Abstract
OBJECTIVE.
The purpose of this article is to compare traditional versus machine learning–based computer-aided detection (CAD) platforms in breast imaging with a focus on mammography, to underscore limitations of traditional CAD, and to highlight potential solutions in new CAD systems under development for the future.
CONCLUSION.
CAD development for breast imaging is undergoing a paradigm shift based on vast improvement of computing power and rapid emergence of advanced deep learning algorithms, heralding new systems that may hold real potential to improve clinical care.
Keywords: artificial intelligence, breast, computer-aided detection, computer-aided diagnosis, mammography, texture analysis
Traditional computer-aided detection (CAD) in mammography is familiar to breast radiologists. Early concepts of using computers to automate detection of mammographic abnormalities date back to the 1960s and were originally intended to help combat human fatigue and error while optimizing the search for subtle cancers against a complex background of breast tissue on mammograms [1]. The early version of CAD for mammography was approved by the U.S. Food and Drug Administration in 1998 but was not fully disseminated until 2002 when the use of CAD was approved for reimbursement by the Centers for Medicare & Medicaid Services. This approval overlapped with the transition from film-screen to digital mammography, particularly after the publication of the Digital Mammographic Imaging Screening Trial in 2005 [2], and with the concurrent integration of PACS into radiology, which together enabled the rapid adoption of CAD into mammographic interpretation. CAD was used in 74% of all screening mammograms by 2008 [3] and in 92% of all screening mammograms by 2016 [4]. It remains an integral part of mammographic screening to date, despite questions about its efficacy in its current form.
CAD has taken on a broader meaning more recently, notably in the context of renewed interest in artificial intelligence in medical imaging. There has been an abundance of research in machine learning in the last decade and in deep learning (DL), a subtype of machine learning, since 2012, which is made possible by advanced algorithms that are powered by faster computers, greater storage capabilities, and availability of big data [5]. In this setting, CAD research has expanded across modalities and beyond breast imaging, and CAD may refer to any computer-assisted algorithm in diagnostic imaging using machine learning. Research in the newer DL-based CAD platforms continues to focus on mammography, which offers simple binary outcomes (cancer vs not cancer), leverages big datasets (large number of screening examinations), and has maximum tangible clinical impact (improved mortality). The new DL-based CAD platforms in mammography will differ from the traditional CAD in important ways, with the potential to overcome the limitations of its predecessor, and is expected to better enhance radiologist accuracy.
Computer-Aided Detection Versus Computer-Aided Diagnosis
There are two components to consider: computer-aided detection (CADe), which focuses on detection, and computer-aided diagnosis (CADx), which focuses on classification. Traditional CAD in mammography emphasizes detection of abnormalities, such as masses, asymmetries, calcifications, and architectural distortions and is limited to CADe. The computer uses recognition techniques based on manually crafted features tailored to specific tasks to delineate and mark sites that appear distinct from normal structures in the breast [6] (Fig. 1). The radiologist then decides whether these findings warrant a patient recall, determining their clinical significance. Traditional CAD was intended to be a second reader in the setting of double reading, which represents the standard of care in European countries and was widely known to increase cancer detection and decrease recall rates, albeit doubling the workload for radiologists and limiting cost-effectiveness [7]. Unlike the second radiologist, however, CADe is beholden to the opinion of the primary radiologist, which, in part, explains why it has been challenging to accurately assess the efficacy of the current CAD system in place. CADx is therefore the natural next step, which aims to automate classification of abnormalities. Although CADx development began with CADe in the early 1990s [8–10], computer algorithms were limited at that time, and CADx technology was not ready to enter the clinical sphere.
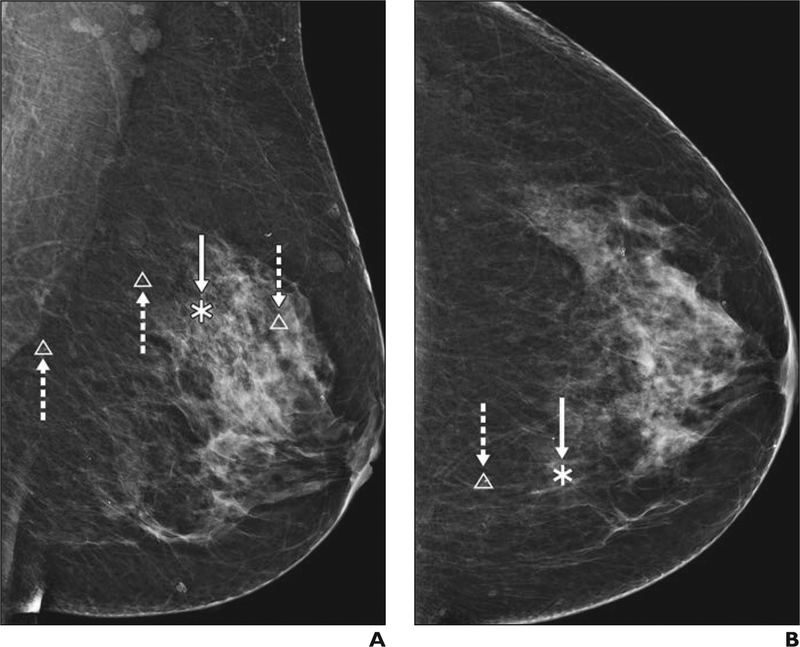
Fig. 1—
46-year-old woman with heterogeneously dense breast tissue. A and B, Digital mammograms of left breast in mediolateral oblique (A) and craniocaudal (B) views show traditional computer-aided detection markings highlighting masses in asterisks (solid arrows) and calcifications in triangles (dashed arrows).
Compared with traditional CAD, the next-generation systems will provide both CADe and CADx, and strive for a fully automated end-to-end process, thus better facilitating machine learning [11–18]. The field has transformed since a few decades ago, now with convergence of large computing resources and increased electronic access to data, making it possible to train convolutional neural networks (CNNs) not only to recognize abnormalities but also to assess them on the basis of continuous learning via back-propagating corrective signals, therefore enhancing performance in ways not previously possible. This learning of multilayered artificial neural networks, also known as DL, is behind the early successes of CAD systems currently under development for mammography [12–21] and offers the potential of improved outcomes over traditional approaches. The range of possible CADx utility has also expanded beyond such tasks as lesion classification or risk stratification for breast cancer now into the realms of radiomics (i.e., converting image pixels into minable data) and radiogenomics (i.e., imaging data combined with genetic information), with the ultimate goal being individualized prognostication and tailored therapy, to derive maximum clinical value from imaging data [5].
Traditional Computer-Aided Detection
The use of traditional CAD in mammography has been controversial. Although early findings were promising in smaller retrospective series and reader studies [22–26], benefits were not reproduced in larger population-based clinical trials once CAD was widely implemented [27–29]. The original intent of early CAD systems was to catch subtle cancers that radiologists might otherwise have missed. Indeed, in one early study using prior mammograms as hindsight in known cases of cancer, CAD marked 86% (30/35) of missed calcifications and 73% (58/80) of missed masses [22]. There was also evidence that CAD may even be able to pick up signs of cancer yet imperceptible to the human eye in some studies, where CAD marked 40–42% of mammographic findings retrospectively deemed occult or nonspecific by the radiologist but that developed into cancers [25], potentially accelerating cancer detection by up to 2–12 months [30]. Even so, these results have not been borne out in the clinical environment, because the radiologist still determines the significance of any given finding flagged by CAD, and if a finding is so subtle as to fall below the threshold of human perception, it may be deemed a false-positive [31]. Nevertheless, early studies found CAD to be beneficial, performing as well as, if not better than, double reading by two radiologists in larger trials from the United Kingdom [32, 33]. Although early CAD often did not identify all cancers the radiologist detected, it heightened sensitivity for small subtle lesions easily missed by the radiologist, overall increasing sensitivity but incurring decreased specificity [34].
To maximize the benefit of traditional CAD, one necessarily had to recall all CAD marked lesions not otherwise seen by the radiologist. For example, in a large study using Breast Cancer Surveillance Consortium data (429,345 mammograms in 222,135 women), approximately 157 women would have to be recalled and 15 women would undergo biopsy owing to CAD, to detect one additional cancer [28]. This is not possible in the clinical setting, where the radiologist must practice within recommended American College of Radiology benchmark parameters (cancer detection rate, ≥ 2.5/1000; recall rate, 5–12%) to avoid unacceptably high false-positives and unnecessary biopsies [35]. For this reason, the extent to which early CAD was used to its full potential depended in no small part on the skill and experience level of the radiologist. Although it added little to already seasoned radiologists with high sensitivity, it helped less-experienced radiologists to a greater extent, although it occasionally lulled others into a false sense of security, ultimately lowering their accuracy [31, 36]. In highlighting subtle findings, traditional CAD also did not see all lesions equally. Although early CAD had high sensitivity for microcalcifications (up to 99%) [23], it performed less well with masses (75–89%) [23, 37, 38] and least well with architectural distortions (38%) [39], the latter of which are most challenging to the radiologist, therefore supporting the notion that CAD may be limited in meeting the clinical needs of radiologists [39, 40]. This is also consistent with later findings that CAD preferentially increased the detection of in situ cancers (calcifications) and did not alter the detection of invasive cancers (masses and architectural distortions) [27], thereby improving early diagnosis but offering limited prospects for potential mortality benefit.
For all these reasons, later larger community-based studies could not reproduce the benefits of CAD seen in earlier trials [27–29]. The largest of these studies from 2015 [27] compared the performance of 271 radiologists across 66 facilities in the Breast Cancer Surveillance Consortium, evaluating a total of over 600,000 mammograms with and without CAD and adjusting for radiologist learning curve and patients’ age, race or ethnicity, breast density, menopausal status, and time since prior mammogram, among other potential confounders, and found no improvement in screening performance with CAD. With and without CAD, the cancer detection rate (4.1/1000), sensitivity (85.3% vs 87.3%), and specificity (91.6% vs 91.4%) were unchanged. In a subset of 107 radiologists who interpreted both with and without CAD, sensitivity was significantly lower when using CAD (83.3% with vs 89.6% without) [27]. This echoed findings of a 2007 study [28] earlier in the CAD experience (429,345 mammograms) that found no change in cancer detection rate with and without CAD (4.2 vs 4.15 per 1000), a nonsignificant increase in sensitivity (84% vs 80.4%), but a significant decrease in specificity (87.2% vs 90.2%), resulting in a nearly 20% increase in the biopsy rate and lower overall accuracy (AUC, 0.871 vs 0.919). In addition, along with the unchanged cancer detection rate, CAD-detected invasive cancers were not associated with more favorable stage, size, or lymph node status [29]. In the end, the clinical environment proved to be far more complex than the laboratory setting, and as found by the large reader study arm of the Digital Mammographic Imaging Screening Trial, practicing radiologists rarely altered their diagnostic decisions on the basis of the addition of CAD [41].
Technical Limitations
The clinical failings of early CAD resulted from several key technical limitations. Because prior development of traditional CAD relied on limited computing resources (i.e., slower central processing units and lack of parallel processing), trained on smaller datasets (i.e., limited availability of digital mammograms at the time of transition from film-screen mammography), and used examinations with poor image quality (i.e., digitized films), the resulting application was not as robust as it could have been. Insufficient computer processing power also precluded CAD from fully functioning to assess multiple mammographic views or integrate prior studies, thus further limiting its role in the clinical setting [42]. Perhaps the truly missed opportunity is that, because traditional CAD is not designed for continuous feedback and learning, it does not independently improve its performance, despite amassing mammographic cases over time. Improvement of traditional CAD is possible but limited by periodic software upgrades, which are difficult to vet. The most important limitation is that traditional CAD relies on human expertise as a reference standard, which is not always a consistent surrogate to the ground truth (i.e., whether a lesion is truly cancer). This expert-dependent feature design in early CAD comes with inherent human bias, limiting its performance. On the other hand, machine-learning algorithms will similarly contain human bias if they are trained to replicate human decisions [43, 44].
Redefining Computer-Aided Detection
New CAD platforms will differ from traditional CAD in several important ways (Table 1). Recent approaches in developing CAD based on DL no longer require manual feature design and minimize human interference. DL algorithms learn discerning features that are best predictive of outcomes independently and may be able to identify novel features not previously known [45]. The capacity for continuous feedback and learning will allow DL-based CAD to improve over time. In theory, DL algorithms can be trained to pattern recognize image data (pixel-related information), correlate it to tumor registry data (the truth), and risk assess when it recognizes a similar pattern (predict likelihood of cancer). Further feedback into the system of whether that prediction is correct (based on truth) will improve its performance in the future. New CAD systems will thus eventually be able to identify novel features associated with more relevant cancers by incorporating patient- and tumor-level variables, that will extend beyond immediate human recognition [14, 46]. This design has the potential to maximize the mortality benefit of breast cancer screening and to address the issues of overdiagnosis and overtreatment.
TABLE 1:
Comparison of Traditional and Deep-Learning (DL)–Based Computer-Aided Detection (CAD) Systems
| Variable | Traditional CAD | Future DL-Based CAD |
|---|---|---|
| Function | Second reader | Stand-alone |
| CADe | CADe plus CADx | |
| Mechanism | Manual feature design (known features) | Deep neural networks (known and novel features) |
| Capacity | Slow CPUs, limited storage | Fast GPUs, large storage |
| Limited digital data | Increasing digital data | |
| Clinical utility | Limited | Potentially unlimited |
| Detects DCIS more than IDC | May detect more invasive cancers | |
| Potential | Radiologist dependent | Radiologist independent |
| No feedback or learning | Learns and improves |
Note—CADe = computer-aided detection, CADx = computer-aided diagnosis, CPU = central processing unit, GPU = graphics processing unit, DCIS = ductal carcinoma in situ, IDC = invasive ductal carcinoma.
This shift in CAD design is made possible by recent breakthroughs in computer technology, data science, and algorithm development. The computer processing speed and memory have increased exponentially, owing to faster graphics processing units and parallel processing. By 1997, the IBM Deep Blue was already capable of evaluating 2 million chess positions per second [47]. By 2011, the IBM Watson could process 500 gigabytes of information per second, the equivalent of 1 million books [43, 48]. Powerful computing makes DL feasible, but data are needed. It is no coincidence that DL gained success only more recently, as digital health data (PACS and electronic medical records) became more readily available. The Digital Mammography DREAM challenge in 2017 [49], which used crowd-sourcing and distilled the three best designs from 1150 coders from around the world, yielded CAD algorithms with high AUCs (up to 0.9), allowing a glimpse of what is possible and thus catalyzing the field. The more recent move by the U.S. Food and Drug Administration in June 2018 to reclassify medical image analyzers (i.e., CAD software) from class III to class II devices, thereby reducing regulatory burdens and obviating premarket approval, reflects a renewed optimism for CAD programs and anticipation of increasing numbers of artificial intelligence–based submissions in the near future.
New Computer-Aided Detection Platforms
Just as traditional CAD systems based on manually hard-coded rules are outdated, more-advanced CAD systems from the pre-DL era also had serious limitations. The shortcomings of simpler machine-learning models, such as logistic regression or decision trees, lie in how they arrive at the prediction. What these methods have in common is that, although the decision process might be complex, they operate directly in the original space of features. They do not learn any intermediate representations of the data. This implies that these methods can work well only if the input features presented to them are very predictive to begin with. In some applications, where the input features indeed have a very well-defined meaning, this is sufficient to achieve high accuracy; however, this is often not the case in classifying medical images such as mammograms. Deep neural networks, on the other hand, break up the task of classification into a sequence of easier learning subtasks. This allows them to find a good representation for the data that is tailored to the final task.
A type of deep neural network particularly suitable to computer vision tasks is a CNN (also known as “convnet”) [50–53]. CNNs have a special connectivity structure in their hidden layers that is implemented by convolutional layers and pooling layers, making them largely invariant to shifting, scaling, and rotation of input images. This means that CNNs are high-fidelity systems resilient to small variations in the input image capable of producing consistent final predictions. Another desirable property of CNNs is that the entire pipeline from input to prediction can be expressed mathematically as one function. For this reason, the parameters of CNNs can be conveniently optimized in a single training loop, greatly accelerating the speed of new developments. This setup also allows the identification of predictive features in the input, even though the original training data only had examination-level labels. Research in this direction in medical imaging is still in its infancy, but some work in natural images has been done using the same principle [54–57].
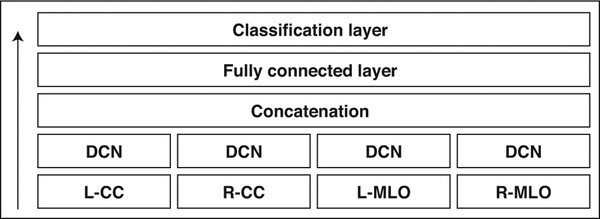
To illustrate the aforementioned concepts, we describe a deep convolutional network used for BI-RADS classification in screening mammography as an example [19]. In this case, the input to the network are four mammographic images displayed in standard views (left and right craniocaudal and left and right mediolateral oblique) (Fig. 2). Each image is first processed independently in a series of convolutional and pooling layers (Fig. 3), then information from all views is concatenated, transformed with one fully connected layer, and finally, this joint representation of the four input images is fed to a simple classification layer, which ultimately produces a prediction. Because the final prediction of this network is a differentiable function of the input images, we can identify the input pixels whose changes influence the change in the confidence of the network the most. This is illustrated in two example cases (Fig. 4). The new CAD platforms under development will build on similar algorithms and concepts and increasingly integrate non–pixel-related factors, such as patient level, tumor level, and population level information.
Fig. 2—
Overview of multiview deep convolutional network (DCN) [19]. DCN refers to series of convolutional and pooling layers applied separately to each mammographic view. It is described in detail in Figure 3. Arrow indicates direction of information flow. L = left, R = right, CC = craniocaudal, MLO = mediolateral oblique.
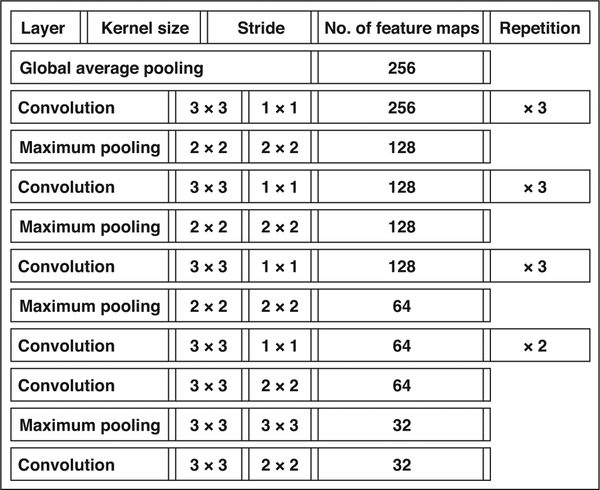
Fig. 3—
Description of subnetwork processing single view. It transforms one mammographic view into fixed size vector that can be concatenated with other vectors for remaining views. For convolutional layers, kernel size denotes size of pattern that layer is extracting. For pooling layers, it denotes size of area over which average or maximum is computed. Stride denotes what is space between applications of convolution or pooling. Number of feature maps indicates how many different patterns network is extracting within each layer. Global average pooling is also type of pooling layer but it acts on entire image. Its purpose is to reduce size of feature maps into single vector.
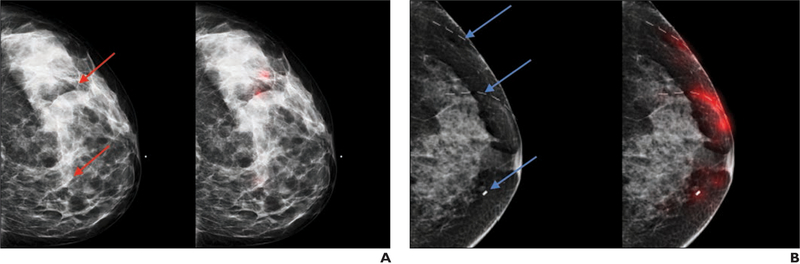
Fig. 4—
Examples of visualization of decisions made by network. A and B, 42-year-old woman (BI-RADS category 0, A) and 51-year-old woman (BI-RADS category 2, B). Both left-hand images show breast with possible suspicious findings (red arrows, A; blue arrows, B). Both right-hand images are corresponding images of same breast with regions of images (highlighted in red) that influence confidence of predictions of neural network. Dashed lines in B denote scar markers overlying left breast.
New Challenges
Developing new DL-based CAD systems is not without challenges. DL requires large databases, which can be costly to compile. As opposed to supervised learning (where image labeling can be labor intensive), unsupervised learning (where the machine must discern intrinsic variables based on unlabeled images) requires high-quality raw data to maximize yield; thus, full-resolution mammographic images, which generate huge data files and storage requirements, are preferred. Techniques such as transfer learning and data augmentation have been used to tailor needs for training data [45]; however, validation and testing datasets currently lack standardization, which makes replicating published results and comparing algorithms difficult [18]. This is also hampered by the limited ability to share and pool data across institutions because of patient privacy considerations mandated by HIPAA, which pertain both to imaging data and clinical data. The benefit of open science shown through the success of the DREAM Challenge underscores the importance of curating large, anonymized, high-quality, sharable, and generalizable datasets [58].
An important pitfall in training deep neural networks is overfitting, which refers to when a model learns idiosyncratic variations of a training dataset, but does not grasp the broadly predictive features of a given problem. This will be manifest as good performance on the training set, but poor performance on testing or validation sets, and is liable to happen when an algorithm is excessively complex relative to the amount of data available [59]. Current DL algorithms typically comprise 30–150 layers, and the complex inner workings of how results are derived are not always apparent, hence the term “black box” [45]. Methods to assess learned parameters within a neural network and to understand its method are needed to provide transparency and accountability [5]. Challenges such as regulatory and medi-colegal considerations, as well as patient privacy and data security, will increasingly play out as DL approaches technical maturity to enter clinical care and will require a unifying vision and oversight by agencies such as the U.S. Food and Drug Administration and the American College of Radiology Data Science Institute [60].
Clinical Applications
The clinical applications of new CAD systems will depend both on performance and implementation. It is likely that future CAD will be implemented both for screening and diagnostic imaging. Although current research emphasizes the development of task-specific tools, careful evaluation of how to best integrate these tools will be needed. We have learned from traditional CAD that a better understanding of radiologist-computer interaction is essential to allow meaningful artificial intelligence assistance. The current status quo of CAD in screening must be disrupted to overcome similar prior challenges. Experts have proposed moving away from automatic CAD overlays, making CAD more interactive and on-demand only when help is needed, possibly flagging cases only when there is a missed finding that CAD deems highly probable to be abnormal, making CAD more evidence-based to provide similar cases with known outcomes and making CAD more personalized and providing radiologists with individual feedback [61]. Ultimately, whether it is possible to rebuild confidence in CAD will depend on successful integration of DL algorithms into real-life clinical workflow.
DL-based CAD systems under development have shown highly promising results in preliminary testing (AUC, ≈ 0.9) [12–21] (Table 2), with work being done in full-field digital mammography (FFDM), digital breast tomosynthesis (DBT), and contrast-enhanced mammography. However, careful validation on large population-based datasets is required. In FFDM, current DL-based CAD systems focus on both cancer detection and tissue quantification. For example, a new CAD system developed to detect and classify malignant versus benign masses and calcifications based on a faster regional CNN trained on the public Digital Database for Screening Mammography performed extremely well when applied to the public INbreast database [12]. Another new CAD system to detect and classify masses and architectural distortions based on an OxfordNetlike CNN trained on the Digital Database for Screening Mammography, integrating not only local information but also context, symmetry, and relationship between two views of the same breast, significantly outperformed traditional CAD, particularly in achieving higher specificity [18]. While some newly proposed CAD schemes improve on prior machine weaknesses, such as detection of architectural distortion (e.g., a bidimensional empirical mode decomposition–based system) [62], other frameworks (e.g., an ROI-based CNN called YOLO [You Only Look Once]) tackle human blind spots such as dense regions or posterior locations [13]. Perhaps most important, work is being done to develop DL-based CAD systems that can directly comment on clinical outcomes. A recently proposed CAD scheme suggests that novel quantitative image markers based on false-positive mammograms could predict short-term breast cancer risk [63]. Other DL-based CAD systems focus on risk assessment. Breast density on mammography, for example, is an established risk marker but is often inconsistently categorized by radiologists. Not only are CNNs performing comparably to human readers in breast density classification [64], they may also enhance clinical assessment of the most difficult-to-distinguish breast density categories (i.e., scattered vs heterogeneously dense) [65]. Further application of DL-based CAD in personalized breast cancer risk assessment is also being explored [66].
TABLE 2:
Emerging Deep Convolutional Neural Network–Based CAD Frameworks for Mammography and Outcomes
| Reference | Deep Convolutional Neural Network | Databases |
|---|---|---|
| Kim et al. [15] | DIB-MG | Multiinstitutional |
| Ribli et al. [12] | Faster R-CNN | DDSM, INbreast |
| Al-Masni et al.[13] | You Only Look Once | DDSM |
| Chougrad et al. [16] | VGG, ResNet50 | DDSM, INbreast, BCDR |
| Inception version 3 | MIAS | |
| Teare et al.[17] | Inception version 3 | DDSM, ZMDS |
| Kooi et al. [18] | VGG | DDSM |
| Geras et al. [19] | Multiview VGG | Institutional |
| Lotter et al. [20] | ResNet | DDSM |
| Zhu et al. [21] | Multiinstance AlexNet | INbreast |
Note—DIB-MG = Data-driven Imaging Biomarker in Mammography, R-CNN = regional convolutional neural network, DDSM = Digital Database for Screening Mammography, VGG = Visual Geometry Group, ResNet 50 = Residual Network 50, BCDR = Breast Cancer Digital Repository, MIAS = Mammographic Image Analysis Society database, ZMDS = Zebra Mammography Dataset.
New CAD systems for DBT have a slightly different focus. As synthetic mammograms increasingly replace 2D FFDM images in combination analysis, CAD software programs have been updated to work with synthetic images from vendors such as Hologic (C-view) and GE (V-preview), although these are not DL-based systems. Work on DL-based CAD-enhanced synthetic mammography is emerging, which accentuates relevant abnormal findings and increases diagnostic accuracy [67]. In addition, rapid integration of DBT into clinical practice has created greater time and concentration demands on the radiologist, who is increasingly prone to fatigue and error [68]. New CAD systems for DBT have reported 23.5–29.2% reduction in reading time with noninferior radiologist performance [69–71]. Because DBT is a relatively new technology, data availability is limited for training neural networks, but work is being done to possibly circumvent this limitation by using FFDM studies to enrich training with some success [72]. Finally, DL-based CAD is also being developed in contrast-enhanced digital mammography as a natural extension of work in mammography, capitalizing on advantages of functional imaging using contrast enhancement, with studies showing new CAD significantly outperforming traditional CAD and improving specificity as compared with human readers [73, 74].
Conclusion
The field of breast imaging has again found itself at the cutting edge of CAD development in the age of renewed interest in artificial intelligence. DL has profoundly transformed our approach to computer-aided imaging analysis, distinguishing it fundamentally from traditional CAD methods. Where traditional CAD in mammography fell short, new CAD platforms have great potential to succeed. However, careful and rigorous training, validation, and testing on external datasets and large population-based datasets are necessary to ensure clinical success of these programs. Although machine-learning based platforms will face unique challenges ahead, as well as the same challenges encountered by traditional CAD in clinical implementation, they present an unprecedented opportunity to better derive clinical value from imaging data and will certainly reshape the way we care for our patients.
References
- 1.Winsberg F, Elkin M, Macy J, et al. Detection of radiographic abnormalities in mammograms by means of optical scanning and computer analysis. Radiology 1967; 89:211–215 [Google Scholar]
- 2.Pisano ED, Gatsonis C, Hendrick E, et al. Diagnostic performance of digital versus film mammography for breast-cancer screening. N Engl J Med 2005; 353:1773–1783 [DOI] [PubMed] [Google Scholar]
- 3.Rao VM, Levin DC, Parker L, Cavanaugh B, Frangos AJ, Sunshine JH. How widely is computer-aided detection used in screening and diagnostic mammography? J Am Coll Radiol 2010; 10:802–805 [DOI] [PubMed] [Google Scholar]
- 4.Keen JD, Keen JM, Keen JE. Utilization of computer-aided detection for digital screening mammography in the United States, 2008–2016. J Am Coll Radiol 2018; 15(1 Pt A):44–48 [DOI] [PubMed] [Google Scholar]
- 5.Giger ML. Machine learning in medical imaging. J Am Coll Radiol 2018; 15(3 Pt B):512–520 [DOI] [PubMed] [Google Scholar]
- 6.Hupse R, Karssemeijer N. Use of normal tissue context in computer-aided detection of masses in mammograms. IEEE Trans Med Imaging 2009; 28:2033–2041 [DOI] [PubMed] [Google Scholar]
- 7.Taylor-Phillips S, Jenkinson D, Stinton C, Wallis MG, Dunn J, Clarke A. Double reading in breast cancer screening: cohort evaluation in the CO-OPS trial. Radiology 2018; 287:749–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jiang YN, Nishikawa RM, Wolverton DE, et al. Malignant and benign clustered microcalcifications: automated feature analysis and classification. Radiology 1996; 198:671–678 [DOI] [PubMed] [Google Scholar]
- 9.Cho HC, Hadjiiski L, Sahiner B, et al. Similarity evaluation in a content-based image retrieval (CBIR) CADx system for characterization of breast masses on ultrasound images. Med Phys 2011; 38:1820–1831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta S, Chyn PF, Markey MK. Breast cancer CADx based on BI-RADS descriptors from two mammographic views. Med Phys 2006; 33:1810–1817 [DOI] [PubMed] [Google Scholar]
- 11.Arevalo J, González FA, Ramos-Pollán R, Oliveira JL, Guevara Lopez MA. Representation learning for mammography mass lesion classification with convolutional neural networks. Comput Methods Programs Biomed 2016; 127:248–257 [DOI] [PubMed] [Google Scholar]
- 12.Ribli D, Horváth A, Unger Z, Pollner P, Csabai I. Detecting and classifying lesions in mammograms with deep learning. Sci Rep 2018; 8:4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Al-Masni MA, Al-Antari MA, Park JM, et al. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput Methods Programs Biomed 2018; 157:85–94 [DOI] [PubMed] [Google Scholar]
- 14.Shi BG, Grimm LJ, Mazurowski MA, et al. Prediction of occult invasive disease in ductal carcinoma in situ using deep learning features. J Am Coll Radiol 2018; 15(3 Pt B):527–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim EK, Kim HE, Han K, et al. Applying data-driven imaging biomarker in mammography for breast cancer screening: preliminary study. Sci Rep 2018; 8:2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chougrad H, Zouaki H, Alheyane O. Deep convolutional neural networks for breast cancer screening. Comput Methods Programs Biomed 2018; 157:19–30 [DOI] [PubMed] [Google Scholar]
- 17.Teare P, Fishman M, Benzaquen O, Toledano E, Elnekave E. Malignancy detection on mammography using dual deep convolutional neural networks and genetically discovered false color input enhancement. J Digit Imaging 2017; 30:499–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kooi T, Litjens G, van Ginneken B, et al. Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 2017; 35:303–312 [DOI] [PubMed] [Google Scholar]
- 19.Geras KJ, Wolfson S, Shen Y, et al. High-resolution breast cancer screening with multi-view deep convolutional neural networks. arXiv website. arxiv.org/abs/1703.07047. Published March 21, 2017. Accessed September 21, 2018
- 20.Lotter W, Sorensen G, Cox D. A multi-scale CNN and curriculum learning strategy for mammogram classification In: Cardoso J, Arbel T, Carneiro G, et al. , eds. Deep learning in medical image analysis and multimodal learning for clinical decision support. Basel, Switzerland: Springer, 2017:169–177 [Google Scholar]
- 21.Zhu W, Lou Q, Vang YS, Xie X. Deep multi-instance networks with sparse label assignment for whole mammogram classification. bioRxiv website. www.biorxiv.org/content/biorxiv/early/2016/12/20/095794.full.pdf. Published December 18, 2016. Accessed September 21, 2018
- 22.Birdwell RL, Ikeda DM, O’Shaughnessy KF, Sickles EA. Mammographic characteristics of 115 missed cancers later detected with screening mammography and the potential utility of computer-aided detection. Radiology 2001; 219:192–202 [DOI] [PubMed] [Google Scholar]
- 23.Warren Burhenne LJ, Wood SA, D’Orsi CJ, et al. Potential contribution of computer-aided detection to the sensitivity of screening mammography. Radiology 2000; 215:554–562 [DOI] [PubMed] [Google Scholar]
- 24.Ciatto S, Del Turco MR, Risso G, et al. Comparison of standard reading and computer aided detection (CAD) on a national proficiency test of screening mammography. Eur J Radiol 2003; 45:135–138 [DOI] [PubMed] [Google Scholar]
- 25.Ikeda DM, Birdwell RL, O’Shaughnessy KF, Sickles EA, Brenner RJ. Computer-aided detection output on 172 subtle findings on normal mammograms previously obtained in women with breast cancer detected at follow-up screening mammography. Radiology 2004; 230:811–819 [DOI] [PubMed] [Google Scholar]
- 26.Helvie MA, Hadjiiski L, Makariou E, et al. Sensitivity of noncommercial computer-aided detection system for mammographic breast cancer detection: pilot clinical trial. Radiology 2004; 231:208–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lehman CD, Wellman RD, Buist DS, et al. Diagnostic accuracy of digital screening mammography with and without computer-aided detection. JAMA Intern Med 2015; 175:1828–1837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fenton JJ, Taplin SH, Carney PA, et al. Influence of computer-aided detection on performance of screening mammography. N Engl J Med 2007; 356:1399–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fenton JJ, Abraham L, Taplin SH, et al. Effectiveness of computer-aided detection in community mammography practice. J Natl Cancer Inst 2011; 103:1152–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gur D, Sumkin JH. CAD in screening mammography. AJR 2006; 187:1474. [DOI] [PubMed] [Google Scholar]
- 31.Krupinski EA. Computer-aided detection in clinical environment: benefits and challenges for radiologists. Radology 2004; 231:7–9 [DOI] [PubMed] [Google Scholar]
- 32.Gilbert FJ, Astley SM, McGee MA, et al. Single reading with computer-aided detection and double reading of screening mammograms in the United Kingdom National Breast Screening Program. Radiology 2006; 241:47–53 [DOI] [PubMed] [Google Scholar]
- 33.Gilbert FJ, Astley SM, Gillan MG, et al. Single reading with computer-aided detection for screening mammography. N Engl J Med 2008; 359:1675–1684 [DOI] [PubMed] [Google Scholar]
- 34.Henriksen EL, Carlsen JF, Vejborg IM, Nielsen MB, Lauridsen CA. The efficacy of using computer-aided detection (CAD) for detection of breast cancer in mammography screening: a systematic review. Acta Radiol 2018. January 1 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 35.Sickles EA, D’Orsi CJ. ACR BI-RADS Follow-up and outcome monitoring In: D’Orsi CJ, Sickles EA, Mendelson EB, et al. , eds. ACR BI-RADS Atlas, Breast Imaging Reporting and Data System. Reston, VA: American College of Radiology, 2013 [Google Scholar]
- 36.Ko JM, Nicholas MJ, Mendel JB, Slanetz PJ. Prospective assessment of computer-aided detection in interpretation of screening mammography. AJR 2006; 187:1483–1491 [DOI] [PubMed] [Google Scholar]
- 37.Petrick N, Sahiner B, Chan HP, Helvie MA, Paguerault S, Hadjiiski LM. Breast cancer detection: evaluation of a mass-detection algorithm for computer-aided diagnosis—experience in 263 patients. Radiology 2002; 224:217–224 [DOI] [PubMed] [Google Scholar]
- 38.Malich A, Marx C, Facius M, Boehm T, Fleck M, Kaiser WA. Tumour detection rate of a new commercially available computer-aided detection system. Eur Radiol 2001; 11:2454–2459 [DOI] [PubMed] [Google Scholar]
- 39.Baker JA, Rosen EL, Lo JY, Gimenez EI, Walsh R, Soo MS. Computer-aided detection (CAD) in screening mammography: sensitivity of commercial CAD systems for detecting architectural distortion. AJR 2003; 181:1083–1088 [DOI] [PubMed] [Google Scholar]
- 40.Destounis SV, Arieno AL, Morgan RC. CAD may not be necessary for microcalcifications in the digital era, CAD may benefit radiologists for masses. J Clin Imaging Sci 2012; 2:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cole EB, Zhang Z, Marques HS, Edward Hendrick R, Yaffe MJ, Pisano ED. Impact of computer-aided detection systems on radiologist accuracy with digital mammography. AJR 2014; 203:909–916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Giger ML, Chan HP, Boone J. Anniversary paper: history and status of CAD and quantitative image analysis—the role of Medical Physics and AAPM. Med Phys 2008; 35:5799–5820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dilsizian SE, Siegel EL. Artificial intelligence in medicine and cardiac imaging: harnessing big data and advanced computing to provide personalized medical diagnosis and treatment. Curr Cardiol Rep 2014; 16:441. [DOI] [PubMed] [Google Scholar]
- 44.Lee CS, Nagy PG, Weaver SJ, Newman-Toker DE. Cognitive and system factors contributing to diagnostic errors in radiology. AJR 2013; 201:611–617 [DOI] [PubMed] [Google Scholar]
- 45.Erickson BJ, Korfiatis P, Kline TL, Akkus Z, Philbrick K, Weston AD. Deep learning in radiology: does one size fit all? J Am Coll Radiol 2018; 15(3 Pt B):521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sutton EJ, Huang EP, Drukker K, et al. Breast MRI radiomics: comparison of computer- and human-extracted imaging phenotypes. Eur Radiol Exp 2017; 1:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.IBM. IBM 100: Deep Blue. IBM website. www-03.ibm.com/ibm/history/ibm100/us/en/icons/deepblue/. Accessed September 21, 2018
- 48.Pearson T IBM Watson: how to replicate Watson hardware and systems design for your own use in your basement. IBM website. www.ibm.com/developerworks/mydeveloperworks/blogs/Inside-SystemStorage/entry/ibm_watson_how_to_build_your_own_watson_jr_in_your_basement7?lang=en. Published February 18, 2011. Updated 2014. Accessed September 21, 2018
- 49.Trister AD, Buist DSM, Lee CI. Will machine learning tip the balance in breast cancer screening? JAMA Oncol 2017; 3:1463–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.LeCun Y, Boser BE, Denker JS, et al. Handwritten digit recognition with a back-propagation network. NIPS proceedings website. papers.nips.cc/paper/293-handwritten-digit-recognition-with-aback-propagation-network. Published 1989. Accessed September 21, 2018
- 51.Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. NIPS proceedings website. papers.nips.cc/paper/4824-imagenet-classification-with-deep-convolutional-neural-networks. Published 2012. Accessed September 21, 2018
- 52.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. IEEE Xplore Digital Library website. ieeexplore.ieee.org/document/7780459/. Published 2016. Accessed September 21, 2018
- 53.Huang G, Liu Z, van der Maaten L, Weinberger KQ. Densely connected convolutional networks. IEEE Xplore Digital Library website. ieeexplore.ieee.org/document/8099726/. Published 2017. Accessed September 21, 2018
- 54.Simonyan K, Vedaldi A, Zisserman A. Deep inside convolutional networks: visualising image classification models and saliency maps. arXiv website. arxiv.org/abs/1312.6034. Published December 20, 2013. Accessed September 21, 2018
- 55.Fong R, Vedaldi A. Interpretable explanations of black boxes by meaningful perturbation. arXiv website. arxiv.org/abs/1704.03296. Published April 11, 2017. Updated January 10, 2018. Accessed September 24, 2018
- 56.Dabkowski P, Gal Y. Real time image saliency for black box classifiers. arXiv website. arxiv.org/abs/1705.07857. Published May 22, 2017. Accessed September 24, 2018
- 57.Zolna K, Geras KG, Cho K. Classifier-agnostic saliency map extraction. arXiv website. arxiv.org/abs/1805.08249. Published May 21, 2018. Accessed September 24, 2018
- 58.Lee RS, Gimenez F, Hoogi A, Miyake KK, Gorovoy M, Rubin DL. A curated mammography data set for use in computer-aided detection and diagnosis research. Sci Data 2017; 4:170177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chartrand G, Cheng PM, Vorontsov E, et al. Deep learning: a primer for radiologists. RadioGraphics 2017; 37:2113–2131 [DOI] [PubMed] [Google Scholar]
- 60.American College of Radiology (ACR). ACR Data Science Institute blog. www.acrdsi.org. Accessed June 11, 2018
- 61.Nishikawa RM, Bae KT. Importance of better human-computer interaction in the era of deep learning: mammography computer-aided diagnosis as a use case. J Am Coll Radiol 2018; 15(1 Pt A):49–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zyout I, Togneri R. A computer-aided detection of the architectural distortion in digital mammograms using the fractal dimension measurements of BEMD. Comput Med Imaging Graph 2018. April 3 [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 63.Mirniaharikandehei S, Hollingsworth AB, Patel B, Heidari M, Liu H, Zheng B. Applying a new computer-aided detection scheme generated imaging marker to predict short-term breast cancer risk. Phys Med Biol 2018; 63:105005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu N, Geras KG, Shen Y, et al. Breast density classification with deep convolutional neural networks. arXiv website. arxiv.org/abs/1711.03674. Published November 10, 2017. Accessed September 24, 2018
- 65.Mohamed AA, Berg WA, Peng H, Luo Y, Jankowitz RC, Wu S. A deep learning method for classifying mammographic breast density categories. Med Phys 2018; 45:314–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Li H, Giger ML, Huynh BQ, Antropova NO. Deep learning in breast cancer risk assessment: evaluation of convolutional neural networks on a clinical dataset of full-field digital mammograms. J Med Imaging (Bellingham) 2017; 4:041304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.James JJ, Giannotti E, Chen Y. Evaluation of a computer-aided detection (CAD)-enhanced 2D synthetic mammogram: comparison with standard synthetic 2D mammograms and conventional 2D digital mammography. Clin Radiol 2018; 73:886–892 [DOI] [PubMed] [Google Scholar]
- 68.Stec N, Arie D, Moody AR, Krupinski EA, Tyrrell PN. A systematic review of fatigue in radiology: is it a problem? AJR 2018; 210:799–806 [DOI] [PubMed] [Google Scholar]
- 69.Balleyguier C, Arfi-Rouche J, Levy L, et al. Improving digital breast tomosynthesis reading time: a pilot multi-reader, multi-case study using concurrent computer-aided detection (CAD). Eur J Radiol 2017; 97:83–89 [DOI] [PubMed] [Google Scholar]
- 70.Benedikt RA, Boatsman JE, Swann CA, Kirkpatrick AD, Toledano AY. Concurrent computer-aided detection improves reading time of digital breast tomosynthesis and maintains interpretation performance in a multireader multicase study. AJR 2018; 210:685–694 [DOI] [PubMed] [Google Scholar]
- 71.Morra L, Sacchetto D, Durando M, et al. Breast cancer: computer-aided detection with digital breast tomosynthesis. Radiology 2015; 277:56–63 [DOI] [PubMed] [Google Scholar]
- 72.van Schie G, Wallis MG, Leifland K, Danielsson M, Karssemeijer N. Mass detection in reconstructed digital breast tomosynthesis volumes with a computer-aided detection system trained on 2D mammograms. Med Phys 2013; 40:041902. [DOI] [PubMed] [Google Scholar]
- 73.Danala G, Patel B, Aghaei F, et al. Classification of breast masses using a computer-aided diagnosis scheme of contrast enhanced digital mammograms. Ann Biomed Eng 2018; 46:1419–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Patel BK, Ranjbar S, Wu T, et al. Computer-aided diagnosis of contrast-enhanced spectral mammography: a feasibility study. Eur J Radiol 2018; 98:207–213 [DOI] [PubMed] [Google Scholar]