in the period between early 2017 and the middle of 2018, which we associate with the increased sampling effort by our study. We undertook two analyses of Re, one not including the novel viruses identified in this study (figure 3A ) and the other including the full curated dataset (figure 3B). By omitting the new sequences, the Re of MERS-CoV seems to decrease over time, highlighting that inclusion of new sequences is important to maintain an accurate picture of viral spread. Notably, this result is subject to previous assumptions made in our analyses.
Figure 3.
Reconstruction of reproductive number of circulating virus based on strains sampled in the Arabian Peninsula
(A) Analysis based on a dataset that excludes the novel viruses contributed by the present study. (B) Analysis including full dataset. Solid lines are estimates, with Bayesian 95% confidence limits indicated by shaded areas. Full data on cohort are in the appendix (pp 7–9).
Discussion
The present study provides further insight into the diversity of MERS-CoV. By analysing a large number of strains of MERS-CoV imported into Saudi Arabia from major African trade ports, we showed that Arabian strains of the virus are isolated from African strains and identify African MERS-CoV clades that were not as clearly distinguished in previous studies. Although all African strains of the virus share common ancestors in east Africa, only clade C1 presently appears in west and north Africa. Even if all African clades are represented in east Africa, no single place sampled so far in east Africa represents the whole diversity of MERS-CoV variants. Phylogeographical and social network analyses suggest that the present distribution of African clades is not predominantly shaped by transregional exchange but mainly reflects the phylogenetic tree structure. Camel trade, such as via the Sahel route connecting east and west Africa, corresponds with the present location of C1.1 strains but does not seem to cause a bidirectional exchange of strains between east and west Africa. Only for the clustering of Moroccan with west African strains and some strains from Kenya and Djibouti, the geographical distance between sites exceeds that of the attributed genetic distance, suggesting recent exchange through trade routes. Our phylogenetic tree structure shows that only parts of the diversity of clades C1 and C3 are represented in Sudan. Recombination between these clades is not observed, despite co-occurrence in Sudan. Therefore, Sudan appears to be a frequent recipient of viral lineages from other regions although we did not identify any strictly Sudan-specific lineages.
For African strain of the virus, our data confirm that the genome regions coding for the ORF4b and ORF3 regions have genetic instability. ORF4b encodes a phosphodiesterase that degrades 2′-5′-oligoadenylate and thereby prevents the activation of RNase L, an important cellular antiviral effector.28, 29 The importance of this function for the spread of MERS-CoV and its ability to infect organisms under natural conditions is of interest. In-vitro studies suggest an attenuation of replication in viruses whose ORF4b region has undergone deletions.8 One could speculate that slightly deleterious mutants can sustain themselves better in so-called sink populations, whereas in source populations they would typically be out-competed by non-deleted strains that have higher reproductive fitness. Our data in combination with those of Chu and colleagues8 suggest that sink populations of MERS-CoV are hosted by west African camels and the possible geographical origin of all African MERS-CoV clades, thus the so-called source population is found in east Africa. The lesser density of camels in west Africa than in east Africa and the Arabian Peninsula supports the idea of an evolutionary dead end for MERS-CoV in that region. Particularly, Nigeria could be considered a dead end or sink area given the reduced density of camels towards the tropical zone of west Africa.
For the strains of the virus detected in Saudi Arabia, our study suggests that Arabian and African viruses have been separated for a time that exceeds the present observation period in all studies (ie, viruses have been separated since before 2012). Despite the steady import of clade C viruses, these African lineages do not appear to establish themselves in Saudi Arabia. We were surprised to see a similar age structure of imported compared with local virus-positive camels, suggesting that age-related contact barriers in husbandry would not explain the absence of transmission; however, notably, approximately 15% of sampled animals were of unknown age. Future studies to understand barriers to transmission should examine local camel populations in direct contact with imported camels, including their susceptibility based on serological testing.
Our analysis of population dynamics suggests that Arabian viruses can maintain endemic status without introduction of additional lineages. This finding corresponds with the observed isolation of Arabian virus lineages; however, these models are influenced by our prior assumptions, and therefore should be taken as a qualitative comparison of the contribution of present data rather than an exact quantitative assessment of R. Also, the point in time since Arabian strains of the virus have been isolated is impossible to infer because of our sample was restricted to virus samples from Africa and the Arabian Peninsula. Future work with a wider sampling timeframe would be able to involve more refined clock models. Nevertheless, our results provide a reminder that continuous surveillance is necessary to trace and potentially discover changes in endemic activity.
Our study had several limitations including restricted geographical coverage in terms of camels studied in Africa and Saudi Arabia, an absence of age-related data in 15% of camels, and an absence of data on local MERS-CoV circulating strains in Sudan. Thus, some of our interpretations might need to be updated based on intensified sampling efforts in the future.
All MERS-CoV strains we isolated in Saudi Arabia belong to clade NRC circulating since 2014.17 Previously, clades in camels have subsequently been replaced by other clades, corresponding to a pattern of enzootic acute infections with short waves of prevailing individual subtypes.17, 30 Although our data are not representative for all of Saudi Arabia, the evidence we collected by sampling from two sites as remote from each other as Jeddah and Riyadh indicates prolonged circulation of the recombinant MERS-CoV clade as a dominant strain. The increased detection rate of MERS-CoV RNA in camels from the Arabian Peninsula reported in this and previous studies17, 23 points towards differences in transmission dynamics and selection pressure. Further studies across a wider geographical area should be undertaken to understand potential changes in virulence and transmissibility associated with this strain. These studies should include direct comparisons between African and Arabian strains.
Acknowledgments
Acknowledgments
The work was supported by the German Ministry of Research and Education (grant number 01KI1723A) and the EU Horizon 2020 project Compare. EIA thanks the King Fahd Medical Research Center and the King Abdullah University of Science and Technology (KAUST) for support. AZ and CD are members of the Pan-African Network on Emerging and Re-emerging Infections and thank the European and Developing Countries Clinical Trials Partnership for support under EU Horizon 2020, the EU's Framework Programme for Research and Innovation. AZ has received a National Institutes of Health Research senior investigator award.
Contributors
EIA, CD, VMC, SAE-K, and AZ conceived, designed, and coordinated the study. AAA, GAA, and ANA were involved in collection of samples from camels and data collection. EIA, SBAM, AMT, SAE-K, AMH, MAM, VMC, and TB did virological testing and sequencing. CD and VMC did phylogenetic analyses. All authors reviewed the data. CD, VMC, EIA, SAE-K, and AZ developed the first draft of the manuscript. All authors contributed to writing and finalising the manuscript and agreed to submit for publication.
Declaration of interests
All authors have an academic interest in zoonotic diseases. We declare no competing interests.
Contributor Information
Christian Drosten, Email: christian.drosten@charite.de.
Esam I Azhar, Email: eazhar@kau.edu.sa.
Supplementary Material
References
- 1.Mehand MS, Al-Shorbaji F, Millett P, Murgue B. The WHO R&D Blueprint: 2018 review of emerging infectious diseases requiring urgent research and development efforts. Antiviral Res. 2018;159:63–67. doi: 10.1016/j.antiviral.2018.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.WHO . World Health Oragnization; Geneva: 2019. Middle East respiratory syndrome coronavirus (MERS-CoV)https://www.who.int/emergencies/mers-cov/en/ [Google Scholar]
- 3.Kelly-Cirino C, Mazzola LT, Chua A, Oxenford CJ, Van Kerkhove MD. An updated roadmap for MERS-CoV research and product development: focus on diagnostics. BMJ Glob Health. 2019;4(suppl 2) doi: 10.1136/bmjgh-2018-001105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Müller MA, Meyer B, Corman VM. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis. 2015;15:559–564. doi: 10.1016/S1473-3099(15)70090-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alraddadi BM, Watson JT, Almarashi A. Risk factors for primary Middle East respiratory syndrome coronavirus illness in humans, Saudi Arabia, 2014. Emerg Infect Dis. 2016;22:49–55. doi: 10.3201/eid2201.151340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Saqib M, Sieberg A, Hussain MH. Serologic evidence for MERS-CoV infection in dromedary camels, Punjab, Pakistan, 2012–2015. Emerg Infect Dis. 2017;23:550–551. doi: 10.3201/eid2303.161285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Müller MA, Corman VM, Jores J. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis. 2014;20:2093–2095. doi: 10.3201/eid2012.141026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chu DKW, Hui KPY, Perera RAPM. MERS coronaviruses from camels in Africa exhibit region-dependent genetic diversity. Proc Natl Acad Sci USA. 2018;115:3144–3149. doi: 10.1073/pnas.1718769115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kiambi S, Corman VM, Sitawa R. Detection of distinct MERS-Coronavirus strains in dromedary camels from Kenya, 2017. Emerg Microbes Infect. 2018;7:195. doi: 10.1038/s41426-018-0193-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Corman VM, Ithete NL, Richards LR. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297–11303. doi: 10.1128/JVI.01498-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Younan M, Bornstein S, Gluecks IV. MERS and the dromedary camel trade between Africa and the Middle East. Trop Anim Health Prod. 2016;48:1277–1282. doi: 10.1007/s11250-016-1089-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer B, Juhasz J, Barua R. Time course of MERS-CoV infection and immunity in dromedary camels. Emerg Infect Dis. 2016;22:2171–2173. doi: 10.3201/eid2212.160382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abdallah HR, Faye B. Typology of camel farming system in Saudi Arabia. Emir J Food Agric. 2013;25:250–260. [Google Scholar]
- 14.Corman VM, Müller MA, Costabel U. Assays for laboratory confirmation of novel human coronavirus (hCoV-EMC) infections. Euro Surveill. 2012;17 doi: 10.2807/ese.17.49.20334-en. [DOI] [PubMed] [Google Scholar]
- 15.Corman VM, Eckerle I, Bleicker T. Detection of a novel human coronavirus by real-time reverse-transcription polymerase chain reaction. Euro Surveill. 2012;17 doi: 10.2807/ese.17.39.20285-en. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Liu D, Shi W. Origin and possible genetic recombination of the Middle East respiratory syndrome coronavirus from the first imported case in China: phylogenetics and coalescence analysis. MBio. 2015;6:e01280–e01315. doi: 10.1128/mBio.01280-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sabir JS, Lam TT, Ahmed MM. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science. 2016;351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 18.Suchard MA, Lemey P, Baele G, Ayres DL, Drummond AJ, Rambaut A. Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 2018;4 doi: 10.1093/ve/vey016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lemey P, Rambaut A, Drummond AJ, Suchard MA. Bayesian phylogeography finds its roots. PLOS Comput Biol. 2009;5 doi: 10.1371/journal.pcbi.1000520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bielejec F, Baele G, Vrancken B, Suchard MA, Rambaut A, Lemey P. SpreaD3: interactive visualization of spatiotemporal history and trait evolutionary processes. Mol Biol Evol. 2016;33:2167–2169. doi: 10.1093/molbev/msw082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: detection and analysis of recombination patterns in virus genomes. Virus Evol. 2015;1 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stadler T, Kühnert D, Bonhoeffer S, Drummond AJ. Birth-death skyline plot reveals temporal changes of epidemic spread in HIV and hepatitis C virus (HCV) Proc Natl Acad Sci USA. 2013;110:228–233. doi: 10.1073/pnas.1207965110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dudas G, Carvalho LM, Rambaut A, Bedford T. MERS-CoV spillover at the camel-human interface. eLife. 2018;7 doi: 10.7554/eLife.31257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu DK, Oladipo JO, Perera RA. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Nigeria, 2015. Euro Surveill. 2015;20 doi: 10.2807/1560-7917.ES.2015.20.49.30086. [DOI] [PubMed] [Google Scholar]
- 25.Chu DK, Poon LL, Gomaa MM. MERS coronaviruses in dromedary camels, Egypt. Emerg Infect Dis. 2014;20:1049–1053. doi: 10.3201/eid2006.140299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau SK, Wernery R, Wong EY. Polyphyletic origin of MERS coronaviruses and isolation of a novel clade A strain from dromedary camels in the United Arab Emirates. Emerg Microbes Infect. 2016;5:e128. doi: 10.1038/emi.2016.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamers MM, Raj VS, Shafei M. Deletion variants of Middle East respiratory syndrome coronavirus from humans, Jordan, 2015. Emerg Infect Dis. 2016;22:716–719. doi: 10.3201/eid2204.152065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Comar CE, Goldstein SA, Li Y, Yount B, Baric RS, Weiss SR. Antagonism of dsRNA-Induced innate immune pathways by NS4a and NS4b accessory proteins during MERS coronavirus infection. MBio. 2019;10:e00319–e00419. doi: 10.1128/mBio.00319-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thornbrough JM, Jha BK, Yount B. Middle East respiratory syndrome coronavirus NS4b protein inhibits host RNase L activation. MBio. 2016;7 doi: 10.1128/mBio.00258-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drosten C, Muth D, Corman VM. An observational, laboratory-based study of outbreaks of Middle East respiratory syndrome coronavirus in Jeddah and Riyadh, Kingdom of Saudi Arabia, 2014. Clin Infect Dis. 2015;60:369–377. doi: 10.1093/cid/ciu812. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.